Abstract
The Golgi is an ancient and fundamental eukaryotic organelle. Evolutionary cell biological studies have begun establishing the repertoire, processes, and level of complexity of membrane-trafficking machinery present in early eukaryotic cells. This article serves as a review of the literature on the topic of Golgi evolution and diversity and reports a novel comparative genomic survey addressing Golgi machinery in the widest taxonomic diversity of eukaryotes sampled to date. Finally, the article is meant to serve as a primer on the rationale and design of evolutionary cell biological studies, hopefully encouraging readers to consider this approach as an addition to their cell biological toolbox. It is clear that the major machinery involved in vesicle trafficking to and from the Golgi was already in place by the time of the divergence of the major eukaryotic lineages, nearly 2 billion years ago. Much of this complexity was likely generated by an evolutionary process involving gene duplication and coevolution of specificity encoding membrane-trafficking proteins. There have also been clear cases of loss of Golgi machinery in some lineages as well as innovation of novel machinery. The Golgi is a wonderfully complex and diverse organelle and its continued exploration promises insight into the evolutionary history of the eukaryotic cell.
The Golgi appeared almost 2 billion years ago, before divergence of the major eukaryotic lineages. Comparative genomics reveals the gene duplication and coevolution of protein specificity that generated its complex trafficking machinery.
Poised at the crossroads of the endocytic and exocytic pathways, the Golgi is a crucial transport nexus in eukaryotic cells. This organelle acts as a site for protein modification and it is here that proteins are selected for traffic to the endocytic organelles, to the cell surface either for presentation or for secretion, or indeed for retrieval back to the ER (Glick 2000; Lorente-Rodriguez and Barlowe 2011). With this central role in membrane trafficking, it is not surprising that disruption of Golgi function is linked to human pathology (Freeze and Ng 2011), whether more rare (e.g., Batten's disease) (Kremmidiotis et al. 1999) or common (e.g., Alzheimer's disease) (Muhammad et al. 2008). For more extensive discussions of all these aspects of the Golgi, readers are directed to the various accompanying articles in this collection.
Golgi are not restricted to human cells, of course, but rather are a general feature of eukaryotes—yeast, plants, algae, and parasites. Indeed, the Golgi appears as one of the key defining features of eukaryotic cells and a landmark that separates us from prokaryotes at a cellular level. As such, the Golgi may be presumed to be ancient, inviting us to explore its origins and history. In this article, we will examine current thought on the history of the Golgi and the endomembrane system in general. We will use comparative genomics to examine which proteins, involved in Golgi-associated vesicular traffic or in Golgi structure, are recent innovations and which represent ancient machinery. This will not only address the evolution of the Golgi, but also provide guidelines for experimental design of such analyses, hopefully illustrating the utility of an evolutionary cell biological approach. Finally, we will briefly discuss attempts to look back further into the events and processes that built the Golgi into the complex organelle we know today.
EVOLUTION OF THE GOLGI: HOW AND WHEN
Perhaps the most prominent feature distinguishing prokaryotic cells from eukaryotic ones is the ubiquitous presence in the latter of compartmentalization into specialized organelles (Stanier 1970). These diverse organelles are thought to have been derived by one of two possible mechanisms. The first mechanism, endosymbiotic acquisition, is clearly responsible for the origins of mitochondria and chloroplasts from α-proteobacteria and cyanobacteria, respectively (Gray and Doolittle 1982). The second mechanism is autogenous origin, that is, derivation from factors present in the pre-eukaryotic ancestor, without large-scale influence from a single endosymbiotic event. Although the origins of some organelles, such as the peroxisomes and even the nucleus, are still under debate, the organelles of the endomembrane system have been relatively noncontroversial, representing perhaps the most solidly established examples of organelles derived via an autogenous mechanism (Dacks and Field 2007). More controversial has been the question of when in eukaryotic evolution the Golgi arose.
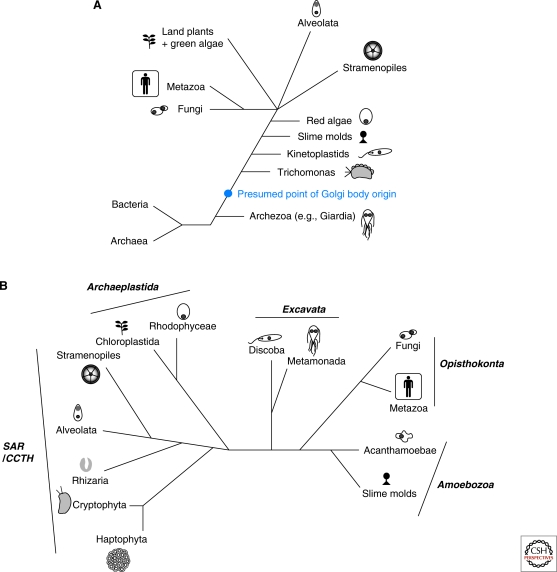
The reason for this is that, to the surprise of many cell biologists, not all eukaryotic cells possess readily visible Golgi when examined microscopically. Several parasites such as Giardia, Entamoeba, and microsporidia, as well as free-living organisms such as oxymonads, lack the hallmark stacked cisternal membrane compartments (Walker et al. 2011). On this basis, it was proposed nearly 30 years ago that these organisms did not possess Golgi and, furthermore, had never possessed Golgi (Cavalier-Smith 1987). Under the paradigmatic Archezoa hypothesis (Cavalier-Smith 1987) these organisms, many of which also lacked identifiable mitochondria and peroxisomes, were held to be lineages that had evolved very early in the history of eukaryotes, before the innovation of the Golgi (Fig. 1A). Initial molecular phylogenetic analyses were consistent with the cytologically simple parasites emerging at the base of the eukaryotic tree (Sogin 1991; Hashimoto et al. 1994). These organisms were proposed to have speciated, producing independent lineages and eventually living ancestors today lacking these key eukaryotic features. Under this theory, at some later time the Golgi arose in the stem lineage leading to the rest of eukaryotes (Fig. 1A).
Figure 1.
Relationships of the major eukaryotic lineages. (A) The Archezoa hypothesis. This diagram, modified and redrawn from Dacks and Doolittle (2001) (reprinted with permission from Elsevier © 2001) shows some of the major eukaryotic groups sampled at the time and their proposed relationships based on phylogenetic analyses derived from ssu rDNA. The cytologically simple Archezoa, here represented by Giardia, were thought to be basal, after which the remaining lineages emerged in a ladder-like fashion culminating in a crown radiation of animals (metazoa), fungi, plants, stramenopiles, and alveolates. The deduced origin of the Golgi after the speciation of the Archezoa is shown as a blue dot. (B) The six-supergroup taxonomic scheme of eukaryotic diversity. This diagram shows the current thinking on eukaryotic relationships based on concatenated gene phylogenies and accommodating ultrastructural evidence. Because of sampling unavailable at the time of the Archezoa hypothesis, this diagram includes many more taxa. Additionally, the following organisms or lineages are now placed as follows: Giardia and Trichomonas are metamonads, kinetoplastids are discoba, red algae are rhodophyceae, land plants, and green algae are chloroplastida. The taxonomic divisions are derived from evidence described in Walker et al. (2011) among others.
