Abstract
The combination of ion mobility separation with mass spectrometry is an emergent and powerful structural biology tool, capable of simultaneously assessing the structure, topology, dynamics and composition of large protein assemblies within complex mixtures. An integral part of the ion mobility-mass spectrometry measurement is the ionization of intact multiprotein complexes and their removal from bulk solvent. This process, during which a substantial portion of protein structure and organization is likely to be preserved, imposes a foreign environment on proteins that may cause structural rearrangements to occur. Thus, a general means must be identified to stabilize protein structures in the absence of bulk solvent. Our approach to this problem involves the protection of protein complex structure through the addition of salts in solution prior to desorption/ionization. Anionic components of the added salts bind to the complex either in solution or during the electrospray process, and those that remain bound in the gas phase tend to have high gas phase acidities. The resulting ‘shell’ of counter-ions is able to carry away excess energy from the protein complex ion upon activation and can result in significant structural stabilization of the gas-phase protein assembly overall. By using ion mobility-mass spectrometry, we observe both the dissociation and unfolding transitions for four tetrameric protein complexes bound to populations of twelve different anions using collisional activation. The data presented here quantifies, for the first time, the influence of a large range of counter-ions on gas-phase protein structure and allows us to rank and classify counter-ions as structure stabilizers in the absence of bulk solvent. Our measurements indicate that tartrate, citrate, chloride and nitrate anions are amongst the strongest stabilizers of gas-phase protein structure identified in this screen. The rank order determined by our data is substantially different when compared to the known Hofmeister salt series in solution. While this is an expected outcome of our work, due to the diminished influence of anion and protein solvation by water, our data correlates well to expected anion binding in solution and highlights the fact that both hydration layer and anion-protein binding effects are critical for Hofmeister-type stabilization in solution. Finally, we present a detailed mechanism of action for counter-ion stabilization of proteins and their complexes in the gas-phase, which indicates that anions must bind with high-affinity, but must dissociate readily from the protein in order to be an effective stabilizer. Anion-resolved data acquired for smaller protein systems allows us to classify anions into three categories based on their ability to stabilize protein and protein complex structure in the absence of bulk solvent.
Keywords: Ion Mobility-Mass Spectrometry, Structural Biology, Collision Induced Unfolding, Gas-phase Protein Structure
INTRODUCTION
Characterizing the structures and functions of multiprotein complexes on a global level within living organisms is a far-reaching scientific goal for the field of structural genomics 1–3. While significant progress has been made in this field over the past few years, it is becoming increasingly apparent that no single analytical tool has the capacity to completely describe the structural complexity of even the simplest proteome. This fact is evident in recent reports where multiple structural datasets have been integrated in order to produce models of large multiprotein systems 4–11. For example, a structural model of the yeast nuclear pore complex was recently determined by integrating the distance constraints derived from multiple datasets, including: affinity chromatography, size exclusion separations, sedimentation coefficients, electron microscopy, and chemical cross-linking mass spectrometry 5. In this and other experiments in integrative structural biology, the orthogonality of the tools employed translates into a structural picture that none of the technologies could produce in isolation.
Mass spectrometry (MS) has played a significant role in such integrative structural biology ventures, as MS has the ability to assess the composition, stoichiometry, and dynamics of multiprotein complexes of low abundance 6,7,12,13. These attributes have allowed MS to provide organizational diagrams, or protein-protein contact maps, for a number of assemblies in advance of atomic resolution structures from X-ray diffraction or nuclear magnetic resonance datasets 14–18. In some cases, MS data have reported on multi-protein systems that exhibit high degrees of polydispersity and complexity 19,20, and are excellent examples of the unique role that MS can play in defining the structure and function of multiprotein complexes. In addition, recent experiments that demonstrate the ability of MS to interrogate the organization of membrane protein systems identifies MS as a technology at the forefront of structural biology research 21,22.
Recently, the utility of ion mobility (IM) separation in tandem with MS for the analysis of protein complexes has generated considerable interest, because early studies have indicated that the quaternary structure of protein complexes can be retained in the absence of solvent 23–26. Originally applied to problems in chemical physics 27–29, trace detection30,31, and used for the analysis of small biomolecules for well-over a decade 32–37, IM separates ions based on their ability to traverse a chamber filled with inert neutrals under the influence of a weak electric field. In a process roughly analogous to a gas-phase version of electrophoretic separation in solution, ions that are large undergo a greater number of collisions with neutral molecules and thus take more time to elute from the chamber than smaller, more compact ions. Ion size in the form of an orientationally-averaged collision cross-section (CCS) is, therefore, the primary information content of IM separation and established computational approaches can be used in conjunction with this information to assign the structure of small biomolecules with a high degree of precision38.
IM-MS experiments have been used to interrogate the structure of a number of multiprotein systems and have provided information that is able to refine topology maps 39–41, establish cavity regions within a protein complex 25,42, and identify conformational changes upon ligand binding 18,43. In addition, protein complexes have been demonstrated to undergo structural collapse and unfolding upon activation with energetic collisions in the gas phase 25,44,45. Current evidence points to a mechanism for collision induced unfolding where a single subunit within the assembly unfolds, inhabiting a number of intermediate structures that are relatively stable on the timescale of the IM measurement44,46,47. In parallel with these experiments aimed at the controlled disruption of protein structure in the gas phase, several reports have highlighted the uncontrolled distortion of protein structure in the absence of solvent, including both the general compaction of protein size and structural rearrangements that may occur upon desolvation and transfer to the gas phase 40,48. While simple normalization procedures have been used extensively to analyze IM-MS data for protein complexes, and remove the influence of protein compaction in order to construct topology models 39,40,49, such rearrangements prompt the development of general strategies aimed at the protection of protein structure, at every level, in the absence of bulk solvent. Such strategies would have far reaching implications in characterizing the structures of gas-phase biomolecules and relating such data to analogues in solution including IM-MS, ion spectroscopy 50, and gas-phase H/D exchange 51.
In solution, the identity and concentration of salts can drastically influence the structure and stability of proteins and their assemblies. Recognized initially in pioneering work by Hofmeister 52, a robust rank-order of the ability of different salts to act as structural stabilizers or denaturants has been developed based on datasets accumulated over decades of biochemical observations 53,54. While the mechanistic details that lead to this rank order are currently an active area of research, recent work points to the importance of direct salt-protein interactions and the local water structure that surrounds both interacting partners as the driving forces behind the Hofmeister series 55. In the electrospray ionization (ESI) of large protein complexes, buffer components and charge carriers can condense and adhere to the assembly during the final stages of desolvation, and previous data indicate that components added in solution can influence the stability of the ions generated 56. While the use of Hofmeister-type salts is potentially a compelling approach for preventing protein unfolding in the absence of bulk solvent, the quantitative influence and mechanism of such stabilizing counter-ions is largely unknown.
In this report, we screen a series of anions for their ability to increase the structural stability of multiprotein complexes in the absence of bulk solvent using IM-MS. Specifically, we use data from four different tetrameric protein complexes, spanning a 100 kDa mass range, where the threshold energies necessary for achieving collision induced dissociation (CID) and collision induced unfolding (CIU) are measured as a function of specific anions added to ammonium acetate-based buffers in solution. In some cases, the residual counter-ions screened here are found to substantially stabilize protein quaternary structure, in other cases gas-phase protein stability is decreased measurably upon addition of the anions in solution. We use all of this data to generally classify anions in terms of their ability to stabilize protein structure in the absence of bulk solvent. In addition, we consider the mechanism of protein stabilization observed in our experiments. By analyzing smaller protein systems and generating adduct-resolved ion populations, we are able to observe that bound counter-ions act as the primary agent of gas-phase protein stabilization under our conditions. Counter-ion binding takes place in solution or during the nano-ESI (nESI) process and the data indicate that the main driver of this binding is the relative acidity of the anions added. Those that bind in large numbers and then evaporate from the assembly upon collisional activation, carrying away excess vibrational energy from the system, provide the greatest stability enhancement for gas-phase proteins and their complexes. In other cases, where the gas-phase acidities of counter-ions are very high, counter-ions may bind with high-affinity to the protein and remain bound after collisional activation resulting in no measurable increase in structural stability for the gas-phase complex. Furthermore, low-acidity anions exhibit very little protein binding affinity, and thus show no significant ability to stabilize protein structure in the absence of bulk solvent. Finally, we consider the relevance of our dataset on Hofmeister stabilization effects observed in solution.
