Abstract
Background/Aims
Gastric epithelial dysplasia is considered a precancerous lesion with a variable clinical course. There is disagreement, however, regarding histology-based diagnoses, which has led to confusion in choosing a therapeutic plan. New objective markers are needed to determine which lesions progress to true malignancy. We measured LINE-1 methylation levels, which have been reported to strongly correlate with the global methylation level in gastric epithelial dysplasia and intramucosal cancer.
Methods
A total of 145 tissue samples were analyzed by two histopathologists. All tissues were excised by therapeutic endoscopic mucosal resection and paired with adjacent normal tissue samples. A modified long interspersed nucleotide elements-combined bisulfite restriction analysis (COBRA-LINE-1) method was used.
Results
Gastric epithelial dysplasia and intramucosal cancer tissues had significantly lower levels of LINE-1 methylation than adjacent normal gastric tissues. High-grade dysplasia and intramucosal cancer were distinguishable from low-grade dysplasia based on LINE-1 methylation levels. Furthermore, the distinction could be determined with high sensitivity and specificity, as shown by the receiver operating characteristic (ROC) curve (AUC, 0.82; 95% confidence interval, 0.74 to 0.88).
Conclusions
LINE-1 methylation levels may provide a diagnostic tool for identifying high-grade dysplasia and intramucosal cancer.
Keywords: LINE-1 methylation, Gastric epithelial dysplasia, Intramucosal cancer
INTRODUCTION
Gastric epithelial dysplasia is considered to be a precursor lesion of gastric cancer, and has various clinical courses. Some progress to adenocarcinoma, whereas others persist unchanged for many years.1,2 Moreover, without uniform and definite criteria for the diagnosis of gastric epithelial dysplasia, treatment is inconsistent and often results in over-treatment of gastric epithelial dysplasia by surgical resection.3-5 In an attempt to resolve this problem, the revised Vienna classification has been proposed,6,7 providing a consensus on guidelines for clinical management. The removal of category 3 (low grade dysplasia) lesions may not be necessary, whereas the removal of category 4 lesions (high grade dysplasia and intramucosal cancer) must be obligatory. But this classification is not perfect and there are frequently cases that do not easily fit into the diagnostic categories of the revised Vienna classification. Approximately 15% to 30% of low grade dysplasia progress to high grade lesions and/or adenocarcinomas,5,8 so this classification system needs a complementary marker. Moreover, the inter-observer and intra-observer variation of histopathologists should be never neglected in the diagnosis of gastric epithelial dysplasia and intramucosal cancer.
The goal of this study was to investigate the level of LINE-1 methylation in gastric epithelial dysplasia and intramucosal cancer samples as well as the level of LINE-1 methylation in the adjacent mucosa. The results could potentially be of clinical interest and we hoped to provide the pathologist with an objective marker to distinguish between low grade dysplasia and high grade dysplasia or between high grade dysplasia and intramucosal cancer.
MATERIALS AND METHODS
All tissues were excised by therapeutic endoscopic mucosal resection. The diagnosis of tissue sample was confirmed by two different histopathologists according to the revised Vienna classification; when they disagreed, the tissue sample was excluded from the study. The lesions were histopathologically assigned to divide into 3 groups according to the revised Vienna classification system: low grade dysplasia (category 3), high grade dysplasia (category 4.1) and intramucosal cancer (categories 4.2, 4.3, and 4.4).
All normal tissues had grossly intact mucosa and were at least 1 cm from the mucosal lesion; they were obtained by gastric biopsy just after an endoscopic mucosal resection. The microscopic examination showed no evidence of malignant cells. Each patient was classified as Helicobacter pylori (H. pylori) positive or negative according to the histological results. In the present study, the resection specimen and gastric biopsy of surrounding mucosa were stained with hematoxylin and eosin and silver stains. To assess H. pylori state accurately, two biopsies were taken both from antrum and corpus after 4 weeks of the endoscopic resection. We evaluated with CLO test or histological examination.