This elegant and logical idea turned out to be false. The initial molecular phylogenetic analyses supporting the Archezoa hypothesis have been discounted as artifact laden (Philippe et al. 2000) and replaced with a taxonomy of eukaryotes comprising six supergroups (Fig. 1B) based on multiple lines of genetic and ultrastructural evidence (Walker et al. 2011). More critically, the major application of the Archezoa hypothesis had to do with mitochondria, and indeed its major downfall was the discovery in each of these organisms of genes and later organelles of mitochondrial origin, the hydrogen producing hydrogenosomes and minuscule mitosomes (reviewed in van der Giezen 2009). Similar evidence for Golgi presence in organisms putatively lacking the organelle was rallied from the presence of genes encoded in the nuclear genomes that are known in model systems to act primarily or exclusively at the Golgi (Dacks et al. 2003; Mowbrey and Dacks 2009). Further evidence comes from cell biological studies in a few lineages identifying Golgi homologs where once there was thought to be none (Ghosh et al. 1999; Marti et al. 2003). Together these studies strongly suggest that the Golgi is a fundamental feature of the eukaryotic membrane-trafficking system and therefore likely coevolved with the other endomembrane compartments.
EVOLUTIONARY CELL BIOLOGY AND THE EUKARYOTIC MEMBRANE: TRAFFICKING SYSTEM
Our evolutionary understanding of the membrane-trafficking system has grown in recent years, because of advances in cell biology and increased availability of genome sequences from microbial eukaryotes. Indeed, the past decade has seen the emergence of a new biological discipline: evolutionary cell biology. If cell biology is defined by its subject and scope (i.e., the study of components, interactions, and processes at the cellular, rather than the molecular or organismal level), then evolutionary cell biology is the study of how such components and complexity emerged. It borrows techniques from both cellular and evolutionary biology weaving genomic, phylogenetic, and functional information about cellular components across a taxonomic framework. Patterns are thus inferred about origins, expansion, and loss of various organelles and cellular systems.
Although a great deal of excellent information in evolutionary cell biology is obtained from detailed phylogenetic work on individual protein families or detailed experimental work on non-model organisms, a strategy that has been highly successful is a comparative genomic approach. Here, sets of proteins are defined as markers for the presence of key organelles or processes within a given cellular system. These proteins are then searched against a battery of genome databases spanning the taxonomic breadth of organisms relevant to the question of interest. From the pattern of presence and absence of the various components, it is possible to infer the timing of both acquisition and reduction of system complexity. This approach is amenable at evolutionary scales ranging from the most ancient (e.g., the ancestor of all life) to the more recent (e.g., the ancestor of all yeasts or of all vertebrates).
Frequently, the goal of such studies is to deduce the degree of complexity of a given cellular system in the last eukaryotic common ancestor or LECA. The LECA is a hypothetical construct, a reconstruction of the ancestor that gave rise to all of the currently existing eukaryotic lineages. Because of the lack of a clear eukaryotic root (Roger and Simpson 2009), the most robust approach to deducing cellular information in the LECA is to take a consensus approach, identifying commonalities in organisms from the diversity of eukaryotes. This approach not only allows for the deduction of evolutionary information but also has the benefit of assessing the applicability of mechanistic models of cell biology beyond their experimental context. It is a simple extension of studying familiar model systems (e.g., Saccharomyces cerevisiae or Caenorhabditis elegans), but in this case writ large across eukaryotes.
This comparative genomic approach has been applied to the membrane-trafficking system with relative success. An initial study performed in 2001 (Dacks and Doolittle 2001) and expanded in 2004 (Dacks and Field 2004) addressed the question of what major membrane-trafficking machinery, as defined by experimental work in mammalian and yeast cells, is present in organisms spanning the breadth of eukaryotes. It was found that the major proteins families involved in vesicle formation (Arf GTPases, coat complexes such as COPI, COPII, clathrin, and adaptins) and vesicle fusion (Rab GTPases, SM proteins, NSF ATPases, and SNARE proteins; see below for further description of the various protein functions) were encoded in genomes from a broad taxonomic range of eukaryotes. Rather than a primitive or simple cellular makeup, as had been predicted by the Archezoa hypothesis, the LECA possessed a complex set of trafficking components. Phylogenetic analyses of SNAREs (Dacks and Doolittle 2002; Dacks and Doolittle 2004), Rabs (Pereira-Leal 2008), oligomeric tethering complexes (Koumandou et al. 2007), and cargo adaptors (Dacks et al. 2008) have delved even more deeply and enhanced the idea of trafficking complexity in this ancient eukaryotic ancestor.
The story is not only one of building complexity. Looking in more detail, a comparative genomic analysis of trafficking proteins acting specifically within the endocytic system showed a more complicated pattern (Field et al. 2007). Many aspects of the system were already established in the LECA such as the endocytic SNARE syntaxin 16, the lysosomal Rab 7, and clathrin. However, many other systems well characterized in mammalian and/or yeast systems, such as caveolins or the ESCRT 0 subcomplex of the multivesicular machinery, turned out to be much later evolutionary inventions (Field et al. 2007; Kirkham et al. 2008; Leung et al. 2008). This means that these components cannot be included in models of membrane trafficking meant to encompass all eukaryotic cells. At the same time, some ancestral features of the endocytic system have been lost, often multiple times independently. For example, Rab 4 appears to have been lost in plants and stramenopiles (Field et al. 2007). Comparative genomics of the endocytic system has revealed a pattern not only of ancient complexity but also of evolutionary plasticity with innovation and reduction playing significant roles.
Turning our attention to the Golgi, we know that the vast majority of our knowledge about the mechanism of transport and structure maintenance in the Golgi is based on experimental work in yeast and mammalian model organisms (Glick 2000). This begs the evolutionary cell biological questions of what aspects of our current models are valid outside of yeast and mammals and when did the components of this model evolve? We can use comparative genomics to ask: How applicable, how ancient?
WHERE DO WE LOOK: EUKARYOTIC DIVERSITY
A comparative genomic study is essentially a series of simple yes/no questions. Is a homolog of my gene of interest found in the genome in which I am searching? Each iteration of this question, though, has three components: Where am I looking, what am I looking for, and how am I looking? We will start with the “where.”
It is critical, when designing a comparative genomic analysis, to choose a taxonomic range for sampling that is at the same time broad enough to capture the full diversity of relevant taxa and evenly enough sampled so as not to have taxonomic overrepresentation. Because the Golgi is an ancient and ubiquitous feature of eukaryotic cells, here this translates into sampling genomes from across the diversity of eukaryotes.