EXPERIMENTAL METHODS
Materials
The proteins avidin tetramer (egg white), transthyretin tetramer (TTR, human), concanavalin A tetramer (ConA, jack bean), alcohol dehydrogenase tetramer (ADH, yeast), cytochrome c monomer (CYC, equine), β-Lactoglobulin A dimer (BLA, bovine), and salts (ammonium cation with acetate, fluoride, chloride, nitrate, citrate, thiocyanate, bicarbonate, tartrate, iodide, hydrogen phosphate, sulfate and perchlorate counter-anions) were purchased from Sigma (St. Louis, MO, USA). All protein samples were buffer exchanged into 100 mM ammonium acetate at pH 7 using Micro Bio-Spin 6 columns (Bio-Rad, Hercules, CA) and prepared to a final concentration of 20 µM (BLA), 10 µM (ADH, ConA, CYC) or 5 µM (avidin, TTR). In order to study the influence of different salts on protein stability without significantly altering buffer capacity or solution pH, the salts were prepared as stock solutions in 100 mM ammonium acetate at a concentration of 20 mM, each of which was then added to the protein solution. Final solutions contained added salt concentrations of 5 mM for Avidin and TTR, 10 mM for ConA and ADH, 1 mM for CYC, and 0.2 mM for BLA samples. The total salt and protein concentrations listed above were chosen primarily to avoid nESI-based ion suppression effects 57. Although previous data, and data acquired during our experiments (Figure S1), suggests that the stabilization effects observed have a salt concentration dependence, the general classifications, mechanistic details and rank orders of stability presented are not similarly dependant 56.
Ion Mobility-Mass Spectrometry
Typically, an sample aliquot (~5 µL) was analyzed using our quadrupole-ion mobility-time-of–flight mass spectrometry (Q-IM-ToF MS) instrument (Synapt G2 HDMS, Waters, Milford MA, USA) 58,59. Protein ions were generated using a nESI source. The capillary of the nESI source was typically held at voltages between 1.2 and 2.0 kV (avidin 1.6 kV; TTR 1.2 kV; ConA 1.8 kV; ADH 1.7 kV; BLA 2.0 kV; CYC 1.5 kV), with the source operating in positive mode. The sample cone was operated as follows: avidin 50 V; TTR 80 V; ConA 10 V; ADH 100 V; BLA 30 V; CYC 30 V. The instrument settings were optimized to allow transmission of intact protein complexes and to preserve non-covalent interactions 49,60,61. The trap traveling-wave ion guide was pressurized to contain 3.3 × 10−2 mbar of argon gas. The ion trap was run in an accumulation mode and ion lifetimes in the trap prior to IM analysis range from 0–50 ms in our experiments. The traveling-wave ion mobility separator was operated at a pressure of ~ 3.5 mbar, and employed a series of DC voltage waves (40 V wave height traveling at 800–1000 m/s) to generate ion mobility separation. The ToF-MS was operated over the m/z range of 800–15000 and at a pressure of 1.6 ×10−6 mbar.
Collision Induced Unfolding and Dissociation
Collisional activation in the ion trap traveling-wave ion guide prior to the ion mobility separator was used for CIU and CID of protein complexes in order to investigate the gas-phase stability of protein ions in the presence of different salts. This work was all performed in tandem-MS (Quad selection) mode. Ions were selected in the quadrupole mass filter at an m/z corresponding to the 16+ charge state of Avidin, 20+ of ConA, 14+ of TTR, 24+ of ADH tetramers and 11+ BLA dimers. Charge states were chosen based on their intensity across each solution state interrogated, and control IM arrival time data were screened for evidence of overlapping non-tetrameric ions at the same m/z value. Each of these mass-selected ions were activated by increasing the trap collision voltage (Trap CE, as indicated in the instrument control software) which acts as a bias voltage between the quadrupole and ion trap traveling-wave ion guide to accelerate ions to increased kinetic energies for CIU and CID experiments. For all protein-salt systems investigated here, energy-dependant arrival-time distribution profiles (CIU ‘fingerprints’) were constructed using 5 V step-wise increments (2 V increments for CYC data) in the trap CE. Upper voltage limits were identified as those where no further dissociation was observed.
Data analysis
All mass spectra were calibrated externally using a solution of cesium iodide (100 mg ml−1) and were processed with Masslynx 4.1 software (Waters, UK). Spectra are shown with minimal smoothing and without background subtraction. The relative abundance of mass-selected tetrameric ions (Itet) was calculated as a percentage of the total intensity of all the signals observed in the mass spectra corresponding to either intact protein complex or its corresponding fragment ions (i.e., monomer or trimer). The relative abundance of the compact form observed for tetrameric ions separated by ion mobility (If) was calculated as a percentage of the total intensity of the peaks in the arrival time distribution observed at a selected m/z value corresponding to intact tetramer (dimer for BLA, or monomer for CYC):
The average relative standard deviation for the determination of either Itet (%) or If (%) is 2–4% (see Supporting Information for a sample calculation). The data shown in Figures 3, 4 and 5 include axes labeled in collision energy (units of eV*). The axis is a normalized version of ion kinetic energy that takes into account both the charge on the ion and reduced mass of the ion-neutral collision complex, for making comparisons across large mass ranges. Although the conversion used here is identical to center-of-mass energy conversions used elsewhere in the literature 62, we do not use this term in this report, as the definition of this quantity has clear implications for ion internal energy and these claims may not extend to the large ions studied here. We use the conversion only as a means of normalizing kinetic energies for CIU and CID comparisons across broad mass ranges.
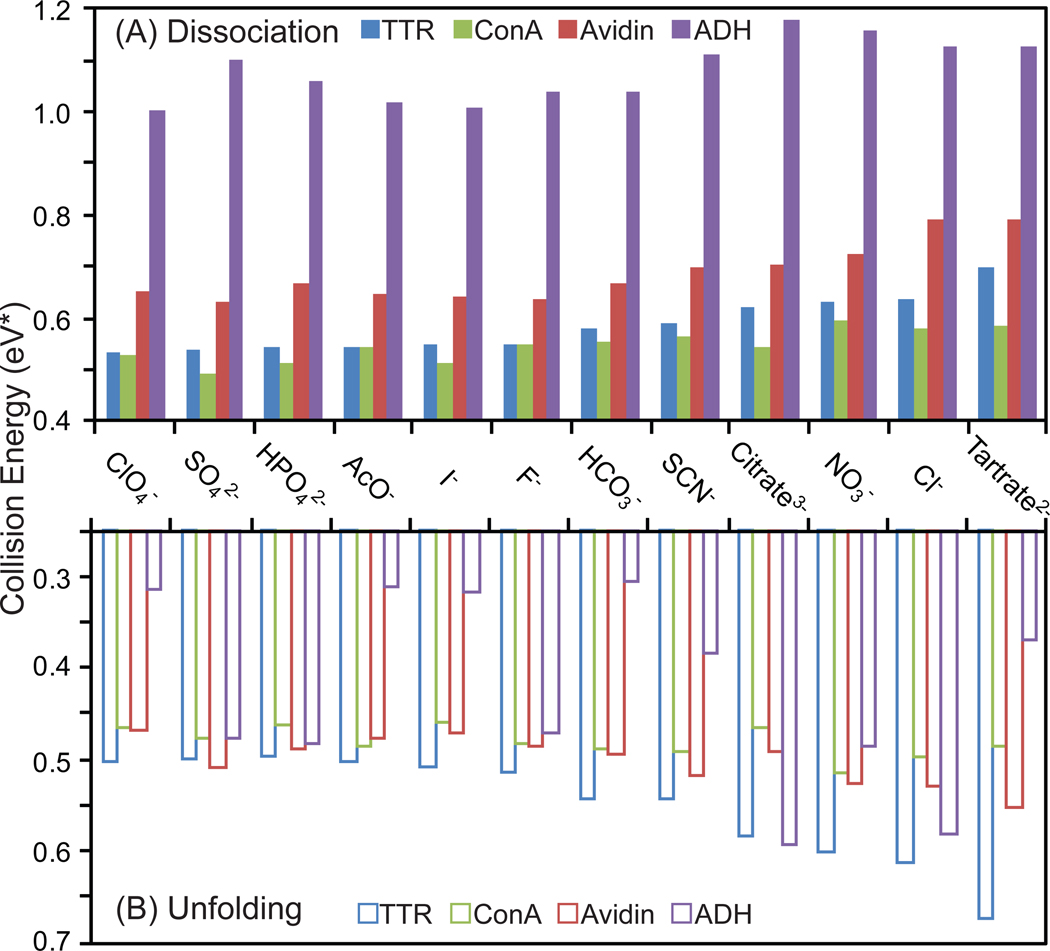
Figure 3.
