Abstract
Background/Aims
Krüppel-like factor 4 (KLF4) is an epithelial-specific transcription factor primarily expressed in the gastrointestinal tract that mediates growth arrest in the colonic epithelium. We tried to find whether KLF4 expression is associated with the progression and differentiation of colorectal cancer.
Methods
We detected KLF4 expression in 109 colorectal specimens (40 normal appearing mucosa, 7 adenomas, and 62 carcinomas) by immunohistochemistry using a tissue microarray. Western blot and RT-PCR analyses were also performed.
Results
The upregulation of KLF4 expression in carcinoma tissue was statistically significant (p<0.05) when compared to normal appearing mucosa. The negative and weak positive staining rates in normal appearing mucosa, adenoma, and carcinoma were 42.5%, 71.4%, and 82.3%, respectively, indicating a decreased degree of KLF4 expression over the course of progressive transformation of normal cells into malignant derivatives. KLF4 protein levels showed no correlation with sex, age, or metastatic state (p>0.05), while KLF4 protein expression correlated with the diagnostic stage (p<0.05). Furthermore, strong KLF4 staining was detected in 22.9% (11/48) and 0% (0/14) of well/moderately and poorly differentiated colorectal cancers, respectively. Our results clearly indicate that KLF4 protein expression significantly correlates with the degree of differentiation in colorectal cancers (p<0.05). KLF4 expression in RKO cells is also upregulated by butyrate, an inducer of differentiation.
Conclusions
Downregulation of KLF4 expression may lead to more poorly differentiated tumors.
Keywords: Krüppel-like factor 4, Differentiation, Colorectal cancer, Immunohistochemisty
INTRODUCTION
Colon and rectum cancers (CRC) accounted for about 1.23 million new cases in 2008 (9.7% of the world total). In terms of incidence, colorectal cancers rank third in frequency.1 At present time, with the development of living conditions and changes of life behaviors, CRC has become more and more frequent in China. Several lines of evidence indicated that tumorigenesis is a multistep process and these steps reflect genetic alterations that drive the progressive transformation of normal human cells into highly malignant derivatives.2 Investigators have made significant progress in understanding the molecular mechanisms that lead to colorectal cancer. In recent days, more studies were focused on the role of transcription factor in regulating abnormal cell growth.
Krüppel-like factor 4 (KLF4), first identified as a gut epithelial enriched gene,3 is a member of the KLF family of conserved zinc-finger-containing eukaryotic transcription factors. It encodes a transcription factor with three C2H2 zinc fingers in the carboxyl terminus.4 KLF4, formerly known as gut-enriched Krüppel-like factor (GKLF) or epithelial zinc finger (EZF), is expressed in the epithelia of skin, lung, gastrointestinal tract and several other organs.
KLF4 is an important regulator of cell cycle in vitro. It controls the G1/S cell cycle checkpoint, and is also required to prevent the G2/M transition after DNA damage. KLF4 is associated with both active and repressive genes that are involved in cell-cycle regulation and differentiation.5 Taken together, these studies point to a wide-ranging and crucial effect of KLF4 in maintaining the integrity of the cell cycle. The observation that KLF4 exerts an important checkpoint function suggested that KLF4 may have a tumor suppressor effect.
A survey of the tissue distribution in adult mice revealed that KLF4 was highly expressed in terminally differentiated, post-mitotic epithelial cells of the intestinal tract.3 Disorder of the expression of KLF4 has been associated with some kinds of carcinoma, for example, KLF4 was increased during the progression of breast cancer6 and dysplastic oral squamous epithelium,7 while it was decreased in colorectal8 and esophageal cancer,9 etc.
Many researches are focused in colorectal cancer in vivo and in vitro. Some studies show the lower expression of KLF4 in colorectal cancer, but there is no any report on this gene expression in colorectal cancer in Chinese population. Here, we detected the expression of KLF4 in 109 colorectal specimens (40 normal appearing mucosa, 7 adenoma, and 62 carcinoma) to study the relationship between KLF4 expression and clinicopathological parameters, such as sex, age, TNM stage, metastasis state and differentiation. We tried to find the evidence in vivo that if KLF4 is associated with the progression of colorectal cancer and whether the expression of KLF4 is related to the differentiation state.
