SUMMARY
Group II introns are mobile ribozymes that self-splice from precursor RNAs to yield excised intron lariat RNAs, which then invade new genomic DNA sites by reverse splicing. The introns encode a reverse transcriptase that stabilizes the catalytically active RNA structure for forward and reverse splicing, and afterwards converts the integrated intron RNA back into DNA. The characteristics of group II introns suggest that they or their close relatives were evolutionary ancestors of spliceosomal introns, the spliceosome, and retrotransposons in eukaryotes. Further, their ribozyme-based DNA integration mechanism enabled the development of group II introns into gene targeting vectors (“targetrons”), which have the unique feature of readily programmable DNA target specificity.
Group II introns are mobile bacterial ribozymes that invade genomes by reverse splicing and reverse transcription. They may represent ancestors of eukaryotic spliceosomes and retrotransposons.
1. INTRODUCTION
Group II introns are mobile genetic elements that are found in bacterial and organellar genomes and are thought to be ancestors of spliceosomal introns and retrotransposons in eukaryotes. They consist of a catalytically active intron RNA (“ribozyme”) and an intron-encoded protein (IEP), whose combined activities enable intron proliferation within genomes. The group II intron RNA catalyzes its own splicing via transesterification reactions that are the same as those of spliceosomal introns, yielding spliced exons and an excised intron lariat RNA. The IEP is a multifunctional reverse transcriptase (RT), which is related to non-LTR-retrotransposon RTs and assists splicing by stabilizing the catalytically active RNA structure. It then remains bound to the excised intron RNA in an RNP that invades DNA sites. DNA invasion occurs by a remarkable mechanism in which the intron RNA uses its ribozyme activity to reverse splice directly into a DNA strand, after which it is reverse transcribed back into DNA by the IEP. Cycles of RNA splicing and reverse splicing enable the introns to proliferate to new DNA sites, while minimally impairing gene expression.
The characteristics of group II introns, including their splicing and mobility mechanisms, active-site structure, and naturally occurring variants that are split into two or more functionally reassociating segments, suggest an evolutionary scenario for the origin of introns, the spliceosome, and retrotransposons in eukaryotes. An evolutionary relationship between group II and spliceosomal introns appears increasingly plausible in light of newly obtained structural information, and indeed, a recent hypothesis asserts that group II intron invasion was the major driving force for the emergence of eukaryotes (Martin and Koonin 2006). Group II introns have also found practical applications as novel gene targeting vectors (“targetrons”), whose ribozyme-based DNA-integration mechanism enables their ready reprogramming to insert into desired DNA sites with high efficiency and specificity.
Here, we present an overview of group II intron structure and their splicing and mobility mechanisms, including recent insights from X-ray crystal structures of a group II intron RNA. We incorporate what has been learned into an evolutionary framework that considers the origin of group II introns, their structural variations, and how they may have evolved into spliceosomal introns and retrotransposons in eukaryotes.
2. CHARACTERISTICS OF GROUP II INTRONS
2.1. Phylogenetic Distribution
Group II introns have been found in bacteria and in the mitochondrial (mt) and chloroplast (cp) genomes of fungi, plants, protists, and an annelid worm (Belfort et al. 2002; Lambowitz and Zimmerly 2004; Toro et al. 2007; Vallès et al. 2008). Group II introns are rare in archaea, with the few found there likely acquired from eubacteria by relatively recent horizontal transfers (Rest and Mindell 2003). In eubacteria, group II introns are present in ∼25% of sequenced genomes, generally in small numbers, and typically as active retroelements with functional ribozyme and RT components. By contrast, group II introns in organelles frequently have degenerate RNA structures and either lack ORFs or encode degenerate IEPs that no longer promote intron mobility (Michel and Ferat 1995; Bonen 2008; Barkan 2009). Such immobile group II introns are inherited vertically and rely on host-encoded splicing factors (see later). Group II introns have not been found in the nuclear genomes of eukaryotes, but their hypothesized descendants, spliceosomal introns and retrotransposons, are highly abundant in eukaryotes, together comprising more than half of the human genome.
2.2. Group II Intron RNAs
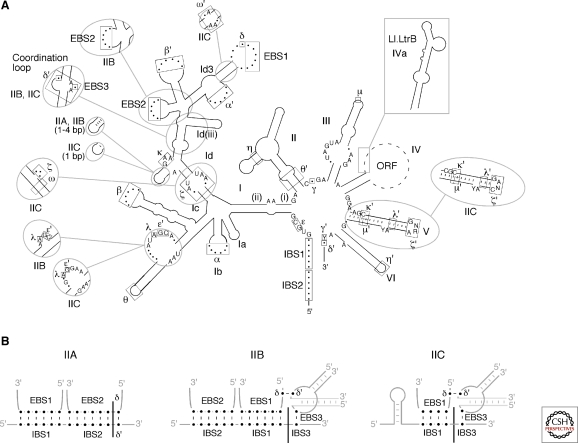
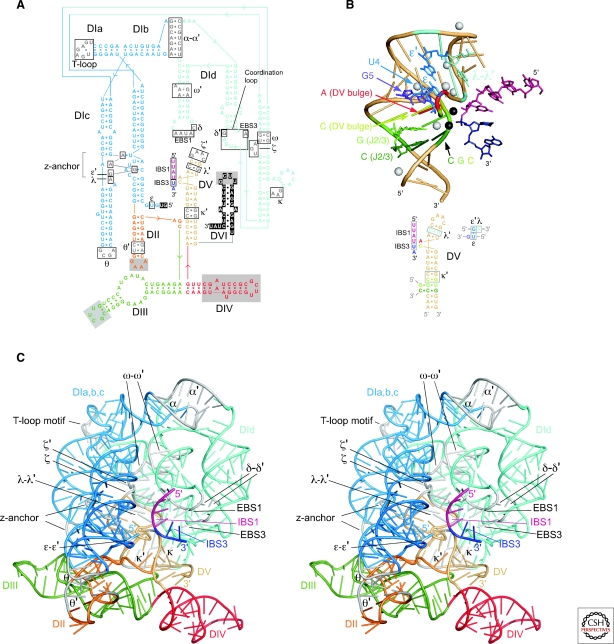
Group II intron RNAs are characterized by a conserved secondary structure, which spans 400-800 nts and is organized into six domains, DI-VI, radiating from a central “wheel” (Fig. 1) (Michel and Ferat 1995; Qin and Pyle 1998; Pyle and Lambowitz 2006). These domains interact to form a conserved tertiary structure that brings together distant sequences to form an active site. The active site binds the splice sites and branch-point nucleotide residue and uses specifically bound Mg++ ions to activate the appropriate bonds for catalysis. DV is the heart of the active site and contains the so-called catalytic triad AGC and an AY bulge, both of which bind catalytically important Mg++ ions. DI is the largest domain, with upper and lower halves separated by the κ and ζ motifs. The lower half contains the ε′ motif, which is associated with the active site, while the upper half contains sequence elements that bind the 5′ and 3′ exons at the active site. DVI contains the branch-point nucleotide, generally a bulged A. DII and III are smaller domains that contribute structurally, with DIII also acting as a “catalytic effector” to accelerate the catalytic step (Fedorova et al. 2003). The DIV loop encodes the IEP, with subdomain DIVa near its 5′ end containing a high-affinity binding site for the IEP. Group II intron RNAs also have conserved 5′- and 3′-end sequences, GUGYG and AY, respectively, which resemble those of spliceosomal introns (GU… AG) and which are bound at the active site by the ε–ε′and γ–γ′ interactions.
Figure 1.
Group II intron RNA secondary structure. (A) Structure of a representative bacterial IIA1 intron (not to scale), with notable variations in IIB and IIC introns shown in circles. Boxes indicate sequences involved in tertiary interactions (Greek letters, EBS, IBS). The “loop” of DIV, which encodes the IEP, is depicted by dashed lines, with a box showing the location and structure of DIVa of the Lactococcus lactis Ll.LtrB intron, a high-affinity binding site for the IEP. Subdomains discussed in the text are labeled, with base pairs (dashes) shown only for DV and the κ-stem-loop. Compared to IIA introns, major differences in other subgroups include structural features of DV (IIC introns); different ε′ motifs (IIB, IIC); the number of base pairs in the κ-stem-loop (IIC); a coordination loop containing EBS3 and δ′ (IIB, IIC); the absence of the DId(iii) stem-loop (IIB, IIC); the absence of a stem in the EBS2 motif (IIB, IIC); a unique ζ–ζ′ motif (IIC); and the ω–ω′ interaction (IIC, some IIB). (B) Base-pairing interactions used by IIA, IIB, and IIC introns to bind the exons at the active site. EBS, exon-binding site; IBS, intron-binding site.
Critical for the folding of group II intron RNAs are a series of conserved motifs involved in long-range tertiary interactions (Greek letters, EBS (exon-binding site), or IBS (intron-binding site) (Fig. 1). Such interactions have been identified by systematic covariation, genetic, and biochemical analyses. Some involve Watson-Crick base pairing (IBS1-EBS1, IBS2-EBS2, IBS3-EBS3, α–α′, β–β′, δ–δ′, ϵ–ϵ′, and γ–γ′), whereas others are tetraloop-receptor interactions (ζ–ζ′, θ–θ′, and η–η′) or other types of non-Watson-Crick interactions (κ–κ′, λ–λ′, and μ–μ′) (Qin and Pyle 1998; Costa et al. 2000; Boudvillain et al. 2000; Fedorova and Pyle 2005). The tertiary interactions between domains make it possible for group II intron RNAs to be split readily into different trans-splicing segments (Belhocine et al. 2008; Glanz and Kück 2009) and for some domains (DIc, DIII, DV, DI/II/III/IV) to act in trans to promote the splicing of introns lacking them (Jarrell et al. 1988a; Goldschmidt-Clermont et al. 1991; Suchy and Schmelzer 1991). Such fragmented group II introns and trans-acting segments occur naturally and underlie evolutionary scenarios for the origin of snRNAs (see later).