Histogram plots charting collision energy (eV*) required to dissociate (A) and unfold (B) 50% of the tetrameric protein ion population for avidin (red), transthyretin (blue), concanavalin A (green), and alcohol dehydrogenase (purple) are shown for a range of anion additives. Control datasets, without added ammonium salts, are marked on the plot.
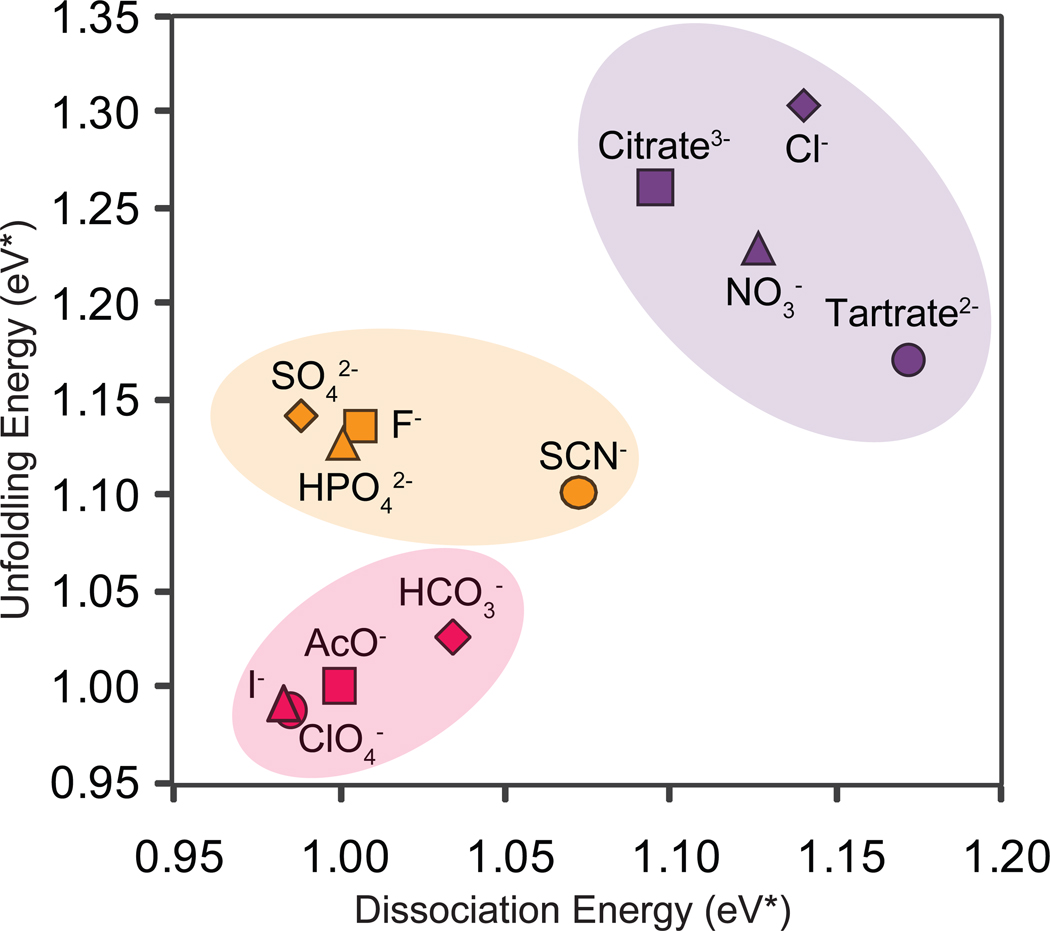
Figure 4.
A plot of collision energy (eV*) averaged over four protein complexes studied herein. The ability of anion additives to affect unfolding and dissociation of protein complexes is expressed by a collision energy axis normalized to the control dataset. The plot reveals the anions can be categorized into three distinct groups, according to their ability to stabilize protein complexes relative to the control dataset.
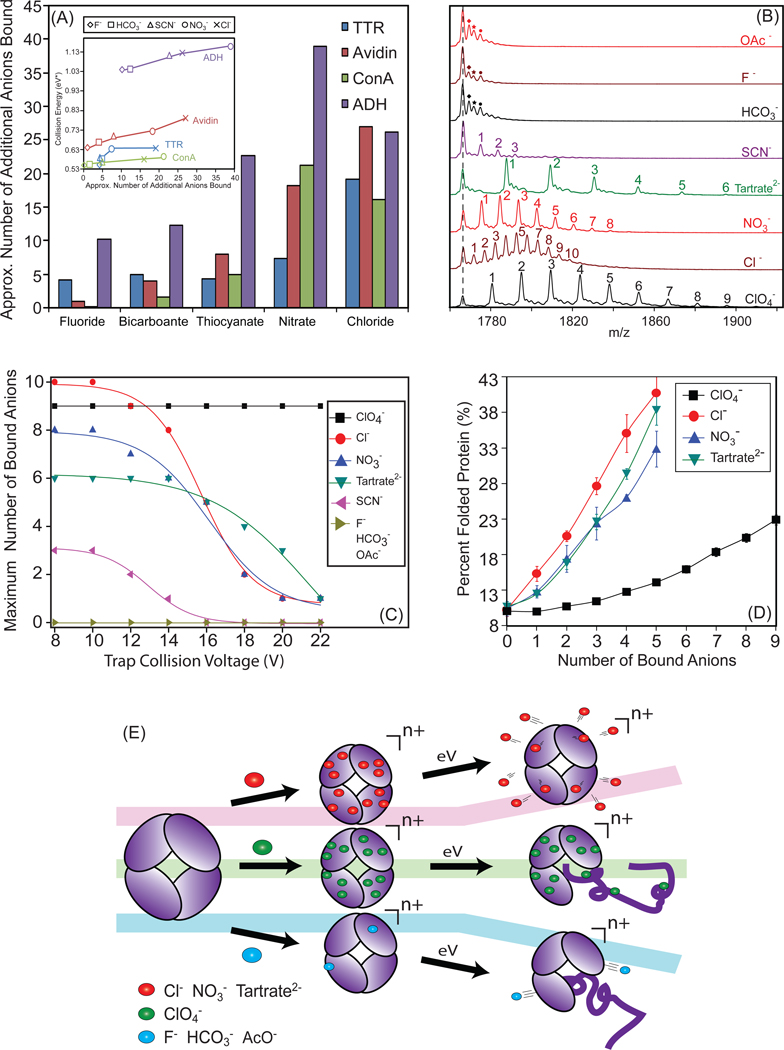
Figure 5.
