Abstract
The newly discovered bacterial phosphothreonine lyases perform a post-translational modification of host cell signaling proteins employing a novel catalytic mechanism that irreversibly removes the phosphate group from a phosphorylated threonine via β-elimination. This “eliminylation” reaction is shown by ab initio QM/MM studies to proceed via the E1cB-like pathway, in which the carbanion intermediate is stabilized by an enzyme oxyanion hole provided by Lys104 and Tyr158 of SpvC.
Post-translational modifications (PTMs), such as methylation and phosphorylation, covalently modify amino acid residues in peptides,1 altering structural properties of the peptides and their interaction with other molecules. PTMs play an important role in cell signaling and gene expression, and their malfunction is implicated in many diseases. This Communication is concerned with a newly discovered PTM process, namely eliminylation,2 which leads to irreversible removal of the phosphate group from a phosphorylated threonine residue. Interestingly, the strategy used by phosphothreonine lyases, which catalyze eliminylation, is quite novel and differs from the well-known phosphoryl transfer mechanism used by protein phosphatases. Instead of cleaving the POγ bond, these enzymes catalyze the breaking of the Cβ-Oγ bond and forming of a Cα=Cβ double bond via β-elimination.3-4 The resulting π system presents an easy target for Michael addition, thus eliminating the possibility of future phosphorylation of the threonine residue.
While eliminylation has yet to be identified in eukaryotic cells, its usage by bacteria is well documented.2 Many bacteria have been found to evade host immune responses,5-6 often by injecting into the host cell protein effectors that disrupt host signal transduction pathways.7-8 One class of such bacterial effectors is phosphothreonine lyase, which has recently been found in Salmonella4, Shigella,3,9 and Pseudomonas syringae.10 The other example of eliminylation is found in bacterial biosynthesis of lantibiotics, which are macrocyclic peptides with thioether cross-links. The lanthionine synthetase catalyzes the removal of the OH group from a serine/threonine residue by first phosphorylating it with a kinase domain, followed by β-elimination of the phosphate group with a lyase domain, in the same fashion as the phosphothreonine lyase. The addition of a terminal Cys residue to the π bond completes the macrocyclization.11 Although no crystal structure is known, the lyase domain of the enzyme shares some active-site residues with phosphothreonine lyases.12
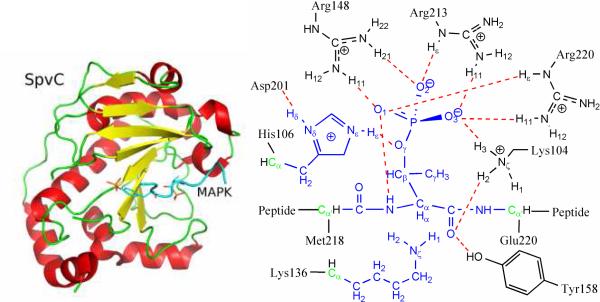
The crystal structure of the phosphothreonine lyase SpvC from S. typhimurium has recently been solved,4,13 and its novel α/β fold shown in Fig. 1 differs from all known folds. The peptide substrate contains the signature pThr-X-pTyr motif of mitogen-activated protein kinases (MAPKs), which is recognized by SpvC. Specifically, pTyr is bound with Lys160, Lys134, and Arg80, while pThr inserts its phosphate group to a pocket formed by Arg220, Arg213, Arg148, and Lys104. The binding of the sub strate induces a conformational change in the loop containing Arg220, which seals the positive pocket surrounding the pThr phosphate. The active-site arrangement of SpvC is depicted in Fig. 1. All phosphothreonine lyases share two critical residues implicated in catalysis by mutagenesis experiments, kinetic data, and their proximity to the substrate in the X-ray structure.4 The putative base for the β-elimination is Lys136 in SpvC, which abstracts the α-proton from pThr in the substrate. A conserved His106 is thought to protonate the phosphate leaving group. Despite this general description, however, the detailed mechanism by which the β-elimination is catalyzed is still unresolved. Information of the microscopic reaction pathway, such as the structure and charge distribution of the transition state(s), might be helpful for designing agents to regulate such PTM events.
Figure 1.
The overall structure of SpvC (left panel) and the active-site arrangement (right panel). The QM region includes those atoms colored in blue and boundary atoms colored in green. Hydrogen bonds are colored red.