Eukaryotic diversity can be classified into six major divisions, or supergroups (Fig. 1B). The familiar model organisms of yeast and mammals are found within a single one, the Opisthokonta. This supergroup is named from the Greek Opisthos (rear-facing) and konta (pole, i.e., flagella) for the propensity of its constituent organisms to have posteriorly directed flagella (Adl et al. 2005). It encompasses animals (e.g., Homo sapiens, Drosophila melanogaster, and Nematostella vectensis), fungi (e.g., Saccharomyces cerevisiae and Batrachochytrium dendrobatidis), and their protozoan relatives (e.g., Monosiga brevicollis). The opisthokonts are most closely related to organisms in the supergroup Amoebozoa (Baldauf et al. 2000; Bapteste et al. 2002). This assemblage of amoeboid organisms (Adl et al. 2005) contains parasites such as Acanthamoeba, the causative agent of keratitis, and the free-living, and sometimes multicellular, Dictyostelium. On the other side of the tree lies a mega-assemblage of three supergroups. The best known of these are the Archaeplastida (Adl et al. 2005). This is an umbrella term for red algae (e.g., Galdieria sulphuraria), glaucocystophyte algae, green algae (e.g., Chlamydomonas reinhardtii and Ostreococcus tauri), as well as land plants (e.g., Arabidopsis thaliana). The other two supergroups are more controversial. Molecular phylogenetic data clearly unites the stramenopiles (e.g., diatoms like Thalassiosira pseudonana, and oomycetes such as the crop pathogen Phytophthora ramorum) with the alveolates (e.g., dinoflagellates that cause red tide, ciliates like Tetrahymena thermophila, and apicomplexans like Toxoplasma gondii, which infects ∼1/3 of the global population and is associated with fatalities in immunocompromised patients) (Rodriguez-Ezpeleta et al. 2007). Both the stramenopiles and alveolates possess a chloroplast of red algal descent (Gould et al. 2008). Surprisingly, the Rhizaria (e.g., Bigelowiella natans), with no hint of photosynthetic ancestry, are related to these groups as well (Burki et al. 2007; Rodriguez-Ezpeleta et al. 2007). This new group has been named the SAR clade and is provisionally treated as a supergroup. At the same time organisms once thought to be closely related to stramenopiles and alveolates are classed in a group of their own. The cryptophyte algae, and nonphotosynthetic telonemids are housed now in the CCTH group (Burki et al. 2009), which also includes the haptophyte algae (e.g., Emiliania huxleyi), capable of producing algal blooms 100, 000s of square kilometers across. Between the Amoebozoa/Opisthokonta on one side and the mega assemblage of (mostly) photosynthetic groups on the other lies the final supergroup, the Excavata (Hampl et al. 2009). This group of organisms is united by possession of a feeding groove supported by an intricate arrangement of cytoskeletal roots and fibers (Simpson 2003). Excavates include Trypanosoma brucei, the agent of African sleeping sickness, and Trichomonas vaginalis, the most globally prevalent nonviral sexually transmitted disease.
In designing our comparative genomic search we chose at least two genomes from each supergroup, when possible, including the first ever representatives of Rhizarian and CCTH genomes. We also chose slightly higher sampling within the opisthokonts, as the supergroup from which most data regarding Golgi function was collected. It is also important to exclude organisms that are highly divergent (have undergone genome reduction, or living in extreme environments such as high temperature or acidity), as long as there are other, more canonical, genomes from that same taxonomic group available. In our case, because we wanted to ask questions about the level of conservation of Golgi proteins, we sampled genomes of 19 organisms for which obvious stacked Golgi have been reported (Table 1).
Table 1.
Taxa used for analysis
| Organism | Supergroup | Subgroup | URL |
|---|---|---|---|
| Homo sapiens | Opisthokonta | Metazoa | http://www.ncbi.nlm.nih.gov/BLAST |
| Nematostella vectensis | Opisthokonta | Metazoa | http://www.jgi.doe.gov/genome-projects |
| Drosophila melanogaster | Opisthokonta | Metazoa | http://www.ncbi.nlm.nih.gov/BLAST |
| Dictyostelium discoideum | Amoebozoa | Slime molds | http://www.ncbi.nlm.nih.gov/BLAST |
| Acanthamoeba castellanii | Amoebozoa | Acanthamoebae | http://blast.hgsc.bcm.tmc.edu/bcm/blast/ |
| Arabidopsis thaliana | Archaeplastida | Chloroplastida | http://www.ncbi.nlm.nih.gov/BLAST |
| Chlamydomonas reinhardtii | Archaeplastida | Chloroplastida | http://www.jgi.doe.gov/genome-projects |
| Ostreococcus tauri | Archaeplastida | Chloroplastida | http://www.jgi.doe.gov/genome-projects |
| Galdieria sulphuraria | Archaeplastida | Rhodophyceae | http://genomics.msu.edu/galdieria |
| Toxoplasma gondii | SAR | Alveolata | http://www.ncbi.nlm.nih.gov/BLAST |
| Tetrahymena thermophila | SAR | Alveolata | http://www.ncbi.nlm.nih.gov/BLAST |
| Thalassiosira pseudonana | SAR | Stramenopiles | http://www.jgi.doe.gov/genome-projects |
| Phytophthora ramorum | SAR | Stramenopiles | http://www.jgi.doe.gov/genome-projects |
| Bigelowiella natans | SAR | Rhizaria | J.M. Archibald, Dacks, Klute, JGl unpublished results |
| Guillardia theta | CCTH | Cryptophyta | J.M. Archibald, Dacks, Klute, JGl unpublished results |
| Emiliania huxleyi | CCTH | Haptophyta | http://www.jgi.doe.gov/genome-projects |
| Trypanosoma brucei | Excavata | Discoba | http://www.ncbi.nlm.nih.gov/BLAST |
| Trichomonas vaginalis | Excavata | Metamonada | http://www.ncbi.nlm.nih.gov/BLAST |
This table provides information for genome sequences searched by organism, supergroup, subgroup (as in Fig. 1B), and URL for the genome database accessed.
WHAT DO WE LOOK FOR: REPRESENTATIVE GOLGI PROTEINS
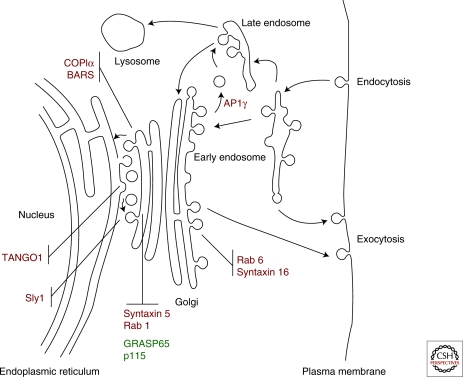
In the design of a comparative genomic survey, the choice of proteins/genes to search for is influenced by similar criteria as the choice of where to look. Just as one wants to evenly sample the full breadth of taxonomic diversity, one wants to look across the breadth of the system of interest, whether breadth is defined as the range of tasks performed or as the range of possible cellular locations. In our case, we chose proteins that act as carriers to and from the Golgi, both at the cis- and trans-faces of the Golgi. We chose matrix proteins implicated in Golgi morphology, as well as proteins involved in trafficking (Fig. 2).
Figure 2.
Eukaryotic membrane-trafficking system and Golgi proteins. This cartoon shows the canonical eukaryotic membrane-trafficking organelles. The 11 proteins examined by comparative genomics in this article (see text for details) are shown at their major location of action with trafficking proteins in red and structural proteins in green. (Modified from Morgan et al. 2002; reprinted with permission from Elsevier © 2002.)
Nine proteins involved in Golgi trafficking were selected (Table 2). COPI is the coat involved in retrograde Golgi to ER trafficking, as well as intra-Golgi trafficking, and is composed of seven subunits (α, β′, β, γ, δ, ε, ζ) (Waters et al. 1991). Because of its large size, the COPIα subunit was chosen for this study. Brefeldin-A ADP-ribosylated substrate (BARS) is a COPI-associated protein that is critical for the fission step of COPI vesicle formation (Yang et al. 2005). Adaptor proteins (adaptins) complex with clathrin to mediate trafficking between the TGN, endosomes, and the plasma membrane, with the clathrin-AP1 complex involved in TGN to endosome trafficking (Robinson 2004). The AP1 complex is a heterotetramer (composed of β, γ, μ, σ subunits) and, again because of its large size, the AP1γ subunit was chosen for this study. Sec1/Munc18 (SM) proteins regulate SNARE complex–dependent trafficking (Toonen and Verhage 2003), with Sly1 being involved in traffic between the ER and Golgi (Ossig et al. 1991; Grabowski and Gallwitz 1997). SNAREs (soluble N-ethylmaleimide sensitive factor attachment protein receptors) are essential for vesicle-target membrane fusion and contribute to organelle identity (Sollner et al. 1993). The two SNAREs chosen for this study were syntaxin 5 (localized to the cis-Golgi) (Rowe et al. 1998; Wooding and Pelham 1998) and syntaxin 16 (localized to the TGN) (Abeliovich et al. 1998). Rab GTPases play multiple roles in the formation and selective docking of transport vesicles (Stenmark 2009). Rab1 is localized to the cis-Golgi and is involved in ER to Golgi transport (Segev et al. 1988). Rab6 is localized to the Golgi, including the trans-Golgi network, and is involved in retrograde intra-Golgi transport and transport to the ER (Goud et al. 1994; Opdam et al. 2000). TANGO1 (components required for transport and Golgi organization) is an integral membrane protein at ER exit sites that facilitates cargo loading into COPII vesicles for transport to the Golgi (Saito et al. 2009).
