Abstract
Background/Aims
Cluster differentiation 44 standard isoform (CD44s) is a transmembrane glycoprotein. CD44s is a known prognostic factor in various cancers, due to its involvement in tumor cell growth, invasion and metastasis. Its prognostic role, however, is debated because it can be a positive or negative prognostic factor depending on tumor type and is still an ambiguous prognostic indicator in other cancers, especially hepatocellular carcinoma (HCC). We investigated the relationship between CD44s expression and survival in HCC patients.
Methods
A total of 260 HCC samples were collected to generate a tissue microarray. Staining of the arrays with a primary mouse CD44s monoclonal antibody was followed by evaluation of the relationship between CD44s expression and tumor differentiation. The effect of CD44s expression on patient survival was analyzed.
Results
CD44s protein expression correlated with histological grade (most and worst Edmondson grade) of the HCC (p=0.029 and p=0.039, respectively) and adversely affected the disease free survival period based on univariate and multivariate analyses (p=0.038 and p=0.077, respectively).
Conclusions
High CD44s protein expression correlates with shorter disease free survival and poorly differentiated HCC. CD44s-targeted therapy may be efficacious for HCC treatment in the future.
Keywords: Hepatocellular carcinoma, CD44s antigen, Protein microarrays
INTRODUCTION
Hepatocellular carcinoma (HCC) is ranked third in prevalence, behind stomach and lung, and mortality rate behind lung and stomach in the world.1 There are some contributing factors to prohibit better prognosis such as difficulty in early detection and frequent recurrent rate.2 Therefore identification of predictive marker for recurrence is still remained as critical subject.
CD44 glycoprotein is integral membrane glycoprotein that recognizes and binds to hyaluronan, a component of extracellular matrix glycan.3 CD44s is known to play a role in the migration of some non-neoplastic cells such as lymphocytes and facilitates their homing to lymphoid organs.2 There are two types of CD44 glycoprotein family-CD44s, the standard isoform4 and multiple spliced variants, CD44v.5 CD44 has been known for having diverse effects on cellular function especially migration, adhesion and invasion and sometimes depending on site of tumor.6,7 In addition, CD44s expression appears to be an indicator of poor prognosis and clinical recurrence of many kinds of cancers, such as renal,8 thyroid,9 and colorectal cancer.10 There are reports about relationship between expressions of CD44s protein and differentiation of tumor in other organs.11,12 In cases of HCC, however, highly expressed CD44s protein has been correlated with poor prognostic factor and poorly differentiated tumor only in few reports.11,13,14 In the present study, we studied relationship between expression of CD44s and tumor differentiation with Edmondson grade systems (most and worst). In addition, expression of CD44s was investigated using tissue microarray and prognostic effect of CD44s protein on HCC.
MATERIALS AND METHODS
1. Patients and samples
Paraffin block tumor and non-neoplastic liver tissues were obtained from the resected specimens of 260 hepatocellular carcinoma patients who underwent hepatectomy or segmentectomy at the Hepatobiliary Department of Surgery of the Korean Cancer Center Hospital, Seoul, Korea between January 1, 1990 and December 25, 2003. Paraffin sections from 10 metastatic colorectal carcinomas in liver were used as immunohistochemical study of CD44s expression as control.
2. Clinicopathological data
Tissue sections (5-µm thick) were deparaffinized in xylene, rehydrated in graded alcohol solution, and stained with hematoxylin and eosin solution. Histological examination of resected specimens was performed by a pathologist who was blinded to the CD44s expression result. Tumor differentiation was classified into four grades, from I to IV, according to criteria proposed by Edmondson and Steiner.15 We estimated most and worst Edmondson grade and categorized them into two groups, high (higher than grade 3) and low grade (less than grade 2). The cut-off value of high alpha-fetoprotein (AFP) designates serum AFP level above 200 ng/mL. Age group was divided into two groups, high and low group, which above 50 years old and below 50 years old, respectively. The measurement of tumor size was done by gross examination directly or photographs. Histologic and cell types were categorized by World Health Organization classification of liver. Cell type was divided into two group, classic type and non-classic type. Histologic type was divided into trabecular group, which including microtrabecular and macrotrabecular type, and non-trabecular groups, which including pseudo-glandular and compact pattern. If there were more than one tumor cell clusters in portal vein, then we counted positive of portal invasion. All patients were regularly followed in a hepatobiliary clinic and all clinicopathological and follow-up data were collected prospectively in a computerized database.