MATERIALS AND METHODS
1. Tissue procurement
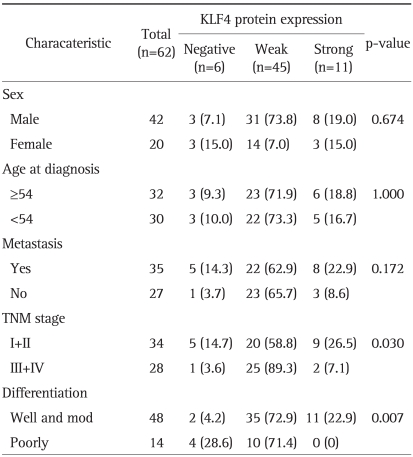
Archived paraffin blocks of formalin-fixed surgical specimens corresponding to 109 cases of colorectal tissues, diagnosed between 2001 and 2003, were obtained from the first affiliated hospital of Anhui Medical University. Fresh colorectal tumor and adjacent normal-appearing colorectal mucosa were also obtained from the hospital. The specimens were neither treated by radiotherapy nor chemotherapy. Patient characteristics were summarized in Table 1 (The ages of the patients ranged from 17 to 78 years).
Table 1.
KLF4 Expression Characteristics of Patients
Data are presented as number (%).
Fisher's exact probabilistic method is done to determine the statistic significance of the relationship of KLF4 expression of various variables.
2. Tissue microarray (TMA)
According to the H&E stain of pathological section, the tissue blocks were selected and be marked with the typical areas in the section by two pathologists. The TMAs were assembled using a tissue-arraying instrument, consisting of thin-walled stainless steel biopsy needles and stylets used to empty and transfer the needle content out of the paraffin block (recipient block). A large diameter stylet (1.5 mm) was used for sampling, and non-necrotic areas of the blocks were routinely sampled with two replicated core samples of tumor and normal (if present) regions from each donor block. To combine donor cores with the recipient block, the paraffin wax was reheated for 1 hour at 55℃. Forty-two tissues were included in each tissue array block. Sections of 4 µm were cut from the resulting microarray one day before immunostaining.
3. Immunohistochemistry
TMAs were deparaffinized, rehydrated and immersed in 3% hydrogen peroxide methanol solution for 10 minutes at room temperature. The sections was incubated with H-180 polyclonal rabbit antibody against human KLF4 (Santa Cruz Biotechology, Santa Cruz, CA, USA) at 4℃ for 14 hours, and sequentially incubated with biotinylated goat anti-rabbit IgG and streptavidin-horseradish peroxidase. Streptavidin-peroxidase method was routinely used.
4. Immunoscoring
All slides were examined and scored independently by two pathologists. A positive reaction was indicated by a reddish-brown precipitate in the cells. Depending on the percentage of positive cells and staining intensity, KLF4 staining was classified into three groups: negative, weak positive, and strong positive. Specifically, the percentage of positive cells was divided into five grades (percentage scores): <10% (0), 10-25% (1), 25-50% (2), 50-75% (3), and >75% (4). The intensity of staining was divided into four grades (intensity scores): no staining (0), light brown (1), brown (2), and dark brown (3). KLF4 staining positivity was determined by the formula: overall scores=percentage score×intensity score. The overall score of ≤3 was defined as negative, of >3-≤6 as weak positive, and of >6 as strong positive.10
5. Statistical analysis
Statistical tests were performed using Stata 8.0 software (Stata Co., College Station, TX, USA). Proportions were compared by χ2 test and Fisher's exact test.
6. Cell culture
Human colorectal cancer cell line RKO was cultured in DMEM medium (high glucose) supplemented with 10% (v/v) bovine serum. KLF4 expression level was detected in RKO cells treated with 0 to 3 mM butyrate for 4 hours.
7. RT-PCR
RNA extractions and PCR were carried out with kit (TaKaRa RNA PCR Kit (AMV) ver.3.0; TaKaRa, Shiga, Japan), according to the manufacturer's instructions. For detecting the expression of KLF4 mRNA, primers which amplify the coding sequence of KLF4 cDNA (+864 to +1298) were used. The forward primer was 5'-CCACCGGCCGGCTGCACACGACT-3', The reverse primer was 5'-TCATCTGAGCGGGCGAATTTCCATCCACA-3'.9 The positive control was β-actin, while the negative control was water. Each PCR reaction was hot started at 94℃ for 5 minutes, and then amplified for 30 cycles (94℃ for 30 seconds, 56℃ for 40 seconds, and 72℃ for 30 seconds). PCR products were visualized on a 2% agarose gel stained by ethidium bromide.