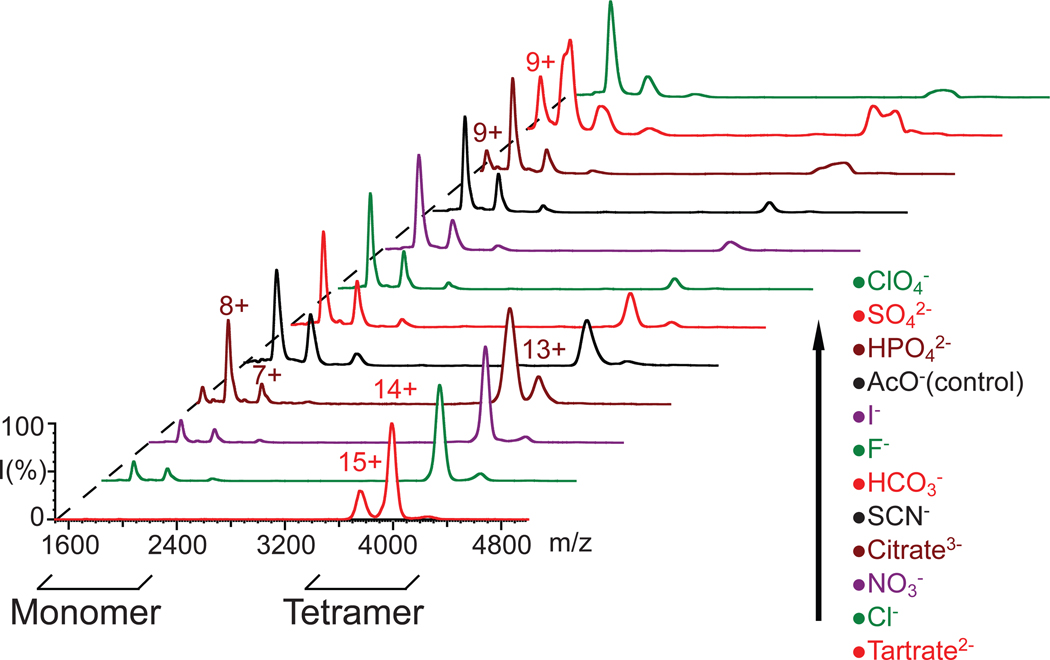
Ion mobility-mass spectrometry reveals a mechanism of protein complex stabilization through anion attachment. (A) A plot of the number of residual anions that remain bound to the protein complex at a trap collision voltage of 40 V, for transthyretin (blue), avidin (red), concanavalin A (green), and alcohol dehydrogenase (purple). The inset shown plots the normalized collision energy required to dissociate 50% of complex ions against the calculated number of additional anions bound to the assembly. A positive correlation is observed for all complexes. (B) Mass spectra of 7+ cytochrome c monomers generated from solutions containing anion additives reveal a distribution of adducts resolved by MS at trap collision voltage of 16 V. Peaks corresponding to adducts arising from sodium-binding are marked with diamonds for the control (acetate), fluoride and bicarbonate anions. (C) Plots of the largest number of additive counter-ions bound to cytochrome c observed as a function of trap collision voltage for a range of additive anions. (D) A plot of compact protein ion signal intensity (If) against the number of anion adducts bound reveals a positive correlation for four high-affinity anions, with perchlorate adducts generating a significantly more-shallow slope than other additives shown here. (E) A mechanistic diagram of protein structure stabilization through bound counter-ions that summarizes our current dataset. Three tracks are shown: a track where anions bind the protein in high affinity and are released upon dissociation leading to high protein structural stability in the gas-phase (red), a high-affinity binding track where anions do not dissociate from the protein, leading to relatively low protein structural stability (green), and a low-affinity binding track that does not produce measureable increases in protein stability in the absence of bulk solvent (blue).
RESULTS & DISCUSSION
Measuring the stability of protein complexes via collision induced unfolding and dissociation
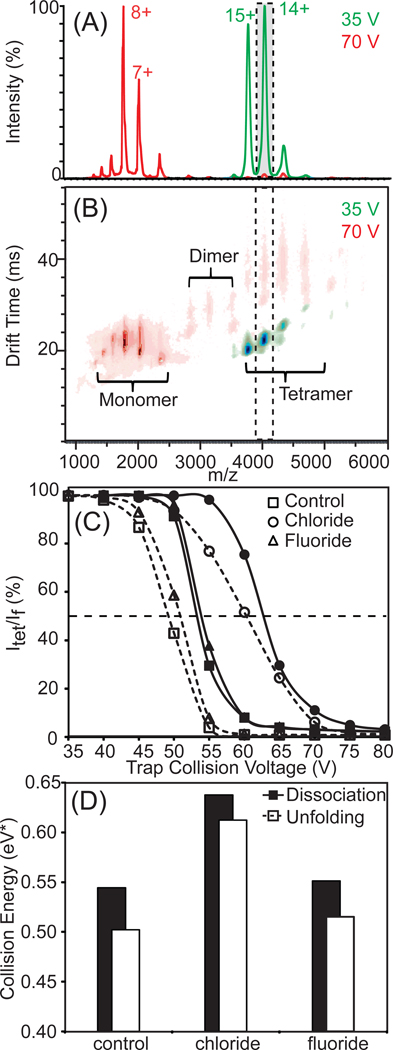
To investigate the influence of anions on the stability of protein complexes in the absence of bulk solvent, we developed a combined CIU / CID approach that allows us to assess the relative stability of proteins and complexes by monitoring their unfolding and dissociation profiles. Figure 1 illustrates this work-flow using TTR, a 55kDa tetrameric protein as an example. The mass spectra of TTR in ammonium acetate buffer (100 mM), acquired at trap collision voltage of 35 V (green), shows peaks corresponding to 13+, 14+, and 15+ charge states of the tetramer exclusively (Figure 1A). By contrast, a large fraction of the tetramer ion population dissociates into monomer and trimer at a trap collision voltage of 70 V (red). The charge state distribution of monomeric TTR spans from 6+ to 11+, with 8+ (1732 m/z) and 7+ (1980 m/z) as the two most intense monomer ion signals.
Figure 1.
Addition of anions in solution alters the dissociation and unfolding profiles of a protein assembly. (A) Mass spectra of TTR acquired at instrument conditions that preserve the tetrameric assembly (trap collision voltage: 35 V, blue), or that activate the protein ion (70 V, red). Peaks corresponding to 13–15+ charge states of tetramer and 6–10+ charge states of monomer are shown. (B) Contour plots of m/z versus drift time acquired at a trap collision voltage of 35 V (green) and 70 V (red). A narrow window that contains the peak corresponding to the 14+ ion of tetrameric TTR indicates the populations of compact and extended tetrameric TTR generated under different instrument conditions (dashed box). (C) Plots of the relative intensities of TTR tetramer 14+ ions (Itet, solid lines), and the relative intensities of compact TTR tetramer 14+ ions (If, dashed lines) are shown as a function of trap collision voltage. TTR ion was generated with solutions containing either chloride (circle), fluoride (triangle), or acetate anion (control, square). The energy at which the relative intensity reduces to 50 % is marked with a horizontal dashed line. (D) A histogram showing the 50% dissociation yield (black) and unfolding yield (white) for TTR tetramers generated from solutions with various additives is shown.
Drift times for the ions were acquired under the same conditions as above and are shown in a plot of drift time versus m/z (Figure 1B). At a collision voltage of 35 V (green), the plot shows a number of peaks resolved in the drift time dimension corresponding to the charge state series for TTR, with 3980 m/z assigned to the 14+ charge state of tetramer (dashed box). At a trap collision voltage of 70 V (red) the majority of the tetrameric ion current is converted into monomer. The peaks corresponding to the tetramer, however, extend to longer drift times, indicating that the remaining tetramers exist in a range of structural states at elevated internal energies. These data are consistent with previous reports where TTR was observed to occupy a number of partially folded intermediate states that are stable on the millisecond timescale 44,63. Previous results have also indicated that the stability of TTR can be enhanced through specific binding of small molecule ligands43.
For quantitative measurements of TTR stability, the trap collision voltage at which ions undergo CIU and CID was monitored, and plots of collision voltage versus the intensity observed for compact (If) and intact (Itet) tetramer ions are recorded (Figure 1C). Based on these plots, a simplified descriptor of tetramer stability is constructed by plotting the normalized collision energy (eV*) at which the intact/compact tetramers (Itet / If respectively) decrease to 50% of their initial values (Figure 1D). For example, a plot of If and Itet for the 14+ charge state of TTR incubated in three different buffer compositions (control/100% 100 mM ammonium acetate, 100 mM ammonium acetate with 5 mM added ammonium fluoride and 100 mM ammonium acetate with 5 mM added ammonium chloride) as a function of trap collision voltage is shown in Figure 1C. Both If and Itet are shown to decrease as the collision voltage is increased, and If decreases at lower voltages than Itet for ions generated from all three buffer compositions. These results indicate that protein complexes dissociate only after tetramer precursor ions have undergone significant unfolding in the absence of the bulk solvent. Importantly, when comparing a plot of If (open, dashed) and Itet (closed, solid), we observe that the addition of ammonium chloride to the sample solution (circles) increases the voltage values at which If and Itet are observed to decrease, and that this change is significantly greater than that observed for the addition of ammonium fluoride (triangles), relative to the measurements made in 100% ammonium acetate based control experiments (squares).
