Abstract
Membrane proteins have evolved to fold and function in a lipid bilayer, so it is generally assumed that their stability should be optimized in a natural membrane environment. Yet optimal stability is not always in accord with optimization of function, so evolutionary pressure, occurring in a complex membrane environment, may favor marginal stability. Here we find that the transmembrane helix dimer, glycophorin A (GpATM) is actually much less stable in the heterogeneous environment of a natural membrane than it is in model membranes and even common detergents. The primary destabilizing factors are electrostatic interactions between charged lipids and charged GpATM side chains, and non-specific competition from other membrane proteins. These effects overwhelm stabilizing contributions from lateral packing pressure and excluded volume. Our work illustrates how evolution can employ membrane composition to modulate protein stability.
Introduction
α-helical membrane proteins fold and assemble via lateral helix-helix interactions in bilayer membranes1. In addition to specific interactions within the protein, the membrane environment itself can play a major role in structuring membrane proteins2. Cell membranes are a highly heterogeneous medium in lipid and protein composition3. For example, E. coli inner membranes contain three major lipid species with different headgroups and the E. coli genome codes for more than a thousand putative α-helical membrane proteins4,5. Thus, there are a wide variety of potential interactions that could modulate transmembrane helix association through specific or nonspecific lipid-protein and protein-protein interactions. It is largely unknown how the complex lipid and protein environment of natural membranes influences the folding and stability of membrane proteins.
Bacterial and eukaryotic cell membranes are overall negatively charged and contain a significant fraction of nonbilayer lipids6. The negative charges of biological membranes help determine the topology of integral membrane proteins by electrostatic interactions with the charged residues7–9. The nonbilayer lipids such as phosphatidylethanolamine (PE) and lyso-phospholipid form inverted hexagonal (HII) and hexagonal (HI) phases, respectively, and thus strongly modulate the curvature stress and lateral pressure profile of the lipid bilayers10. These elastic properties can be important modulators of the structure, function, and topogenesis of many integral membrane proteins2,11,12.
In addition to the wide variety of lipids, membrane proteins occupy more than a half of total membrane mass13. The high concentration of proteins may perturb the structure and dynamics of the surrounding lipid molecules and reduce the surface area accessible for diffusion14,15. It is also possible that embedded membrane proteins can compete with specific protein-protein interactions in cell membranes.
Here, we investigated the effects of natural lipid and protein components on the stability of a specific transmembrane (TM) helix-helix interaction in reconstituted bilayer systems. TM helix interactions are prevalent in cell membranes and are involved in assembly, signaling, and subcellular localization of membrane proteins16,17. They have also served as an important model system for studying the lateral transmembrane helix–helix interactions that occur in the folding of α-helical membrane proteins18,19. As our model, we chose the well characterized glycophorin A transmembrane domain, GpATM, which forms a stable sequence-specific dimer in lipid environments. Extensive mutagenesis and thermodynamic studies have been carried out in detergent micelles and lipid bilayers to identify the driving forces determining the sequence specificity of the dimerization20,21. GpATM is sufficiently stable that it remains dimeric in SDS solution20,22,23. We recently found that GpATM is so stable in palmitoyloleoyl phosphatidylcholine (C16:0C18:1c9PC) model bilayers that it should be almost completely dimeric in vivo24. Nevertheless, the results of Duong et al. suggests that GpATM is mostly monomeric in E. coli inner membranes25. Chen et al. also reported that the wild-type GpATM is predominantly monomeric in bleb vesicles derived from mammalian cells26. These results suggest that there may be something different about natural membranes that destabilizes GpATM interactions relative to a pure C16:0C18:1c9PC bilayer.
Here we investigate the physico-chemical origin of this destabilization. Our results reveal several new aspects of lipid-protein and protein-protein interactions that can modulate the stability of transmembrane helix-helix interactions.
Results
The steric trap method
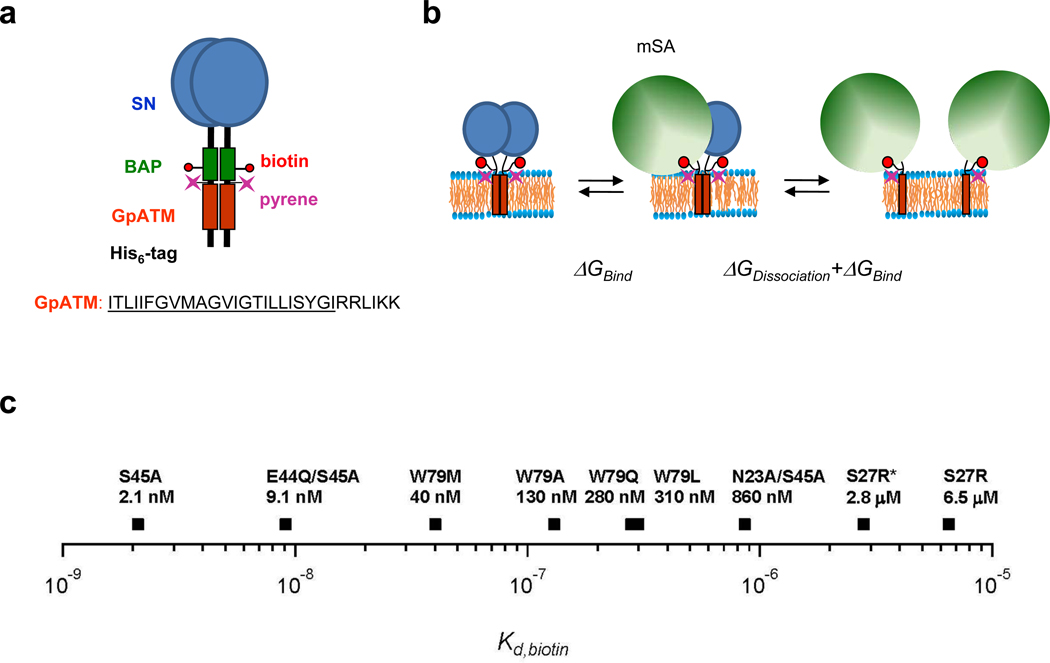
We used our recently developed steric trap method to measure the dimer stability of a staphylococcal nuclease-GpATM fusion, SNGpATM, in lipid bilayers24,27. The method, shown in Figure 1a and 1b has been described previously24. We attach two biotin tags that are sufficiently close in the dimer that only one biotin can be bound by a large monovalent streptavidin (mSA) molecule. Because of steric overlap, the second biotin tag can only be bound when the dimer falls apart to a monomer. Thus, the affinity of the second mSA binding is coupled to dissociation and therefore reflects dimer affinity. Binding is observed by monitoring the fluorescence of a pyrene label on SNGpATM. The method is applicable to high affinity interactions because dissociation can be thermodynamically driven by the high affinity of mSA and by its concentration. The affinity of mSA must be tuned to the affinity of the dimer interaction being examined. We therefore previously generated a series of monovalent streptavidin mutants whose biotin affinity range from ~10−9 M to ~10−6 M (Figure 1c)24. Employing a mSA variant with the appropriate affinity, biphasic binding is observed, corresponding to the high affinity binding of the first mSA, followed by SNGpATM dissociation and binding of the second mSA occurring at lower affinity. If the mSA affinity is too high, only one binding phase is observed because dimer affinity cannot compete with mSA binding.
Figure 1.
The steric trap method. (a) Design of the construct used for steric trapping24. GpATM: orange, SN: blue (staphylococcal nuclease fused to the N-terminus), and BAP: green (biotin acceptor peptide). A unique cysteine was labeled with a pyrene fluorescent probe for the detection of binding/dissociation. The amino acid sequence of human GpATM is composed of the hydrophobic transmembrane segment (underlined) followed by a series of positively charged residues in the C-terminus. (b) Reaction scheme of the steric trap method. A single monovalent streptavidin (green sphere) can bind to one of the biotin tags in the dimer with its intrinsic binding affinity (ΔGbind). Because of steric overlap in the dimeric form, however, a second streptavidin can only bind to the dissociated form. Thus, the affinity of the second streptavidin is modulated by the stability of the dimer, yielding an overall free energy of binding equal to ΔGdissociation + ΔGbind. (c) If the affinity of mSA is too high, binding is insensitive to the contribution from dissociation. If the affinity is too low, impractically high concentrations of mSA are required to drive dissociation. We therefore employ a library of mSA mutants with various intrinsic biotin binding affinities (Kd biotin)24. mSA-S27R (asterisk) has a stronger biotin binding affinity in the negatively charged membranes than in neutral lipid environments (see text).