8. Western blot analysis
Protein samples were isolated from fresh specimens of cancerous or noncancerous portions. Standard western blot was performed with the H-180 polyclonal rabbit antibody against human KLF4 (Santa Cruz Biotechnology).
RESULTS
1. KLF4 protein expression of colorectal normal mucosa, adenoma, and carcinoma
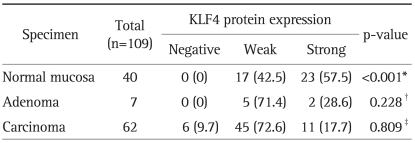
KLF4 protein expression was detected in 40 normal appearing mucosa, 7 adenoma, and 62 carcinoma (Table 2). The negative/weak positive staining of KLF4 was 42.5% in normal mucosa, while the rate was up to 71.4% and 82.3% in adenoma, and carcinoma. A decreased tendency was evident, though not significant, in KLF4 expression in the course of normal, adenoma, and carcinoma. Scoring carefully, the strong positive rate of normal appearing mucosa, adenoma, and carcinoma were 57.5%, 28.6%, and 17.7%. The staining patterns were significant different between normal mucosa and carcinoma (p<0.01).
Table 2.
KLF4 Protein Expression in Colorectal Normal Mucosa and Carcinoma
Data are presented as number (%).
Fisher's exact test is done with the STATA software program to determine the statistical significance of the level of expression of KLF4 in different type tissue specimens.
*Normal mucosa versus carcinoma; †Normal mucosa versus adenoma; ‡Adenoma versus carcinoma.
2. Characteristics of the patients and the KLF4 protein expression
To investigate what properties of colorectal carcinoma are associated with KLF4 expression, we identified 109 surgical specimens which were well characterized for clinicopathological parameters, including sex, age at diagnosis, metastasis state, TNM stage11 and differentiation (Table 1). Obviously, the level of KLF4 had no correlation with the sex, age, and metastasis state (p>0.05). Meanwhile, strong KLF4 staining was detected higher in well and moderately differentiated colorectal cancer than that in poorly differentiated colorectal cancer (22.9% vs 0%, p<0.01). In this case, the expression of KLF4 protein was significantly correlated with the differentiation in the colorectal cancer (p<0.01). Loss of expression of KLF4 or loss of its expression pattern may lead to more poorly differentiation. The KLF4 expressions of specimens of stage I or II were also differed from the specimens that are diagnosis in the stage of III or IV (p<0.05).
3. Immunohistochemistry for KLF4 in colon and rectum
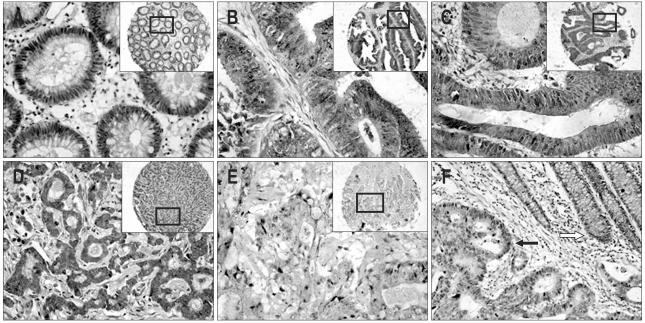
KLF4 expression was examined in TMAs collected from histology sections of normal colon, adenomatous polyp, and colon cancer. The immunostain patterns differed in different types of specimens. Strongly KLF4 immunostains were predominantly cytoplasmic at normal crypt epithelium, and mainly distributed around the nucleus, which showed obvious cell polarity (Fig. 1A). In colonic adenoma (Fig. 1B), cell polarity was also observed, while the localization of the KLF4 immunostains were cytoplasmic and nuclear in the crypt epithelium. Downregulation of KLF4 expression and loss of cell polarity were detected in the course of malignant transformation from normal to carcinoma (Fig. 1F).
Fig. 1.