This result is clearly illustrated through a comparison of the normalized collision energy at which If and Itet decrease to 50% of their original intensity (Figure 1D). This result implies that the presence of added chloride anions in solution causes the stability of TTR to increase relative to control, while such effects are negligible when fluoride anions are added in solution. Thus, the collective approach described in Figure 1 provides us with a basis for quantitatively comparing the stability of protein complexes in terms of both their unfolding (CIU) and dissociation (CID) profiles.
Ion mobility-mass spectrometry reveals the differential stability of gas-phase protein complexes
In order to assess the influence of anions on the stability of protein complexes in general, we examined a range of anions whose relative ability to stabilize proteins in solution is recognized. Figure 2 shows a series of tandem mass spectra of the 14+ charge state of TTR acquired at a trap collision voltage of 60 V. For each spectrum, all instrument parameters were kept constant; only the composition of the buffer additives was altered and their effect on protein complex dissociation (Itet) monitored. We find that the peaks corresponding to tetramer and monomer are produced in substantially different abundances, clearly demonstrating the influence of anion identity on CID yields. Of the anions examined here, the addition of tartrate in solution confers the greatest stability to TTR, as evidenced by a lack of peaks corresponding to monomer at low m/z in the mass spectrum (Figure 2). The MS spectra also reveal the appearance of a minor peak corresponding to the 15+ charge state of TTR due to the loss of negatively charged counter ions from the 14+ charge state 64, and 13+ tetramer ion signals corresponding to positive charge stripping 65. Note that such signals are prominent even in the mass spectrum acquired from the control, and are likely enhanced by the long life-times of tetramer ions in the ion trap region of the IM-MS instrumentation used in our experiments. While the charge state distribution of the monomer ions produced from all tetramer ions remains relatively constant, we note that a peak corresponding to the 9+ monomer is observed primarily when relatively destabilizing salts, such as hydrogen phosphate and sulfate, are added. The tetramer dissociation yield, measured from the data shown in Figure 2, increases in the following rank order: tartrate, chloride, nitrate, citrate, thiocyanate, bicarbonate, fluoride, iodide, acetate (control), hydrogen phosphate, sulfate and finally perchlorate. The difference in the stability conferred to the tetrameric protein ions can be quantified by comparing the signal intensity of monomer ions relative to tetramer, as previously described.
Figure 2.
The mass spectra of TTR incubated with a range of anions reveals different extents of dissociation. The 14+ charge state of TTR ions selected by the quadrupole mass filter was subjected to a trap collision voltage of 60 V in the trapping region between the quadrupole mass analyzer and ion mobility separator. Major peaks from the charge state series corresponding to monomeric and tetrameric TTR are labeled. The anions are arranged by their ability to limit dissociation of TTR.
To further investigate these effects, we determined the stability afforded to three additional tetrameric protein complexes (avidin, ConA and ADH) in the gas-phase upon addition of the same 12 anions in solution prior to ion desolvation. Each protein/anion pairing was assessed by examining a single charge state isolated in the quadrupole mass filter, and this same charge state (one for each complex) was assessed across all solution compositions in our CIU and CID experiments. Both If and Itet follow typical sigmoid-type decay curves as a function of the trap collision voltage for all systems studied 43. Figure 3A shows a histogram plot of the energy necessary to reduce Itet to 50% of its original value as a function of anion identity for each of the four protein complexes studied here. The plot reveals a surprisingly wide range of values for the energy necessary to reduce tetramer intensity by 50%, and a comparison among the datasets shown here reveals several features regarding the stability of multiprotein complexes. We first note that, in general, several anions added in solution have a universally stabilizing influence on Itet. These include tartrate, chloride, nitrate, citrate, and thiocyanate. Conversely anions such as perchlorate and iodide have a destabilizing influence on protein complexes relative to control (ammonium acetate). In addition, we note a significant difference in the energy required to dissociate the four different protein complexes studied here. The stability order of protein complexes relative to CID in our dataset indicates that the ADH tetramer is the most stable, followed by avidin, TTR, and finally ConA, (Figure 3A). In a similar manner, Figure 3B tracks the changes in If through CIU experiments for each of the same 48 anion/protein pairs shown in Figure 3A. In contrast to the dissociation data, TTR is the most stable relative to unfolding, followed by avidin, Con A, and ADH is the most prone to unfolding by CIU measurements. Counter-ions can be grouped similarly using CIU data as they are in the CID data above. The stabilizing anions identified relative to the unfolding transition probed in Figure 3B are identical to those identified in Figure 3A (e.g. tartrate, chloride and nitrate). On the other hand, anions that act as destabilizing agents for protein structure in the gas-phase are less clearly distinguished from stabilizers by CIU measurements when compared with CID data, suggesting that bound agents can act to decouple the two processes as observed previously 43.
Our CIU and CID datasets reveal a number of general points regarding protein complex stability in the gas phase. Firstly, that different bound counter-ions influence protein stability differentially. For example, the increases in CID stability relative to control experiments range from a 10% increase for ConA to a 30% percent increase for TTR. CIU data shows a substantially greater amount and range of stabilization, spanning between a 13% increase in stability for ConA and a 90% increase for ADH. In general, the protein complexes studied here are observed to undergo CIU at lower energies relative to CID, as reported previously 43,44. For each protein complex, however, the two processes are energetically separated to different degrees, from 0.7 eV* separating the two transitions in ADH to 0.03 eV* for TTR. Such wide variations in the collision energies between CIU and CID processes across the protein tetramers studied here highlights the importance of assessing the stability of protein complexes in terms of both unfolding and dissociation, especially for IM-MS experiments where proteins and complexes are activated in order to obtain higher mass measurement accuracy and resolving power 66.
A comparison of the CID and CIU data also highlights that anions added in solution modulate protein stability to different extents for different protein complexes For example, although measurable differences in the energy required to deplete 50% of the tetramer population and 50% of the folded population is observed for most TTR CIU and CID datasets, unfolding and dissociation are isoenergetic processes when TTR is bound to highly stabilizing anion populations (tartrate, chloride, or nitrate). In contrast, ADH is observed to undergo CIU at drastically lower energies relative to all other protein complexes interrogated here, and bound anions seem to influence this process in a somewhat different manner when compared to the other three complexes shown in Figure 3.
Classifying and ranking the influence of anions on protein stability
In order to develop a classification system that allows us to generally rank anions in terms of the stability afforded to gas-phase protein complexes through their addition in solution, we first normalized our stability measurements to our control experiments and then combined our CIU and CID datasets to derive a consensus measure of gas-phase protein complex stability. This data is shown in Figure 4, and reveals the presence of at least three general levels of protein ion stabilization in our CID and CIU datasets. Remarkably, despite the differences apparent in Figure 3, both CIU and CID datasets generally correlate well. As noted previously, many anions provide little or no stability to protein complex ions upon addition in solution, resulting in dissociation and unfolding threshold values similar to control experiments in pure ammonium acetate buffer systems. These anions cluster together within the same region of Figure 4 (light pink). The two other clusters of anions shown (orange and purple) exhibit enhanced stability relative to control (light pink) in both datasets. For example, tartrate, chloride, citrate, and nitrate are among the most efficient stabilizers of protein unfolding, as well as increasing the energy required for protein dissociation. Thus, these anions populate a ‘highly-stabilizing’ cluster shown in purple within Figure 4. All of the remaining anions exhibit mid-range values for CID and CIU stabilization when bound to protein complexes in the gas-phase, and form a final ‘medium-stabilizing’ cluster of counter-ions (orange, Figure 4). Overall, these results suggest that anions influence the unfolding and dissociation processes of protein complexes in concert, rather than independently. It is worth noting that there are several exceptions to this general statement within our dataset, where unfolding or dissociation is stabilized preferentially relative to the other. For example, tartrate and sulfate anions show significantly different rank orders when unfolding and dissociation are considered separately, indicating that these salts may influence the tertiary and quaternary structure of protein complex differentially.