Although all group II introns have similar overall secondary structures, three major subgroups, denoted IIA, IIB, and IIC, and further subdivisions (A1, A2, B1, B2) are distinguished by specific variations (Fig. 1A; Michel et al. 1989; Toor et al. 2001; Dai et al. 2008). Unlike group I introns, in which subgroups differ mainly in peripheral structures, the differences between group II intron subgroups extend to the active site. One defining difference involves the interactions that bind the exons at the active site, which are critical for both RNA splicing and DNA target site recognition during intron mobility (Fig. 1B). IIA and IIB introns recognize their flanking exons via three base-pairing interactions (IBS1/EBS1 and IBS2/EBS2 for the 5′ exon and δ–δ′ (IIA) or IBS3/EBS3 (IIB) for the 3′ exon), while IIC introns use only two of these interactions (IBS1/EBS1 and IBS3/EBS3) and may also recognize a stem-loop derived from a transcription terminator or attC site in the 5′ exon (Fig. 1B) (Toor et al. 2006). Group II intron subgroups also differ in DV and in tertiary interaction motifs (see Fig. 1 legend). The latter include the “coordination loop” containing EBS3 and δ′, which is present in IIB and IIC but not IIA introns. The coordination loop may also position the branch-point A at the active site (Hamill and Pyle 2006), but this proposal has been questioned (Michel et al. 2009). A notable subfamily of IIB introns contains two additional stem-loops between DVI and the 3′-splice site (Stabell et al. 2009).
2.3. Degenerate, Twintron, and Trans-Splicing Group II Introns
Many organellar group II introns have structural defects that impair ribozyme activity. Plant mt and cp DNAs, for example, each encode about twenty group II introns, none of which is self-splicing or mobile (Barkan 2004). Their structural deviations include mispairs, insertions and deletions in DV and DVI, the absence of the bulged A in DVI, and subdomains that are unrecognizable or absent (Michel et al. 1989; Bonen 2008). The most extreme examples are found in Euglena cp DNA, which contains ∼150 introns lacking various domains. The smallest, referred to as group III introns, are ∼100 nt and contain only DI-like and DVI-like structures, lacking even DV, which is catalytically essential (Copertino and Hallick 1993). The splicing of these highly degenerate introns presumably requires trans-acting RNAs or proteins that compensate for the missing RNA structures.
Group II introns sometimes form twintrons in which one intron has inserted into another, reflecting their activity as mobile elements (Copertino and Hallick 1993). In some cases, the invading intron disrupts a critical feature of the outer intron and must be spliced first (Drager and Hallick 1993), while in other cases, the invading intron need not be spliced before the outer intron (Nakamura et al. 2002). Some twintrons form large nested arrays by repeated insertion of one group II intron into another (Dai and Zimmerly 2003).
Finally, trans-splicing group II introns are variants caused by genomic rearrangements that split an intron into two or more separately transcribed segments (Glanz and Kück 2009). These segments reassociate via tertiary contacts between group II intron domains and trans-splice the exons to produce a functional mRNA. Trans-splicing group II introns split at different locations have arisen naturally many times, particularly in plant mitochondria and chloroplasts (Qiu and Palmer 2004; Bonen 2008). One example is mt nad1-I4, which is continuous in some flowering plants, but trans-splicing in others, the only significant difference in the intron being its division into two independently transcribed segments (Bonen 1993). More extreme examples are cp psaA-I2 in Chlamydomonas spp. and mt nad5-I3 in some angiosperms, which are transcribed in three segments, with the middle segment beginning in DI and ending in DIV (Goldschmidt-Clermont et al. 1991; Knoop et al. 1997).
2.4. Group II Intron-Encoded Proteins
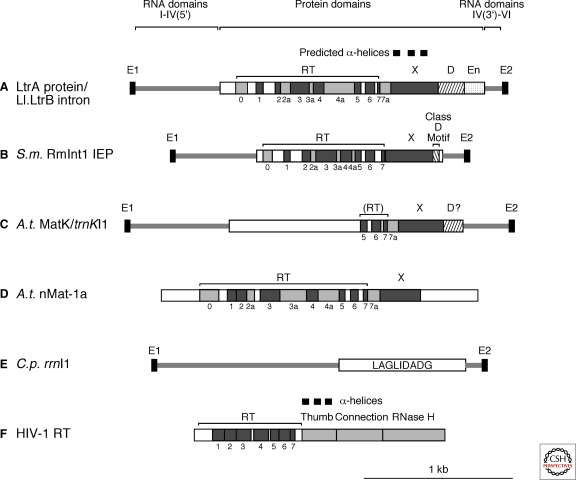
Most group II introns in bacteria and about half in mitochondria and chloroplasts encode an IEP in the loop of DIV. The best characterized IEP is the LtrA protein encoded by the Lactococcus lactis Ll.LtrB intron (Fig. 2A). LtrA has four domains: RT, reverse transcriptase; X/thumb; D, DNA binding; and En, DNA endonuclease. The RT domain is characterized by seven conserved sequence blocks (RT1-7) that form the fingers and palm regions of retroviral RTs, with RT5 containing the highly conserved sequence YADD that is part of the RT active site (Xiong and Eickbush 1990). Although presumably having a similar fold, group II intron and non-LTR-retrotransposon RTs are larger than those of retroviral RTs because of an amino-terminal extension (RT0) and “insertions” between the RT sequence blocks (RT2a, 3a, 4a, 7a in LtrA; cf., HIV-1 RT in Fig. 2F). Some of these insertions have conserved structural features and may be functionally important (Malik et al. 1999; Blocker et al. 2005).
Figure 2.
Group II intron IEPs and related proteins. (A) LtrA protein encoded by the L. lactis Ll.LtrB intron. (B) IEP lacking an En domain encoded by the Sinorhizobium meliloti RmInt1 intron, which belongs to bacterial lineage D (see Fig. 3). The “class D motif” at the carboxy-terminus is a conserved sequence that is required for splicing and mobility functions in lineage D IEPs (Molina-Sánchez et al. 2010). (C) MatK protein encoded by the Arabidopsis thaliana trnKI1 intron. MatK proteins retain conserved sequence blocks RT5-7 and domain X, but their amino-terminal halves have diverged from those of canonical group II IEPs, and they lack an En domain (Mohr et al. 1993). (D) nMat-1a protein encoded by a nuclear gene in Arabidopsis thaliana. nMat-1 proteins contain complete RT and X domains, but have mutations expected to inhibit RT activity; nMat-2 proteins (not shown) also contain an En domain, but with mutations expected to inhibit En activity (Mohr and Lambowitz 2003). (E) LAGLIDADG protein encoded by Cryphonectria parasitica rrnI1. (F) HIV-1 RT. Schematics of introns and ORFs are to scale. Insertions between RT sequence blocks are denoted 2a, 3a, 4a, and 7a. The locations of the three-predicted α-helices characteristic of thumb domains are shown above domain X in LtrA (cf. with HIV-1 RT in panel F).
Domain X is sometimes referred to as the “maturase domain” because it was identified as a site of mutations affecting RNA splicing activity (Mohr et al. 1993; Moran et al. 1994). It is characterized by conserved sequences and three predicted α-helices that are structurally analogous to the thumb domain of retroviral RTs (Blocker et al. 2005). In addition to reverse transcription, the RT and X/thumb domains function together in specific binding of the intron RNA, which both promotes formation of the active ribozyme structure and positions the protein to initiate cDNA synthesis (Saldanha et al. 1999; Wank et al. 1999; Cui et al. 2004).
The carboxy-terminal D and En domains interact with the target DNA during intron mobility. Domain D contributes to DNA binding, whereas the En domain is a Mg++-dependent DNA endonuclease of the H-N-H family that cleaves a target DNA strand to generate the primer for reverse transcription (San Filippo and Lambowitz 2002). Some bacterial group II introns, the best studied of which is Sinorhizobium meliloti RmInt1, encode IEPs that lack an En domain and use a different mechanism to prime reverse transcription (Fig. 2B) (see later).
Many group II IEPs have lost conserved sequences required for RT activity, but continue to function in RNA splicing, which is essential for gene expression after intron insertion. Some of these degenerate IEPs have small changes (e.g., mutations in the conserved YADD motif at the RT active site), whereas others are highly degenerate. The latter include two prominent examples of plant IEPs that have evolved to splice multiple organellar group II introns: cp MatK proteins (Fig. 2C) (Mohr et al. 1993) and nuclear-encoded nMat proteins (nMat-1a, -1b, -2a, -2b) (Fig. 2D) (Mohr and Lambowitz 2003) (see later).
Finally, a small group of fungal mt group II introns encode IEPs belonging to a family of DNA endonucleases characterized by the conserved motif LAGLIDADG (Fig. 2E) (Toor and Zimmerly 2002). These enzymes are often encoded in group I introns, where they function to promote intron homing by double-strand-break stimulated homologous recombination and sometimes act as maturases to promote RNA splicing (Belfort et al. 2002). Whether they function similarly in group II intron splicing and mobility is unknown.
2.5. Group II Intron Lineages
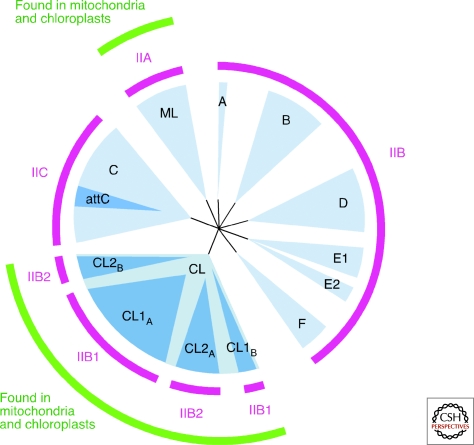
Group II intron ribozymes and IEPs function together as RNPs, with each IEP binding specifically to the intron RNA that encodes it. As a result, the intron RNAs and IEPs have coevolved over long times to form phylogenetic lineages of mobile introns (Fontaine et al. 1997; Toor et al. 2001). This situation again contrasts with that of group I introns, whose IEPs generally act independently as homing endonucleases and are frequently exchanged among introns (Belfort et al. 2002). Phylogenetic analyses identified eight lineages of group II intron IEPs, termed bacterial classes A-F, ML (mitochondrial-like) and CL (chloroplast-like), the latter because they are the major lineages in mitochondria and chloroplasts, respectively (Zimmerly et al. 2001; Simon et al. 2008) (Fig. 3). Each IEP lineage is associated with a specific RNA subgroup: ML with IIA, bacterial class C with IIC, and the remainder with IIB RNAs. CL IEPs are associated with IIB1 and IIB2 RNAs, while bacterial A, B, D, E, and F IEPs are associated with less typical IIB structures (Simon et al. 2009). Notably, bacteria contain all group II intron lineages, while mitochondria and chloroplasts contain both ML and CL introns but not other lineages. This distribution may reflect that ML and CL introns were present in bacterial endosymbionts that colonized eukaryotes and then exchanged between the two organelles.
Figure 3.