Affinity of SNGpATM dimer is low in E. coli lipid membranes
To provide a lipid environment mimicking natural cell membranes, we measured the dissociation constant of a GpATM dimer in lipid bilayers composed of E. coli phospholipid extracts. E. coli inner membranes have a simpler lipid composition than eukaryotic cell membranes4,28. Due to the lack of cholesterol and sphingolipids, a complex lipid phase behavior in model membranes can be avoided29,30. More importantly, dimerization of GpATM has been extensively studied in E. coli inner membranes in vivo using the TOXCAT assay so that our results can be directly compared with the previous reports25,31.
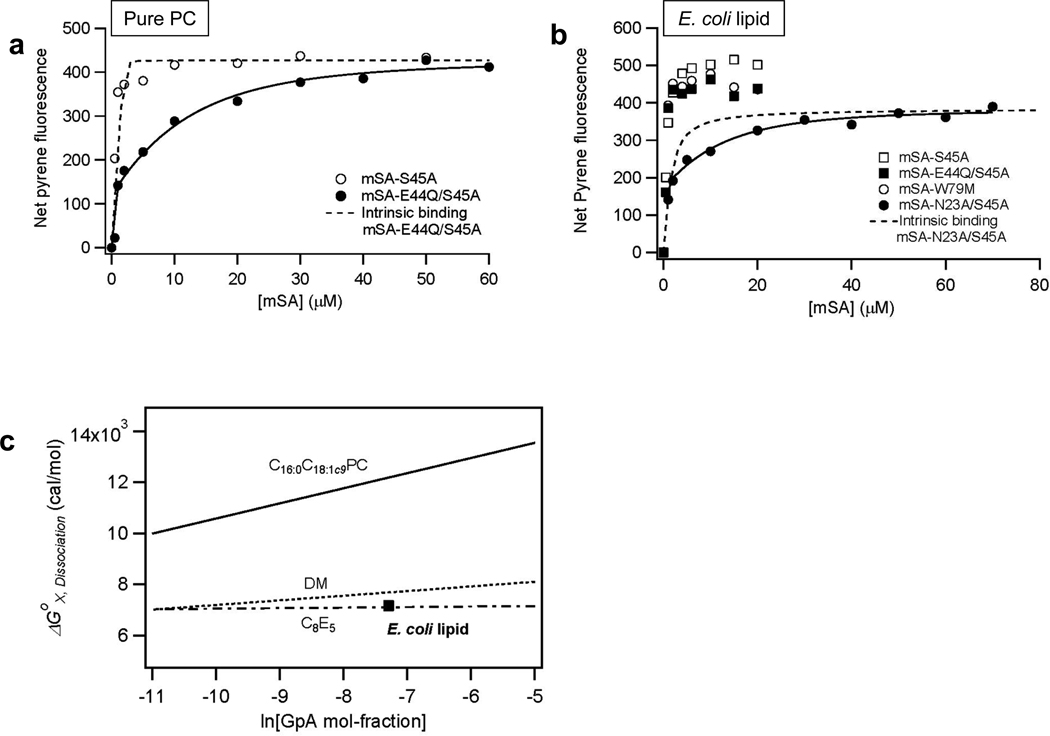
SNGpATM in E. coli lipid membranes was titrated with a series of mSA mutants with different intrinsic biotin binding affinities (Kd,biotin) (Figure 2b). Only a single tight binding phase was observed with the stronger biotin binding mSA mutants mSA-S45A (Kd,biotin=2.1×10−9 M), mSA-E44Q/S45A (Kd,biotin=9.1×10−9 M), and mSA-W79M (Kd,biotin=4.5×10−8 M), indicating that the affinity of these mSA variants was too high to effectively measure the SNGpATM dissociation constant. The characteristic two phase binding curve could be obtained with a weaker biotin binding mSA-N23A/S45A (Kd,biotin=8.6×10−7 M) (for the detailed procedure, see Supporting Figure 2). The fitted dissociation constant (Kd,GpA) of the GpATM dimer was 1.0±0.3×10−8 M. By comparison, the dissociation constant in pure C16:0C18:1c9PC bilayers was 1.5±0.6×10−12 M and required the use of higher affinity mSA variant, mSA-E44Q/S45A (Kd,biotin=9.1×10−9 M) (Figure 2a). Thus, the affinity of GpATM dimer was severely weakened in E. coli lipid membranes relative to C16:0C18:1c9PC membranes by ΔΔGox,Dissociation =~5 kcal/mol using a mol-fraction scale32. Surprisingly, the affinity is not much different than in detergent micelle solutions (Figure 2c)32,33.
Figure 2.
Comparison of of SNGpA dimer in pure PC and E. coli lipid bilayers. (a) mSA binding curves in pure C16:0C18:1c9PC. The affinity of mSA-S45A is too high under these conditions to observe a second binding phase from dissociation, but the mSA-E44Q/S45A mutant (Kd,biotin=9.1×10−9 M) allows the dissociation phase to be observed. The fitted Kd,GpA of GpATM dimer was 1.5±0.6×10−12 M at L/P=1000 with mSA-E44Q/S45A. (b) Selection of a mutant mSA with an optimal Kd,biotin to extract the dissociation constant (Kd,GpA) of SNGpATM dimer in E. coli lipid membranes. In contrast to pure PC bilayers, the mSA-E44Q/S45A variant has too high affinity to measure dissociation in E. coli lipid. Instead, a much lower affinity variant was required, mSA-N23A/S45A (Kd,biotin=8.6×10−7 M), indicative of the weaker dimerization of SNGpATM in E. coli lipids. For the mSA-N23A/S45A binding curve, we obtain Kd,GpA = 1.0±0.3×10−8 M at a lipid-to-protein molar ratio (L/P) of 1500. All measurements were performed in 20 mM MOPS (pH 7.4), 200 mM NaCl buffer solutions with mSA-accessible SNGpATM concentrations of 1.8 µM. (c) Comparison of the dissociation free energy (ΔGX,Dissociation) of GpATM dimer in E. coli lipid membranes relative to pure C16:0C18:1c9PC bilayers24 (solid line), DM micelles33 (dotted line), and C8E5 micelles32 (dashed line). ΔGX,Dissociation of GpATM dimer is expressed in mol-fraction scale32 as a function of molar ratio of GpATM to total detergents or lipid concentrations. The lines indicating the concentration dependence of dissociation free energies in various environments were obtained previously24,32,33.
Host-guest system to test the effects of individual lipid components
To dissect the bilayer forces that contribute to the destabilization of the GpATM dimer in E. coli lipid membranes, we set out to test the effects of the major lipid components of E. coli inner membranes which are composed of 70~75 % of zwitterionic phophatidylethanolamine (PE), 15~20% of negatively charged phosphatidylglycerol (PG), and 5~10 % of negatively charged cardiolipin (CL) lipids. The major hydrocarbon composition is 30~45 % of 16:0, 30~45 % of 16:1c9, and 15~30 % of 18:1c114.
We designed a host-guest lipid system, which is composed of purely synthetic phospholipids. C16:0C18:1c9PC was chosen as a host lipid, and the guest lipids, which have a various headgroup structure but the same hydrocarbon composition (C16:0C18:1c9), were incorporated into the lipid bilayers to an appropriate ratio. The hydrocarbon composition was fixed to C16:0C18:1c9 because E. coli lipids have a similar phase behavior to palmitoyloleoyl phosphatidylethanolamine (C16:0C18:1c9PE)4. We chose naturally abundant C16:0C18:1c9PE, C16:0C18:1c9PG, lyso-phosphatidylcholine (C18:1c9PC), palmitoyloleoyl phosphatidylserine (C16:0C18:1c9PS), and cardiolipin (tetra-C18:1c9CL) as guest lipids. Thus, in the lipid bilayers with different lipid compositions, the variations in the hydrophobic thickness were minimized so that the dominant headgroup effects could be studied.