Table 2.
Queries used for analysis
| H. sapiens | S. cerevisiae | |
|---|---|---|
| COPIα | NP_001091868.1 | NP_010136.1 |
| AP1γ | NP_001025178.1 | NP_015354.1 |
| Syntaxin 5 | NP_003155.2 | NP_013126.1 |
| Syntaxin 16 | NP_001001433.1 | NP_014624.1 |
| Sly1 | NP_057190.2 | NP_010475.1 |
| Rab1 | NP_112243.1 | NP_116615.1 |
| Rab6 | NP_002860.2 | NP_013363.1 |
| P115 | NP_003706.1 | NP_010225.1 |
| GRASP65 | NP_114105.1 | NP_010805.1 |
| BARS | NP_001319.1 | – |
| TANGO1 | NP_940953.2 | – |
This table provides information for proteins used as queries listed by protein name and the Homo sapiens and Saccharomyces cerevisiae accession number where relevant.
Two Golgi structural proteins were selected (Table 2). GRASP65 (Golgi reassembly and stacking protein) is required for connectivity of the Golgi complex ribbon structure (Puthenveedu et al. 2006). p115, a golgin, functions at the cis-Golgi as an extensively coiled-coil tether (Ramirez and Lowe, 2009). p115 has been shown to function in COPII vesicle tethering to the Golgi (Allan et al. 2000) and has been detected on COPI vesicles participating in intra-Golgi trafficking (Seemann et al. 2000). Together, the GRASPs and golgins form a Golgi matrix that maintains Golgi structure and function (reviewed in Ramirez and Lowe 2009).
HOW TO FIND IT: A BRIEF INTRODUCTION TO HOMOLOGY SEARCHING
The final component to a comparative genomic experiment is the methodology used to identify potential homologs in a variety of organisms. The aim is to establish the presence in the genome of the organism of interest (e.g., a plant) of a gene that is the direct equivalent of a gene originally discovered or characterized in a different organism (e.g., humans). Such homologous genes are the retained descendent products of a gene that was present in the ancestor of the two organisms in question (e.g., humans and plants).
This assessment is determined using homology searching algorithms. The most commonly used, and that used here, is the Basic Local Alignment Search Tool or BLAST (Altschul et al. 1990). BLAST searches for regions of local similarity between two sequences and makes alignments from these locations. Each BLAST result has an expect (E) value that predicts the false-positive rate (Johnson et al. 2008), with the value representing the number of alignments that would be expected by random chance of the same identity/similarity as the one found. The cutoff that we used was 0.05, that is, that there was only a 1 in 20 probability of this match being because of random chance.
When possible we searched predicted protein databases. This allows for sensitivity across greater evolutionary distances, because of the increased informational content, and lower chance of sequence convergence, of 20 amino acid states rather than four nucleotide states. As well, because predicting proteins is a later step in the production of a genome sequence, the availability of a predicted protein database is a general indication of genome project completeness. However, in the case of G. theta, only an expressed sequence tag (EST) database was available. Although it was sampled to allow for inclusion of this important taxon in our analysis, EST databases have key limitations. First, ESTs often represent portions of a gene, and, second, they only represent the mRNA that was transcribed at a given time rather than the cells entire coding capacity. ESTs can be reliably used to confirm that a protein is definitively present, but not to establish its absence.
The membrane-trafficking machinery is often composed of proteins families with different family members performing mechanistically similar functions but at discrete and characteristic locations within the cell (Bonifacino and Glick 2004). Because many of our queries are Golgi-specific versions of such proteins families in species X (e.g., SNAREs, Rabs, and adaptins from S. cerevisiae or H. sapiens), we ensured that the candidate homologs in species Y were true orthologs (or same member of a gene family) by a “reciprocal best hit” criterion. Briefly, to be considered a valid ortholog, a candidate homolog in species Y is used as a query in a BLAST search either into the genome of species X from which the query was derived and into the nonredundant (nr) protein database at NCBI. These “reciprocal” BLAST searches had to retrieve the original query with an E-value of less than 0.05.
WHAT WE FOUND
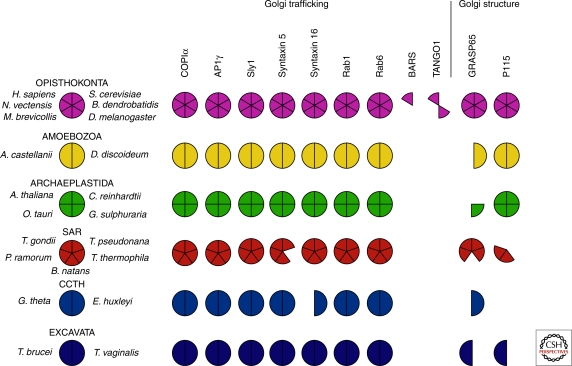
Our analyses yielded examples of the three basic comparative genomic patterns (Fig. 3): near ubiquitous distribution, patchy distribution or lineage-specific distribution. Details of individual results are available from the authors by request.
Figure 3.
Comparative genomic survey results of Golgi-associated protein machinery across diverse eukaryotes. Most Golgi-associated machinery is ancient. This Coulsen plot shows the presence or absence distribution of the 11 Golgi-associated proteins examined across 21 eukaryotic genomes. The presence of a homolog, confirmed by reciprocal BLAST analysis, is shown by a filled pie sector. Nine of the factors show a wide eukaryotic distribution, with clear losses in the cases of both GRASP and p115. BARS and TANGO1 show restricted distribution and are likely recent innovations in animals.
Consistent with past analyses of the respective protein families, the Golgi-specific homologs of adaptin (AP1γ; Dacks et al. 2008), Syntaxins (Syn5 and Syn16; Dacks and Doolittle 2004; Yoshizawa et al. 2006), SM proteins (Sly1; Koumandou et al. 2007), COPIα (Neumann et al. 2010), and Rabs (Rab1 and Rab 6; Pereira-Leal 2008) were identified in almost all of the taxa examined. This study significantly extends the previous analyses, most critically through the inclusion of data from the E. huxleyi, G. theta, and B. natans databases. All six eukaryotic supergroups have now been included for the first time, representing the broadest sampling ever achieved for these Golgi-associated proteins.
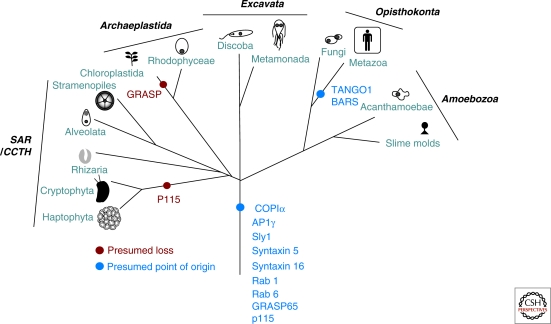
The identification of these various proteins in taxa from the full span of eukaryotic diversity implies first that this machinery is ancient, likely having been present in the LECA (Fig. 4). Second, because these factors are found in organisms beyond the narrow taxonomic span in which they were characterized, it supports the conclusion that such components are likely to be involved in their respective processes in those taxa. The overall model of Golgi trafficking that includes such factors may be now extrapolated to the full breadth of eukaryotes.