Immunostaining of human colorectal cancers with anti-KLF4 antibody (original magnification, ×200). Staining is indicated as a brown precipitate. Unstained nuclei appear blue from the hematoxylin counterstain. (A) Normal appearing colonic mucosa. KLF4 immunostains are predominantly cytoplasmic in the normal crypt epithelium and primarily distributed around the nucleus, a pattern clearly indicative of cell polarity. (B) Colonic villous adenoma. KLF4 immunostains are cytoplasmic and nuclear in the crypt epithelium. Cell polarity is observed. (C) Tubular adenocarcinoma (well-differentiated). KLF4 staining is strong in the cytoplasm and clearly observed in the nuclei of some cells. (D) Papillary adenocarcinoma (moderately differentiated). KLF4 immunostaining is diffusely distributed in the cytoplasm. Several nuclear yellow stains are also observed. (E) Mucous adenocarcinoma (poorly differentiated). KLF4 immunostaining is predominantly cytoplasmic and is weakly positive. Most cells demonstrates a loss of cell polarity. (F) Another specimen illustrates the difference in KLF4 protein between poorly and well-differentiated cells; the yellow stain is darker in the well-differentiated crypt (white arrow) than in the poorly differentiated portion (black arrow).
KLF4 expression also differed among different types of carcinoma. Poorly differentiated carcinomas (Fig. 1E) were characterized with lighter yellow staining in cytoplasm compared to well/moderately differentiated carcinoma (Fig. 1C and D). This was coincident with the positive percentage of the KLF4 staining (Table 2). And the localization of KLF4 immunostaining was mainly in cytoplasm, while yellow-stained nucleus also was detected in some part of the cells.
4. Expression of KLF4 transcript in colon and rectum
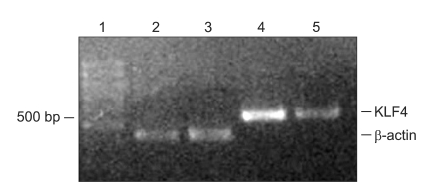
RT-PCR was performed on total mRNA isolated from fresh specimens of cancerous and noncancerous portions. The PCR product was designed to be 435 bp. The positive control was β-actin. The levels of KLF4 mRNA were lower in the colorectal mucosa than in normal colon (Fig. 2).
Fig. 2.
RT-PCR analysis of KLF4 transcript expression in colonic tissues. KLF4 mRNA is highly expressed in normal tissue and down-regulated in colorectal cancer tissue. Lane 1, molecular marker; Lanes 2 and 4, β-actin and KLF4 PCR products in normal mucosa; Lanes 3 and 5, β-actin and KLF4 PCR products in colorectal cancer specimens.
5. Expression of KLF4 protein in colon and rectum
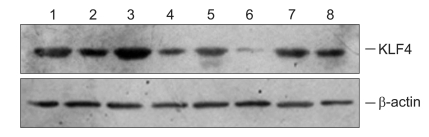
As showed in Fig. 3, the expression of KLF4 protein was lower in colorectal tumor than that in normal-appearing colorectal mucosa.
Fig. 3.
Western bolt analysis of KLF4 protein expression in colorectal tissue. High KLF4 expression is detected in normal colorectal and well-differentiated tissues. Moreover, a downregulation of expression is observed in poorly differentiated colorectal cancer tissues. Lanes 1 and 3 show KLF4 expression in normal-appearing tissues, while the other lanes show expression in cancerous specimens.
6. Expression of KLF4 protein in RKO cells
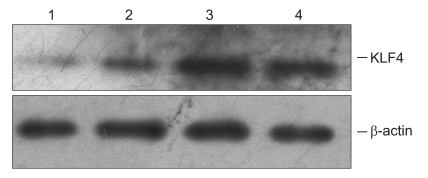
As showed in Fig. 4, the expression of KLF4 protein was elevated in RKO treated with butyrate in a dose-dependent manner.
Fig. 4.
Western blot analysis of KLF4 expression in RKO cells. KLF4 protein expression is elevated in RKO cells treated with butyrate in a dose-dependent manner. Lanes 1-4 show results from cells treated with 0, 1, 2, and 3 mM butyrate, respectively.