Bilayer lateral pressure modulates transmembrane helix-heix interaction
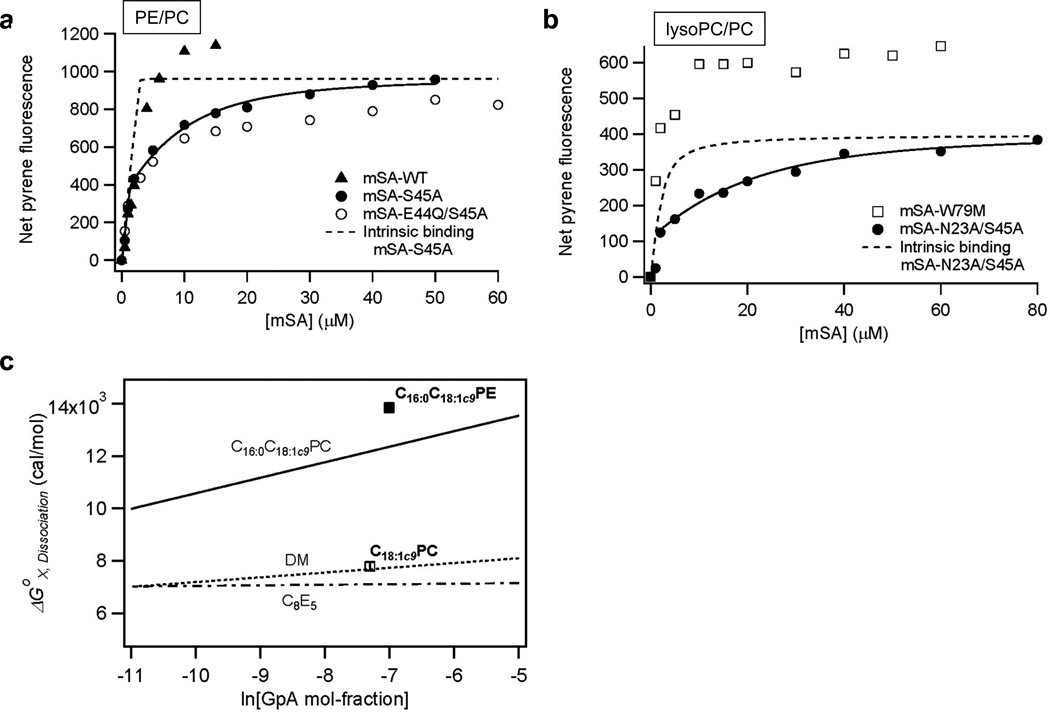
First, we tested the effect of PE lipids on the dimerization of GpATM. PE lipids with longer acyl chains and cis-double bonds form an inverted hexagonal phase (HII) above the liquid-hexagonal phase transition temperature 34. When incorporated into a bilayer, they induce an intrinsic negative curvature in each monolayer leaflet and increase the lateral pressure in the central region of the bilayer35,36. Although highly stable in pure C16:0C18:1c9PC bilayers (Kd,GpA=1.5±0.6 ×10−12 M at lipid-to-protein molar ratio (L/P)=1000, Figure 2a), the GpATM dimer became even more stable in C16:0C18:1c9PE/PC (40/60 in mol-%) bilayers (Kd,GpA=2.1±0.5 ×10−13 M at L/P=1100). At a comparable L/P ratio, the net stabilization effect by 40 % C16:0C18:1c9PE was ΔΔGox,Dissociation =−1.5 kcal/mol relative to pure C16:0C18:1c9PC bilayers.
A complication with the incorporation of the small headgroup PE lipid into PC bilayers is that it increases not only the lateral pressure but also the hydrophobic thickness of the bilayers37. Thus, dimer stability was tested in 100 % C18:0C18:1c9PC bilayers (Supporting Figure3). C18:0C18:1c9PC has a thicker hydrocarbon thickness (dhc=27.6 Å37) than C16:0C18:1c9PC (dhc=26.5 Å38) and has a similar gel-to-liquid phase transition temperature (Tm=13 °C39) as C16:0C18:1c9PC/PE (60/40) mixed bilayers (Tm=~12 °C40). The Tm and dhc of pure C16:0C18:1c9PE are 24 °C40 and 30.1 Å37, respectively. Increasing the hydrocarbon thickness from C16:0C18:1c9PC to C18:0C18:1c9PC destabilized the GpATM dimer. Thus, the net stabilization effect by PE lipid likely originates from the increase in the bilayer lateral pressure rather than the increase in thickness.
To further test the lateral pressure effect on the stability of the GpATM dimer, the micelle-forming lyso-phosphatidylcholine (C18:1c9PC) was perfused into the C16:0C18:1c9PC-SNGpATM proteoliposomes to a final 20 mol-% of the total lipids. In constrast to PE lipids, lysoPC induces a positive intrinsic monolayer curvature and reduces the lateral pressure in the bilayer center2. In the bilayers containing C18:1c9PC, the dimer stability was dramatically reduced by ~4.5 kcal/mol (Figure 3b and 3c) relative to pure C16:0C18:1c9PC bilayers. These results again suggest that packing pressure has a significant effect on dimer stability. The presence of PE in E. coli membranes, which increases lateral packing pressure, is therefore not likely to be the origin of the destabilization of the SNGpATM dimer.
Figure 3.
Effects of bilayer lateral pressure profile on the stability of GpATM dimer. (a) mSA-binding curves in C16:0C18:1c9PE/PC (40/60) lipid bilayers. Kd,GpA=2.1±0.6×10−13 M was obtained at L/P=1100 with mSA-S45A (Kd,biotin=2.1×10−9 M). (b) mSA binding curves in C18:1c9PC/ C16:0C18:1c9PC (20/80) bilayers. Kd,GpA=6.0±2.4×10−9 M was obtained at L/P=1500 with mSA-N23A/S45A (Kd,biotin=8.6×10−7 M). (c) Summary of changes in ΔGX,Dissociation induced by 40 mol-% C16:0C18:1c9PE (filled square) and 20 mol-% C18:1c9PC (empty square) relative to pure C16:0C18:1c9PC bilayers. The lines indicating the concentration dependence of dissociation free energies in various environments were obtained previously24,32,33.
Strong destabilization of TM helix interactions in negatively charged membranes
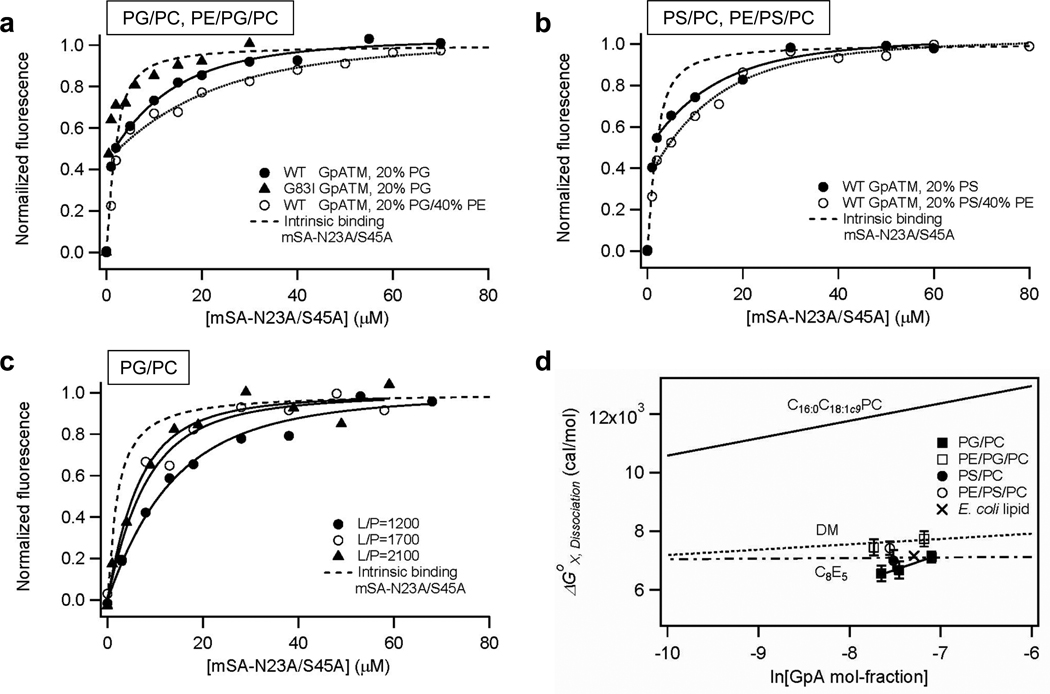
The effects of the negatively charged lipids PG and PS on the dimerization of GpATM were tested in C16:0C18:1c9PG/PC (20/80) and C16:0C18:1c9PS/PC (20/80) bilayers. PG is the major negatively charged lipid in E. coli inner membranes while PS is the major negatively charged lipid in eukaryotic cell membranes. As shown in Figure 4, the stability of the GpATM dimer dramatically decreased in the negatively charged membranes by ΔΔGox,Dissociation= 4.5~5 kcal/mol relative to pure C16:0C18:1c9PC bilayers (Figure 4a, and 4b filled circles, also see Supporting Figure 4). Moreover, the chemical structure of the lipid headgroup is not a factor since the incorporation of either PG and PS lipids decreased the stability of the GpATM dimer to a similar level as E. coli lipids (Figure 4d). Incorporation of 5 mol-% of cardiolipin (tetra-C18:1c9CL) also destabilized the dimer, but only to a moderate extent (Supporting Figure 5).