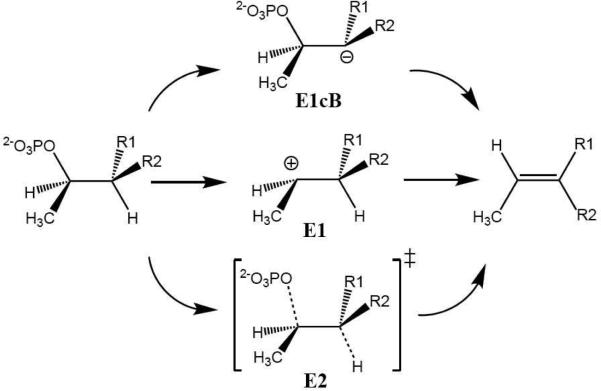
As shown in Scheme 1, there are three possible mechanistic scenarios for a β-elimination reaction. The E1cB mechanism envisions a carbanion intermediate created by the abstraction of the α-proton. The E1 mechanism, on the other hand, assumes a carbocation intermediate generated by the unimolecular loss of inorganic phosphate. Finally, there is the concerted, or E2, mechanism where the two processes occur simultaneously. To identify the correct molecular mechanism, we have investigated the reaction using a quantum mechanical/molecular mechanical (QM/MM) approach, a proven method for studying enzyme catalysis.14-16 While the simulation protocols are described in details in Supporting Information (SI), we note here that the QM system, which is colored blue in Fig. 1, includes the phosphothreonine and the side chains of both Lys136 and His106, totaling 52 atoms. Based on pH-titration curves of wild type and mutants of SpvC,4 Lys136 and His106 were assigned to be deprotonated and protonated, serving as the general base and general acid, respectively. The effects of several important secondary residues, such as Asp201, Tyr158, and Lys104, were approximated via the MM force field. The QM region was treated at the B3LYP/6-31G* level of theory with the pseudobond formalism.17-18 The non-empirical treatment of the QM region is important for a reliable description of the chemistry.
Scheme 1.
Three possible reaction mechanisms for β-elimination.
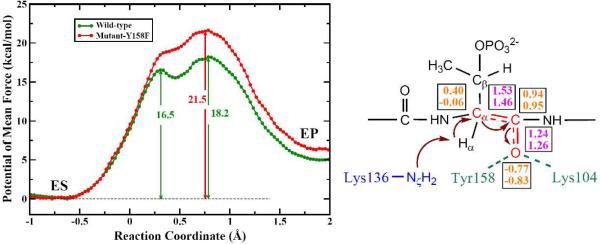
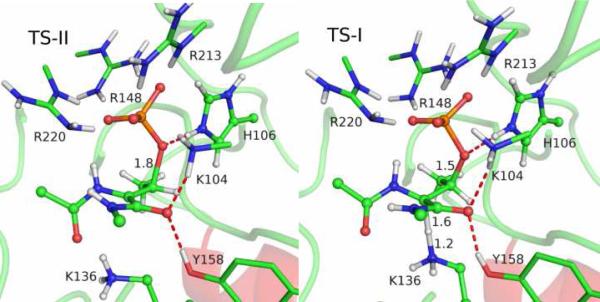
To address the mechanistic questions on the catalyzed reaction, we first calculated the QM/MM minimal energy surface in both the α proton abstraction coordinate and the Cβ-Oγ bond breaking coordinate. The resulting two-dimensional surface, as shown in Fig. S1 in SI, strongly suggests that the proton abstraction occurs prior to the Cβ-Oγ bond cleavage. Using the reaction coordinate, RC = dCβ-Oγ - dHα-Nζ, the potential of mean force (PMF) was then calculated for the wild-type of SpvC using the same QM/MM Hamiltonian. The free-energy profile displayed in Fig. 2 indicates a step-wise E1cB-like reaction mechanism with a shallow minimum flanked by two transition states. As shown in Fig. 3, the first transition state (TS-I) is primarily for the abstraction of the α proton by Lys136, while the second and rate-limiting transition state (TS-II) is for Cβ-Oγ bond cleavage. The shallow minimum in the PMF represents a carbanion intermediate (INT). The overall barrier of 18.2 kcal/mol, which is in reasonably good agreement with the experimental data of 16.0 kcal/mol, estimated from the kcat of 10.60 s-1.4
Figure 2.
Left: the calculated PMFs for both the wild-type (green) and Y158F mutant (red) of SpvC. Right: Charge stabilization scheme for the carbanion intermediate. The atomic charges are given in orange, while the bond lengths in purple. The first and second numbers are for ES and INT, respectively.
Figure 3.
Structures of TS-I and TS-II for the wild-type SpvC. The hydrogen bonds of interest are represented in red dash lines and some key distances are given.