3. Tissue microarray and immunohistochemistry
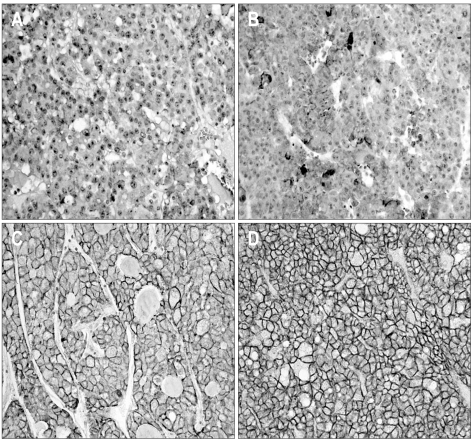
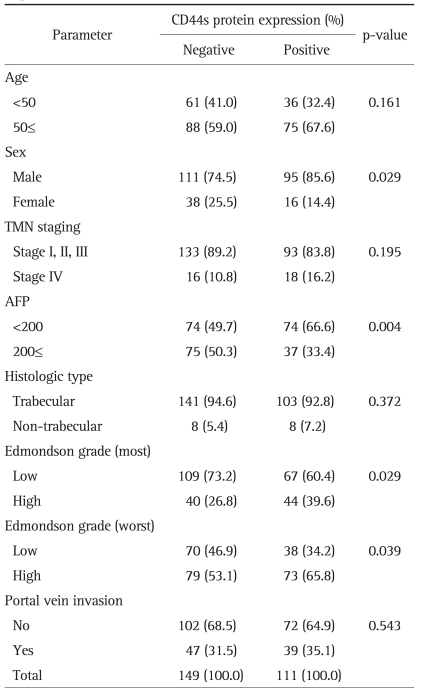
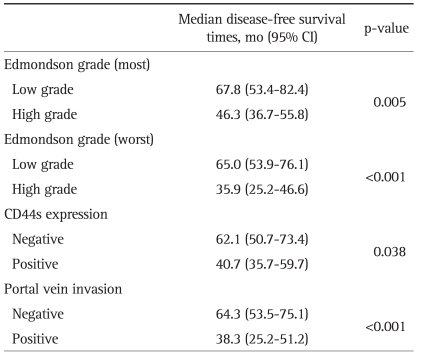
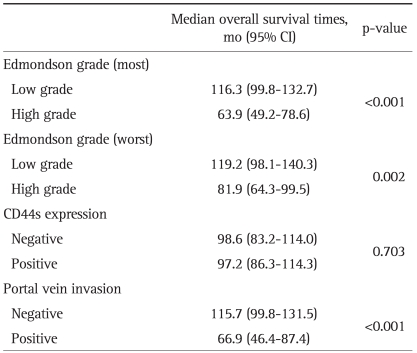
For each cancer case, all of the histologically well preserved target tissue (cancer and normal) was circled on the guide slides using different colored markers. No effort was made to distinguish between specific areas of available tumor (center versus periphery, depth of invasion, etc.). The formalin-fixed, paraffin-embedded tissues corresponding to the histological sections were sampled from these marked regions using a specialized manual arraying instrument (Model MTA 1; Beecher Instruments, Sun Prairie, WI, USA). Using this device, the marked slides were aligned with the original tissue block, and a needle was used to remove a cylindrical core (3 mm in diameter) from the original blocks (one to four cores of tumor were arrayed per original block); the core samples were then placed into an empty paraffin block. The tissue microarray (TMA) consisted of 59 malignant and 1 normal cores from HCC and normal control cases. Non-malignant tissues (normal liver parenchyma of metastatic colorectal carcinoma in liver) were placed as controls for orientation at the top of each TMA block. In addition to the primary TMA set of 7 blocks, 4 duplicate sets were also constructed by additional sampling of each original block and positioning the tissues at identical coordinates. After the arraying was completed, TMA blocks were completely sectioned at 5-µm thickness, yielding 20 slides from each block and immunohistochemical staining was performed. Non-specific binding was blocked with 1% bovine serum albumin (BSA) in 0.01 mol/L citrate buffer (pH=6.0) for 30 minutes before exposure to the primary antibody for CD44s. After incubation at room temperature for 30 minutes with biotinylated anti-mouse IgG (Dako, Golstrup, Denmark) sections were incubated with avidin-biotin peroxidase (1:200, Vectastain ABC kit; Vector Laboratories, Burlingame, CA, USA) for 60 minutes and rinsed again with PBS. They were counterstained with Mayer's hematoxyline and mounted. The antibody used was primary mouse CD44s monoclonal antibody (1:250, Zymed Lab Inc., San Francisco, CA, USA). The slides were evaluated in a double-blind manner by two pathologists (H.S.R. and S.H.P.). CD44s immunoreactivity along the cytoplasmic membrane tumor cells was only considered as positive reaction then Immunostaining was scored based on the percentage of stained cells into 4 categories: no expression (Score 0, 0% of stained cells), low expression (score 1, 1-10% of stained cells), moderate expression (score 2, 11-50%) and high expression (score 3, >50% of stained cells). For statistical purposes, staining scores were dichotomized as two groups on a scale of negative and positive scoring (Fig. 1).