Figure 4.
The effect of negatively charged lipids on SNGpATM dimer affinity. (a) mSA binding curves of wild-type and non-dimerizing G83I GpATM mutant in C16:0C18:1c9PG/PC (20/80) and wild-type GpATM in C16:0C18:1c9PE/PG/PC (40/20/40) bilayers at L/P=1200~1300. Kd,GpA=1.4±0.4×10−8 M and Kd,GpA=5.1±2.2×10−9 M were obtained in C16:0C18:1c9PG/PC and C16:0C18:1c9PE/PG/PC, respectively, using a weaker biotin binding mSA-N23A/S45A (Kd,biotin=8.6×10−7M). (b) mSA binding curves of wild-type GpATM in C16:0C18:1c9PS/PC (20/80) and C16:0C18:1c9PE/PS/PC (40/20/40) bilayers at L/P=1800~1900. Kd,GpA=2.5±1.5×10−8 M and Kd,GpA=1.0±0.4×10−8 M were obtained in C16:0C18:1c9PS/PC and C16:0C18:1c9PE/PS/PC, respectively, using mSA-N23A/S45A (Kd,biotin=8.6×10−7M). (c) Dilution effects of GpATM in the negatively charged C16:0C18:1c9PG/PC (20/80) membranes. The fitted Kd,GpA’s were 1.4±0.4×10−8 M at L/P=1200, 3.2±1.6×10−8 M at L/P=1700, and 4.7±2.1×10−8 M at L/P=2100. (d) Summary of the effects of negatively charged lipids. C16:0C18:1c9PG/PC (20/80) and C16:0C18:1c9PS/PC (20/80) destabilize the SNGpATM dimer relative to pure C16:0C18:1c9PC bilayers. A modest stabilization by the introduction of C16:0C18:1c9PE can be seen by the increase of dissociation free energies in C16:0C18:1c9PE/PG/PC (40/20/40) and C16:0C18:1c9PE/PS/PC (40/20/40) bilayers. The stability of GpATM dimer in E. coli lipid membranes is also indicated for comparison. The lines indicating the concentration dependence of dissociation free energies in various environments were obtained previously24,32,33.
It is possible that the negatively charged membranes increased streptavidin binding affinity, thereby falsely reporting a weakened GpATM dimerization affinity. We therefore tested the intrinsic binding affinity of mSA to a monomeric GpATM mutant, GpATM-G83I mutant, so that dimerization and mSA binding affinity could be separated. In C16:0C18:1c9PC/PG (20/80) bilayers (Figure 4a, filled triangles), the overall binding curve closely overlapped the intrinsic biotin binding curve of mSA-N23A/S45A, as expected for binding to a monomeric variant. In addition, dilution of SNGpATM by a higher concentration of C16:0C18:1c9PG/PC lipids also shifted the second phase binding curve to lower mSA concentration region, consistent with destabilization of the dimer (Figure 4c). Therefore, we could conclude that the specificity of dimer formation is maintained in the negatively charged membranes. The intrinsic biotin binding affinity of mSA mutants did not change in the negatively charged membranes except for mSA-S27R, whose intrinsic affinity increased by only 2.3 times in the C16:0C18:1c9PG/PC bilayers (Supporting Figure 1).
We next tested if the stability enhancing effect by the nonbilayer PE lipid and the stability reducing effect by the negatively charged PG or PS are additive when these two lipid species were incorporated at the same time in lipid bilayers. The resulting lipid compositions (C16:0C18:1c9PE/PG/PC or PS) are more similar to those of E. coli inner membrane or eukaryotic cell membranes. As before (see above), the addition of 40 mol-% C16:0C18:1c9PE to 20 mol-% PG or 20 mol-% PS mildly increased the dimer stability by ΔΔGox,Dissociation=−0.5±0.4 kcal/mol in PS-containing membranes and ΔΔGox,Dissociation=−0.6±0.3~−0.9±0.4 kcal/mol in PG-containing membranes, respectively (Figure 4a and 4b, open circles). The stability enhancement, however, was smaller than in neutral C16:0C18:1c9PE/PC (40/60) bilayers, however (ΔΔGox,Dissociation=−1.5 kcal/mol, Figure 3c).
Electrostatic interaction between negatively charged lipids and the positive charge residues of TM helix is responsible for the destabilization
What is the origin of the strong destabilization of GpATM dimer induced by the negatively charged lipids? GpATM contains four positively charged residues, Arg96, Arg97, Lys100, and Lys101 (ITLIIFGVMAGVIGTILLISYGIRRLIKKLG: TM domain underlined, and Arg and Lys residues in bold.) in the C-terminal region of the transmembrane domain23. In an erythrocyte membrane, these residues are likely to be located in the membrane-cytoplasm interfacial region. We therefore hypothesized that electrostatic interactions between the interfacial positively charged residues and the negatively charged lipid headgroups may affect the dimerization equilibrium of GpATM.
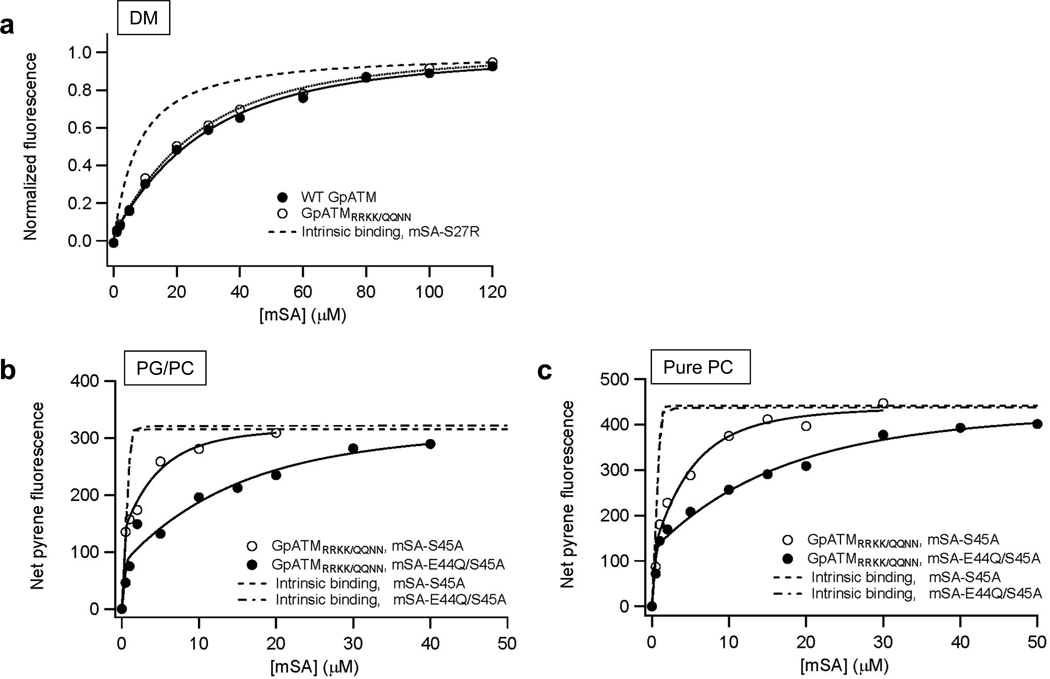
To address this question, a quadruple mutant R96Q/R97Q/K100N/K101N (GpATMRRKK/QQNN) was generated, in which all the C-terminal positively charged residues were replaced by neutral polar residues. This construct showed the similar dimer stability to wild-type SNGpATM in non-denaturing decyl malotoside (DM) micelles (Figure 5a). When the stability of the GpATMRRKK/QQNN dimer was measured in the negatively charged C16:0C18:1c9PG/PC (20/80) and the neutral C16:0C18:1c9PC bilayer, the destabilization effect of the negatively charged lipids was completely eliminated (Figure 5b and 5c). The dissociation constants of the GpATMRRKK/QQNN dimer in the neutral and negatively charged bilayers (Kd,GpA =5.8±2.4×10−13 M in C16:0C18:1c9PC and Kd,GpA =7.3±4.1×10−13 M in C16:0C18:1c9PG/PC) were indistinguishable and close to the dissociation constant of wild-type GpATM dimer in neutral C16:0C18:1c9PC bilayers (Kd,GpA =1.5±0.6×10−12 M). This result strongly suggests that the specific electrostatic interaction between the interfacial positively charged residues of GpATM and the negatively charged membranes strongly reduce the stability of the GpATM dimer.