Figure 4.
The evolution of Golgi-associated machinery in the six eukaryotic supergroups. This diagram shows the diverse eukaryotic supergroups with gains (blue) and losses (red) of Golgi-associated factors across the various lineages. Proposed losses are only those with multiple taxa in a lineage, to be more confident in ascribed absence. Note that the majority of examined machinery was already present in the LECA, with BARS and TANGO likely being metazoa-specific innovations. (Modified from Field et al. 2007; reprinted with express permission of the authors from Springer © 2007.)
Both p115 and GRASP, the representative proteins chosen for the Golgi matrix, showed a patchy distribution. That is, whereas homologs of these proteins were found in most eukaryotic supergroups, several genomes distributed across many supergroups lacked recognizable homologs. To be conservative, we specifically use the term “not identified” because there are a number of technical reasons why there may be a failure to identify a homolog in a given genomic database. In cases in which the database is incomplete (e.g., EST datasets or genomic databases with low coverage), the sequence may not be present because of sampling error. In cases in which the gene sequences in the taxon of interest are highly divergent, then the homolog of interest may be so different in sequence from the query as to be unrecognizable by the homology-searching algorithm. In this case use of more sensitive homology-searching algorithms such as PSI-BLAST (Altschul et al. 1997) or HMMer (http://hmmer.janelia.org/) may be successful. Nonetheless, reductive evolution is a prevalent phenomenon and so proteins not identified might, in fact, not be present. One can increase the confidence of absence if there is a consistent pattern, either a single protein missing in multiple genomes from related group organisms or multiple subunits of a complex missing in a given genome.
In the case of the Golgi structure factors in question, the most conservative interpretation of the data is that p115 and GRASP are likely to have been present in the LECA, as suggested previously (Short et al. 2005). This interpretation is supported by the positive identification of a GRASP homolog in each of the eukaryotic supergroups and of p115 in all but the CCTH representatives. Several of the “not identified” observations may well be because of technical failures of the search algorithm, particularly the absence of both factors from the highly divergent T. vaginalis genome. On the other hand, the absence of p115 from both CCTH representatives could well represent a loss in that lineage (Fig. 4). Similarly, it has been reported that GRASP is lacking from plants (Struck et al. 2008). We extend this observation to green algae but interestingly can pinpoint the loss in the viridiplantae stem lineage, as we identified a red algal GRASP protein.
Of the genes that we examined, both TANGO1 and BARS were identified only in a limited taxonomic distribution, protostome and deuterostome animals. Because of this finite and coherent distribution, the most consistent explanation is that these factors represent recent, lineage-specific innovations (Fig. 4) and that they may not be validly incorporated into general models of Golgi mechanism. This in no way undermines their importance in the organisms in which they are found. The TANGO1 protein may well be present in more taxa than were observed, because homology searching failed to identify even the functionally characterized TANGO1 found in D. melanogaster, with the TANGO1 candidate shown in Figure 3 based on the independent functional characterization performed by Saito et al. (2009).
The BARS protein illustrates a potential pitfall of comparative genomics and the importance of multiple lines of evidence when assessing homology. Homology-searching algorithms are powerful and so conserved functional domains in a protein may cause misinterpretation as an ortholog. BARS includes a dehydrogenase domain, and so candidate homologs were identified by our homology searches in several genomic databases. On reciprocal BLAST into the human genome database, several of the candidate proteins retrieved BARS protein as the best-scoring homolog with moderate E-values. This resulted in assignment of BARS homologs in several, taxonomically diverse, genomic databases. However, on searching with the same queries in the nr database, clear matches to other dehydrogenase proteins were retrieved with much higher statistical significance, E-values of 40–50 higher orders of magnitude in some cases. Consequently, it seems much more likely that these candidate BARS homologs are homologs of other dehydrogenase families instead. Proteins with conserved domains can be difficult to assess and require particular care when being incorporated into comparative genomic analyses. Based on these criteria BARS, also named CTBP1, was initially found restricted to humans in our comparative genomic survey. However, a protein termed CTPB identified in D. melanogaster had similar BLAST values to both CTBP1 and a second gene CTBP2, suggesting that the duplication giving rise to CTBP1 and 2 had not yet occurred for the D. melanogaster version.
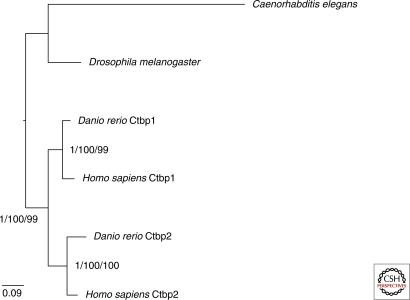
To clarify the evolution of BARS and its closely related proteins, we searched in the Danio rerio (fish) and Caenorhabditis elegans (worm) genomes for CTPB homologs and performed a phylogenetic analysis (Fig. 5). Consistent with previous analyses (Kim et al. 2002; Nicholas et al. 2008), CTBP1 and CTBP2 represent a vertebrate-specific duplication. The CTBP homologs found in D. melanogaster and C. elegans can be counted as legitimate homologs, but not orthologs. However, as only CTBP1 and not CTBP2 is involved in Golgi function, it is not clear that the CTBP preduplicates in the invertebrates (Mani-Telang and Arnosti 2007; Nicholas et al. 2008) share functional as well as evolutionary homology with BARS.
Figure 5.
Evolution of BARS in animals. This phylogenetic analysis shows that the duplication of CTBP1 and CTBP2 predates the vertebrate lineage, as both fish and humans have CTBP1 and CTBP2 homologs. Sequences were aligned using MUSCLE (Edgar 2004), and only unambiguously homologous positions were used for analysis. The best Bayesian tree topology, generated by the program Mr. Bayes (Ronquist and Huelsenbeck 2003), is shown with posterior probability values at each node based on 106 MCMC generations and burnin determined by eliminating all trees prior to a graphically defined plateau. The analysis incorporated a mixed model of sequence evolution with a γ correction for rate among sites. Additionally, bootstrap values are given at each node for maximum-likelihood bootstrap analysis by PhyML (Guindon and Gascuel 2003) and RAxML (Stamatakis 2006) with JTT + G model and PROTCATWAG models applied, respectively. Sequences included are as follows: Caenorhabditis elegans-accession NP_508983.2; Drosophila melanogaster-accession NP_524336.2; Homo sapiens CTBP1-accession NP_001319.1; Homo sapiens CTBP2-accession NP_001077383.1, Danio rerio CTBP1-accession NP_001035480.1; Danio rerio CTBP2-accession NP_571790.1.