1. DNA extraction
Four-micrometer-thick tissue sections from the dysplasia/cancer and normal tissues were placed on a glass slide and stained with hematoxylin and eosin. The diagnosis of the tissue samples was confirmed by two different histopathologists. Prior to DNA extraction, all the microdissected tumor sites were checked for the tumor cell contents ≥70% using a stereomicroscope under a ×40 magnification.
Two ten-micrometer-thick tissue sections from cancer samples and normal tissues were placed on glass slides. The tissue sections were added by xylene (1 mL), incubated for 10 minutes and repeated 3 times. These were then dehydrated in graded ethanol solutions (100% ethanol, 1 mL), dried without a cover glass for 10 minutes and repeated three times. The DNA was extracted from the tissues with 20 uL of extraction buffer (100 mmol/L Tris-HCl; 2 mmol/L ethylene diamine tetraacetic acid [EDTA], pH 8.0; 400 ug/mL of proteinase K) at 55℃ overnight. The tubes were boiled for 7 minutes to inactivate the proteinase K, and cooling on ice. The solution was added by 20 uL (Phenol:Chloloform:Isoamyl alcohol, 25:24:1) and centrifuged at 12,000 rpm, 4℃ for 5 minutes. This process was de-proteinization to extract DNA of tissue. The supernatant was added by ethanol (1 mL, 100%) and the tube was gently inverted. It was incubated at -20℃ for 10 minutes and centrifuged at 12,000 rpm, 4℃ for 5 minutes. DNA pellet was made by washing with ethanol (1 mL, 75%) and centrifuged at 14,000 rpm for 5 minutes. The pellet was dissolved by adding 20 uL of dextrose water, and then 1 uL of this extract was used for each polymerase chain reaction (PCR) amplification.
2. Assessment of LINE-1 methylation status
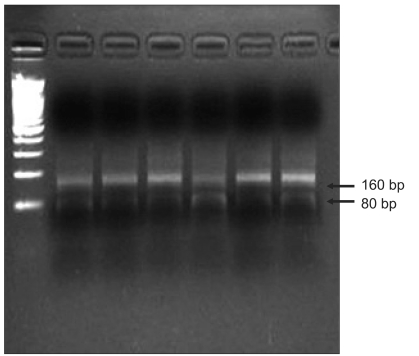
A modified long interspersed nucleotide elements-combined bisulfite restriction analysis (COBRA LINE-1) method was used to analyze LINE-1 methylation status of the cancers and normal mucosa.9-11 This method is based on the principle that cytosine in DNA is converted to uracil when DNA is treated with sodium bisulfite, whereas methylated cytosine is protected from the conversion. Thus, the methylated and unmethylated cytosine could be distinguished by digestion with a restriction enzyme that recognizes sequences containing CpG. The extracted DNA was treated with sodium bisulfite, and isolated using the EZ DNA methylation kit (Zymo Research, Orange, CA, USA). Bisulfite-treated DNA was amplified by 40 cycles of PCR with two primers, LINE3 (5V-GYGTAAGGGGTTAGGGAGTTTTT) and LINE4 (5V-AACRTAAAACCCTCCRAACCAAATATAAA), at an annealing temperature of 50℃. The PCR products were digested with the TaqI restriction enzyme, which recognizes TCGA, for 1 hour at 65℃, and then were separated by electrophoresis on 2% agarose gels. The densities of the digested and undigested bands were obtained by scanning with Gel Doc XR (Bio-Rad, Hercules, CA, USA) and scoring with Quantity One Software (Bio-Rad). The ratio of the digested fragments (80 bp) derived from the methylated DNA divided by the sum of the digested fragments and the undigested fragments (160 bp) derived from the unmethylated DNA represents the fractional methylation (expressed as a percentage) at the LINE TaqI site.