It is interesting to note that the QM/MM results differ from our earlier density functional theory study with a truncated active-site model, which suggested a concerted mechanism.19 This discrepancy is likely due to the small size of the truncated active-site model used in our earlier work. An analysis of charges along the reaction path shows that as α proton is extracted, Cα develops a negative charge. However, this charge can be stabilized via a resonance structure featuring an enolate form of the adjacent backbone carbonyl, as illustrated in Fig. 2. This hypothesis is supported by the fact that the charge of the carbonyl O changes from -0.77 ± 0.04 at the enzyme-substrate (ES) complex to -0.83 ± 0.06 at INT. The development of a negative charge at the carbonyl O is accompanied by the shortening of the Cα-C bond from 1.53 ± 0.07 Å (ES) to 1.46 ± 0.07 Å (INT), and by the elongation of the carbonyl bond from 1.24 ± 0.05 Å (ES) to 1.26 ± 0.05 Å (INT). The spread of the negative charge is facilitated by an oxyanion hole provided by the enzyme. Indeed, the hydrogen bonds of the carbonyl to Lys104 and Tyr158 strengthen as the negative charge develops at the enolate O, as indicated by the shortening of the hydrogen bond lengths from 1.95±0.19 Å and 1.81±0.16 Å (ES) to 1.83±0.12 Å and 1.79±0.15 Å (INT), respectively. These two residues were not included in our earlier model.
To further confirm our hypothesis, we have carried out PMF calculations for the mutant Y158F, which abolishes the hydrogen bond with the carbonyl O. However, the hydrogen bond involving Lys104 is still maintained in all stationary states except EP, where the phosphate group is the sole strong interaction. The removal of the hydrogen bond in Y158F mutant increases the overall barrier by 3.3 kcal/mol while the shape remains roughly the same, as shown in Fig. 2. This increase of barrier is consistent with muta-genesis data, which showed that the Y158F mutant retains only 5% catalytic activity of the wild-type.4 To further understand the role played by secondary residues and the enzyme itself, we have computed the non-bonded contributions of all MM residues to the barriers. As discussed in SI, these results are also consistent with the mutagenesis data, providing additional support of the proposed reaction mechanism.
While the strategy used by the phosphothreonine lyase SpvC is unique and novel, it shares some features with those used by several well-studied enzymes. The enolase superfamily, for example, is known to abstract the α protons of carboxylic acids, although the subsequent events may differ significantly for different members of the superfamily.20-21 Much discussion has been devoted to the acidity of the α proton and the mechanism of the catalysis.22-24 Convincing evidence based on kinetic isotope effects (KIEs) indicated that the reaction is stepwise and involves a carbanion intermediate.25 This conclusion was confirmed by QM/MM calculations,26-28 which suggested that the negative charge developed on Cα is spread to an enolate oxygen stabilized by metal ions (e.g. Mg2+). Phosphothreonine lyases differ in several aspects. First, the α proton is from the backbone of a peptide substrate, instead of carboxylic acid. Second, no metal ion was involved. Instead, the oxyanion hole is stabilized by hydrogen bond interactions. Finally, the β-elimination removes a phosphate group, rather than H2O in the case of enolases. In the last aspect, SpvC is perhaps more closely related to methylglyoxal synthase (MGS), which catalyzes the elimination of the phosphate group of dihydroxyace-tone phosphate (DHAP) initiated by proton abstraction.29
In conclusion, our ab initio QM/MM calculations reveal that eliminylation catalyzed by SpvC utilizes an E1cB-like mechanism, in which the reaction is initiated by α proton abstraction by Lys136. A metastable enolate intermediate is identified, in which the negative charge developed on Cα is spread via a conjugated resonance to the backbone carbonyl oxygen. The carbanionic intermediate is stabilized by an enzyme oxyanion hole consisting of Tyr158 and Lys104. The second and rate-liming elimination step in this enzymatic reaction is catalyzed by His106, which protonates the leaving group. In addition, several cationic residues, namely Arg220, Arg213, Arg148 and Lys104, provide further electrostatic stabilization of the phosphate leaving group. Their catalytic roles are consistent with structural and mutagenesis data.
Supplementary Material
Acknowledgements
This work was supported by NIH (R01-GM079223 to YZ, R03-AI071992 to HG). The calculations were performed at the National Center for Supercomputing Applications (NCSA) and the New Mexico Center for Computational Applications. We thank Alvan Hengge for several useful discussions.