Our searches did retrieve the ANGUSTIFOLIA protein, the named CTBP homolog in Arabidopsis (Kim et al. 2002). However, on use as a query for BLAST into the nr database, the E-value for retrieval of the CTBP homologs was e-28, whereas the E-value for other dehydrogenases was e-26. The evolutionary scenarios for the presence of a protein shared between multicellular plants and animals to the exclusion of all other eukaryotes would involve either extensive gene loss or horizontal gene transfer between two lineages with dedicated germlines. Neither scenario is particularly likely. Given the presence of a conserved functional (dehydrogenase) domain, the equivocal E-values between the CTBP and other dehydrogenases, and the demonstration of dissimilar functional properties between ANGUSTIFOLIA and CTBP homologs (Stern et al. 2007), we suggest that perhaps the resemblance of these two protein families may be because of convergent evolution rather than direct orthology. Either way, it has been shown that ANGUSTIFOLIA does not have an effect on the Golgi (Stern et al. 2007), and so BARS per se remains a vertebrate-specific innovation.
PRE-LECA HINTS ABOUT GOLGI
A complex organelle, with machinery that acts distinctively at the cis-Golgi and the TGN, appears to have been present in the ancestor of eukaryotes. This result is consistent with past studies that have addressed overlapping subsets of these proteins or have confirmed that the Golgi was present as a separate organelle from other membrane-trafficking compartments at this point in time (Dacks and Field 2007 inter alia). This complexity in the LECA, however, simply pushes back in time our most fundamental questions about the evolution and origin of the Golgi: What processes allowed the evolutionary differentiation, from the other endomembrane organelles, of the Golgi, and its the subsequent functional complexity giving rise to the cis-, intermediate, and trans-faces, as well as the TGN?
These questions are now squarely in the investigative realm of the prokaryote to eukaryote transition. The LECA is quite a different hypothetical construct than the first eukaryotic common ancestor, or FECA. This point of evolutionary reconstruction is a hypothetical organism presumably with some traits rendering it recognizably eukaryotic rather than prokaryotic but likely not possessing full cellular complexity. Understanding the evolutionary events involved in building from the FECA to the LECA is a much more nebulous task than concrete reconstructions from comparative complements of extant eukaryotic genomes. It essentially gets at fundamental mechanisms of organellogenesis, in the case of the Golgi and the other membrane-trafficking organelles, autogenous organelle evolution.
Based on the facts (1) that much of the machinery encoding trafficking specificity and organelle identity is composed of paralogous gene families, (2) that these organelle-specific families were already present in the LECA, and (3) that multiple such families together encoded this specificity through their combinatorial interactions, the organelle paralogy hypothesis (OPH) was proposed as a mechanism of autogenous organelle evolution (Dacks and Field 2007). This mechanism contends that the various eukaryotic membrane-trafficking organelles are derived from a single primordial endomembrane compartment through a process of gene duplication and coevolution of the various protein families that encode organelle specificity. Although initially incorporating the Rabs, SNAREs, and vesicle coats (Dacks and Field 2007), the hypothesis explicitly encompasses all specificity encoding machinery that shows a pattern of paralogous expansion and are derived before the LECA. As well, although it was proposed to cover the membrane-trafficking organelles, it could theoretically be applied to all homologous autogenous organelles. This wider application dovetails well with the proto-coatomer hypothesis (Devos et al. 2004). Here, structural analyses have revealed homology between the membrane-deformation machinery at play in the COPI, COPII, and clathrin vesicle coats, as well as the nuclear pore complex (Devos et al. 2004). Recent evidence also implicates the intraflagellar transport machinery (Jin et al. 2010; Jekely and Arendt 2006). The proto-coatomer hypothesis contends that a single ancestral protein gave rise to nuclear pore and intraflagellar transport machinery along with the ER, endosomes, plasma membrane, and the Golgi.
One important implication of both hypotheses is that if the order of paralogous expansions could be unraveled, then the evolutionary order of how the organelles themselves evolved would be revealed. This principle was implemented in the first major test of the OPH, whereby phylogenetic analyses of the Rabs, SNAREs, and β-adaptin subunits were performed (Dacks et al. 2008). In each case the endocytic organelle-specific homologs showed precisely the pattern predicted, if the family was evolving as described by the OPH, and if these organelles were the most recent to evolve, being caught in the process midstream (Dacks et al. 2008).
The application of this idea to the Golgi has thus far been limited, primarily because of the technical issues surrounding phylogenetic analysis of membrane-trafficking machinery. Although most analyses can reconstruct the organelle-specific families with confidence, the relationship between these families has been elusive. Nonetheless, we can speculate as to the nature of some of the events of machinery expansion that must have surrounded the emergence of the Golgi. The gene duplications giving rise to the AP complexes and the COPI F-subcomplex, as separate entities, likely contributed to the establishment of the cis-Golgi from the TGN. Likewise, the duplications producing the various Golgi-associated SNARE, Rab, and Arf-GAP families from the ER and endosomal versions should tell us about the emergence of both the organelle itself and its internal complexity. Because of the multiplicity of trafficking pathways that exist in the cells, and particularly at the Golgi, the patterns may not be as straight-forward as all Golgi paralogs grouping together. The possibility exists that complex trafficking pathways, perhaps optimizing recycling or providing cellular redundancy, were connected after the emergence of simpler organellar systems. These results, however, should provide penetrating insight into the history of the Golgi, the membrane-trafficking system, and indeed the eukaryotic cell itself.
CONCLUSIONS
We are still very much in the early stages of applying the tools and approaches of evolutionary cell biology to questions of the origin and history of the Golgi. Nonetheless, we can say with some certainty, that there was some form of Golgi in the last eukaryotic common ancestor and that this organelle had begun the differentiation process into cis-Golgi and TGN. Indeed, most of the machinery that we examined, whether vesicle trafficking or structural, appears conserved across the broadest diversity of eukaryotes sampled to date. This implies that the machinery in question is ancient and that the models of Golgi function, characterized in a small subset of eukaryotic organisms, is applicable well beyond its original derivation. Importantly, it also tells us about Golgi biology in organisms that are currently of limited experimental tractability but of ecological or medical relevance.
Nonetheless, some of the Golgi machinery examined was derived recently and appears functionally relevant to only a narrow subset of organisms. This not only serves as a caution about extrapolating utility of findings beyond sampled taxonomic points, it should whet our appetite for the fascinating biology that remains to be discovered about the Golgi in non-model eukaryotes. The existence of such biology is assured, not only from our comparative genomic data, but from the diversity of form and function in eukaryotes, whether the ring Golgi in the malaria parasite Plasmodium falciparum (Struck et al. 2005), the highly mobile Golgi of plants (Faso et al. 2009), or the production of intricate algal scales in post-Golgi secretory vesicles/cisternae (Hawkins and Lee 2001). Although comparative genomics can begin to shed light on currently intractable organisms, it also underlines the value of the experimental model organisms currently being established in the various supergroups: Toxoplasma in the SAR clade; Trypanosoma in the excavates; Dictyostelium in the Amoebozoa; and Chlamydomonas in the Archaeplastida. As cell biology in these systems matures, sequence homology can be paired with functional homology to weave a much richer tapestry of understanding for the Golgi, both mechanistic and historical.
The purpose of this article was not only to examine the origin and emergence of the Golgi, but also to act as a general primer for evolutionary cell biological analyses, hopefully convincing you the reader of the value and tractability of such an approach. The methods and rationales used here can be applied to a wide variety of cell biological questions—complexes, pathways, and organelles. As the tools improve in the coming years in their sensitivity and accuracy and as genome sequence sampling takes yet another giant leap forward because of next-generation sequencing technology, it will become increasingly feasible to examine organelle origins and evolutionary mechanisms, addressing the fundamental question: “How did the eukaryotic cell arise?”