Figure 5.
Stability of the neutral GpATMRRKK/QQNN in various lipid environments. (a) Binding curves of mSA-S27R to 2 µM wild-type SNGpATM (Kd,GpA=1.7±0.3×10−7 M) and SNGpATMRRKK/QQNN (Kd,GpA=2.6±0.6×10−7 M) in 40 mM DM micelles. (b) mSA binding curves of SNGpATMRRKK/QQNN in the negatively charged C16:0C18:1c9PG/PC (20/80) bilayers at L/P=2100. The stability of the dimer was probed using mSA-S45A (Kd,GpA=4.8±2.5×10−13 M) and mSA-E44Q/S45A (Kd,GpA=7.3±4.1×10−13 M). (c) mSA binding curves of SNGpATMRRKK/QQNN in neutral C16:0C18:1c9PC bilayers at L/P=1900. The stability of the dimer was probed using mSA-S45A (Kd,biotin=2.1×10−9 M, Kd,GpA=4.0±2.2×10−13 M) and mSA-E44Q/S45A (Kd,biotin=9.1×10−9 M, Kd,GpA=5.8±2.4×10−13 M).
Incorporation of E. coli inner membrane proteins leads to a significant destabilization of specific TM helix-helix interactions in membranes
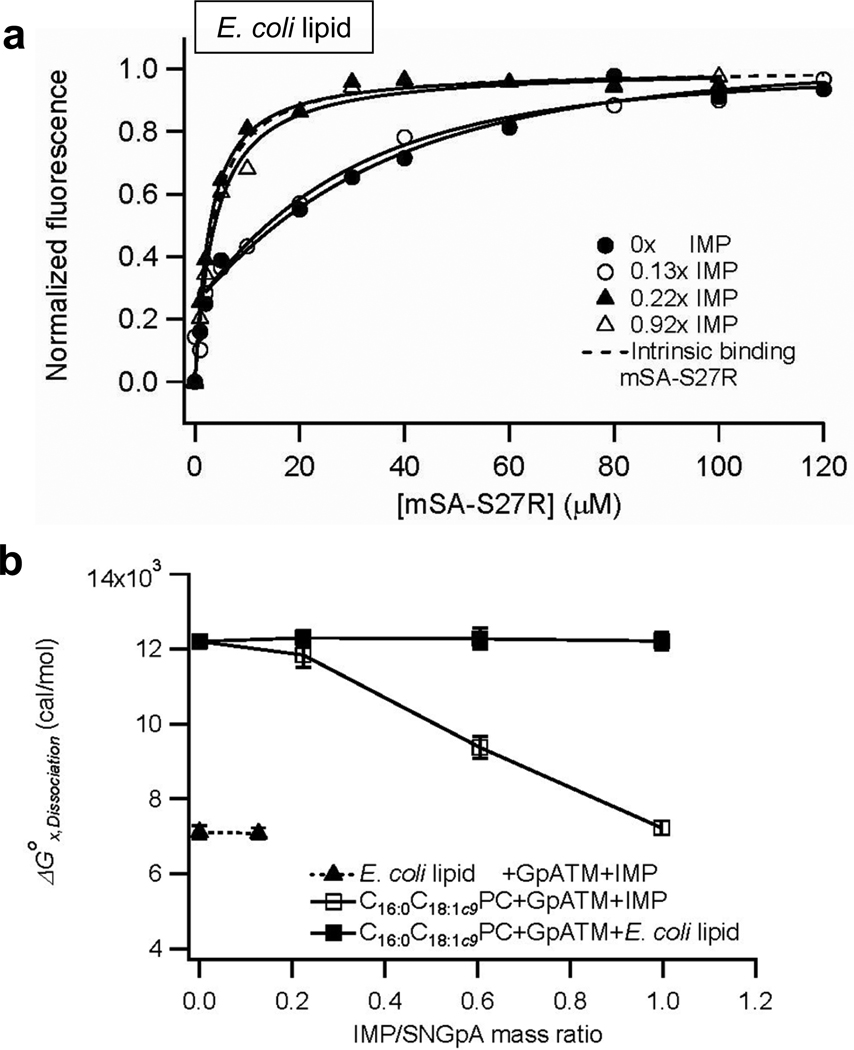
To investigate the effects of heterogeneous membrane proteins on the specific dimerization of TM helices, the total inner membrane proteins (IMP) of E. coli cells were solubilized, and reconstituted in E. coli lipid membranes with SNGpATM (Supporting Figure 6a and 6c). As shown in Figure 6a, the incorporation of IMPs dramatically decreased the SNGpATM dimer affinity. The overall mSA-binding curve reached an intrinsic biotin binding regime of mSA-S27R (Kd,biotin =2.8×10−6 M) at the IMP-SNGpATM mass ratio of 0.22. Therefore, in E. coli lipid membranes, the presence of a relatively small amount of general E. coli inner membrane proteins led to a large destabilization of the GpATM dimer to the level where the dissociation constant (Kd,GpA) of the dimer was so weak that it could not be measured by the steric trapping. The mass ratio of the total reconstituted membrane proteins (SNGpATM plus IMPs) to phospholipids was approximately 1:7.0 at the highest IMP content in the proteoliposomes, which is considerably more dilute than the total protein-to-lipid mass ratio of 2.8:1 of the solubilized E. coli inner membranes.
Figure 6.
Effects of total E. coli inner membrane protein extracts (IMP) on the stability of SNGpATM dimer. (a) Binding curves of mSA-S27R to 2.1~2.7 µM SNGpATM with an increasing amount of IMPs in E. coli lipid membranes at L/P=1500. The fitted Kd,GpA’s were 1.1±0.4×10−8 M and 1.4±0.3×10−8 M at the IMP to SNGpATM mass ratios of 0 and 0.13, respectively. At higher mass ratios, the dissociation constant was too weak to measure, even with our lowest affinity mSA variant. (b) Changes in the dissociation free energies (ΔGX,Dissociation) of the SNGpATM dimer as a function of the mass ratio of IMPs to SNGpATM in E. coli lipid and C16:0C18:1c9PC membranes. For the detailed experimental design and data, see the Supporting Figure 7.
To quantitatively assess the destabilization effect by IMPs at higher IMP-SNGpATM mass ratio, SNGpATM and IMPs were reconstituted in pure C16:0C18:1c9PC bilayers, in which the GpATM dimer is highly stable (Figure 6b and Supporting Figure 6d). The high stability in C16:0C18:1c9PC bilayers allows a wider range of the destabilizing effects to be quantitatively assessed than is possible in E. coli lipid membranes. As the protein to lipid mass ratio increased from 0 to 1.0, the dissociation constants increased from Kd,GpA =3.0×10−12 M to Kd,GpA =9.2±2.9×10−9 M or ~3 orders of magnitude at L/P=1400~1600 (Supporting Figure 7). At the highest IMP-SNGpATM mass ratio tested, the net destabilization free energy of the dimer amounted to remarkable ΔΔGox,Dissociation=4~5 kcal/mol.