Fig. 1.
Representative photomicrographs showing CD44s expression. (A) Negative reactions for CD44s protein in normal hepatocytes. (B-D) Positive reactions for CD44s protein expression in grade 1 (B), grade 2 (C) and grade 3 (D) hepatocellular carcinomas, respectively (×200). CD44s staining is observed along the cytoplasmic membrane.
4. Statistical analysis
Statistical analysis for the evaluation of relationship between CD44s immunostaining and clinicopathologic parameters was performed using Pearson's chi-square test. We analyzed correlation between CD44s expression of immunohistochemical staining and several clinicopathologic parameters, including tumor cell grade of HCC (most and worst Edmondson-Steiner grade), age, sex, TNM stage, AFP, tumor histologic type, and portal vein invasion. The influence of prognostic factors on tumor-related over all and disease free survival was assessed by Kaplan-Meier estimates and subgroups were compared by the log-rank test for univariate analysis. The multivariate Cox proportional hazard model was applied using a stepwise forward method to detect independent predictors of survival. Statistical analysis was carried out using SPSS for Windows (version 16.0; SPSS Inc., Chicago, IL, USA). Two-tailed p values of 0.05 or less were considered to be statistically significant.
RESULTS
Overall, 260 patients (median age, 52.1 years) were included in the study. Their clinical characteristics are given in Table 1. The male to female ratio was 3.7. Data for disease-free and overall survival were available in all patients. Median follow-up was 26±19 months (range, 2 to 137 months) for all patients together (n=260). Of them, 119 patients (45.7%) recurred and 37 patients (14.2%) died of recurrent HCC, 22 patients (8.4%) died with hepatic failure, 4 patients (1.5%) due to respiratory failure, and 5 patients (1.7%) died with metastasis.
Table 1.
Clinicopathologic Parameters of HCC according to CD44s Expression (n=260)
1. Relationship between protein expression and clinicopatholgic parameters
There was one or two significant relationship between each biological marker and clinicopathologic parameters. CD44s expression was associated with serum AFP level (p=0.004) and both most and worst Edmondson grades (p=0.029 and p=0.039). As both of Edmondson grade was getting higher, CD44s protein expression was increased. However, there was no significant correlation between CD44s expression and remaining clinicopathologic values (Table 1).