DISCUSSION
Cancer cells differ from normal cells in many aspects, including hyperproliferation and loss of differentiation. The recent concept of colonic carcinogenesis begins with damage to normal cellular genes, such as the activation of oncogenes, or the loss of the tumor suppressor genes. The loss of chromosome 9q, in which KLF4 resides, is observed in 25% to 50% of sporadic CRC.12 Zhao et al.13 found evidence for loss of heterozygosity (LOH) in two of eight surgically resected CRC specimens, and further studies in colorectal cancer cell lines also confirmed that in five of six exhibited LOH while other cell line RKO was found to be hemizygous deletion in the KLF4 gene. Krüppel like factor 4, a eukaryotic transcription factor expressed extensively in the gastrointestinal tract, was turned out to be a potential tumor suppressor gene in the colorectal cancer.
In this report, we analyzed the expression patterns of KLF4 in Chinese sporadic colorectal cancers to study the relationship between KLF4 expression and clinicopathological parameters. Our studies showed that the negative and weak positive rate of normal appearing mucosa, adenoma, and carcinoma were 42.5%, 71.4%, and 82.3%, and the expression level of KLF4 in carcinoma, as compared to normal appearing mucosa, is statistically significant (p<0.05), which sames that the downregultaion of KLF4 is a precancer lesion in colon and rectum. Western blot and RT-PCR were also performed to confirm the results. This data was also supported by the result of northern blot analysis in precious studies.6,13 All those results meant the downregulation of KLF4 expression in the course of malignant transformation, and indicated that KLF4 is closely correlated with differentiation and may be a potential tumor suppressor gene in colorectal cancer.
We also found that the expression patterns of KLF4 differed according to differentiation type of colorectal cancer. Higher percentage of strong positive expression of KLF4 was detected in well and moderately differentiated colorectal cancer than in the poorly differentiated ones. (22.9% vs 0%) (p<0.01). This result was different from Choi et al.14 The distinct point here is that KLF4 expression patterns differed in different stage of the specimens (p<0.05). Strong KLF4 expression was found in 26.5% of stage I or II, and it was down to 7.1% of stage III or IV. The reasons were not clear. It could be due to the different genetic background or the reasonable sampling error. It was coincident with recent study of Patel et al.15 that they showed an obvious trend of a decreased odds ratio at higher stages compared with stage I cancer. Those suggested that the loss of KLF4 or its expression pattern leads to poor differentiation.
KLF4 expression was correlated with the differentiation in CRC in vivo, more and more studies were performed in vitro to support this. Sodium butyrate, a differentiation promoting agent of colon cancer,16 could induce KLF4 mRNA expression and stimulated KLF4 promoter activity in a dose-and time-dependent manner in HT-29 cells.17 In our study, the protein levels of KLF4 were elevated in the colorectal cancer cell line RKO treated with 0 to 3 mM butyrate in a dose-dependent manner. RKO cells induced to overexpression of KLF4 demonstrated a corresponding dose-dependent increase in intestinal alkaline phosphatase (IAP) expression, which is known as enterocyte differentiation marker.18 This notion is further supported by the identification of that KLF4 plays an important role in colonic epithelial cell differentiation, and it is a goblet cell specific differentiation factor. KLF4-/- mice displayed marked decrease in the number of goblet cells and abnormal goblet cell morphology.19 Loss of KLF4 in mice also caused altered differentiation and precancerous changes in the adult stomach.20 This reminded the critical role of KLF4 in regulating the differentiation of colorectal cancer. Further studies are needed to analyze the role of KLF4 in the regulation of cell cycle in the colorectal cancer cell.
But it is still premature to say that the upregulation of KLF4 is the cause of differentiation. In the recent study of stem cells, it is demonstrated that induction of pluripotent stem cells from mouse embryonic or adult fibroblasts by introducing four factors which including KLF4.21 KLF4 was also downregulated at early stage of mouse embryonic stem cells differentiation.22 These studies showed the dual role of KLF4 in cell differentiation. The expression patterns differed in different cells.
We did not found the level of KLF4 had correlation with the sex, age, pathology type, and metastasis state (p>0.05), which was consistent with findings from Choi et al.14
In conclusion, our results showed the downregulation of KLF4 expression in the course of progressive malignant transformation, and indicated that KLF4 was significantly correlated with differentiation and may be a potential tumor suppressor gene in colorectal cancer.