3. Statistical analysis
For the quantitative variables, the mean and its standard error were calculated. For the qualitative variables, the percent and its 95% confidence interval (CI) were calculated. We used the χ2 test to analyze the association between the H. pylori status and other baseline characteristics. For comparison of age and the level of LINE-1 methylation we used the unpaired t test and one way ANOVA. The diagnostic yield of the level of LINE-1 methylation, for the prediction of premalignant or early cancer lesions, was calculated using the area under the receiver operating characteristic (ROC) curve. The area under the ROC curve (AUC) is the measure of separation of two probability distributions: excellent for AUC values greater than 0.9, good for 0.8 to 0.9, fair for 0.7 to 0.8, poor for 0.6 to 0.7, and failure for values less than 0.6. We used the SPSS statistical package (version 12.0.1; SPSS Inc., Chicago, IL, USA) for all analyses.
RESULTS
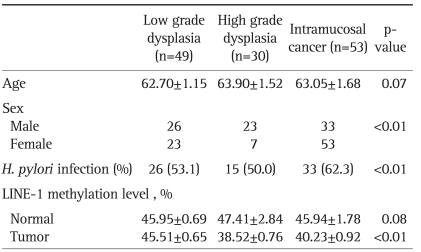
Total 145 tissue samples were examined and thirteen tissue samples were excluded because of disagreement on diagnosis and inappropriate preparation of tissue. The concordance rate between the two histopathologists was 91.0% and 132 patients were analyzed. Seventy nine cases of gastric epithelial dysplasia and 53 intramucosal carcinoma tissue samples were paired with adjacent normal mucosal tissues. Forty-one patients (51.9%) with gastric epithelial dysplasia and 33 patients (62.3%) with intramucosal carcinoma had associated H. pylori infection (p=0.24). When the gastric epithelial dysplasia was divided according to the revised Vienna classification, 49 tissues were low grade and 30 were high grade dysplasia.
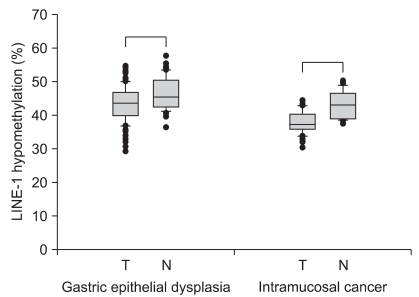
The level of LINE-1 methylation in the gastric epithelial lesions and adjacent normal mucosa, from the same subject, was examined by a modified COBRA LINE-1 method (Fig. 1). The gastric epithelial dysplasia and cancer tissues had significantly lower levels of LINE-1 methylation than the adjacent normal tissues (Fig. 2). In gastric epithelial dysplasia, the dysplasia lesion showed a significantly lower LINE-1 methylation level (42.85±0.63%) compared to normal epithelium (46.35±0.69%) (p<0.01). In intramucosal cancer, the cancer lesion showed a significantly lower LINE-1 methylation level (40.23±0.92%) compared to normal epithelium (45.94±1.78%) (p<0.01). When the dysplastic lesions with H. pylori infection compared to the lesions without infection, there was no significant difference (42.46±0.88 and 43.22±0.89, p=0.27).
Fig. 1.
Assessment of LINE-1 hypomethylation status using the COBRA-LINE-1 method. Calculations are based on the ratio of the digested bands divided by the sum of the digested and undigested bands, as described in the materials and methods section.
Fig. 2.
LINE-1 hypomethylation levels in gastric epithelial dysplasia and intramucosal cancer. When compared to adjacent normal mucosa, gastric epithelial dysplasia and cancer tissues have significantly lower LINE-1 methylation levels. Box plots illustrate median values, 25th and 75th percentiles, and outliers on a linear scale. The unpaired t-test is applied for nonparametric statistical analysis, and a p value of less than 0.05 is considered significant.
N, normal; T, tumor.