ACKNOWLEDGMENTS
We wish to acknowledge the many genome projects that make their data freely available. We thank J.M. Archibald, M.W. Gray, P.J. Keeling, G.I. McFadden, and C.E. Lane for access to preliminary genome sequence data from Guillardia theta and Bigelowiella natans, which was produced by the Joint Genome Institute's Community Sequencing Program (http://www.jgi.doe.gov/). The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. This work in this article was supported by an NSERC discovery grant to J.B.D.
Footnotes
Editors: Graham Warren and James Rothman
Additional Perspectives on The Golgi available at www.cshperspectives.org
REFERENCES
- Abeliovich H, Grote E, Novick P, Ferro-Novick S 1998. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J Biol Chem 273: 11719–11727 [DOI] [PubMed] [Google Scholar]
- Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, et al. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryotic Microbiol 52: 399–451 [DOI] [PubMed] [Google Scholar]
- Allan BB, Moyer BD, Balch WE 2000. Rab1 recruitment of p115 into a cis-SNARE complex: Programming budding COPII vesicles for fusion. Science 289 444–448 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290: 972–977 [DOI] [PubMed] [Google Scholar]
- Bapteste E, Brinkmann H, Lee JA, Moore DV, Sensen CW, Gordon P, Durufle L, Gaasterland T, Lopez P, Muller M, et al. 2002. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc Nat Acad Sci 99: 1414–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS 2004. The mechanisms of vesicle budding and fusion. Cell 116: 153–166 [DOI] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Minge M, Skjaeveland A, Nikolaev SI, Jakobsen KS, Pawlowski J 2007. Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE 2: e790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Inagaki Y, Brate J, Archibald JM, Keeling PJ, Cavalier-Smith T, Sakaguchi M, Hashimoto T, Horak A, Kumar S, et al. 2009. Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, Telonemia and Centroheliozoa, are related to photosynthetic chromalveolates. Gen Biol Evol 1: 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T 1987. Eukaryotes with no mitochondria. Nature 326: 332–333 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Doolittle WF 2001. Reconstructing/deconstructing the earliest eukaryotes: How comparative genomics can help. Cell 107: 419–425 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Doolittle WF 2002. Novel syntaxin gene sequences from Giardia, Trypanosoma and algae: Implications for the ancient evolution of the eukaryotic endomembrane system. J Cell Sci 115: 1635–1642 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Doolittle WF 2004. Molecular and phylogenetic characterization of syntaxin genes from parasitic protozoa. Mol Biochem Parasitol 136: 123–136 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Field MC 2004. Eukaryotic cell evolution from a genomic perspective: The endomembrane system. In Organelles, genomes and eukaryote phylogeny: An evolutionary synthesis in the age of genomics (ed. Hirt RP, Horner DS), pp. 309–334 CRC Press, London [Google Scholar]
- Dacks JB, Field MC 2007. Evolution of the eukaryotic membrane-trafficking system: Origin, tempo and mode. J Cell Sci 120: 2977–2985 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Davis LA, Sjogren AM, Andersson JO, Roger AJ, Doolittle WF 2003. Evidence for Golgi in proposed “Golgi-lacking” lineages. Proc Biol Sci 270 Suppl 2: S168–S171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacks JB, Poon PP, Field MC 2008. Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc Natl Acad Sci 105: 588–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol 2: e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faso C, Boulaflous A, Brandizzi F 2009. The plant Golgi: Last 10 years of answered and open questions. FEBS Lett 583: 3752–3757 [DOI] [PubMed] [Google Scholar]
- Field MC, Gabernet-Castello C, Dacks JB 2007. Reconstructing the evolution of the endocytic system: Insights from genomics and molecular cell biology. Adv Exp Med Biol 607: 84–96 [DOI] [PubMed] [Google Scholar]
- Freeze H, Ng BG 2011. Golgi glycosylation and human inherited diseases. Cold Spring Harb Perspect Biol 3: a005371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh SK, Field J, Frisardi M, Rosenthal B, Mai Z, Rogers R, Samuelson J 1999. Chitinase secretion by encysting Entamoeba invadens and transfected Entamoeba histolytica trophozoites: Localization of secretory vesicles, endoplasmic reticulum, and Golgi. Infect Immun 67: 3073–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS 2000. Organization of the Golgi. Curr Opin Cell Biol 12: 450–456 [DOI] [PubMed] [Google Scholar]
- Goud B, Yang C, Roa M, Martinez O, Slepnev V 1994. Study of Rab6, a Ras-like GTP-binding protein associated with the Golgi complex. Ann NY Acad Sci 733: 340–343 [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI 2008. Plastid evolution. Annu Rev Plant Biol 59: 491–517 [DOI] [PubMed] [Google Scholar]
- Grabowski R, Gallwitz D 1997. High-affinity binding of the yeast cis-Golgi t-SNARE, Sed5p, to wild-type and mutant Sly1p, a modulator of transport vesicle docking. FEBS Lett 411: 169–172 [DOI] [PubMed] [Google Scholar]
- Gray MW, Doolittle WF 1982. Has the endosymbiont hypothesis been proven? Microbiol Rev 46: 1–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AG, Roger AJ 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups.” Proc Nat Acad Sci 106: 3859–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Nakamura Y, Nakamura F, Shirakura T, Adachi J, Goto N, Okamoto K, Hasegawa M 1994. Protein phylogeny gives a robust estimation for early divergences of eukaryotes: Phylogenetic place of a mitochondria-lacking protozoan, Giardia lamblia. Mol Biol Evol 11: 65–71 [DOI] [PubMed] [Google Scholar]
- Hawkins EK, Lee JJ 2001. Architecture of the Golgi of a scale-forming alga: Biogenesis and transport of scales. Protoplasma 216: 227–238 [DOI] [PubMed] [Google Scholar]
- Jekely G, Arendt D 2006. Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. Bioessays 28: 191–198 [DOI] [PubMed] [Google Scholar]
- Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV 2010. The conserved Bardet–Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141: 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL 2008. NCBI BLAST: A better web interface. Nucleic Acids Res 36: W5–W9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Shoda K, Tsuge T, Cho KH, Uchimiya H, Yokoyama R, Nishitani K, Tsukaya H 2002. The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J 21: 1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M, Nixon SJ, Howes MT, Abi-Rached L, Wakeham DE, Hanzal-Bayer M, Ferguson C, Hill MM, Fernandez-Rojo M, Brown DA, et al. 2008. Evolutionary analysis and molecular dissection of caveola biogenesis. J Cell Sci 121: 2075–2086 [DOI] [PubMed] [Google Scholar]
- Koumandou VL, Dacks JB, Coulson RM, Field MC 2007. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol 7: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremmidiotis G, Lensink IL, Bilton RL, Woollatt E, Chataway TK, Sutherland GR, Callen DF 1999. The Batten disease gene product (CLN3p) is a Golgi integral membrane protein. Hum Mol Genet 8: 523–531 [DOI] [PubMed] [Google Scholar]
- Leung KF, Dacks JB, Field MC 2008. Evolution of the multivesicular ESCRT machinery; retention across the eukaryotic lineage. Traffic 9: 1698–1716 [DOI] [PubMed] [Google Scholar]