Towards a mechanistic understanding of gas-phase protein stabilization through bound anions
Insight into the mechanism by which protein complexes are stabilized through the addition of salt in buffer solutions prior to nESI-IM-MS analysis can be obtained by observing the intact mass of protein complex ions generated as a function of buffer composition. Although resolved adduct populations cannot be resolved by MS for these large complexes, it is reasonable to assume that the excess mass, relative to the ammonium acetate control, arises from binding of additional anions added in solution and this excess mass can be converted to an estimated average number of additional anions bound in our tetramer data. This is shown in Figure 5A, where we measured the mass of the protein complexes incubated with several salt additives under identical instrument conditions (Trap CE = 40V). Such an analysis is limited to those cases where the protein ions produced from solutions containing salt additives generate MS data having resolved MS features under identical conditions. While this limits the panel of anions that can be tested significantly from our original pool of 12, the trend observed in this data is clear. As the excess mass relative to the protein sequence alone increases, the structural stability afforded the assembly also increases. This suggests that the amount of bound counter-ions carried with the complex from solution or the nESI process is the determining factor in the stability enhancements observed in our CID and CIU datasets. The inset shown in Figure 5A indicates a clear positive correlation between the amount of excess mass observed bound to protein complex ions in our gas-phase measurements and the structural stability of those ions relative to both dissociation and unfolding, further indicating a role for bound anions in the stability enhancements observed here.
In order to test the hypothesis that bound buffer material is the main cause of the stability enhancement observed for gas-phase protein complexes in our data, we broadened our initial dataset to include smaller protein systems, including both CYC monomer and BLA dimer datasets (Figure S2). Example data from the 7+ charge state of CYC monomer (12 kDa) is shown in Figure 5B, where the ions are generated from solutions with additives which are identical to those used to generate the protein complex datasets shown in this report. The data clearly demonstrate that the increase in mass observed in Figure 5A is due to anion binding and not other solution components. Resolved adduct populations corresponding to [H-‘anion’] type adducts where the ‘anion’ is chloride, nitrate, tartrate, perchlorate, or thiocyanate, are all observed in our monomer dataset. The maximum number of adducts bound to the CYC monomer (5–10 for high-affinity counter-ions) scales well with respect to our tetramer data shown in Figure 5A, where the excess mass recorded for protein tetramers corresponds to 5–10 adducts per monomer in the cases of those anion additives with high apparent protein affinity. Critically, despite the significant structural differences between the protein complexes measured in Figure 5A and the monomeric CYC, the observed relative protein binding affinity of the anions remains similar.
These results also agree well with previous data 67,68 where counter-ion adduction, principally to basic sites on the surface of the protein, is observed by ESI-MS from solutions containing added salts while changes to the ionic strength were not observed to alter the charge state distribution of the protein ions significantly 69. These previous results also cite a strong correlation between the number of protein-bound counter-ions and the relative gas-phase acidity of the anions studied 67,68. Our data corroborates these general findings for a wider panel of anions and proteins than reported previously (Figure S3, Table S1). We note that all additives that do not lead to appreciable adduct formation correlate well with those anions identified in Figures 3 and 4 as having little influence on protein stability; a list that includes: acetate, bicarbonate, and fluoride salts. Conversely, many of the anions observed to bind in large numbers to CYC correlate well with those that confer significant stability to protein complex structure in the absence of bulk solvent (e.g. tartrate, chloride, and nitrate).
Although the data in Figure 5B clearly shows a correlation between protein-anion binding, either in solution or during the nESI process, and the stability enhancements observed in our CIU and CID datasets for protein complexes, a limited number of salt additives appear to follow a different trend. For example, perchlorate anions bind in large numbers to CYC (Figure 5B), but the resultant gain in stability for the gas-phase protein ion is minimal. To elucidate the difference between the action of anions like perchlorate and other anions that lead both to large numbers of adducts and an appreciable increase in protein stability, we recorded MS data for CYC-anion complexes as a function of activation in the ion trap region of the instrument (prior to IM separation). Most anions that bind with high affinity, including chloride, nitrate, and tartrate, readily dissociate from the protein as the activation voltage is increased (Figure 5C). In contrast, perchlorate-based adducts do not readily dissociate from the protein over the range of voltages shown in Figure 5C (black squares). This data, then, suggests a second condition for the effective stabilization of gas-phase protein ions through counter-ion attachment. Anions must bind with high-affinity, but must also dissociate readily from the protein in order to be an effective stabilizer. This balance between protein-anion binding in solution and gas-phase dissociation is reflected in the relative acidities, both in solution and in the gas-phase, for those anions that effectively stabilize protein structure in the absence of bulk solvent. For singly-charged anions, where protein-binding is simplified, there is a strong correlation between those anions that stabilize protein structure and a narrow range of acceptable acidity values (Figure S3).
The correlation between the ability of adducts to dissociate from the complex and gas-phase structural stability enhancements are further corroborated by adduct-resolved CIU experiments performed for CYC-anion complexes (Figure 5D). The data shown for stabilizing counter-ions (chloride, nitrate, and tartrate) display positive correlations between the relative amounts of compact protein observed and the number of counter-ions bound, while a significantly shallower trend is observed for perchlorate-bound CYC. These data indicate that while perchlorate binds in larger numbers to proteins and their complexes when compared to other anion additives, the bound adducts do not preserve the protein in a compact configuration, as indicated by the low percentage of folded protein recorded as a function of the number of perchlorate adducts bound. In contrast, tartrate, chloride and nitrate-bound protein ions all retain a significantly greater folded percentage per-unit adducted anion. The trend-lines observed for all three stabilizing counter-ions are relatively similar, having similar slopes and intercept values, further indicating a common mechanism of protein stabilization.
We have summarized the observations discussed above into a schematic diagram (Figure 5E) that illustrates our current understanding of the molecular mechanism of protein complex ion structural stabilization through counter-ion attachment and binding. Our mechanism delineates and classifies anions into three categories based on both their protein binding affinity and their ability to dissociate from proteins and their complexes following activation in the gas-phase. Counter-ions that exhibit a strong stabilizing influence on protein structure (red track, Figure 5E) both bind with high affinity and readily dissociate from the protein surface. The observed dissociation of anion-based adducts as neutrals appears to be the key metric that allows protein-adduct ion populations to retain compact, native like gas-phase conformations under conditions where protein ions produced from solutions containing more-volatile additives unfold and dissociate. It is reasonable to assume that the dissociation of adducts acts to carry away excess rotational and vibrational energy from gas-phase protein ions, thus abating any dramatic internal energy increases for the protein and allowing it to retain in a compact, native-like structure. A second class of anions bind to the protein, but are not easily dissociated from the complex in the gas-phase upon activation (green track, Figure 5E). Typically comprised of anions that exhibit greater proton affinities than those highly-stabilizing anions described in the red track, this class of anions demonstrates a poor ability to stabilize the gas-phase structure of protein complex ions. Similarly, those anions that do not bind with high-affinity to proteins in solution, or during the nESI process, also do not stabilize gas-phase protein structure (blue track, Figure 5E). It is important to point out that these three anion classes, while related, are not the same as those presented in Figure 4. Anions that populate the blue and green tracks in Figure 5E are likely to represent the weak stabilizers observed in Figure 4 (light pink), whereas the mild and strong stabilizers observed in that data (orange and purple) can all be placed on the red track in Figure 5E to various degrees. As noted above, there is a strong correlation between anion acidity, both in the gas-phase and in solution, and the three classes of stabilizing anions described in Figure 5E. This correlation is readily apparent when singly-charged anions are considered (Figure S3, Table S1), where anions with low acidity fall into the blue track, those with intermediate acidity in the red track, and those with the highest acidity in the green track. Multiply-charged anions are more difficult to classify due to likely multi-dentate interactions with basic sites on the protein surface. Anion additives that stabilize protein structure in the absence of solvent should, therefore, possess sufficient affinity for basic sites on the protein sufrace to drive initial binding, but also have a low enough binding strength to allow for adduct dissociation and effective ‘evaporative cooling’ of the protein complex in the gas-phase.