Group II intron lineages. The major lineages of group II intron IEPs, denoted CL (chloroplast-like), ML (mitochondrial-like), and bacterial classes A-F, are shown as blue sectors. Notable sublineages, including four subdivisions of CL and a subclass of IIC introns that inserts after attC sites, are shown as darker blue sectors within the major lineages. RNA structural subgroups that correspond to IEP lineages are shown in magenta. All group II intron lineages and RNA types are found in bacteria. Lineages and RNA types also found in organelles are delineated in green (outer circle). Note that there may be limited exceptions to the overall pattern of co-evolution within the CL group, with different sublineages possibly having exchanged IIB RNA structures (Simon et al. 2009). An alternate nomenclature for group II lineages has been proposed, which does not distinguish between IEP and ribozyme lineages or take into account exceptions to their coevolution (Toro et al. 2002).
3. REACTIONS CATALYZED BY GROUP II INTRON RNAs
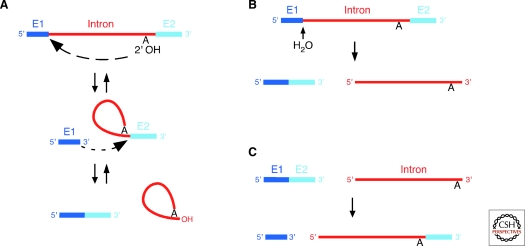
Group II ribozymes catalyze their own splicing via two sequential transesterification reactions (Fig. 4A). In the first step, the 2′ OH of the bulged A in DVI acts as the nucleophile to attack the 5′-splice site, producing an intron lariat/3′-exon intermediate. In the second step, the 3′ OH of the cleaved 5′ exon is the nucleophile and attacks the 3′-splice site, resulting in exon ligation and excision of an intron lariat RNA. Some group II introns self-splice in vitro, but the reaction is generally slow (kobs= 0.2 − 1.0 × 10−2 /min) and requires nonphysiological conditions—e.g., high concentrations of monovalent salt and/or Mg++ (Jarrell et al. 1988b; Daniels et al. 1996; Hiller et al. 2000), reflecting that proteins are needed to help fold group II intron RNAs into the catalytically active structure for efficient splicing. An important variation of the splicing reaction, termed “hydrolytic splicing,” involves the use of water as the nucleophile for the first transesterification reaction, rather than the 2′ OH of the bulged A of DVI (Fig. 4B) (van der Veen et al. 1987; Jarrell et al. 1988b). Some group II introns can splice exclusively via this pathway in vivo (Podar et al. 1998a; Bonen 2008).
Figure 4.
Reactions catalyzed by group II introns RNAs. (A) Forward and reverse splicing. (B) Hydrolytic splicing. The initial step is hydrolytic cleavage at the 5′ splice site. The second step leading to exon ligation (not shown) is the same as for splicing via lariat formation (panel A). (C) Partial reverse splicing by linear intron RNA, leading to ligation of the 3′ end of the intron RNA to the 5′ end of the 3′ exon. Intron RNA, red; 5′ and 3′ exons (E1 and E2), dark and light blue, respectively.
The active site for the splicing reaction contains at least two specifically bound Mg++ ions, which appear to be associated with the AGC triad and AY bulge in DV based on thio substitution/rescue and metal ion cleavage experiments (Chanfreau and Jacquier 1994; Sigel et al. 2000; Gordon and Piccirilli 2001; Gordon et al. 2007). For group I introns, Rp- and Sp-phosphorothioate substitutions at the splice sites have opposite effects on the two transesterification steps, which are simple reversals of each other at the same active site (McSwiggen and Cech 1989). By contrast, group II and spliceosomal introns show strong sensitivity to Rp but not Sp substitutions for both steps (Moore and Sharp 1993; Padgett et al. 1994; Podar et al. 1995; Podar et al. 1998b). These and other findings suggest that group II introns either use separate active sites with different Mg++ ions to catalyze the two splicing steps or a single active site, which is rearranged between steps (discussed in Pyle and Lambowitz (2006)). The active sites for the two steps at least partially overlap, as all of the reacting groups for both steps are in close proximity prior to splicing (de Lencastre et al. 2005) and mutations to key residues block both steps (Chanfreau and Jacquier 1994). A conformational change that occurs after the first step involves the formation of the η−η′ interaction between the distal loops of DII and DVI and is thought to remove the branch-point A from the active site (Chanfreau and Jacquier 1996). Other conformational changes may be linked to exon binding and release (Costa and Michel 1999).
Because transesterification reactions are reversible and energetically neutral, excised group II intron RNAs can reverse splice into ligated exons, guided by the same EBS/IBS and δ–δ′ base pairing interactions between the intron and flanking exon sequences used for RNA splicing. Lariat RNAs can carry out both reverse splicing steps, resulting in insertion of the intron RNA between the 5′ and 3′ exons (Fig. 4A, reverse arrows), while linear intron RNAs can carry out only the first step, ligation of the 3′ end of the intron to the 3′ exon (Fig. 4C) (Mörl and Schmelzer 1990; Mörl et al. 1992; Roitzsch and Pyle 2009). Reverse splicing of lariat or linear introns occurs into DNA as well as RNA exons, with a relatively small decrease of reactivity, reflecting a modest contribution of the 2′ OHs of the RNA exons (Mörl et al. 1992; Griffin et al. 1995). The ability to reverse splice efficiently into DNA underlies group II intron mobility.
Group II intron RNAs can carry out a number of other reactions in vitro, but their biological significance is unclear. These include: spliced-exon reopening (SER) in which the lariat intron hydrolytically cleaves the exon junction in RNA or DNA (Jarrell et al. 1988b; Mörl et al. 1992); RNA cleavage after 5′-exon sequences (Müller et al. 1988); reverse branching in which excised lariat ligates itself to 5′ exon RNA sequence through the reverse of the first splicing step (Chin and Pyle 1995); and circle formation in which an intron's 3′ end attacks the 5′ exon–intron junction, releasing the 5′ exon and an intron RNA cyclized by a 2′-5′ bond (Murray et al. 2001). Circular group II intron RNAs have also been reported in vivo (Li-Pook-Than and Bonen 2006; Molina-Sánchez et al. 2006).
4. THREE-DIMENSIONAL STRUCTURE OF GROUP II INTRON RIBOZYMES
A recent milestone in understanding group II ribozymes is the 3.1-Å X-ray crystal structure of the catalytic core of an Oceanobacillus iheyensis IIC intron (Toor et al. 2008a). The construct used for crystallization was a 412-nt RNA that was deleted for the distal stems of DII, III and VI, as well as the ORF of DIV (Fig. 5A). The intron was crystallized after hydrolytic splicing in vitro and hence is not branched. The 5′ and 3′ termini and DVI were not visible in the structure, and DVI was later found to be degraded (Toor et al. 2010). In a follow-up structure, what was initially thought to be a 6-nt exon RNA bound at the active site (Toor et al. 2008b) was found to be an unidentified RNA fragment, possibly derived from the degraded DVI (Toor et al. 2010).
Figure 5.
Crystal structure of the Oceanobacillus iheyensis group IIC intron. (A) Sequence and secondary structure of the crystallized RNA. Boxes indicate motifs involved in tertiary interactions. Solid gray boxes in DII, DIII, and DIV indicate regions deleted from the crystallization construct and replaced with sequences not present in the wild-type intron. Nucleotide residues in DI and DVI shown as white letters on a black background are not visible in the crystal structure. (B) Structure of the active-site region, with a corresponding color-coded secondary structure below. DV is a beige tube helix, with a bound RNA modeled as the 5′ and 3′ exons (pink and indigo, respectively; Toor et al. 2008b; Toor et al. 2010). The triple interactions in the triple helix stack between the CGC triad, the CG of J2/3, and the C of the AC bulge are shown in dark green, green and yellow-green. The three-base stack consisting of the A of the AC bulge, G5, and U4 of the ε–ε′ interaction are in red, purple and blue, respectively, while the λ–λ′ interaction is cyan. Metal ions bound to DV in the crystal are indicated by spheres, with black spheres representing the proposed active-site Mg++ ions, which were identified by binding of Yb3+ in the crystal derivatives. (C) X-ray crystal structure. A stereoview is shown, with domains colored as in (A) and regions involved in tertiary interactions colored gray (Toor et al. 2008a,b). Note added in proof: The conserved single base pair in the κ stem-loop, which we noted was missing in the original structure (Toor et al. 2008a), is present in the recently corrected and refined structure (Toor et al. 2010).
The structures provide critical insights into the nature of the active site (Fig. 5B). The catalytic center, DV, whose structure in isolation had been determined by crystallography and NMR (Zhang and Doudna 2002; Sigel et al. 2004), assumes a very different structure in the context of the ribozyme. The two helices of DV, rather than stacking coaxially, are bent severely to bring the AC bulge near the CGC triad, thus juxtaposing the phosphate backbones of the most conserved DV sequences. Nine potential Mg++-binding sites were assigned in or near DV, of which two bound Yb3+ ions in heavy atom derivatives. These two metal ions are coordinated to the phosphate backbones of the CGC triad and AC bulge, which were identified as catalytic metal-binding sites in other group II introns (see above), and are separated by a distance consistent with a two-metal-ion model of catalysis (Steitz and Steitz 1993).
The folded structure of DV is stabilized by the close packing of other conserved elements (Fig. 5B,C). The two bases just upstream of the γ-nucleotide in J2/3 form major groove base triples with the CG of the catalytic triad. The C of the AC bulge stacks on top of these two triple pairs to form a third triple base pair with the final C of the CGC triad. The A of the AC bulge protrudes in a different direction and stacks below G5 and U4 of the ε–ε′ interaction, which is in turn positioned by the λ–λ′ interaction between DIc and the upper stem of DV.
The remainder of the structure is dominated by DI, which along with DV comprises the minimal catalytic core (Fig. 5C). The two halves of DI wrap around and cap DV, anchored by the ζ–ζ′ and κ–κ′ interactions. On the opposite side of DV, the α–α′ pairing connects the halves. One half of DI, which consists of DI(i,ii)/Ia,/Ib/Ic, is a structural unit that positions the ε–ε′ pairing at the core of the molecule. Key to this positioning is the z-anchor, a zig-zagging series of four bases in the ε′ motif, which alternately form pairings with ε and the bulge loop of DI(i). Another key organizing feature is the 5-way junction formed by the loop of DIa interacting with the helices of DI(ii), DIb, DIc and DId, similar to the T-loop in tRNAs (Michel et al. 2009).