DISCUSSION
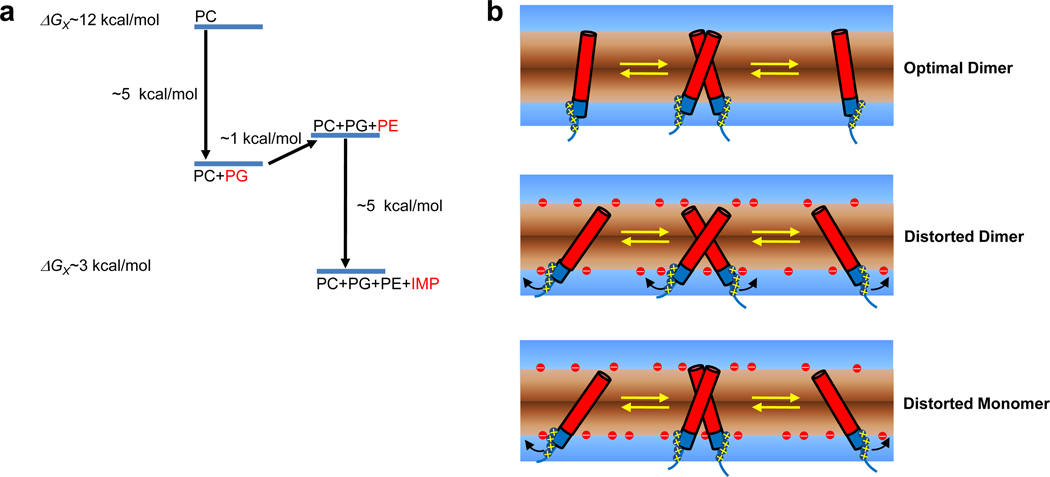
Here we showed that individual lipid components differentially affect the dimerization equilibrium of GpATM and that generic membrane proteins extracted from the E. coli inner membranes significantly destabilized the TM helix-helix interaction in lipid bilayers. The combination of these environmental factors imposes strong destabilizing forces on the GpATM dimer. Our results are summarized in Figure 7a. In pure C16:0C18:1c9PC bilayers, the dimer stability is ΔGx ~12 kcal/mol. The addition of the negatively charged lipid C16:0C18:1c9PG in the proportion seen in E. coli membranes destabilizes the dimer about ~5 kcal/mol due to electrostatic interactions between the membrane and the protein. The E. coli lipid C16:0C18:1c9PE increases stability by about ~−1 kcal/mol through an increase in lateral packing pressure. Finally, the addition of E. coli IMP, which recreates a facsimile of a natural E. coli membrane environment, destabilized the dimeric form by roughly ~5 kcal/mol to a level of ΔGx ~ 3 kcal/mol. This value is remarkably similar to the values estimated in membrane blebs by Chen et al. 26. Our results provide a clearer picture of the physico-chemical origin of the relatively weak GpATM dimerization seen in these natural membranes.
Figure 7.
Summary of the effects of membrane components on GpATM dimer stability. (a) Major environmental forces which contribute to the weak TM helix-helix interactions in natural cell membranes. Relative to neutral fluid C16:0C18:1c9PC bilayers, the negatively charged lipids such as PG and PS dominate the lipid effects, destabilizing the dimer by ~5 kcal/mol. The increased elastic stiffness by the inverted hexagonal phase forming PE lipids mildly stabilizes the dimer. The addition of non-specific proteins to the membranes can further destabilize the specific helix-helix interactions. (b) Possible models for the destabilization of GpATM in the negatively charged membranes. In neutral membrane vesicles, the dimer is stabilized by the restricted lateral and angular motions (top). In the negatively charged membranes, however, interfacial positively charged Arg and Lys residues of TM helices are strongly attracted to the negatively charged lipid headgroups. The resulting adjustment of each monomeric TM helix relative to bilayer normal may cause the distortion of the dimer interface from the optimal side-chain packing destabilizing the dimer (middle). It is also possible that the optimal dimer packing is maintained, but the significant free energy cost is required to form a dimer from a structurally altered, electrostatistically stabilized monomer.
Membrane proteins occupy a significant fraction of the total membrane surface, reducing the area accessible to the free diffusion of the embedded proteins15. Grasberger et al. suggests that the excluded volume effect of inert proteins should increase the dimerization equilibrium constant by about 2.7 kcal/mol when the fractional occupancy of the inert proteins increases up to 0.5 41. We find the opposite effect, however. At a mass ratio of ~1, the GpATM dimer is destabilized by 4~5 kcal/mol. It seems likely that this effect is due to competition for the binding surface with other potential protein partners in the crowded membrane. Certainly, the GxxxG dimerization motif of GpATM is not uncommon in other membrane proteins42. We cannot exclude the possibility that the added membrane proteins may alter lipid structure, however14. Although the relative concentration of IMP in our experimental condition is less than in real E. coli cell membranes, the destabilization effect already exceeds the expected stabilization effect of excluded volume, indicating the importance of nonspecific interactions in determining the stability of helix-helix interactions cell membranes. The potential for non-specific contributions could be a significant complication when evaluating the effects of mutations using in vivo systems or predicting in vivo effects from in vitro studies.
We have made the first direct thermodynamic measurements on the role of the bilayer lateral pressures in the TM helix-helix associations of α-helical membrane proteins (Figure 3). Increasing the lateral pressure by 40 % inverted hexagonal phase forming PE increased the dimer stability by ΔΔGx = ~−1.5 kcal/mol while decreasing lateral pressure with 20 % micellar lysoPC exerted the opposite effect (ΔΔGx=~4.5kcal/mol destabilization). Many studies have demonstrated the profound effects of bilayer elastic properties on the structure, function, and folding of integral membrane proteins2. For example, the open states of several mechanosensitive channels are stabilized by addition of lysoPCs43–45. Electron paramagnetic resonance (EPR) studies showed that the incorporation of lysoPC induces a dramatic structural rearrangement of MscL44 and MscS45. The effects of the lateral pressure induced by PE lipids are manifested in opposite ways for two channel forming antimicrobial peptides, alamethicin46 and gramicidin47. The incorporation of PE lipids increases the conductance level and open lifetime of alamethicin, a transmembrane peptide, by facilitating the lateral association of the peptides. In contrast, PE lipids decrease the channel activity of gramicidin, which spans half of a full bilayer thickness. The modification of lateral pressure profile also significantly changes the refolding yield and kinetics of α-helical membrane proteins48,49 and the thermodynamic stability of β-barrel membrane proteins50. It is still not clear how and why the lateral pressure affects the conformation of membrane proteins. In general, the higher lateral pressure condition seems to favor the association and packing of the transmembrane secondary structure elements. Exposure of the protein surfaces of the associated and folded state of the membrane proteins may be more energetically costly under high lateral pressure. In addition, the hourglass shapes of most integral membrane proteins of known structures may relieve the negative monolayer curvature strains induced by PE lipids stabilizing the folded state50,51. In the case of GpATM, the helices splay from the central GxxxG packing interface52,53, so that the volume of the dimer should be higher at the edges of the bilayer—a structure that would be favored by the positive pressure in the center and negative pressure at the edges.
We were able to reproduce degree of destabilization by E. coli lipids in entirely synthetic lipid membranes by introducing negatively charged lipids. The electrostatic interactions between the negatively charged lipid headgroups and the positively charged residues in the cytosolic side of the GpATM played a prominent role in this phenomenon. As illustrated in Figure 7b, the interfacial charge interactions could either alter the structure of the dimer or the monomer, thereby reducing the net stability of the dimeric form. All biological membranes are essentially negatively charged and the positively charged residues of membrane proteins are significantly enriched near the bilayer-water interface in the cytoplasmic side of the membranes54. This biased charge distribution, called ‘the positive-inside rule’ serves as a strong determinant of membrane protein topology. Although the polar lipid headgoups mostly comprise the interfacial region, this region is not fully hydrated55. Therefore, electrostatic interactions can be much stronger than in the bulk water phase. It has been frequently observed that the hydrocarbon region of Lys or Arg side chains in membrane proteins are buried in the bilayer core and the polar charged group is anchored in the water-bilayer interface56. This snorkeling effect may help the stabilization and precise positioning of α-helical segments of membrane proteins in lipid bilayers57. Our results strongly suggest that the electrostatic interactions between the interfacial Lys and Arg residues and the negatively charged membranes 58, not only determine the topology but also may strongly affect the energetics of the TM helix-helix associations in membrane proteins or their complexes.
The relatively non-specific environmental factors examined in this work highlight a tool evolution can employ to adjust membrane protein interactions to a level appropriate for function. Indeed, evolutionary pressure does not generally favor optimal stability. Random mutations tend to drive proteins toward marginal stability and functional constraints can also demand low stability59. Protein stabilization involves a complex balance of forces involving interactions within the protein itself and between the protein and the environment1. Our work illustrates how evolution can employ the relatively structured membrane environment to modulate stability in accord with evolutionary pressure.
MATERIALS and METHODS
Cloning, expression and purification of SNGpATM with biotin acceptor peptide tag
The pET11a vector encoding staphylococcal nuclease-glycorphorinA transmembrane domain fusion (SNGpATM) was a gift from Karen Fleming (Johns Hopkins University). The detailed procedures for the cloning, expression, and purification of SNGpATM with biotin acceptor peptide (BAP) tag were described previously 60. The gene encoding SNGpATMRRKK/QQNN was generated by mutating Lys100 and Lys101 to Asn100 and Asn101 at the same time using the primer 5’-GGT ATT TCC TCA CTG ATA AAT AAC CTC GAG CAC CAC CAC followed by simultaneously substituting Arg96 and Arg97 for Gln96 and Gln97 with the primer 5’-TTA ATT TCT TAC GGT ATT CAA CAA CTG ATA AAT AAC CTC G.