Footnotes
Supporting Information Available: Details of simulation protocol, minimal energy surface, and analysis of QM-MM interactions. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Wold F. Annu. Rev. Biochem. 1981;50:783. doi: 10.1146/annurev.bi.50.070181.004031. [DOI] [PubMed] [Google Scholar]
- 2.Brennan DF, Barford D. Trends Biochem. Sci. 2009;34:108. doi: 10.1016/j.tibs.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou JM, Shao F. Science. 2007;315:1000. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Y, Li H, Long C, Hu L, Xu H, Liu L, Chen S, Wang DC, Shao F. Mole. Cell. 2007;28:899. doi: 10.1016/j.molcel.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 5.Brodsky IE, Medzhitov R. Nat. Cell Biol. 2009;11:521. doi: 10.1038/ncb0509-521. [DOI] [PubMed] [Google Scholar]
- 6.Ribet D, Cossart P. Cell. 2010;694-702 doi: 10.1016/j.cell.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis GR. Nat. Rev. Microbiol. 2006;4:811. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 8.Galan JE, Wolf-Watz H. Nature. 2006;444:567. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 9.Arbibe L, Kim DW, Batsche E, Pedron T, Mateescu B, Muchardt C, Parsot C, Sansonetti PJ. Nat. Immunol. 2007;8:47. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, Chen S, Tang X, Zhou J-M. Cell Host Microbe. 2007;1:175. doi: 10.1016/j.chom.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Goto Y, Li B, Shi Y, Bibb MJ, van der Donk WA. PLOS Biol. 2010;8:e1000339. doi: 10.1371/journal.pbio.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goto Y, Okesli A, van der Donk WA. Biochem. 2011;50:891. doi: 10.1021/bi101750r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, Wang H, Zhang J, Gu L, Huang N, Zhou J-M, Chai J. Nat. Struc. Mole. Biol. 2008;15:101. doi: 10.1038/nsmb1329. [DOI] [PubMed] [Google Scholar]
- 14.Warshel A. Annu. Rev. Biophys. Biomol. Struct. 2003;32:425. doi: 10.1146/annurev.biophys.32.110601.141807. [DOI] [PubMed] [Google Scholar]
- 15.Gao J, Ma S, Major DT, Nam K, Pu J, Truhlar D. Chem. Rev. 2006;106:3188. doi: 10.1021/cr050293k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senn HM, Thiel W. Angew. Chem. Int. Ed. 2009;48:1198. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Lee T, Yang W. J. Chem. Phys. 1999;110:46. [Google Scholar]
- 18.Zhang Y. J. Chem. Phys. 2005;122:24114. doi: 10.1063/1.1834899. [DOI] [PubMed] [Google Scholar]
- 19.Smith GK, Ke Z, Hengge AC, Xu D, Xie D, Guo H. J. Phys. Chem. B. 2009;113:15327. doi: 10.1021/jp9052677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babbitt PC, Mrachko GT, Hasson MS, Huisman GW, Kolter R, Ringe D, Petsko GA, Kenyon GL, Gerlt JA. Science. 1995;267:1159. doi: 10.1126/science.7855594. [DOI] [PubMed] [Google Scholar]
- 21.Babbitt PC, Hasson MS, Wedekind JE, Palmer DRJ, Barrett WC, Reed GH, Rayment I, Ringe D, Kenyon GL, Gerlt JA. Biochem. 1996;35:16489. doi: 10.1021/bi9616413. [DOI] [PubMed] [Google Scholar]
- 22.Gerlt JA, Gassman PG. J. Am. Chem. Soc. 1992;114:5928. [Google Scholar]
- 23.Guthrie JP, Kluger R. J. Am. Chem. Soc. 1993;115:11569. [Google Scholar]
- 24.Gerlt JA, Gassman PG. J. Am. Chem. Soc. 1993;115:11552. [Google Scholar]
- 25.Anderson SR, Anderson VE, Knowles JR. Biochem. 1994;33:10545. doi: 10.1021/bi00200a041. [DOI] [PubMed] [Google Scholar]
- 26.Alhambra C, Gao J, Corchado JC, Villa J, Truhlar DG. J. Am. Chem. Soc. 1999;121:2253. [Google Scholar]
- 27.Liu H, Zhang Y, Yang W. J. Am. Chem. Soc. 2000;122:6560. [Google Scholar]
- 28.Feierberg I, Aqvist J. Theo. Chem. Acc. 2002;108:71. [Google Scholar]
- 29.Zhang X, Harrison DH, Cui Q. J. Am. Chem. Soc. 2002;124:14871. doi: 10.1021/ja027063x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.