The other half of DI consists of nearly all of DId and is the exon-binding region. An important organizing center here is the ω–ω′ interaction, a ribose zipper that anchors the EBS1 stem near the ζ receptor, thus helping to position the 5′ exon. EBS3 and the coordination loop are positioned under EBS1 near the active site. Completing the structure are DII, which stacks coaxially with DI(i) and helps to dock DIc through the θ−θ′ interaction, and DIII and DIV, which are stacked coaxially along the bottom of the RNA.
The structure contains most of the previously predicted tertiary interactions, including IBS1-EBS1, α–α′, ε–ε′, δ–δ′, θ–θ′, ζ–ζ′, κ–κ′, and λ–λ′. Interactions not observed are either missing from IIC introns (β–β′, IBS2-EBS2), deleted from the crystallized construct (η–η′, μ–μ′), or unresolved (γ–γ′). However, some features of the structure are not consistent with previous data, particularly the position of DIII, which is pointed away from the ribozyme in the crystal structure, but whose conserved internal loop is predicted to be in the core of the molecule in contact with the ε motif, based on both chemogenetic and cross-linking experiments (Dai et al. 2008; Fedorova and Pyle 2008). In addition, the κ stem-loop does not form the single base pair that is phylogenetically conserved in IIC introns (Figs. 1A, 5A), and IBS3 and EBS3, while near each other, are not paired, possibly reflecting binding of a noncomplementary RNA fragment (Toor et al. 2010) (see earlier). Thus, it is not certain that the entire RNA has crystallized in a native conformation. IIA and IIB introns are expected to have active-site structures similar to that determined for the IIC intron, but contain a number of motifs not found in IIC introns and vice versa and thus must stabilize this structure in different ways.
5. THE INVOLVEMENT OF PROTEINS IN GROUP II INTRON SPLICING
As for other large RNAs, proteins play a major role in folding group II introns into their active three-dimensional structure in vivo (Lambowitz et al. 1999). Some of these proteins are RNA splicing factors that stabilize the active structure, while others are RNA chaperones that resolve stable, inactive structures (“kinetic traps”) that limit the rate of productive RNA folding.
5.1. Group II Intron-Encoded RTs are Intron-Specific RNA Splicing Factors
A key splicing factor for mobile group II introns is the IEP, which promotes splicing of the intron that encodes it. This splicing function (maturase activity) was first shown genetically for the yeast mt aI1 and aI2 IEPs (Carignani et al. 1983; Moran et al. 1994) and has been studied biochemically for the L. lactis Ll.LtrB IEP (LtrA protein) (Fig. 2A) (Matsuura et al. 1997; Saldanha et al. 1999). All three of these important model introns are members of subgroup IIA and encode ML lineage proteins. Purified recombinant LtrA can by itself promote splicing of the Ll.LtrB intron at near-physiological Mg++ concentrations (5 mM), at which the intron cannot self-splice efficiently. To promote splicing, LtrA binds tightly and specifically to the intron RNA to stabilize the active RNA structure, which reverts rapidly to the inactive structure when the protein is removed with protease (Matsuura et al. 2001; Noah and Lambowitz 2003; Mohr et al. 2006). Although largely monomeric in solution, LtrA binds the intron RNA with a stoichiometry of 2:1, suggesting that it functions as a dimer, the same active form as HIV-1 RT (Saldanha et al. 1999; Rambo and Doudna 2004).
Biochemical studies showed that LtrA has a high-affinity binding site in DIVa, an idiosyncratic stem-loop structure at the beginning of the LtrA ORF, accounting in part for its intron-specificity (Fig. 1A, inset) (Wank et al. 1999). The binding of LtrA to DIVa involves recognition of specific bases in the terminal loop and helical bulges, as well as their secondary structure context via phosphate-backbone interactions, analogous to classical phage coat-protein stem-loop interactions (Watanabe and Lambowitz 2004). Because DIVa in the Ll.LtrB intron contains the Shine-Dalgarno sequence and start codon of the IEP, the binding of LtrA to DIVa down-regulates its own translation (Singh et al. 2002; Cui et al. 2004).
Anchored by its tight binding to DIVa, LtrA makes additional contacts with conserved core regions of the intron RNA that stabilize the active RNA structure (Wank et al. 1999; Matsuura et al. 2001; Dai et al. 2008). If DIVa is deleted, LtrA can still promote splicing at reduced efficiency by binding directly to these core regions. The yeast aI2 IEP, which is translated as a fusion protein with the upstream exon and then proteolytically processed, also binds both DIVa and catalytic core regions, suggesting a conserved mode of interaction even when DIVa does not contain the start codon for the intron ORF (Huang et al. 2003).
LtrA-binding sites on the intron RNA were identified by RNA-footprinting (Matsuura et al. 2001) and by using circularly permuted intron RNAs to position fluorescence quenching and cross-linking reagents at different sites within the intron (Dai et al. 2008). Mapping of these sites onto a three-dimensional model of the intron RNA revealed a large continuous binding surface, which extends from DIVa across contiguous regions of DI, II, and VI. This binding surface suggests that LtrA promotes splicing by stabilizing interactions between these RNA domains, as well as the folded structure of DI. After splicing, the IEP remains tightly bound to excised intron and presumably uses most or all of the same interactions to stabilize the ribozyme structure for reverse splicing into DNA during intron mobility. Despite the structural differences between IIA, IIB, and IIC introns, modeling suggests that the IEP could bind and act similarly in all three cases (Dai et al. 2008).
Regions of the LtrA protein required for splicing were identified by using a high throughput mutagenesis method and mapped onto a three-dimensional model of the protein (Cui et al. 2004; Blocker et al. 2005). The results suggest that the RNA-binding surface extends across the RT and X/thumb domains and includes the amino-terminal extension, template-primer binding track, and patches on the back of the protein. Biochemical and genetic experiments suggest that LtrA's amino-terminal extension binds DIVa, whereas other regions of the RT and X domains bind the intron core (Cui et al. 2004; Gu et al. 2010). Unlike retroviral RTs, non-LTR-retroelement RTs bind specifically to their template RNAs for initiation of cDNA synthesis. Thus, the splicing function of group II intron RTs may have evolved from interactions that were used initially for RNA template recognition (Kennell et al. 1993).
5.2. Evolution of IEPs to Function in Splicing Multiple Group II Introns
Although most IEPs splice only the intron that encodes them, some have evolved to splice multiple introns, providing a common splicing apparatus. In the simplest cases, several bacterial introns that have proliferated within a genome are spliced by a single IEP, enabling all but one intron to lose its own ORF (Dai and Zimmerly 2003; Meng et al. 2005). Other IEPs, however, have expanded their splicing function to include more distantly related introns. Thus, the cp MatK protein, which is encoded by a trnKI1 intron in land plants and by a free-standing ORF in some nonphotosynthetic plants, is a highly degenerate IEP that has lost mobility functions but acquired the ability to bind and splice multiple cp IIA introns that lack ORFs (Fig. 2C; Ems et al. 1995; Vogel et al. 1999; Zoschke et al. 2010). This more general splicing function presumably reflects the loss of intron-specific interactions, such as those involving DIVa, and increased reliance on interactions with structural features shared by IIA introns. A further development is illustrated by the nMat proteins (nMat-1a, -1b, 2a, and 2b) of flowering plants (Fig. 2D; Mohr and Lambowitz 2003). These proteins, which evolved from mt group II intron IEPs, are encoded by nuclear genes and are transported into organelles to promote the splicing of group II introns that lack ORFs (Nakagawa and Sakurai 2006; Keren et al. 2009). This evolutionary progression in which an IEP loses mobility functions, evolves to splice multiple introns, and is ultimately subsumed into the host genome, limits potentially deleterious intron mobility and facilitates host regulation of RNA splicing in concert with other cellular processes.
5.3. Recruitment of Host-Encoded Proteins to Function in Group II Intron Splicing
Most mt and cp group II introns lack ORFs and rely on host-encoded proteins to promote their splicing. This situation is similar to that for group I introns, where a variety of host proteins, such aminoacyl-tRNA synthetases, have been recruited to function in splicing different introns (Lambowitz et al. 1999). Host-encoded splicing factors have been studied in the greatest detail for plant cp group II introns. Genetic analysis supported by in vivo binding data indicate that these cp group II introns utilize a battery of splicing factors, including proteins belonging to three large plant-specific families of RNA-binding proteins, CRM (chloroplast RNA splicing and ribosome maturation), POROR (plant RNA recognition), and pentatricopeptide repeat (PPR) proteins (Stern et al. 2010). These proteins function combinatorially to promote the splicing of different introns, with most introns requiring multiple splicing factors that associate with the RNA to form large complexes (Till et al. 2001; Kroeger et al. 2009). In the green alga Chlamydomonas reinhardti, the trans-splicing of cp psaA-I1 and -I2 requires a different set of host-encoded proteins, which also appear to function in large complexes, some of which may be membrane bound (Merendino et al. 2006; Perron et al. 1999). The different host-encoded splicing factors for plant and algal cp group II introns presumably reflect that their splicing function evolved relatively recently, after the divergence of plants and green algae.
5.4. DEAD-Box Proteins Function as RNA Chaperones in Group II Intron Splicing
In addition to the splicing factors that stabilize the active RNA structure, the efficient splicing of group II introns requires RNA chaperones to resolve stable inactive or intermediate structures that limit the rate of productive RNA folding (Huang et al. 2005; Köhler et al. 2010). In S. cerevisiae mitochondria, a key protein that plays this role is Mss116p, a member of the DEAD-box family of ATP-dependent RNA helicases, which function in diverse RNA structural rearrangements in all organisms (Séraphin et al. 1989; Huang et al. 2005). MSS116 null mutants are defective in splicing all four mt group II introns. Two of these, aI1 and aI2, are mobile IIA introns that encode RT/maturase proteins that stabilize their active structures, while the other two, aI5γ and bI1, are small IIB introns that do not encode proteins. Significantly, these group II intron splicing defects are leaky, and MSS116 mutants are also defective in splicing all mt group I introns, translation of some mt mRNAs, and other RNA processing reactions. Further, Mss116p can be replaced in group I or group II intron splicing reactions in vitro or in vivo by CYT-19, a N. crassa DEAD-box protein that functions as an RNA chaperone in mt group I intron splicing (Huang et al. 2005) or by other DEAD-box proteins, which do not ordinarily function in group I and II intron splicing (e.g., S. cerevisiae Ded1p and Escherichia coli SrmB; Del Campo et al. 2009 and refs. therein). Thus, Mss116p and these other DEAD-box proteins appear to interact nonspecifically with structurally diverse RNA and RNP substrates, as expected for general RNA chaperones. Biochemical studies confirmed these nonspecific interactions, as well as a correlation with the DEAD-box protein's RNA-unwinding activity in all cases, but leave open the possibility that other DEAD-box protein activities, such as strand annealing or RNA structural stabilization, also contribute to the splicing of some group II introns (Mohr et al. 2006; Solem et al. 2006; Halls et al. 2007; Del Campo et al. 2007, 2009). The involvement of DEAD-box proteins in group II intron splicing is an important parallel to the spliceosome, where DEAD-box proteins and related RNA helicases function at multiple steps to accelerate specific structural transitions.