Preparation of total E. coli inner membrane proteins
E. coli (BL21(DE3)Rp strain, Agilent Techonologies) cells were grown at 37 °C in LB media containing 0.5 mM chloramphenicol to an OD600nm ~ 0.9. The harvested cells were resuspended in 20 ml of 50 mM TrisHCl buffer (pH 8.0) containing 20 % glycerol and lyzed using a French press four times at 8,000 psi. The resuspension was diluted with an equal volume of the resuspension buffer and centrifuged at 4,000 g for 10 minutes. The supernatant was then centrifuged at 40,000 g for 90 minutes in an ultracentrifuge (Optima L-90K, Beckman-Coulter) to obtain the total membrane fraction. The pellet was resuspended and washed with 10 ml of 50 mM TrisHCl buffer (pH 8.0) containing 20 % sucrose and 4 M urea. The resuspension was loaded to the top of a continuous sucrose gradient (30~60 % sucrose) prepared in a Quick-Seal centrifuge tube (38×102 mm, Beckman Instruments) and centrifuged at 27,000 rpm in a 45 K rotor for 16 hours. The red-colored inner membrane fraction layer was collected in a 25 ml volume using a syringe with 18G needle. The collected inner membranes were diluted 5 times with 50 mM TrisHCl (pH 8.0) without sucrose and centrifuged again at 27,000 rpm for 1 hour in the 45 Ti rotor. The pellet was solubilized with 3 ml of 4 % decylmaloside (DM) solution (pH 8.0 50 mM TrisHCl, 200 mM NaCl) and centrifuged at 13,000 rpm for 5 minutes. The total concentrations of phospholipids and proteins in the supernatant were determined by an organic phosphate assay61 and a BioRad Dc assay, respectively. The final stock solution contained 4.2 mM of phospholipids and 8.5 mg/ml of proteins.
Preparation of proteoliposomes
E. coli polar extract lipids and synthetic phospholipids were purchased from Avanti polar lipids in chloroform solutions. Typically, 10 µmol of pure lipids or lipid mixtures dissolved in chloroform were added to glass tubes and dried under a stream of nitrogen gas. The residual solvent was further removed in a vacuum dessiccator for 2 hours. The dried lipid films were hydrated and solubilized in 1 ml of 3 % β-octylglucoside (Anatrace, Anagrade), 20 mM MOPS, 200 mM NaCl (pH 7.5). The SNGpATM stock solution (290~350 µM) in DM was added to the solubilized lipids at a final concentration of 10 µM and incubated at room temperature for 1 hour. SNGpATM was reconstituted into lipid vesicles by dialyzing against 250 sample volumes of 20 mM MOPS, 200 mM NaCl buffer solution (pH 7.5), with three buffer exchanges over 48 hours at 5 °C (Spectra/Por 25 kDa cut-off dialysis membrane, 7.5 mm diameter). The resulting proteoliposomes were passed through Nucleo track etch membrane (Whatman) with 200 nm pore-sizes using a mini-extruder (Avanti polar lipids) 15 times. To prepare the lipid vesicles containing 20 mol-% lysoPC (C18:1c9PC), the dialyzed pure C16:0C18:1c9PC proteoliposomes containing SNGpATM was mixed with the micellar solution of C18:1c9PC, incubated at room temperature for 5 hours, and extruded with the 200 nm pore size membrane. The typical average diameter of proteoliposomes was 110 ±60 nm as measured by dynamic light scattering (DynaPro light-scattering systems, Protein Solutions). The liposomal solutions were stored at 4 °C.
Determination of protein and lipid concentrations in proteoliposomes
The proteolipsomes prepared above were solubilized with an equal volume of 200 mM C8E5 solution (20 mM MOPS, 200 mM NaCl, pH 7.5). To determine SNGpATM concentration, the solubilized sample was titrated with wild-type mSA and incubated overnight. The total concentration of biotinylated pyrene-labeled SNGpATM was determined by fitting the stoichiometric binding data with a fixed parameter, Kd,biotin=10−14 M. The total lipid concentration in proteoliposome samples was measured by an organic phosphate assay61. The orientation distributions of reconstituted GpATM in proteoliposomes were determined by avidin binding assay as described previously24.
Monovalent streptavidin binding measurements in detergent micelles and bilayers
For binding measurements in detergent micelles, 2 µM of biotinylated pyrene-labeled wild-type or mutant SNGpATMs were titrated with monovalent streptavidin (mSA) in buffer solutions containing 40 mM DM (Anatrace, Anagrade), 20 mM 3-(N-morpholino)propanesulfonic acid (MOPS), and 200 mM NaCl (pH 7.5). A total volume of 70 µl of each sample was transferred to a 96-well UV-compatible microplate (BD-Falcon), sealed with a polyolefin tape, and incubated overnight at room temperature. Binding of mSA coupled with the dissociation of the SNGpATM dimer was monitored by pyrene monomer fluorescence at 390 nm with an excitation wavelength of 330 nm using a SpectraMax M5 plate reader (Molecular Devices). At the end of the measurements, excess free biotin was added to a final concentration of 2 mM and incubated more than 1 hour at room temperature to dissociate bound mSA from the biotinylated SNGpATM. The pyrene fluorescence intensity was measured at least five times for the biotin added samples and used as the background. The background-subtracted data were fitted to Eq. (1) to extract the dissociation constants (Kd,GpA) of the SNGpATM dimer.
For binding measurements in lipid vesicle solution, 20~40 µl of proteliposomes was mixed with various compositions of buffer/BSA (bovine serum albumin, Sigma)/mSA mixtures. BSA was added to match the total protein concentrations of the sample solutions which can affect osmolarity. The total molar concentration of BSA and mSA was adjusted to 100 µM in all sample solutions. A total volume of 80 µl of each sample was transferred to a microplate, sealed with a polyolefin tape. When the lipid vesicles were composed of neutral zwitterionic lipids such as PC and PE, the samples were incubated overnight. When the vesicles contained negatively charged lipids such as E. coli polar extract, PG, or PS, the samples were incubated for 2~3 hours. The same instrumental parameters as in the micellar solutions were used. The mSA-induced changes in the pyrene-fluorescence intensity were measured at least 5 times and averaged. At the end of the measurements, excess free biotin was added to a final concentration of 2 mM and incubated more than 6 hours for the samples containing strong biotin binding mSA mutants (mSA-S45A, -E44Q/S45A, -W79M, and -W79L) and for 1 hour for the samples containing weak biotin binding mSA mutants (mSA-N23A/S45A and –S27R) at room temperature to dissociate bound mSA from the biotinylated SNGpATM. The fluorescence data from the biotin-blocked samples, which were averaged from at least 5 times of measurements at least, served as a background. The background-subtracted data were fitted to the Eq. (1) or Eq. (2) to extract Kd,GpA of the SNGpATM dimer24.
| (1) |
| (2) |
, where Fθ is the change in the specific binding signal of mSA, Po is the mSA-accessible SNGpATM concentration, A1 and A2 are the amplitudes of the binding phase and initial fluorescence level, respectively, [mSA] is the total mSA concentration, and Kd,biotin is the intrinsic biotin dissociation constant for mSA. When an ideal two phase binding curve was obtained with a strong biotin binding mSA mutant (mSA-S45A, -E44Q/S45A, -W79M, and -W79L), a simultaneous fitting of the first and the second binding phase (Eq. (1)) was applied. But when a weaker mSA mutant was used (mSA-N23A/S45A and -S27R), the second binding phase was selectively fitted to Eq. (2). The onset of the second binding phase was identified by the breaking point of the first derivative of the overall binding curve24.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH Grants R01GM063919 and R01GM081783 to J.U.B.. H.H. is supported by the Leukemia and Lymphoma Society Career Development Award (Fellow). We thank Karen Fleming lab (Johns Hopkins University) and Alice Ting lab (MIT) for providing plasmids, and Bowie lab members for critically reading the manuscript.