CONCLUSIONS
In general, there are four main conclusions from our work. First, our dataset has greatly expanded the known buffer conditions and additives that are amenable to nESI of protein complexes without complete suppression of usable ion signal. All data shown here included salts at 1–10 mM, however higher concentrations are possible for many of the salts discussed, in some cases extending to ~1 M concentrations for anions having weak protein binding affinity. While lower salt concentrations are preferred for maximum mass accuracy and resolution, our data indicate that higher salt concentrations and tailored salt identity provide maximally-stabilized protein complexes for IM measurements of protein structure.
Secondly, we have ranked and classified twelve anions, for the first time, according to their effectiveness in stabilizing gas-phase protein structure, and rank orders from both CIU and CID experiments are surprisingly similar. This similarity indicates that the interactions between anion and complex are linked to both local protein structure and protein-protein interactions. We were surprised to find that some salts previously reported to be gas-phase protein denaturants, are highly-stabilizing to protein ion structure in our dataset. For example, tartrate-based salts have been used in ‘top-down’ protein fragmentation experiments to increase fragment ion yield 70. While our data suggests that tartrate be classified as a stabilizing salt in most cases, the overall mechanism of gas-phase protein stabilization may involve partially unfolded forms (Figure S4). Clearly, our data indicates that the influence of tartrate and other salts on ‘top-down’ fragmentation efficiency via electron and collision-based activation methods requires further study.
Thirdly, we have elucidated a complete mechanistic picture of protein structure stabilization in the gas-phase achieved through nESI buffer additives. The data shown in Figure 5 clearly indicates that protein stabilization in the gas-phase is correlated with the binding affinity between the anions and proteins studied here. In order to stabilize gas-phase protein structure, anions must bind to the protein, be carried into the gas-phase as protein-anion complexes, and dissociate from the protein upon activation. The final dissociation step acts to siphon excess energy from the protein system and preserve compact gas-phase structures. Anion additives that do not bind, nor dissociate from gas-phase protein ions do not provide significant structural stabilization in CID and CIU datasets. In addition to the tetramer and protein monomer data shown here, further data collected for the BLA dimer also correlates well with our overall mechanism (Figure S2). Small differences in enhanced structural stability can be observed among those counter-ions that provide the greatest stability, and evidence from CIU fingerprinting, where changes in IM drift time are plotted for selected ions against activation voltage, suggest that some anions remain bound to the complex and stabilize partially unfolded structures of the complex even for strong gas-phase protein stabilizers like chloride, tartrate, and nitrate (Figure S4). Overall, the results presented herein enable the direct manipulation of gas-phase protein complex stability by controlling the composition of the nESI sample solution. Therefore, this dataset will likely enable IM-MS by making a greater number of structurally unstable systems amenable for study in the absence of bulk solvent.
The data shown in this report prompts a comparison between the well-known Hofmeister series of anions, describing their influence upon protein stability in solution: SO42− > HPO4− > F− > Acetate− > Citrate3− > HCO3− > Cl− > NO3− > I− > ClO4− > SCN−. While still an active area of research, much is currently known about the molecular mechanism surrounding Hofmeister-type stabilization of solution-phase protein structures. In general, the Hofmeister series depends upon anion hydration entropy, the ability of anions to alter water surface tension, and direct anion-protein binding 71. On first inspection, drastic differences are apparent between protein stabilizing anions in solution and in the gas-phase. For example, nitrate, thiocyanate and chloride are all strong protein stabilizers in the gas-phase, but are relative denaturants in bulk solvent. Our data present a direct measurement of anion-protein binding, as the relative amounts of counter-ions bound in our data correlate well with the relative acidity of those same anions in solution (for singly charged ions). The stability enhancements observed in our data can be viewed as built entirely upon protein-anion interactions. Differences between the two rank orders can be ascribed to the lack of solvation effects in the dataset presented in this report, and highlight the importance of such affects for Hofmiester stabilization of proteins in solution.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Alex Pagliaro for collecting some proof-of-principle data on this project. This work is supported by the National Institutes of Health (1-R01-GM-095832-01 and 1-R01-CA-154455-01) and by University of Michigan startup funds.
Footnotes
SUPPORTING INFORMATION
Concentration dependant stabilization data (S1), Anion-bound protein stability measurements for BLA dimer (S2), Graph correlating acidity values in the gas-phase and in solution for singly charged anions (S3), CIU fingerprinting data showing evidence for stabilizing anions that stay bound to proteins following dissociation (S4), tabular data for gas-phase acidity and ionic radii (Table S1), tabular data for protein stability and structure (Table S2), and example standard deviation calculations from our dataset are included.
REFERENCES
- 1.Burley SK, Almo SC, Bonanno JB, Capel M, Chance MR, Gaasterland T, Lin DW, Sali A, Studier FW, Swaminathan S. Nature Genet. 1999;23:151. doi: 10.1038/13783. [DOI] [PubMed] [Google Scholar]
- 2.Chandonia JM, Brenner SE. Science. 2006;311:347. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 3.Robinson CV, Sali A, Baumeister W. Nature. 2007;450:973. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- 4.Alber F, Dokudovskaya S, Veenhoff LM, Zhang WZ, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Rout MP, Sali A. Nature. 2007;450:683. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 5.Alber F, Dokudovskaya S, Veenhoff LM, Zhang WH, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. Nature. 2007;450:695. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 6.Taverner T, Hernandez H, Sharon M, Ruotolo BT, Matak-Vinkovic D, Devos D, Russell RB, Robinson CV. Accounts Chem Res. 2008;41:617. doi: 10.1021/ar700218q. [DOI] [PubMed] [Google Scholar]
- 7.Heck AJR. Nat. Methods. 2008;5:927. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 8.Alber F, Forster F, Korkin D, Topf M, Sali A. Annu. Rev. Biochem. 2008;77:443. doi: 10.1146/annurev.biochem.77.060407.135530. [DOI] [PubMed] [Google Scholar]