6. GROUP II INTRON MOBILITY
Group II intron mobility occurs by a novel mechanism in which the excised intron RNA uses its ribozyme activity to reverse splice directly into a DNA strand where it is reverse transcribed by the IEP. The introns use different variations of this mechanism both to “retrohome” to specific DNA target sites at frequencies up to 100% and to “retrotranspose” to ectopic sites that resemble the normal homing sites at low frequencies (10−4 to 10−6). Retrohoming was first observed for yeast mt introns, which were found to invade intronless alleles during genetic crosses (Meunier et al. 1990). The high efficiency and specificity of retrohoming underlie the use of group II introns in gene targeting.
6.1. Group II Intron Retrohoming by Reverse Splicing into DNA
The mechanism of group II intron retrohoming was elucidated by studies of the yeast mt aI1 and aI2 and L. lactis Ll.LtrB introns (reviewed in Lambowitz and Zimmerly 2004). Retrohoming is mediated by the RNP that is formed during RNA splicing and consists of the IEP and excised intron lariat RNA (Fig. 6A). RNPs initiate retrohoming by using both the IEP and intron RNA to recognize DNA target sequences. The IEP recognizes specific bases or structural features of the DNA target site, which differ for each intron, and this interaction helps separate the DNA strands, enabling the intron RNA to base pair to the 5′ and 3′ DNA exons. Importantly, this base pairing involves the same subgroup-specific EBS/IBS and δ–δ′ interactions used for RNA splicing (see earlier; Guo et al. 1997; Eskes et al. 1997; Mohr et al. 2000; Jiménez-Zurdo et al. 2003; Robart et al. 2007; Zhuang et al. 2009a). By using the same base-pairing interactions for both RNA splicing and DNA integration, the intron ensures that it inserts only at sites from which it can subsequently excise by RNA splicing.
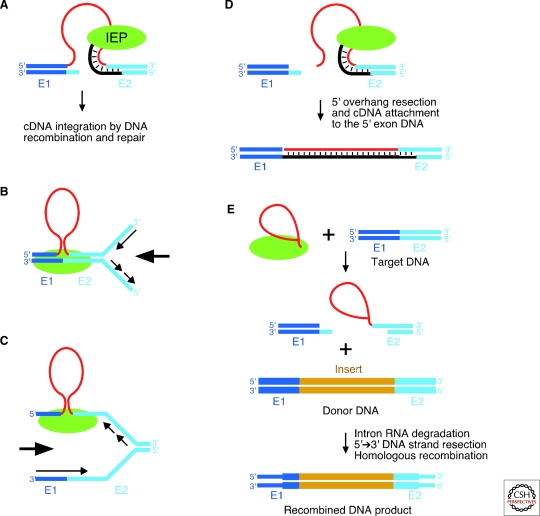
Figure 6.
Group II intron mobility mechanisms. (A) Retrohoming via reverse splicing of the intron RNA into double-stranded DNA. After reverse splicing of the intron RNA into the top strand, the bottom strand is cleaved by the En domain of the IEP, and the 3′ end at the cleavage site is used as a primer for reverse transcription of the inserted intron RNA. The resulting intron cDNA is integrated by cellular DNA recombination and/or repair mechanisms. (B) Reverse splicing of the intron RNA into double-stranded DNA, with priming by the nascent leading strand of the DNA replication fork. (C) Reverse splicing of the intron RNA into single-stranded DNA, with priming by the nascent lagging strand of the DNA replication fork. (D) Retrohoming of linear intron RNA by the first step of reverse splicing, bottom-strand cleavage, reverse transcription, and attachment of the free cDNA end to the 5′ exon DNA likely by NHEJ (Zhuang et al. 2009b). (E) Use of group II introns to introduce a targeted double-strand break that stimulates gene targeting by homologous recombination. The top-strand break by the first step of reverse splicing can be made either by lariat RNA as shown in the figure or by linear intron RNA (not shown; Mastroianni et al. 2008). Recombination results in the precise insertion of a novel DNA sequence (gold) from the donor DNA into the target DNA. The target and donor DNAs are shown with different widths to illustrate the origin of different DNA segments in the recombined DNA product. Intron RNA, red; 5′ and 3′ exons (E1 and E2), dark and light blue, respectively; IEP, green. In (B) and (C), large arrows indicate the direction of the replication fork, and small arrows indicate the direction of DNA synthesis.
After base pairing, the intron reverse splices into the DNA strand, resulting in the insertion of linear intron RNA between the two DNA exons. The En domain of the IEP cleaves the opposite DNA strand a short distance downstream (position +10 for aI1 and aI2 and +9 for Ll.LtrB), and the 3′ OH of the cleaved DNA is used as a primer for reverse transcription of the inserted intron RNA, a process referred to as “target-primed reverse transcription” (TPRT). For yeast aI1 and aI2, cDNA integration generally occurs by a mechanism in which a nascent cDNA initiates recombination with an intron-containing allele, leading to transfer of the intron plus a variable length of the upstream exon (Eskes et al. 1997, 2000). By contrast, cDNA integration for the Ll.LtrB intron in both L. lactis and E. coli occurs by a DNA repair mechanism without coconversion of flanking exon sequences (Cousineau et al. 1998). In E. coli, this repair mechanism appears to involve synthesis of a full-length intron cDNA, removal of the RNA strand by the host RNase H, second-strand synthesis by the host replicative DNA polymerase III, and the use of host DNA ligase to seal nicks (Smith et al. 2005).
6.2. DNA Target Site Recognition By Group II Intron RNPs
The mechanism of DNA target site recognition by group II intron RNPs has been studied in most detail for the L. lactis Ll.LtrB group II intron, taking advantage of an efficient E. coli expression system to obtain large amounts of purified RNPs (Saldanha et al. 1999). Ll.LtrB RNPs bind to DNA nonspecifically and scan for target sites by facilitated diffusion, analogous to site-specific DNA binding proteins (Aizawa et al. 2003). The IEP is thought to first recognize a small number of specific bases in the distal 5′-exon region of the DNA target site via major groove interactions, most critically T-23, G-21, and A-20 (Singh and Lambowitz 2001). These initial base interactions, bolstered by phosphate backbone and possibly IEP interactions in the adjacent minor groove (positions-17 to -13), trigger local DNA melting, enabling the intron RNA's EBS2, EBS1, and δ sequences to base pair to the IBS2, IBS1, and δ′ sequences for reverse splicing. Intron RNA base pairing may occur concomitantly with and help drive DNA melting. Bottom-strand cleavage occurs at position +9 from the insertion site and requires additional interactions between the IEP and the 3′ exon, the most critical being recognition of T+5 (Singh and Lambowitz 2001). Binding of the RNP to the 5′ and 3′ DNA exons bends the target DNA, with the bend angle increasing when the cleaved 3′ end is repositioned from the En to RT active site for initiation of reverse transcription (Noah et al. 2006). Because reverse splicing into the DNA target site is reversible and the equilibrium favors intron excision, the reaction must be driven forward by the initiation of cDNA synthesis, which blocks the 3′-splice site and prevents excision (Aizawa et al. 2003). Other group II introns use the same mode of DNA target site recognition, but the 5′- and 3′-exon sequences recognized by the IEP differ even for closely related introns, suggesting rapid evolution of IEP specificity (Zhuang et al. 2009a and references therein).
6.3. Variations of the Retrohoming Mechanism
Group II introns whose IEPs lack an En domain or whose En activity is inactivated by mutation use a variation of the retrohoming pathway in which a nascent strand at a DNA replication fork primes reverse transcription (Fig. 6B,C; Zhong and Lambowitz 2003; Martínez-Abarca et al. 2004). Group II introns retrohome via this mechanism either by reverse splicing into double-stranded DNA with preferential use of leading strand DNA primers, or by reverse splicing into transiently single-stranded DNA at a DNA replication fork or transcription bubble with preferential use of lagging strand DNA primers. The latter pathway appears to be favored by naturally occurring group II introns whose IEPs lack an En domain (Ichiyanagi et al. 2002), including IIC introns, where single-stranded DNA facilitates formation of the DNA stem-loop of a transcription terminator or attC site that is recognized by the intron RNP (Robart et al. 2007; Léon and Roy 2009).
Linear group II intron RNAs, which may be generated by hydrolytic splicing or debranching of lariats, use another retrohoming variation, which was uncovered by injecting Ll.LtrB RNPs containing linear intron RNA into Xenopus laevis oocyte nuclei or Drosophila melanogaster embryos (Fig. 6D; Zhuang et al. 2009b). Here, the linear intron RNA carries out the first step of reverse splicing into the DNA target, thereby ligating the RNA to the 3′ but not the 5′ exon. The intron RNA is then reverse transcribed by the IEP, and the free end of the cDNA is linked to the 5′ exon DNA, likely via nonhomologous end-joining (NHEJ), a widely studied DNA repair process. Although inefficient, this mechanism could be used for retrohoming of linear RNAs, not only in eukaryotes but also in many prokaryotes, which have analogous NHEJ machinery (Bowater and Doherty 2006).
6.4. Retrotransposition of Group II Introns to New Sites
Retrotransposition to ectopic sites that resemble the normal homing site occurs at low frequency providing a means of group II intron dispersal to new genomic locations. In all cases examined, retrotransposition occurs by reverse splicing of the intron RNA into a DNA site, but with different variations of the pathway favored depending on the organism and growth conditions (Yang et al. 1998; Martínez-Abarca and Toro 2000; Dickson et al. 2001; Ichiyanagi et al. 2002). In L. lactis, retrotransposition of the Ll.LtrB intron occurs primarily by reverse splicing into transiently single-stranded DNA with priming by the nascent lagging strand (Fig. 6C) (Ichiyanagi et al. 2002). In E. coli, however, retrotransposition of this intron occurs both by the above mechanism and by inaccurate reverse splicing into double-stranded DNA, with or without En cleavage of the opposite strand (Fig. 6B,C) (Coros et al. 2005). As expected, pathways using nascent DNA strands as primers are favored under rapid growth conditions, which lead to an increased frequency of replication forks (Coros et al. 2005). Retromobility of the Ll.LtrB intron is also influenced in interesting ways by cellular interactions, host factors, and stress responses (Beauregard et al. 2008; Zhao et al. 2008; Coros et al. 2009).