- Lorente-Rodriguez A, Barlowe C 2011. Entry and exit mechanisms at the cis-face of the Golgi complex. Cold Spring Harb Perspect Biol 3: a005207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani-Telang P, Arnosti DN 2007. Developmental expression and phylogenetic conservation of alternatively spliced forms of the C-terminal binding protein corepressor. Dev Genes Evol 217: 127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti M, Regos A, Li Y, Schraner EM, Wild P, Muller N, Knopf LG, Hehl AB 2003. An ancestral secretory in the protozoan parasite Giardia intestinalis. J Biol Chem 278: 24837–24848 [DOI] [PubMed] [Google Scholar]
- Morgan GW, Hall BS, Denny PW, Field MC, Carrington M 2002. The endocytic of the kinetoplastida. Part II: Machinery and components of the system. Trends Parasitol 18: 540–546 [DOI] [PubMed] [Google Scholar]
- Mowbrey K, Dacks JB 2009. Evolution and diversity of the Golgi. FEBS Lett 583: 3738–3745 [DOI] [PubMed] [Google Scholar]
- Muhammad A, Flores I, Zhang H, Yu R, Staniszewski A, Planel E, Herman M, Ho L, Kreber R, Honig LS, et al. 2008. Retromer deficiency observed in Alzheimer's disease causes hippocampal dysfunction, neurodegeneration, and Aβ accumulation. Proc Natl Acad Sci 105: 7327–7332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann N, Lundin D, Poole AM 2010. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS One 5: e13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas HR, Lowry JA, Wu T, Crossley M 2008. The Caenorhabditis elegans protein CTBP-1 defines a new group of THAP domain-containing CtBP corepressors. J Mol Biol 375: 1–11 [DOI] [PubMed] [Google Scholar]
- Opdam FJ, Echard A, Croes HJ, van den Hurk JA, van de Vorstenbosch RA, Ginsel LA, Goud B, Fransen JA 2000. The small GTPase Rab6B, a novel Rab6 subfamily member, is cell-type specifically expressed and localised to the Golgi. J Cell Sci 113: 2725–2735 [DOI] [PubMed] [Google Scholar]
- Ossig R, Dascher C, Trepte HH, Schmitt HD, Gallwitz D 1991. The yeast SLY gene products, suppressors of defects in the essential GTP-binding Ypt1 protein, may act in endoplasmic reticulum-to-Golgi transport. Mol Cell Biol 11: 2980–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal JB 2008. The Ypt/Rab family and the evolution of trafficking in fungi. Traffic 9: 27–38 [DOI] [PubMed] [Google Scholar]
- Philippe H, Lopez P, Brinkmann H, Budin K, Germot A, Laurent J, Moreira D, Muller M, Le Guyader H 2000. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proc R Soc London B Biol Sci 267: 1213–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD 2006. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol 8: 238–248 [DOI] [PubMed] [Google Scholar]
- Ramirez IB, Lowe M 2009. Golgins and GRASPs: Holding the Golgi together. Semin Cell Dev Biol 20: 770–779 [DOI] [PubMed] [Google Scholar]
- Robinson MS 2004. Adaptable adaptors for coated vesicles. Trends Cell Biol 14: 167–174 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Brinkmann H, Burger G, Roger AJ, Gray MW, Philippe H, Lang BF 2007. Toward resolving the eukaryotic tree: The phylogenetic positions of jakobids and cercozoans. Curr Biol 17: 1420–1425 [DOI] [PubMed] [Google Scholar]
- Roger AJ, Simpson AG 2009. Evolution: revisiting the root of the eukaryote tree. Curr Biol 19: R165–R167 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Rowe T, Dascher C, Bannykh S, Plutner H, Balch WE 1998. Role of vesicle-associated syntaxin 5 in the assembly of pre-Golgi intermediates. Science 279: 696–700 [DOI] [PubMed] [Google Scholar]
- Saito K, Chen M, Bard F, Chen S, Zhou H, Woodley D, Polischuk R, Schekman R, Malhotra V 2009. TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell 136: 891–902 [DOI] [PubMed] [Google Scholar]
- Seemann J, Jokitalo EJ, Warren G 2000. The role of the tethering proteins p115 and GM130 in transport through the Golgi in vivo. Mol Biol Cell 11: 635–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N, Mulholland J, Botstein D 1988. The yeast GTP-binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell 52: 915–924 [DOI] [PubMed] [Google Scholar]
- Short B, Haas A, Barr FA 2005. Golgins and GTPases, giving identity and structure to the Golgi. Biochim Biophys Acta 1744: 383–395 [DOI] [PubMed] [Google Scholar]
- Simpson AG 2003. Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota). Int J Syst Evolutionary Microbiol 53: 1759–1777 [DOI] [PubMed] [Google Scholar]
- Sogin ML 1991. Early evolution and the origin of eukaryotes. Curr Opin Genet Dev 1: 457–463 [DOI] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE 1993. SNAP receptors implicated in vesicle targeting and fusion. Nature 362: 318–324 [DOI] [PubMed] [Google Scholar]
- Stamatakis A 2006. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Stanier R 1970. Some aspects of the biology of cells and their possible evolutionary significance. In Organization and control in prokaryotic and eukaryotic cells (ed. Charles H, Knight B), pp. 1–38 Syndics of the Cambridge University Press, Cambridge [Google Scholar]
- Stenmark H 2009. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10: 513–525 [DOI] [PubMed] [Google Scholar]
- Stern MD, Aihara H, Cho KH, Kim GT, Horiguchi G, Roccaro GA, Guevara E, Sun HH, Negeri D, Tsukaya H, et al. 2007. Structurally related Arabidopsis ANGUSTIFOLIA is functionally distinct from the transcriptional corepressor CtBP. Dev Genes Evol 217: 759–769 [DOI] [PubMed] [Google Scholar]
- Struck NS, de Souza Dias S, Langer C, Marti M, Pearce JA, Cowman AF, Gilberger TW 2005. Re-defining the Golgi complex in Plasmodium falciparum using the novel Golgi marker PfGRASP. J Cell Sci 118: 5603–5613 [DOI] [PubMed] [Google Scholar]
- Struck NS, Herrmann S, Langer C, Krueger A, Foth BJ, Engelberg K, Cabrera AL, Haase S, Treeck M, Marti M, et al. 2008. Plasmodium falciparum possesses two GRASP proteins that are differentially targeted to the Golgi complex via a higher- and lower-eukaryote-like mechanism. J Cell Sci 121: 2123–2129 [DOI] [PubMed] [Google Scholar]
- Toonen RF, Verhage M 2003. Vesicle trafficking: Pleasure and pain from SM genes. Trends Cell Biol 13: 177–186 [DOI] [PubMed] [Google Scholar]
- van der Giezen M 2009. Hydrogenosomes and mitosomes: conservation and evolution of functions. J Eukaryotic Microbiol 56: 221–231 [DOI] [PubMed] [Google Scholar]
- Walker G, Dorrell RG, Schlacht A, Dacks JB 2011. Eukaryotic systematics: A user's guide for cell biologists and parasitologists. Parasitology 15: 1–26 [DOI] [PubMed] [Google Scholar]
- Waters MG, Serafini T, Rothman JE 1991. “Coatomer”: A cytosolic protein complex containing subunits of non-clathrin-coated Golgi transport vesicles. Nature 349: 248–251 [DOI] [PubMed] [Google Scholar]
- Wooding S, Pelham HR 1998. The dynamics of Golgi protein traffic visualized in living yeast cells. Mol Biol Cell 9: 2667–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Lee SY, Spano S, Gad H, Zhang L, Nie Z, Bonazzi M, Corda D, Luini A, Hsu VW 2005. A role for BARS at the fission step of COPI vesicle formation from Golgi membrane. EMBO J 24: 4133–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa AC, Kawashima S, Okuda S, Fujita M, Itoh M, Moriya Y, Hattori M, Kanehisa M 2006. Extracting sequence motifs and the phylogenetic features of SNARE-dependent membrane traffic. Traffic 7: 1104–1118 [DOI] [PubMed] [Google Scholar]