ACKNOWLEDGEMENTS
This study was supported by Natural Science Foundation of Anhui Province (No. 050430705) and National Natural Science Foundation of China (No. 090413116).
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Harguindey S, Orive G, Luis Pedraz J, Paradiso A, Reshkin SJ. The role of pH dynamics and the Na+/H+ antiporter in the etiopathogenesis and treatment of cancer: two faces of the same coin--one single nature. Biochim Biophys Acta. 2005;1756:1–24. doi: 10.1016/j.bbcan.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yet SF, McA'Nulty MM, Folta SC, et al. Human EZF, a Krüppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J Biol Chem. 1998;273:1026–1031. doi: 10.1074/jbc.273.2.1026. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Whitney EM, Gao SY, Yang VW. Transcriptional profiling of Krüppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J Mol Biol. 2003;326:665–677. doi: 10.1016/S0022-2836(02)01449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster KW, Frost AR, McKie-Bell P, et al. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–6495. [PubMed] [Google Scholar]
- 7.Foster KW, Ren S, Louro ID, et al. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 1999;10:423–434. [PubMed] [Google Scholar]
- 8.Shie JL, Chen ZY, O'Brien MJ, Pestell RG, Lee ME, Tseng CC. Role of gut-enriched Krüppel-like factor in colonic cell growth and differentiation. Am J Physiol Gastrointest Liver Physiol. 2000;279:G806–G814. doi: 10.1152/ajpgi.2000.279.4.G806. [DOI] [PubMed] [Google Scholar]
- 9.Wang N, Liu ZH, Ding F, Wang XQ, Zhou CN, Wu M. Down-regulation of gut-enriched Krüppel-like factor expression in esophageal cancer. World J Gastroenterol. 2002;8:966–970. doi: 10.3748/wjg.v8.i6.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Wei D, Huang S, et al. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin Cancer Res. 2003;9:6371–6380. [PubMed] [Google Scholar]
- 11.Sobin LH, Fleming ID. TNM Classification of Malignant Tumors, fifth edition (1997). Union Internationale Contre le Cancer and the American Joint Committee on Cancer. Cancer. 1997;80:1803–1804. doi: 10.1002/(sici)1097-0142(19971101)80:9<1803::aid-cncr16>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 12.Gryfe R, Swallow C, Bapat B, Redston M, Gallinger S, Couture J. Molecular biology of colorectal cancer. Curr Probl Cancer. 1997;21:233–300. doi: 10.1016/s0147-0272(97)80003-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Krüppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23:395–402. doi: 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi BJ, Cho YG, Song JW, et al. Altered expression of the KLF4 in colorectal cancers. Pathol Res Pract. 2006;202:585–589. doi: 10.1016/j.prp.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Patel NV, Ghaleb AM, Nandan MO, Yang VW. Expression of the tumor suppressor Krüppel-like factor 4 as a prognostic predictor for colon cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:2631–2638. doi: 10.1158/1055-9965.EPI-10-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnard JA, Warwick G. Butyrate rapidly induces growth inhibition and differentiation in HT-29 cells. Cell Growth Differ. 1993;4:495–501. [PubMed] [Google Scholar]
- 17.Chen ZY, Rex S, Tseng CC. Krüppel-like factor 4 is transactivated by butyrate in colon cancer cells. J Nutr. 2004;134:792–798. doi: 10.1093/jn/134.4.792. [DOI] [PubMed] [Google Scholar]
- 18.Hinnebusch BF, Siddique A, Henderson JW, et al. Enterocyte differentiation marker intestinal alkaline phosphatase is a target gene of the gut-enriched Krüppel-like factor. Am J Physiol Gastrointest Liver Physiol. 2004;286:G23–G30. doi: 10.1152/ajpgi.00203.2003. [DOI] [PubMed] [Google Scholar]
- 19.Katz JP, Perreault N, Goldstein BG, et al. The zinc-finger transcription factor KLF4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katz JP, Perreault N, Goldstein BG, et al. Loss of KLF4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128:935–945. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 22.Hailesellasse Sene K, Porter CJ, Palidwor G, et al. Gene function in early mouse embryonic stem cell differentiation. BMC Genomics. 2007;8:85. doi: 10.1186/1471-2164-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]