2. Relationship between CD44s protein expression and survival
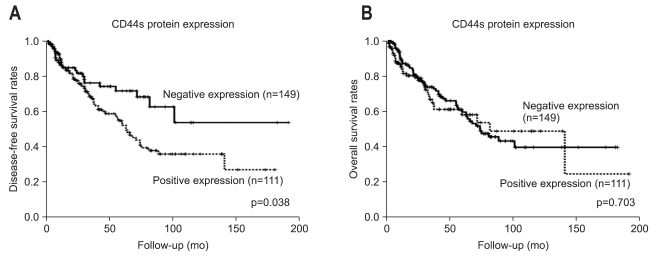
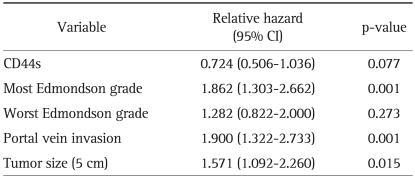
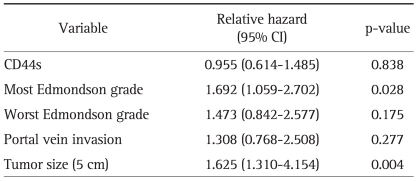
As Edmondson grade system was known for time honored prognostic factor and there was the relationship between CD44s and these classical grades system, we tried to evaluate correlation of CD44s expression with overall and disease-free survival length. According to the result of relationship between Edmondson grade and survival lengths, the patients with low Edmondson grade showed significant longer surviving perio d than those of higher grade (Tables 2 and 3). CD44s also showed shortened disease-free survival period in CD44s expressed patients group than negative expression group (p=0.038, Table 2, Fig. 2A). However no correlation between overall survival length and CD44s expression was identified (Table 3, Fig. 2B). There are 26 patients (10%) who died with no evidence of tumor recurrence and we evaluated survival analysis after exclusion of these patients, which showed similar results in disease-free survival in univariate analysis (p=0.026). In multivariate analysis, most Edmondson grade (p=0.001) and portal vein invasion (p=0.001) were independent predictors of recurrence. CD44s was seen as marginally significant risk factor of recurrence of disease (p=0.077) (Table 4). Tumor size was also revealed as one of significant independent variables for both overall and disease-free survival (p=0.004 and 0.015). Multivariate analysis of overall survival indicated no correlation between CD44s expression of the tumor (p=0.838) (Table 5).
Table 2.
Disease-Free Survival in All Patients Using Kaplan-Meier Estimates
CI, confidence interval.
Table 3.
Overall Survival in All Patients Using Kaplan-Meier Estimates
CI, confidence interval.
Fig. 2.
Disease-free survival curve (A) and overall survival curve (B) according to CD44s protein expression. CD44s expression correlates with a negative prognosis (A), but there is no significant correlation between CD44s protein expression and overall survival (B).
Table 4.
Multivariate Analysis of Significantly Associated Factors and CD44s with Disease-Free Survival
CI, confidence interval.
Table 5.
Multivariate Analysis of Significantly Associated Factors and CD44s with Overall Survival
CI, confidence interval.
DISCUSSION
Up to date, liver cancer stem cell (CSC) has been identified by several cell surface antigens such as CD133, CD90 and CD44.16,17 CSCs are a newly identified subpopulation that possesses stem cell properties and are thought to be the cells that cause resistance to chemotherapy and radiotherapy.16 CD44s was shown to be a part of the CSC in other sites such as breast cancer and was known to contribute to poor prognosis.18
Up to date, we can identify only a few numbers of articles about CD44s in HCC.14,19-21 Endo and Terada14 presented their study of 107 HCC patients, which shows expression of series of CD44 protein correlated with reduced survival period (p<0.005). In addition they concluded CD44s was a single independent prognostic factor in multivariate survival analysis (hazards ratio, 1.95 [1.00 to 3.78]). According to a previous data about CD44s protein expression in HCC, highly expressed CD44s molecule has been shown to be related with increased rate of vascular invasion.13 They presented that increased CD44s expression in 5 out of 6 patients showed vascular invasion opposed to the cases of CD44s negative showing vascular invasion in 5 out of 21. Although number of case involved in the study was small, they resulted in significant difference between CD44s positive and negative groups (p<0.016). The present study was failed to found correlation between CD44s expression and vascular invasion with portal vein. However we identified that this factor was significantly correlated with disease free survival and independent factor in multivariate analysis (p=0.001). Beckebaum et al.19 studied 53 HCC patients and role of CD44s expression in aspect of survival and other factors. They failed to present a significant correlation between patient survival and CD44s expression, however expression of CD44s was shown as a significant predictable marker for lymph node metastasis (p<0.05).
As almost of studies previously reported showed significance of CD44s in aspect of survival, the present study are also led to similar result. According to our result, CD44s was especially correlated with disease free survival of HCC, although we couldn't find statistically significant correlation but marginal relationship between CD44s and disease-free survival (p=0.077).