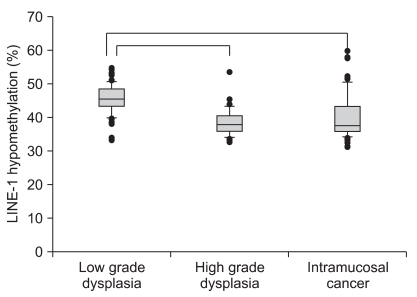
The tissue samples were divided into three groups according to the revised Vienna classification: low grade dysplasia, high grade dysplasia and intramucosal cancer. Forty nine patients with low grade dysplasia, 30 patients with high grade dysplasia and 53 with intramucosal cancer were evaluated (Table 1). The high grade dysplasia and intramucosal cancer (category 4) had lower level of LINE-1 methylation than the low grade dysplasia (category 3) (Fig. 3). However, there was no difference between the high grade dysplasia and the intramucosal cancer with regard to the LINE-1 methylation level (p=0.21).
Table 1.
Basal Characteristics of the Enrolled Patients
Fig. 3.
LINE-1 hypomethylation levels in gastric epithelial neoplasias categorized using the revised Vienna classification. When tissue samples are divided into three groups according to the revised Vienna classification, high-grade dysplasia and intramucosal cancer (category 4) shows significantly lower LINE-1 methylation levels than low-grade dysplasia (category 3). Box plots illustrate median values, 25th and 75th percentiles, and outliers on a linear scale. An unpaired t-test and one way ANOVA are applied for nonparametric statistical analyses, and a p value of less than 0.05 is considered significant.
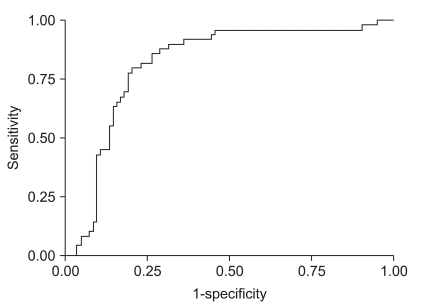
We checked the reproducibility of this method by retest reliability, which measured stability over time. It was administering the same test to the same subjects at two points in time. For continuous data, consensus was measured by intraclass correlation (ICC). We performed the quantitative methylation-specific PCR three times or more, and the data exhibited a high ICC (0.82; 95% CI, 0.74 to 0.88). Also, we evaluated the level of LINE-1 methylation for the assessment of premalignant lesions. ROC curve analysis differentiated the high grade dysplasia/intramucosal cancer (categoty 4) from the low grade dysplasia (category 3). The ROC curve represents the specificity and sensitivity of the diagnostic criteria for high grade dysplasia/intramucosal cancer. The area under the ROC curve (AUC) measures the overall performance of a diagnostic test and is interpreted as the average sensitivity for all possible values of specificity. The results showed that high grade dysplasia and intramucosal cancer were accurately diagnosed using the LINE-1 methylation level (AUC=0.82; 95% CI, 0.74 to 0.88) (Fig. 4).
Fig. 4.
Receiver operating characteristic (ROC) curve for the diagnosis of high-grade dysplasia/intramucosal cancer. Gastric high-grade dysplasia and intramucosal cancer are accurately diagnosed using LINE-1 methylation levels (AUC, 0.82; 95% confidence interval, 0.74 to 0.88).
DISCUSSION
Epigenetics is the study of the modification of gene expression without change of the underlying DNA sequence. The best-known epigenetic marker is DNA methylation.12-14 In cancer cells, DNA methylation changes are characterized by regional CpG island hypermethylation and generalized genomic hypomethylation. Although both kinds of changes are observed simultaneously, these two changes are thought to be independent events.15 Several studies suggest that genome wide hypomethylation generally arises earlier and is strongly linked to chromosomal instability, whereas hypermethylation occurs in promoters and is usually a later event.16-19 Moreover, during the development of a neoplasm, the level of methylation decreases as the lesion progresses from a benign proliferation of cells to an invasive cancer.19-21 In gastric carcinogenesis, changes in human DNA methylation appear to be an early molecular event, and most of the methylation studies have focused on aberrant methylation of tumor associated genes.22-24 However, there is limited data currently available on the level of global DNA methylation in precursor lesions of gastric cancer.25,26 Differential level of global DNA methylation between dysplastic lesion and early cancer in multistep gastric carcinogenesis has not been described in the literature.