6.5. Targetrons
Because group II introns recognize DNA target sites largely by base pairing of the intron RNA to the DNA target sequence, it is possible to retarget them to insert into desired DNA sites simply by modifying the base pairing sequences in the intron RNA (Guo et al. 2000; Karberg et al. 2001). This feature, combined with the high efficiency and specificity of the retrohoming reaction, enabled the development of group II introns into gene targeting vectors (“targetrons”; Perutka et al. 2004). A targetron based on the Ll.LtrB intron is sold commercially and widely used for gene targeting in bacteria. Recently, other group II introns have also been adapted for gene targeting, expanding the range of accessible target sites and providing other useful properties (Zhuang et al. 2009a).
Targetrons are also being developed for use in eukaryotes, where the obstacles include nuclear accessibility of RNPs and suboptimal Mg++ concentrations (Mastroianni et al. 2008). In X. laevis oocyte nuclei and D. melanogaster embryos, microinjected group II intron RNPs retrohome efficiently if additional Mg++ is provided (Mastroianni et al. 2008; Zhuang et al. 2009b). A more general solution is to identify or select introns that function at lower Mg++ concentrations. Importantly, targetrons can be used not only for site-specific DNA integration, but also to generate a targeted double-strand DNA break that stimulates targeted DNA integration by homologous recombination. The double-strand break results from the initial partial reverse splicing and second-strand cleavage reactions of the group II intron RNP (Fig. 6E; Karberg et al. 2001; Mastroianni et al. 2008). The introduction of a recombinogenic double-strand break by a protein endonuclease, such as a Zn-finger nuclease or a meganuclease, is a favored mode of gene targeting in higher organisms (Porteus and Carroll 2005), for which targetrons have an inherent advantage in the ease of retargeting breaks to desired sites.
7. GROUP II INTRON EVOLUTION
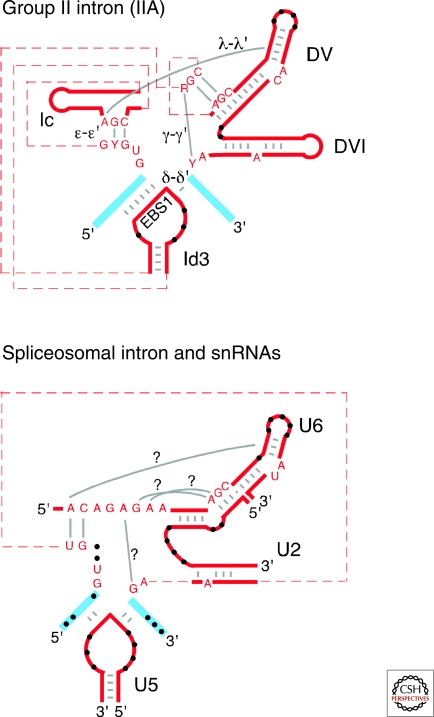
Group II introns are thought to have played a major role in eukaryotic genome evolution as ancestors of both spliceosomal introns and non-LTR-retrotransposons (Sharp 1985; Cech 1986; Zimmerly et al. 1995). An evolutionary relationship between group II and spliceosomal introns is suggested by their identical splicing pathways, similar boundary sequences, and structural similarities between key regions of group II intron domains and spliceosomal RNAs (Madhani and Guthrie 1992; Shukla and Padgett 2002; Keating et al. 2010). The latter include: divalent metal-ion binding sites in DV and U6 snRNA, which may contribute to catalysis; the similar branch site motifs of DVI and the U2-intron pairing, in which an equivalent adenosine is the branch site for the first step of splicing; the similarity between the ϵ–ϵ′ interaction of group II introns and the pairing between the ACAGAGA sequence of U6 and the 5′ region of the intron; and recognition of 5′ and 3′ exons, in which the IBS1-EBS1 and δ–δ′ motifs of IIA introns are analogous to the U5 snRNA stem-loop (Fig. 7). Confidence in these similarities has been bolstered by the recent IIC intron crystal structure (Toor et al. 2008a) and by increasing evidence that snRNAs can catalyze reactions related to RNA splicing (Valadkhan et al. 2009). An evolutionary relationship between group II introns and non-LTR-retrotransposons is suggested by the similarity of their RT sequences (Xiong and Eickbush 1990; Blocker et al. 2005) and TPRT mechanisms, in which a cleaved DNA target site is used as the primer for reverse transcription of a specifically bound RNA template (Luan et al. 1993; Zimmerly et al. 1995).
Figure 7.
Similarities between the active site of group II introns and the putative active site of the spliceosome. Group II intron RNA and spliceosomal snRNA segments are shown in red, and exons are shown in blue. Base-pairing interactions that are similar for group II and spliceosomal introns are shown by gray bars, and unpaired bases at similar positions are shown by black dots. Dashed lines indicate connecting sequence of unspecified length. Question marks indicate hypothetical interactions that may occur in the spliceosome, based on interactions found in group II intron RNAs (Boudvillain et al. 2000; Toor et al. 2008a). The similarity between DId3 and the U5 snRNA is closest for IIA introns, while the ε-ε′ and DV/U6 similarities are closest for IIA and IIB introns (see Fig. 1).
The phylogenetic distribution of group II introns, which are common in eubacteria and eukaryotic organelles but rare in archaebacteria, suggests a scenario in which mobile group II introns originated in eubacteria and were transmitted to eukaryotes, possibly via endosymbiotic bacteria that gave rise to mitochondria and chloroplasts (Cavalier-Smith 1991; Palmer and Logsdon 1991). The idea that group II introns originated as retroelements in bacteria (the “retroelement ancestor hypothesis”) is supported by the observation that bacterial group II introns include all known lineages and generally behave as retroelements, whereas organellar introns belong to only the ML or CL lineages, and frequently lack ORFs and/or have degenerate IEP or RNA structures (Toor et al. 2001). The ancestral eubacterial retroelement might have arisen by invasion of a self-splicing ribozyme by an RT (Wank et al. 1999) or from a retroelement that evolved a self-splicing RNA at its termini to ameliorate damage to the host (Curcio and Belfort 1996). While IIC introns may be the earliest branching class (i.e., most similar to the last common ancestor), statistical support for this position is weak (Rest and Mindell 2003; Simon et al. 2009).
In eukaryotes, group II introns are thought to have invaded the nucleus and proliferated to many genomic sites, after which the ribozyme structure degenerated and fragmented into snRNAs that function in trans in a common splicing apparatus (Sharp 1991). A recent hypothesis suggests that the introduction of group II introns by a bacterial endosymbiont was the driving force for a fundamental step in the evolution of eukaryotes, the formation of the nuclear membrane, which separates transcription from translation and thus helps prevent translation of incompletely spliced RNAs (Martin and Koonin 2006). Regardless of its origin, the separation of transcription and translation by the nuclear membrane prevents immediate access of the IEP to the intron RNA, and necessitates the evolution of splicing factors that function in trans. In flowering plants, host-encoded splicing factors for cp group II introns evolved by the expansion and diversification of different families of RNA-binding proteins, with different proteins required to splice different introns. Such a solution would be impractical for the large numbers of introns in eukaryotic nuclear genomes, which instead evolved snRNAs derived from group II intron domains into a common RNA-based catalytic machinery replacing that in individual introns. These snRNAs continue to recognize the introns via conserved 5′ and 3′ sequences and a branch-point nucleotide similar to those of group II introns and catalyze splicing by the same transesterification reactions. This byzantine RNA-based spliceosomal machinery is arguably the strongest evidence that the eukaryotic splicing apparatus evolved from group II introns or their close relatives rather than de novo.
Acknowledgments
We thank Alice Barkan, Marlene Belfort, Mark Del Campo, Georg Mohr, Sabine Mohr, and Rick Russell for comments on the manuscript. Work in the authors' laboratories was supported by National Institutes of Health grants GM37949 and GM37951 and Welch Foundation grant F-1607 to A.M.L. and CIHR grant MOP-93662 and NSERC grant RGP 2003717-02 to S.Z.