Our data presents another characteristic finding about relationship between CD44s expression and Edmondson grade. Most and worst Edmondson grades had a statistically significant correlation with CD44s protein expression, respectively. Mattew et al.13 also presented similar result as higher grade of tumor is related with upregulation of CD44s expression. Endo and Terada14 also concluded expression of CD44s correlated with poorly differentiated tumor in HCC. However, other studies from extrahepatic organs showed opposite results so there are still controversial in relationship between expression of CD44s molecule and tumor grades.22,23
In this study, AFP had an inverse correlation with CD44s expression. Serum AFP level is widely used as a diagnostic tool or predictive factor in HCC and elevation of AFP level is considered as a poor prognostic factor as CD44s protein. Therefore we could speculate the correlation between two markers. Yang et al.24 reported a similar result that serum level of AFP and CD44s protein expression in immunohistochemistry had an inverse correlation (p<0.001). Although serum AFP level is used as a sensitive marker for diagnosis of HCC, serum AFP level could be not elevated even in patients with obvious HCC.25 Therefore CD44s could be used as a valuable predictive marker for tumor recurrence for the patients with normal serum AFP levels.
Few authors have been tried to inhibit the activity of CD44s in other sites.26,27 Gunia et al.26 studied a function of CD44s by blocking of CD44s protein using anti-CD44s monoclonal antibody in gliosarcoma cell line. They concluded that inhibition of CD44s protein function could be effective in preventing the tumor progression. However there was no data about blocking effect of anti-CD44s antibody on tumor progression in HCC.
In conclusion, expression of CD44s tends to show an adverse prognostic effect in HCC. Also the expression depended on differentiation of tumor cells. We suggested that CD44s could effect on recurrence of HCC and be used as a candidate for targeted therapy of HCC.
References
- 1.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 2.Tölg C, Hofmann M, Herrlich P, Ponta H. Splicing choice from ten variant exons establishes CD44 variability. Nucleic Acids Res. 1993;21:1225–1229. doi: 10.1093/nar/21.5.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naor D, Sionov RV, Ish-Shalom D. CD44: structure, function, and association with the malignant process. Adv Cancer Res. 1997;71:241–319. doi: 10.1016/s0065-230x(08)60101-3. [DOI] [PubMed] [Google Scholar]
- 4.Fox SB, Fawcett J, Jackson DG, et al. Normal human tissues, in addition to some tumors, express multiple different CD44 isoforms. Cancer Res. 1994;54:4539–4546. [PubMed] [Google Scholar]
- 5.Mackay CR, Terpe HJ, Stauder R, Marston WL, Stark H, Günthert U. Expression and modulation of CD44 variant isoforms in humans. J Cell Biol. 1994;124:71–82. doi: 10.1083/jcb.124.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naor D, Nedvetzki S, Golan I, Melnik L, Faitelson Y. CD44 in cancer. Crit Rev Clin Lab Sci. 2002;39:527–579. doi: 10.1080/10408360290795574. [DOI] [PubMed] [Google Scholar]
- 7.Ponta H, Wainwright D, Herrlich P. The CD44 protein family. Int J Biochem Cell Biol. 1998;30:299–305. doi: 10.1016/s1357-2725(97)00152-0. [DOI] [PubMed] [Google Scholar]
- 8.Gilcrease MZ, Guzman-Paz M, Niehans G, Cherwitz D, McCarthy JB, Albores-Saavedra J. Correlation of CD44S expression in renal clear cell carcinomas with subsequent tumor progression or recurrence. Cancer. 1999;86:2320–2326. doi: 10.1002/(sici)1097-0142(19991201)86:11<2320::aid-cncr20>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 9.Böhm JP, Niskanen LK, Pirinen RT, et al. Reduced CD44 standard expression is associated with tumour recurrence and unfavourable outcome in differentiated thyroid carcinoma. J Pathol. 2000;192:321–327. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH711>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 10.Huh JW, Kim HR, Kim YJ, et al. Expression of standard CD44 in human colorectal carcinoma: association with prognosis. Pathol Int. 2009;59:241–246. doi: 10.1111/j.1440-1827.2009.02357.x. [DOI] [PubMed] [Google Scholar]