- 9.Foerster F, Lasker K, Nickell S, Sali A, Baumeister W. Mol. Cell. Proteomics. 2010;9:1666. doi: 10.1074/mcp.R000002-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lasker K, Phillips JL, Russel D, Velazquez-Muriel J, Schneidman-Duhovny D, Tjioe E, Webb B, Schlessinger A, Sali A. Mol. Cell. Proteomics. 2010;9:1689. doi: 10.1074/mcp.R110.000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lasker K, Sali A, Wolfson HJ. Proteins. 2010;78:3205. doi: 10.1002/prot.22845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benesch JLP, Ruotolo BT, Simmons DA, Robinson CV. Chem Rev. 2007;107:3544. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- 13.Sharon M, Robinson CV. Annu. Rev. Biochem. 2007;76:167. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez H, Dziembowski A, Taverner T, Seraphin B, Robinson CV. EMBO Rep. 2006;7:605. doi: 10.1038/sj.embor.7400702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Synowsky SA, van den Heuvel RHH, Mohammed S, Pijnappel W, Heck AJR. Mol. Cell. Proteomics. 2006;5:1581. doi: 10.1074/mcp.M600043-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Zhou M, Sandercock AM, Fraser CS, Ridlova G, Stephens E, Schenauer MR, Yokoi-Fong T, Barsky D, Leary JA, Hershey JW, Doudna JA, Robinson CV. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18139. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharon M, Mao HB, Erba EB, Stephens E, Zheng N, Robinson CV. Structure. 2009;17:31. doi: 10.1016/j.str.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Koschubs T, Lorenzen K, Baumli S, Sandstrom S, Heck AJR, Cramer P. Nucleic Acids Res. 2010;38:3186. doi: 10.1093/nar/gkq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stengel F, Baldwin AJ, Painter AJ, Jaya N, Basha E, Kay LE, Vierling E, Robinson CV, Benesch JLP. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2007. doi: 10.1073/pnas.0910126107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shoemaker GK, van Duijn E, Crawford SE, Uetrecht C, Baclayon M, Roos WH, Wuite GJL, Estes MK, Prasad BVV, Heck AJR. Mol. Cell. Proteomics. 2010;9:1742. doi: 10.1074/mcp.M900620-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Science. 2008;321:243. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 22.Barrera NP, Isaacson SC, Zhou M, Bavro VN, Welch A, Schaedler TA, Seeger MA, Miguel RN, Korkhov VM, van Veen HW, Venter H, Walmsley AR, Tate CG, Robinson CV. Nat. Methods. 2009;6:585. doi: 10.1038/nmeth.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loo JA, Berhane B, Kaddis CS, Wooding KM, Xie YM, Kaufman SL, Chernushevich IV. J Am Soc Mass Spectr. 2005;16:998. doi: 10.1016/j.jasms.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 24.Bernstein SL, Wyttenbach T, Baumketner A, Shea JE, Bitan G, Teplow DB, Bowers MT. J. Am. Chem. Soc. 2005;127:2075. doi: 10.1021/ja044531p. [DOI] [PubMed] [Google Scholar]
- 25.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Science. 2005;310:1658. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 26.Ruotolo BT, Robinson CV. Curr Opin Chem Biol. 2006;10:402. doi: 10.1016/j.cbpa.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 27.Kemper PR, Bowers MT. J. Phys. Chem. 1991;95:5134. [Google Scholar]
- 28.Bowers MT, Kemper PR, Vonhelden G, Vankoppen PAM. Science. 1993;260:1446. doi: 10.1126/science.260.5113.1446. [DOI] [PubMed] [Google Scholar]
- 29.Jarrold MF. J. Phys. Chem. 1995;99:11. [Google Scholar]
- 30.Eiceman GA. Crit. Rev. Anal. Chem. 1991;22:17. [Google Scholar]
- 31.Baumbach JI, Eiceman GA. Appl. Spectrosc. 1999;53:338A. doi: 10.1366/0003702991947847. [DOI] [PubMed] [Google Scholar]
- 32.Vonhelden G, Wyttenbach T, Bowers MT. Science. 1995;267:1483. doi: 10.1126/science.267.5203.1483. [DOI] [PubMed] [Google Scholar]
- 33.Clemmer DE, Jarrold MF. J Mass Spectrom. 1997;32:577. [Google Scholar]
- 34.Valentine SJ, Liu XY, Plasencia MD, Hilderbrand AE, Kurulugama RT, Koeniger SL, Clemmer DE. Expert Rev. Proteomics. 2005;2:553. doi: 10.1586/14789450.2.4.553. [DOI] [PubMed] [Google Scholar]
- 35.McLean JA, Ruotolo BT, Gillig KJ, Russell DH. Int. J. Mass Spectrom. 2005;240:301. [Google Scholar]
- 36.Bohrer BC, Mererbloom SI, Koeniger SL, Hilderbrand AE, Clemmer DE. Annu. Rev. Anal. Chem. 2008;1:293. doi: 10.1146/annurev.anchem.1.031207.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH. J Mass Spectrom. 2008;43:1. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- 38.Tao L, Dahl DB, Perez LM, Russell DH. J Am Soc Mass Spectr. 2009;20:1593. doi: 10.1016/j.jasms.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 39.Pukala TL, Ruotolo BT, Zhou M, Politis A, Stefanescu R, Leary JA, Robinson CV. Structure. 2009;17:1235. doi: 10.1016/j.str.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 40.Politis A, Park AY, Hyung SJ, Barsky D, Ruotolo BT, Robinson CV. Plos One. 2010;5 doi: 10.1371/journal.pone.0012080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorenzen K, Olia AS, Uetrecht C, Cingolani G, Heck AJR. J. Mol. Biol. 2008;379:385. doi: 10.1016/j.jmb.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Duijn E, Barendregt A, Synowsky S, Versluis C, Heck AJR. J. Am. Chem. Soc. 2009;131:1452. doi: 10.1021/ja8055134. [DOI] [PubMed] [Google Scholar]
- 43.Hyung SJ, Robinson CV, Ruotolo BT. Chem. Biol. 2009;16:382. doi: 10.1016/j.chembiol.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Ruotolo BT, Hyung SJ, Robinson PM, Giles K, Bateman RH, Robinson CV. Angew Chem Int Edit. 2007;46:8001. doi: 10.1002/anie.200702161. [DOI] [PubMed] [Google Scholar]
- 45.Benesch JLP. J Am Soc Mass Spectr. 2009;20:341. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 46.Felitsyn N, Kitova EN, Klassen JS. Anal. Chem. 2001;73:4647. doi: 10.1021/ac0103975. [DOI] [PubMed] [Google Scholar]
- 47.Jurchen JC, Williams ER. J. Am. Chem. Soc. 2003;125:2817. doi: 10.1021/ja0211508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jurneczko E, Barran PE. Analyst. 2011;136:20. doi: 10.1039/c0an00373e. [DOI] [PubMed] [Google Scholar]
- 49.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung SJ, Robinson CV. Nat. Protoc. 2008;3:1139. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 50.Oomens J, Sartakov BG, Meijer G, Von Helden G. Int. J. Mass Spectrom. 2006;254:1. [Google Scholar]
- 51.Green MK, Lebrilla CB. Mass Spectrom. Rev. 1997;16:53. doi: 10.1002/(SICI)1098-2787(1997)16:2<53::AID-MAS1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 52.Hofmeister F. Archiv Fur Experimentelle Pathologie Und Pharmakologie. 1908:273. [Google Scholar]
- 53.Baldwin RL. Biophys J. 1996;71:2056. doi: 10.1016/S0006-3495(96)79404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang YJ, Cremer PS. Curr Opin Chem Biol. 2006;10:658. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Cremer PS. Annu Rev Phys Chem. 2010;61:63. doi: 10.1146/annurev.physchem.59.032607.093635. [DOI] [PubMed] [Google Scholar]
- 56.Freeke J, Robinson CV, Ruotolo BT. Int. J. Mass Spectrom. 2010;298:91. [Google Scholar]
- 57.Annesley TM. Clin. Chem. 2003;49:1041. doi: 10.1373/49.7.1041. [DOI] [PubMed] [Google Scholar]
- 58.Zhong Y, Hyung SJ, Ruotolo BT. Analyst. 2011 doi: 10.1039/c0an00987c. [DOI] [PubMed] [Google Scholar]
- 59.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. Int. J. Mass Spectrom. 2007;261:1. [Google Scholar]
- 60.Hernandez H, Robinson CV. Nat. Protoc. 2007;2:715. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 61.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Anal. Chem. 2010;82:9557. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 62.Shukla AK, Futrell JH. J Mass Spectrom. 2000;35:1069. doi: 10.1002/1096-9888(200009)35:9<1069::AID-JMS54>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 63.Pagel K, Hyung SJ, Ruotolo BT, Robinson CV. Anal Chem. 2010;82:5363. doi: 10.1021/ac101121r. [DOI] [PubMed] [Google Scholar]
- 64.Sobott F, McCammon MG, Robinson CV. Int. J. Mass Spectrom. 2003;230:193. [Google Scholar]
- 65.Jurchen JC, Williams ER. J Am Chem Soc. 2003;125:2817. doi: 10.1021/ja0211508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKay AR, Ruotolo BT, Ilag LL, Robinson CV. J. Am. Chem. Soc. 2006;128:11433. doi: 10.1021/ja061468q. [DOI] [PubMed] [Google Scholar]
- 67.Verkerk UH, Kebarle P. J Am Soc Mass Spectr. 2005;16:1325. doi: 10.1016/j.jasms.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 68.Flick TG, Merenbloom SI, Williams ER. Anal. Chem. 2011;83:2210. doi: 10.1021/ac1031012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang GD, Cole RB. Anal. Chem. 1994;66:3702. [Google Scholar]
- 70.Han XM, Jin M, Breuker K, McLafferty FW. Science. 2006;314:109. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 71.Bostrom M, Tavares FW, Finet S, Skouri-Panet F, Tardieu A, Ninham BW. Biophys. Chem. 2005;117:217. doi: 10.1016/j.bpc.2005.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.