Footnotes
Editors: John F. Atkins, Raymond F. Gesteland, and Thomas R. Cech
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Aizawa Y, Xiang Q, Lambowitz AM, Pyle AM 2003. The pathway for DNA recognition and RNA integration by a group II intron retrotransposon. Mol Cell 11: 795–805 [DOI] [PubMed] [Google Scholar]
- Barkan A 2004. Intron splicing in plant organelles. In Molecular biology and biotechnology of plant organelles (eds Daniell H, Chase C), pp. 281–308 Kluwer Academic Publishers, Dordrecht [Google Scholar]
- Barkan A 2009. Genome-wide analysis of RNA-protein interactions in plants. Methods Mol Biol 553: 13–37 [DOI] [PubMed] [Google Scholar]
- Beauregard A, Curcio MJ, Belfort M 2008. The take and give between retrotransposable elements and their hosts. Ann Rev Genet 42: 587–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M, Derbyshire V, Parker MM, Cousineau B, Lambowitz AM 2002. Mobile introns: Pathways and proteins. In Mobile DNA II (eds Craig NL, Craigie R, Gellert M, Lambowitz AM), pp. 761–783 ASM Press, Washington D.C [Google Scholar]
- Belhocine K, Mak AB, Cousineau B 2008. Trans-splicing versatility of the Ll.LtrB group II intron. RNA 14: 1782–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker FJ, Mohr G, Conlan LH, Qi L, Belfort M, Lambowitz AM 2005. Domain structure and three-dimensional model of a group II intron-encoded reverse transcriptase. RNA 11: 14–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L 1993. Trans-splicing of pre-mRNA in plants, animals, and protists. FASEB J 7: 40–46 [DOI] [PubMed] [Google Scholar]
- Bonen L 2008. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8: 26–34 [DOI] [PubMed] [Google Scholar]
- Boudvillain M, de Lencastre A, Pyle AM 2000. A tertiary interaction that links active-site domains to the 5′ splice site of a group II intron. Nature 406: 315–318 [DOI] [PubMed] [Google Scholar]
- Bowater R, Doherty AJ 2006. Making ends meet: Repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genet 2: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carignani G, Groudinsky O, Frezza D, Schiavon E, Bergantino E, Slonimski PP 1983. An mRNA maturase is encoded by the first intron of the mitochondrial gene for the subunit I of cytochrome oxidase in S. cerevisiae. Cell 35: 733–742 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 1991. Intron phylogeny: A new hypothesis. Trends Genet 7: 145–148 [PubMed] [Google Scholar]
- Cech TR 1986. The generality of self-splicing RNA: Relationship to nuclear mRNA splicing. Cell 44: 207–210 [DOI] [PubMed] [Google Scholar]
- Chanfreau G, Jacquier A 1994. Catalytic site components common to both splicing steps of a group II intron. Science 266: 1383–1387 [DOI] [PubMed] [Google Scholar]
- Chanfreau G, Jacquier A 1996. An RNA conformational change between the two chemical steps of group II self-splicing. EMBO J 15: 3466–3476 [PMC free article] [PubMed] [Google Scholar]
- Chin K, Pyle AM 1995. Branch-point attack in group II introns is a highly reversible transesterification, providing a potential proofreading mechanism for 5′-splice site selection. RNA 1: 391–406 [PMC free article] [PubMed] [Google Scholar]
- Copertino D, Hallick R 1993. Group II and group III introns of twintrons: potential relationships with nuclear pre-mRNA introns. Trends Biochem Sci 18: 467–471 [DOI] [PubMed] [Google Scholar]
- Coros CJ, Landthaler M, Piazza CL, Beauregard A, Esposito D, Perutka J, Lambowitz AM, Belfort M 2005. Retrotransposition strategies of the Lactococcus lactis Ll.LtrB group II intron are dictated by host identity and cellular environment. Mol Microbiol 56: 509–524 [DOI] [PubMed] [Google Scholar]
- Coros CJ, Piazza CL, Chalamcharla VR, Smith D, Belfort M 2009. Global regulators orchestrate group II intron retromobility. Mol Cell 34: 250–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Michel F 1999. Tight binding of the 5′ exon to domain I of a group II self-splicing intron requires completion of the intron active site. EMBO J 18: 1025–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Michel F, Westhof E 2000. A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J 19: 5007–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousineau B, Smith D, Lawrence-Cavanagh S, Mueller JE, Yang J, Mills D, Manias D, Dunny G, Lambowitz AM, Belfort M 1998. Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell 94: 451–462 [DOI] [PubMed] [Google Scholar]
- Cui X, Matsuura M, Wang Q, Ma H, Lambowitz AM 2004. A group II intron-encoded maturase functions preferentially in cis and requires both the reverse transcriptase and X domains to promote RNA splicing. J Mol Biol 340: 211–231 [DOI] [PubMed] [Google Scholar]
- Curcio MJ, Belfort M 1996. Retrohoming: cDNA-mediated mobility of group II introns requires a catalytic RNA. Cell 84: 9–12 [DOI] [PubMed] [Google Scholar]
- Dai L, Zimmerly S 2003. ORF-less and reverse-transcriptase-encoding group II introns in archaebacteria, with a pattern of homing into related group II intron ORFs. RNA 9: 14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Chai D, Gu SQ, Gabel J, Noskov SY, Blocker FJ, Lambowitz AM, Zimmerly S 2008. A three-dimensional model of a group II intron RNA and its interaction with the intron-encoded reverse transcriptase. Mol Cell 30: 472–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Michels WJ Jr, Pyle AM 1996. Two competing pathways for self-splicing by group II introns: a quantitative analysis of in vitro reaction rates and products. J Mol Biol 256: 31–49 [DOI] [PubMed] [Google Scholar]
- de Lencastre A, Hamill S, Pyle AM 2005. A single active-site region for a group II intron. Nat Struct Mol Biol 12: 626–627 [DOI] [PubMed] [Google Scholar]
- Del Campo M, Mohr S, Jiang Y, Jia H, Jankowsky E, Lambowitz AM 2009. Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones. J Mol Biol 389: 674–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo M, Tijerina P, Bhaskaran H, Mohr S, Yang Q, Jankowsky E, Russell R, Lambowitz AM 2007. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol Cell 28: 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson L, Huang HR, Liu L, Matsuura M, Lambowitz AM, Perlman PS 2001. Retrotransposition of a yeast group II intron occurs by reverse splicing directly into ectopic DNA sites. Proc Natl Acad Sci 98: 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drager RG, Hallick RB 1993. A complex twintron is excised as four individual introns. Nucleic Acids Res 21: 2389–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ems SC, Morden CW, Dixon CK, Wolfe KH, dePamphilis CW, Palmer JD 1995. Transcription, splicing and editing of plastid RNAs in the nonphotosynthetic plant Epifagus virginiana. Plant Mol Biol 29: 721–733 [DOI] [PubMed] [Google Scholar]
- Eskes R, Liu L, Ma H, Chao MY, Dickson L, Lambowitz AM, Perlman PS 2000. Multiple homing pathways used by yeast mitochondrial group II introns. Mol Cell Biol 20: 8432–8446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskes R, Yang J, Lambowitz AM, Perlman PS 1997. Mobility of yeast mitochondrial group II introns: Engineering a new site specificity and retrohoming via full reverse splicing. Cell 88: 865–874 [DOI] [PubMed] [Google Scholar]
- Fedorova O, Pyle AM 2005. Linking the group II intron catalytic domains: Tertiary contacts and structural features of domain 3. EMBO J 24: 3906–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova O, Pyle AM 2008. A conserved element that stabilizes the group II intron active site. RNA 14: 1048–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova O, Mitros T, Pyle AM 2003. Domains 2 and 3 interact to form critical elements of the group II intron active site. J Mol Biol 330: 197–209 [DOI] [PubMed] [Google Scholar]
- Fontaine JM, Goux D, Kloareg B, Loiseaux-de Goër S 1997. The reverse-transcriptase-like proteins encoded by group II introns in the mitochondrial genome of the brown alga Pylaiella littoralis belong to two different lineages which apparently coevolved with the group II ribozyme lineages. J Mol Evol 44: 33–42 [DOI] [PubMed] [Google Scholar]
- Glanz S, Kück U 2009. Trans-splicing of organelle introns–a detour to continuous RNAs. Bioessays 31: 921–934 [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M, Choquet Y, Girard-Bascou J, Michel F, Schirmer-Rahire M, Rochaix JD 1991. A small chloroplast RNA may be required for trans-splicing in Chlamydomonas reinhardtii. Cell 65: 135–143 [DOI] [PubMed] [Google Scholar]
- Gordon PM, Piccirilli JA 2001. Metal ion coordination by the AGC triad in domain 5 contributes to group II intron catalysis. Nat Struct Biol 8: 893–898 [DOI] [PubMed] [Google Scholar]
- Gordon PM, Fong R, Piccirilli JA 2007. A second divalent metal ion in the group II intron reaction center. Chem Biol 14: 607–612 [DOI] [PubMed] [Google Scholar]
- Griffin EA Jr, Qin Z, Michels WJ Jr, Pyle AM 1995. Group II intron ribozymes that cleave DNA and RNA linkages with similar efficiency, and lack contacts with substrate 2′-hydroxyl groups. Chem Biol 2: 761–770 [DOI] [PubMed] [Google Scholar]
- Gu S-Q, Cui X, Mou S, Mohr S, Yao J, Lambowitz AM 2010. Genetic identification of potential RNA-binding regions in a group II intron-encoded reverse transcriptase. RNA 16: 732–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Karberg M, Long M, Jones JP 3rd, Sullenger B, Lambowitz AM 2000. Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science 289: 452–457 [DOI] [PubMed] [Google Scholar]
- Guo H, Zimmerly S, Perlman PS, Lambowitz AM 1997. Group II intron endonucleases use both RNA and protein subunits for recognition of specific sequences in double-stranded DNA. EMBO J 16: 6835–6848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM 2007. Involvement of DEAD-box proteins in group I and group II intron splicing: Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol 365: 835–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill S, Pyle AM 2006. The receptor for branch-site docking within a group II intron active site. Mol Cell 23: 831–840 [DOI] [PubMed] [Google Scholar]
- Hiller R, Hetzer M, Schweyen RJ, Mueller MW 2000. Transposition and exon shuffling by group II intron RNA molecules in pieces. J Mol Biol 297: 301–308 [DOI] [PubMed] [Google Scholar]
- Huang HR, Chao MY, Armstrong B, Wang Y, Lambowitz AM, Perlman PS 2003. The DIVa maturase binding site in the yeast group II intron aI2 is essential for intron homing but not for in vivo splicing. Mol Cell Biol 23: 8809–8819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS 2005. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci 102: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyanagi K, Beauregard A, Lawrence S, Smith D, Cousineau B, Belfort M 2002. Retrotransposition of the Ll.LtrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol Micoibiol 46: 1259–1272 [DOI] [PubMed] [Google Scholar]
- Jarrell KA, Dietrich RC, Perlman PS 1988a. Group II intron domain 5 facilitates a trans-splicing reaction. Mol Cell Biol 8: 2361–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell KA, Peebles CL, Dietrich RC, Romiti SL, Perlman PS 1988b. Group II intron self-splicing: Alternative reaction conditions yield novel products. J Biol Chem 263: 3432–3439 [PubMed] [Google Scholar]
- Jiménez-Zurdo JI, García-Rodríguez FM, Barrientos-Durán A, Toro N 2003. DNA target site requirements for homing in vivo of a bacterial group II intron encoding a protein lacking the DNA endonuclease domain. J Mol Biol 326: 413–423 [DOI] [PubMed] [Google Scholar]
- Karberg M, Guo H, Zhong J, Coon R, Perutka J, Lambowitz AM 2001. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat Biotech 19: 1162–1167 [DOI] [PubMed] [Google Scholar]