- 11.Heyse TJ, Malcherczyk D, Moll R, et al. CD44: survival and metastasis in chondrosarcoma. Osteoarthritis Cartilage. 2010;18:849–856. doi: 10.1016/j.joca.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Gunia S, May M, Koch S, Dietel M, Erbersdobler A. Expression of CD44s in incidental prostate cancer is more strongly associated with Gleason scores on subsequent radical prostatectomies than conventional prognostic parameters. Pathobiology. 2009;76:286–292. doi: 10.1159/000245894. [DOI] [PubMed] [Google Scholar]
- 13.Mathew J, Hines JE, Obafunwa JO, Burr AW, Toole K, Burt AD. CD44 is expressed in hepatocellular carcinomas showing vascular invasion. J Pathol. 1996;179:74–79. doi: 10.1002/(SICI)1096-9896(199605)179:1<74::AID-PATH531>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 14.Endo K, Terada T. Protein expression of CD44 (standard and variant isoforms) in hepatocellular carcinoma: relationships with tumor grade, clinicopathologic parameters, p53 expression, and patient survival. J Hepatol. 2000;32:78–84. doi: 10.1016/s0168-8278(00)80192-0. [DOI] [PubMed] [Google Scholar]
- 15.Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462–503. doi: 10.1002/1097-0142(195405)7:3<462::aid-cncr2820070308>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 16.Lee TK, Castilho A, Ma S, Ng IO. Liver cancer stem cells: implications for a new therapeutic target. Liver Int. 2009;29:955–965. doi: 10.1111/j.1478-3231.2009.02040.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhu Z, Hao X, Yan M, et al. Cancer stem/progenitor cells are highly enriched in CD133+CD44+ population in hepatocellular carcinoma. Int J Cancer. 2010;126:2067–2078. doi: 10.1002/ijc.24868. [DOI] [PubMed] [Google Scholar]
- 18.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beckebaum S, Chen X, Sotiropoulos GC, et al. Role of osteopontin and CD44s expression for patients with hepatocellular carcinoma undergoing liver transplantation or resection. Transplant Proc. 2008;40:3182–3184. doi: 10.1016/j.transproceed.2008.08.034. [DOI] [PubMed] [Google Scholar]
- 20.Gao C, Guo H, Downey L, Marroquin C, Wei J, Kuo PC. Osteopontin-dependent CD44v6 expression and cell adhesion in HepG2 cells. Carcinogenesis. 2003;24:1871–1878. doi: 10.1093/carcin/bgg139. [DOI] [PubMed] [Google Scholar]
- 21.Zhang P, Chan DW, Zhu Y, et al. Identification of carboxypeptidase of glutamate like-B as a candidate suppressor in cell growth and metastasis in human hepatocellular carcinoma. Clin Cancer Res. 2006;12:6617–6625. doi: 10.1158/1078-0432.CCR-06-1307. [DOI] [PubMed] [Google Scholar]
- 22.Kuncová J, Urban M, Mandys V. Expression of CD44s and CD44v6 in transitional cell carcinomas of the urinary bladder: comparison with tumour grade, proliferative activity and p53 immunoreactivity of tumour cells. APMIS. 2007;115:1194–1205. doi: 10.1111/j.1600-0643.2007.00602.x. [DOI] [PubMed] [Google Scholar]
- 23.Bendardaf R, Algars A, Elzagheid A, et al. Comparison of CD44 expression in primary tumours and metastases of colorectal cancer. Oncol Rep. 2006;16:741–746. [PubMed] [Google Scholar]
- 24.Yang GH, Fan J, Xu Y, et al. Osteopontin combined with CD44, a novel prognostic biomarker for patients with hepatocellular carcinoma undergoing curative resection. Oncologist. 2008;13:1155–1165. doi: 10.1634/theoncologist.2008-0081. [DOI] [PubMed] [Google Scholar]
- 25.Daniele B, Bencivenga A, Megna AS, Tinessa V. Alpha-fetoprotein and ultrasonography screening for hepatocellular carcinoma. Gastroenterology. 2004;127(5 Suppl 1):S108–S112. doi: 10.1053/j.gastro.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 26.Gunia S, Hussein S, Radu DL, et al. CD44s-targeted treatment with monoclonal antibody blocks intracerebral invasion and growth of 9L gliosarcoma. Clin Exp Metastasis. 1999;17:221–230. doi: 10.1023/a:1006699203287. [DOI] [PubMed] [Google Scholar]
- 27.Breyer R, Hussein S, Radu DL, et al. Disruption of intracerebral progression of C6 rat glioblastoma by in vivo treatment with anti-CD44 monoclonal antibody. J Neurosurg. 2000;92:140–149. doi: 10.3171/jns.2000.92.1.0140. [DOI] [PubMed] [Google Scholar]