Footnotes
Supporting Information Available. Supporting Figures 1–7. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Bowie JU. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 2.Andersen OS, Koeppe RE., 2nd Annu Rev Biophys Biomol Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 3.Sachs JN, Engelman DM. Annu Rev Biochem. 2006;75:707–712. doi: 10.1146/annurev.biochem.75.110105.142336. [DOI] [PubMed] [Google Scholar]
- 4.Morein S, Andersson A, Rilfors L, Lindblom G. J Biol Chem. 1996;271:6801–6809. doi: 10.1074/jbc.271.12.6801. [DOI] [PubMed] [Google Scholar]
- 5.Wallin E, von Heijne G. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Kruijff B. Curr Opin Chem Biol. 1997;1:564–569. doi: 10.1016/s1367-5931(97)80053-1. [DOI] [PubMed] [Google Scholar]
- 7.van Klompenburg W, de Kruijff B. J Membr Biol. 1998;162:1–7. doi: 10.1007/s002329900336. [DOI] [PubMed] [Google Scholar]
- 8.von Heijne G. Nat Rev Mol Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 9.Bogdanov M, Xie J, Dowhan W. J Biol Chem. 2009;284:9637–9641. doi: 10.1074/jbc.R800081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seddon JM. Biochim Biophys Acta. 1990;1031:1–69. doi: 10.1016/0304-4157(90)90002-t. [DOI] [PubMed] [Google Scholar]
- 11.Rietveld AG, Koorengevel MC, de Kruijff B. Embo J. 1995;14:5506–5513. doi: 10.1002/j.1460-2075.1995.tb00237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowhan W, Bogdanov M. Annu Rev Biochem. 2009;78:515–540. doi: 10.1146/annurev.biochem.77.060806.091251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raetz CR. Microbiol Rev. 1978;42:614–659. doi: 10.1128/mr.42.3.614-659.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitra K, Ubarretxena-Belandia I, Taguchi T, Warren G, Engelman DM. Proc Natl Acad Sci U S A. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelman DM. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 16.Matthews EE, Zoonens M, Engelman DM. Cell. 2006;127:447–450. doi: 10.1016/j.cell.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Moore DT, Berger BW, DeGrado WF. Structure. 2008;16:991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming KG, Engelman DM. Proc Natl Acad Sci U S A. 2001;98:14340–14344. doi: 10.1073/pnas.251367498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rath A, Tulumello DV, Deber CM. Biochemistry. 2009;48:3036–3045. doi: 10.1021/bi900184j. [DOI] [PubMed] [Google Scholar]
- 20.Lemmon MA, Flanagan JM, Treutlein HR, Zhang J, Engelman DM. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 21.Doura AK, Fleming KG. J Mol Biol. 2004;343:1487–1497. doi: 10.1016/j.jmb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 22.Bormann BJ, Knowles WJ, Marchesi VT. J Biol Chem. 1989;264:4033–4037. [PubMed] [Google Scholar]
- 23.Lemmon MA, Flanagan JM, Hunt JF, Adair BD, Bormann BJ, Dempsey CE, Engelman DM. J Biol Chem. 1992;267:7683–7689. [PubMed] [Google Scholar]
- 24.Hong H, Blois TM, Cao Z, Bowie JU. Proc Natl Acad Sci U S A. 2010;107:19802–19807. doi: 10.1073/pnas.1010348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duong MT, Jaszewski TM, Fleming KG, MacKenzie KR. J Mol Biol. 2007;371:422–434. doi: 10.1016/j.jmb.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Novicky L, Merzlyakov M, Hristov T, Hristova K. J Am Chem Soc. 2010;132:3628–3635. doi: 10.1021/ja910692u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blois TM, Hong H, Kim TH, Bowie JU. J Am Chem Soc. 2009;131:13914–13915. doi: 10.1021/ja905725n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Meer G, Voelker DR, Feigenson GW. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rand RP, Fuller N, Parsegian VA, Rau DC. Biochemistry. 1988;27:7711–7722. doi: 10.1021/bi00420a021. [DOI] [PubMed] [Google Scholar]
- 30.Crane JM, Tamm LK. Biophys J. 2004;86:2965–2979. doi: 10.1016/S0006-3495(04)74347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russ WP, Engelman DM. Proc Natl Acad Sci U S A. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleming K. G. J Mol Biol. 2002;323:563–571. doi: 10.1016/s0022-2836(02)00920-8. [DOI] [PubMed] [Google Scholar]
- 33.Fisher LE, Engelman DM, Sturgis JN. Biophys J. 2003;85:3097–3105. doi: 10.1016/S0006-3495(03)74728-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tate MW, Gruner SM. Biochemistry. 1987;26:231–236. doi: 10.1021/bi00375a031. [DOI] [PubMed] [Google Scholar]
- 35.Cantor RS. Biophys J. 1999;76:2625–2639. doi: 10.1016/S0006-3495(99)77415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gruner SM. Proc Natl Acad Sci U S A. 1985;82:3665–3669. doi: 10.1073/pnas.82.11.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rand RP, Parsegian VA. Biochim Biophys Acta. 1989;988:351–376. [Google Scholar]
- 38.Lewis BA, Engelman DM. J Mol Biol. 1983;166:211–217. doi: 10.1016/s0022-2836(83)80007-2. [DOI] [PubMed] [Google Scholar]
- 39.Op den Kamp JA, Kauerz MT, van Deenen LL. Biochim Biophys Acta. 1975;406:169–177. doi: 10.1016/0005-2736(75)90001-2. [DOI] [PubMed] [Google Scholar]
- 40.Epand RM, Bottega R. Biochim Biophys Acta. 1988;944:144–154. doi: 10.1016/0005-2736(88)90427-0. [DOI] [PubMed] [Google Scholar]
- 41.Grasberger B, Minton AP, DeLisi C, Metzger H. Proc Natl Acad Sci U S A. 1986;83:6258–6262. doi: 10.1073/pnas.83.17.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senes A, Engel DE, DeGrado WF. Curr Opin Struct Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Maingret F, Patel AJ, Lesage F, Lazdunski M, Honore E. J Biol Chem. 2000;275:10128–10133. doi: 10.1074/jbc.275.14.10128. [DOI] [PubMed] [Google Scholar]
- 44.Perozo E, Cortes DM, Sompornpisut P, Kloda A, Martinac B. Nature. 2002;418:942–948. doi: 10.1038/nature00992. [DOI] [PubMed] [Google Scholar]
- 45.Vasquez V, Sotomayor M, Cordero-Morales J, Schulten K, Perozo E. Science. 2008;321:1210–1214. doi: 10.1126/science.1159674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keller SL, Bezrukov SM, Gruner SM, Tate MW, Vodyanoy I, Parsegian VA. Biophys J. 1993;65:23–27. doi: 10.1016/S0006-3495(93)81040-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bezrukov SM. J Membr Biol. 2000;174:1–13. doi: 10.1007/s002320001026. [DOI] [PubMed] [Google Scholar]
- 48.Curran AR, Templer RH, Booth PJ. Biochemistry. 1999;38:9328–9336. doi: 10.1021/bi982322+. [DOI] [PubMed] [Google Scholar]
- 49.Curnow P, Lorch M, Charalambous K, Booth PJ. J Mol Biol. 2004;343:213–222. doi: 10.1016/j.jmb.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 50.Hong H, Tamm LK. Proc Natl Acad Sci U S A. 2004;101:4065–4070. doi: 10.1073/pnas.0400358101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bowie JU. Proc Natl Acad Sci U S A. 2004;101:3995–3996. doi: 10.1073/pnas.0400671101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith SO, Song D, Shekar S, Groesbeek M, Ziliox M, Aimoto S. Biochemistry. 2001;40:6553–6558. doi: 10.1021/bi010357v. [DOI] [PubMed] [Google Scholar]
- 53.MacKenzie KR, Prestegard JH, Engelman DM. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 54.Heijne G. Embo J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krepkiy D, Mihailescu M, Freites JA, Schow EV, Worcester DL, Gawrisch K, Tobias DJ, White SH, Swartz KJ. Nature. 2009;462:473–479. doi: 10.1038/nature08542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chamberlain AK, Lee Y, Kim S, Bowie JU. J Mol Biol. 2004;339:471–479. doi: 10.1016/j.jmb.2004.03.072. [DOI] [PubMed] [Google Scholar]
- 57.Liang J, Adamian L, Jackups R., Jr Trends Biochem Sci. 2005;30:355–357. doi: 10.1016/j.tibs.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 58.Hong M, Su Y. Protein Sci. 2011;20:641–655. doi: 10.1002/pro.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taverna DM, Goldstein RA. Proteins. 2002;46:105–109. doi: 10.1002/prot.10016. [DOI] [PubMed] [Google Scholar]
- 60.Hong H, Joh NH, Bowie JU, Tamm LK. Methods Enzymol. 2009;455:213–236. doi: 10.1016/S0076-6879(08)04208-0. [DOI] [PubMed] [Google Scholar]
- 61.Ames BN, Dubin DT. J Biol Chem. 1960;235:769–775. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.