LINE-1, a highly repeated interspersed human retrotransposon, is ubiquitous and constitutes approximately 17% of the human genome. Its methylation status reflects the genome wide methylation level. LINE-1 has been shown to be responsible for the overall losses of DNA methylation, and COBRA LINE-1 method is suitable for indirectly evaluating whole genome hypomethylation.10,16 The levels of LINE-1 methylation were significantly different among different tissue types. Most carcinomas including breast, colon, bladder, liver, prostate and stomach, revealed a greater level of LINE-1 hypomethylation than their normal tissue counterparts, whereas the levels in thyroid, esophageal tissue and lymphoma did not discriminate between the tumor and normal tissue.10 Since routine formalin-fixed paraffin-embedded tissues are suitable of the COBRA LINE-1 analysis, identification of this molecular event might be a useful adjunct to the standard pathological examination of the excised specimen.
In the present study, we applied COBRA LINE-1 method to efficiently and quantitatively evaluate LINE-1 methylation status of gastric epithelial dysplasia and intramucosal cancer. We evaluated how global hypomethylation evolves during the multistep process of carcinogenesis of the gastric cancer. The results showed that gastric epithelial dysplasia and cancer tissues had significantly lower level of LINE-1 methylation than adjacent normal tissues. In addition, the level of LINE-1 methylation correlated well with gastric lesions defined by the revised Vienna classification. It might substantiate the distinction between the low grade and high grade dysplasia/intramucosal cancer. Moreover, the reproducibility of the test showed relatively high ICC value (0.816; 95% CI, 0.736 to 0.876). These findings were very promising. Next, we plan to determine whether the precancerous lesions have different global DNA methylation level depending on whether there is invasive carcinoma or not. Unfortunately, the sample size of this study was too small and validation study was not performed. Also, it should be necessary to evaluate a methylation profile between low grade dysplasia and reactive changes.
Correa suggested a human model of gastric carcinogenesis and postulated that hyperproliferation caused by H. pylori gastritis is the starting point of a sequence of events that leads to gastric cancer.27 Previously several investigators proposed that aberrant DNA methylation of gastric mucosa infected by H. pylori was possibly associated with gastric cancer risk.28,29 In recent study, the association between methylation abnormality and H. pylori infection is debatable. The levels of LINE-1 methylation decrease from the chronic gastritis to gastric cancer stages, regardless of the status of H. pylori infection.26 Our results showed that lesions with H. pylori infection had a slightly lower level of LINE-1 methylation compared to the lesions without H. pylori infection; however, these differences were not significant. It had several limitations that a small number of patients were enrolled and the controlled group was absent. We did not consider whether the enrolled patients had past exposure to H. pylori.
In summary, the level of LINE-1 methylation might be considered as a useful diagnostic tool for the differentiation of high grade dysplasia/intramucosal cancer, and help to determine the appropriate treatment strategy for patients.
ACKNOWLEDGEMENTS
The authors have declared no conflicts of interest.
Sources of support: None.
References
- 1.Kamiya T, Morishita T, Asakura H, Miura S, Munakata Y, Tsuchiya M. Long-term follow-up study on gastric adenoma and its relation to gastric protruded carcinoma. Cancer. 1982;50:2496–2503. doi: 10.1002/1097-0142(19821201)50:11<2496::aid-cncr2820501140>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 2.Orlowska J, Jarosz D, Pachlewski J, Butruk E. Malignant transformation of benign epithelial gastric polyps. Am J Gastroenterol. 1995;90:2152–2159. [PubMed] [Google Scholar]
- 3.Schlemper RJ, Itabashi M, Kato Y, et al. Differences in diagnostic criteria for gastric carcinoma between Japanese and Western pathologists. Lancet. 1997;349:1725–1729. doi: 10.1016/S0140-6736(96)12249-2. [DOI] [PubMed] [Google Scholar]
- 4.Schlemper RJ, Kato Y, Stolte M. Well-differentiated adenocarcinoma or dysplasia of the gastric epithelium: rationale for a new classification system. Verh Dtsch Ges Pathol. 1999;83:62–70. [PubMed] [Google Scholar]
- 5.Stolte M. Diagnosis of gastric carcinoma: Japanese fairy tales or Western deficiency? Virchows Arch. 1999;434:279–280. doi: 10.1007/s004280050342. [DOI] [PubMed] [Google Scholar]
- 6.Schlemper RJ, Riddell RH, Kato Y, et al. The Vienna classification of gastrointestinal epithelial neoplasia. Gut. 2000;47:251–255. doi: 10.1136/gut.47.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stolte M. The new Vienna classification of epithelial neoplasia of the gastrointestinal tract: advantages and disadvantages. Virchows Arch. 2003;442:99–106. doi: 10.1007/s00428-002-0680-3. [DOI] [PubMed] [Google Scholar]
- 8.Lauwers GY, Riddell RH. Gastric epithelial dysplasia. Gut. 1999;45:784–790. doi: 10.1136/gut.45.5.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalitchagorn K, Shuangshoti S, Hourpai N, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 11.Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holliday R. The inheritance of epigenetic defects. Science. 1987;238:163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 14.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 15.Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr Top Microbiol Immunol. 2006;310:251–274. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert N, Doucet AJ, Bucheton A. Genomic instability associated with human LINE-1 rétrotransposition. J Soc Biol. 2004;198:419–424. [PubMed] [Google Scholar]
- 17.Ehrlich M. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J Nutr. 2002;132(8 Suppl):2424S–2429S. doi: 10.1093/jn/132.8.2424S. [DOI] [PubMed] [Google Scholar]
- 18.Cho NY, Kim BH, Choi M, et al. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007;211:269–277. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 19.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 20.Luczak MW, Jagodziński PP. The role of DNA methylation in cancer development. Folia Histochem Cytobiol. 2006;44:143–154. [PubMed] [Google Scholar]
- 21.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 22.Fang JY, Xiao SD. Alteration of DNA methylation in gastrointestinal carcinogenesis. J Gastroenterol Hepatol. 2001;16:960–968. doi: 10.1046/j.1440-1746.2001.02554.x. [DOI] [PubMed] [Google Scholar]
- 23.Kang GH, Shim YH, Jung HY, Kim WH, Ro JY, Rhyu MG. CpG island methylation in premalignant stages of gastric carcinoma. Cancer Res. 2001;61:2847–2851. [PubMed] [Google Scholar]
- 24.Lee JH, Park SJ, Abraham SC, et al. Frequent CpG island methylation in precursor lesions and early gastric adenocarcinomas. Oncogene. 2004;23:4646–4654. doi: 10.1038/sj.onc.1207588. [DOI] [PubMed] [Google Scholar]
- 25.Cravo M, Pinto R, Fidalgo P, et al. Global DNA hypomethylation occurs in the early stages of intestinal type gastric carcinoma. Gut. 1996;39:434–438. doi: 10.1136/gut.39.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SY, Yoo EJ, Cho NY, Kim N, Kang GH. Comparison of CpG island hypermethylation and repetitive DNA hypomethylation in premalignant stages of gastric cancer, stratified for Helicobacter pylori infection. J Pathol. 2009;219:410–416. doi: 10.1002/path.2596. [DOI] [PubMed] [Google Scholar]
- 27.Correa P. A human model of gastric carcinogenesis. Cancer Res. 1988;48:3554–3560. [PubMed] [Google Scholar]
- 28.Maekita T, Nakazawa K, Mihara M, et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12(3 Pt 1):989–995. doi: 10.1158/1078-0432.CCR-05-2096. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima T, Maekita T, Oda I, et al. Higher methylation levels in gastric mucosae significantly correlate with higher risk of gastric cancers. Cancer Epidemiol Biomarkers Prev. 2006;15:2317–2321. doi: 10.1158/1055-9965.EPI-06-0436. [DOI] [PubMed] [Google Scholar]