- Keating KS, Toor N, Perlman PS, Pyle AM 2010. A structural analysis of the group II intron active site and implications for the spliceosome. RNA 16: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennell JC, Moran JV, Perlman PS, Butow RA, Lambowitz AM 1993. Reverse transcriptase activity associated with maturase-encoding group II introns in yeast mitochondria. Cell 73: 133–146 [DOI] [PubMed] [Google Scholar]
- Keren I, Bezawork-Geleta A, Kolton M, Maayan I, Belausov E, Levy M, Mett A, Gidoni D, Shaya F, Ostersetzer-Biran O 2009. AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15: 2299–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V, Altwasser M, Brennicke A 1997. A tripartite group II intron in mitochondria of an angiosperm plant. Mol Gen Genetics 255: 269–276 [DOI] [PubMed] [Google Scholar]
- Köhler D, Schmidt-Gattung S, Binder S 2010. The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana. Plant Mol Biol 72: 459–467 [DOI] [PubMed] [Google Scholar]
- Kroeger TS, Watkins KP, Friso G, van Wijk KJ, Barkan A 2009. A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc Natl Acad Sci 106: 4537–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S 2004. Mobile group II introns. Ann Rev Genet 38: 1–35 [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Caprara MG, Zimmerly S, Perlman PS 1999. Group I and group II ribozymes as RNPs: Clues to the past and guides to the future. In The RNA world (eds Gesteland RF, Cech TR, Atkins JF), pp. 451–485 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Li-Pook-Than J, Bonen L 2006. Multiple physical forms of excised group II intron RNAs in wheat mitochondria. Nucleic Acids Res 34: 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon G, Roy PH 2009. Group IIC intron mobility into attC sites involves a bulged DNA stem-loop motif. RNA 15: 1543–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell 72: 595–605 [DOI] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C 1992. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817 [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH 1999. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol 16: 793–805 [DOI] [PubMed] [Google Scholar]
- Martin W, Koonin EV 2006. Introns and the origin of nucleus-cytosol compartmentalization. Nature 440: 41–45 [DOI] [PubMed] [Google Scholar]
- Martínez-Abarca F, Toro N 2000. RecA-independent ectopic transposition in vivo of a bacterial group II intron. Nucleic Acids Res 28: 4397–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Abarca F, Barrientos-Durán A, Fernández-López M, Toro N 2004. The RmInt1 group II intron has two different retrohoming pathways for mobility using predominantly the nascent lagging strand at DNA replication forks for priming. Nucleic Acids Res 32: 2880–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroianni M, Watanabe K, White TB, Zhuang F, Vernon J, Matsuura M, Wallingford J, Lambowitz AM 2008. Group II intron-based gene targeting reactions in eukaryotes. PLoS ONE 3: e3121 10.1371/journal.pone.0003121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura M, Noah JW, Lambowitz AM 2001. Mechanism of maturase-promoted group II intron splicing. EMBO J 20: 7259–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura M, Saldanha R, Ma H, Wank H, Yang J, Mohr G, Cavanagh S, Dunny GM, Belfort M, Lambowitz AM 1997. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: Biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev 11: 2910–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen JA, Cech TR 1989. Stereochemistry of RNA cleavage by the Tetrahymena ribozyme and evidence that the chemical step is not rate-limiting. Science 244: 679–683 [DOI] [PubMed] [Google Scholar]
- Meng Q, Wang Y, Liu XQ 2005. An intron-encoded protein assists RNA splicing of multiple similar introns of different bacterial genes. J Biol Chem 280: 35085–35088 [DOI] [PubMed] [Google Scholar]
- Merendino L, Perron K, Rahire M, Howald I, Rochaix JD, Goldschmidt-Clermont M 2006. A novel multifunctional factor involved in trans-splicing of chloroplast introns in Chlamydomonas. Nucleic Acids Res 34: 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier B, Tian G-L, Macadre C, Slonimski PP, Lazowska J 1990. Group II introns transpose in yeast mitochondria. In Structure function and biogenesis of energy transfer systems (eds Quagliariello E, Papa S, Palmieri F, Saccone CC), pp 169–174 Elsevier Scientific Publishers, Amsterdam [Google Scholar]
- Michel F, Ferat JL 1995. Structure and activities of group II introns. Ann Rev Biochem 64: 435–461 [DOI] [PubMed] [Google Scholar]
- Michel F, Costa M, Westhof E 2009. The ribozyme core of group II introns: A structure in want of partners. Trends Biochem Sci 34: 189–199 [DOI] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H 1989. Comparative and functional anatomy of group II catalytic introns—a review. Gene 82: 5–30 [DOI] [PubMed] [Google Scholar]
- Mohr G, Lambowitz AM 2003. Putative proteins related to group II intron reverse transcriptase/maturases are encoded by nuclear genes in higher plants. Nucleic Acids Res 31: 647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr G, Perlman PS, Lambowitz AM 1993. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res 21: 4991–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr G, Smith D, Belfort M, Lambowitz AM 2000. Rules for DNA target-site recognition by a lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes Dev 14: 559–573 [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Matsuura M, Perlman PS, Lambowitz AM 2006. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci 103: 3569–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Sánchez MD, Martínez-Abarca F, Toro N 2006. Excision of the Sinorhizobium meliloti group II intron RmInt1 as circles in vivo. J Biol Chem 281: 28737–28744 [DOI] [PubMed] [Google Scholar]
- Molina-Sánchez MD, Martínez-Abarca F, Toro N 2010. Structural features in the C-terminal region of the Sinorhizobium meliloti RmInt1 group II intron-encoded protein contribute to its maturase and intron DNA-insertion function. FEBS J 277: 244–255 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA 1993. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature 365: 364–368 [DOI] [PubMed] [Google Scholar]
- Moran JV, Mecklenburg KL, Sass P, Belcher SM, Mahnke D, Lewin A, Perlman P 1994. Splicing defective mutants of the COXI gene of yeast mitochondrial DNA: Initial definition of the maturase domain of the group II intron AI2. Nucleic Acids Res 22: 2057–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörl M, Schmelzer C 1990. Integration of group II intron bI1 into a foreign RNA by reversal of the self-splicing reaction in vitro. Cell 60: 629–636 [DOI] [PubMed] [Google Scholar]
- Mörl M, Niemer I, Schmelzer C 1992. New reactions catalyzed by a group II intron ribozyme with RNA and DNA substrates. Cell 70: 803–810 [DOI] [PubMed] [Google Scholar]
- Müller MW, Schweyen RJ, Schmelzer C 1988. Selection of cryptic 5′ splice sites by group II intron RNAs in vitro. Nucleic Acids Res 16: 7383–7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray HL, Mikheeva S, Coljee VW, Turczyk BM, Donahue WF, Bar-Shalom A, Jarrell KA 2001. Excision of group II introns as circles. Mol Cell 8: 201–211 [DOI] [PubMed] [Google Scholar]
- Nakagawa N, Sakurai N 2006. A mutation in At-nMat1a, which encodes a nuclear gene having high similarity to group II intron maturase, causes impaired splicing of mitochondrial NAD4 transcript and altered carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol 47: 772–783 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kaneko T, Sato S, Ikeuchi M, Katoh H, Sasamoto S, Watanabe A, Iriguchi M, Kawashima K, Kimura T, et al. 2002. Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res 9: 123–130 [DOI] [PubMed] [Google Scholar]
- Noah JW, Lambowitz AM 2003. Effects of maturase binding and Mg2+ concentration on group II intron RNA folding investigated by UV cross-linking. Biochemistry 42: 12466–12480 [DOI] [PubMed] [Google Scholar]
- Noah JW, Park S, Whitt JT, Perutka J, Frey W, Lambowitz AM 2006. Atomic force microscopy reveals DNA bending during group II intron ribonucleoprotein particle integration into double-stranded DNA. Biochemistry 45: 12424–12435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett RA, Podar M, Boulanger SC, Perlman PS 1994. The stereochemical course of group II intron self-splicing. Science 266: 1685–1688 [DOI] [PubMed] [Google Scholar]
- Palmer JD, Logsdon JM Jr, 1991. The recent origins of introns. Curr Opinion Genet Dev 1: 470–477 [DOI] [PubMed] [Google Scholar]
- Perron K, Goldschmidt-Clermont M, Rochaix JD 1999. A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J 18: 6481–6490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutka J, Wang W, Goerlitz D, Lambowitz AM 2004. Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J Mol Biol 336: 421–439 [DOI] [PubMed] [Google Scholar]
- Podar M, Perlman PS, Padgett RA 1995. Stereochemical selectivity of group II intron splicing, reverse splicing, and hydrolysis reactions. Mol Cell Biol 15: 4466–4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M, Perlman PS, Padgett RA 1998b. The two steps of group II intron self-splicing are mechanistically distinguishable. RNA 4: 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M, Chu VT, Pyle AM, Perlman PS 1998a. Group II intron splicing in vivo by first-step hydrolysis. Nature 391: 915–918 [DOI] [PubMed] [Google Scholar]
- Porteus MH, Carroll D 2005. Gene targeting using zinc finger nucleases. Nat Biotechnol 23: 967–973 [DOI] [PubMed] [Google Scholar]
- Pyle AM, Lambowitz AM 2006. Group II introns: Ribozymes that splice RNA and invade DNA. In The RNA world, 3rd ed. (eds Gesteland RF, Cech TR, Atkins JF), pp. 469–506 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Qiu YL, Palmer JD 2004. Many independent origins of trans splicing of a plant mitochondrial group II intron. J Mol Evol 59: 80–89 [DOI] [PubMed] [Google Scholar]
- Qin PZ, Pyle AM 1998. The architectural organization and mechanistic function of group II intron structural elements. Curr Opinion Struct Biol 8: 301–308 [DOI] [PubMed] [Google Scholar]
- Rambo RP, Doudna JA 2004. Assembly of an active group II intron-maturase complex by protein dimerization. Biochemistry 43: 6486–6497 [DOI] [PubMed] [Google Scholar]
- Rest JS, Mindell DP 2003. Retroids in archaea: Phylogeny and lateral origins. Mol Biol Evol 20: 1134–1142 [DOI] [PubMed] [Google Scholar]
- Robart AR, Seo W, Zimmerly S 2007. Insertion of group II intron retroelements after intrinsic transcriptional terminators. Proc Natl Acad Sci 104: 6620–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitzsch M, Pyle AM 2009. The linear form of a group II intron catalyzes efficient autocatalytic reverse splicing, establishing a potential for mobility. RNA 15: 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha R, Chen B, Wank H, Matsuura M, Edwards J, Lambowitz AM 1999. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry 38: 9069–9083 [DOI] [PubMed] [Google Scholar]
- San Filippo J, Lambowitz AM 2002. Characterization of the C-terminal DNA-binding/DNA endonuclease region of a group II intron-encoded protein. J Mol Biol 324: 933–951 [DOI] [PubMed] [Google Scholar]
- Séraphin B, Simon M, Boulet A, Faye G 1989. Mitochondrial splicing requires a protein from a novel helicase family. Nature 337: 84–87 [DOI] [PubMed] [Google Scholar]
- Sharp PA 1985. On the origin of RNA splicing and introns. Cell 42: 397–400 [DOI] [PubMed] [Google Scholar]
- Sharp PA 1991. “Five easy pieces”. Science 254: 663. [DOI] [PubMed] [Google Scholar]
- Shukla GC, Padgett RA 2002. A catalytically active group II intron domain 5 can function in the U12-dependent spliceosome. Mol Cell 9: 1145–1150 [DOI] [PubMed] [Google Scholar]
- Sigel RK, Vaidya A, Pyle AM 2000. Metal ion binding sites in a group II intron core. Nature Struct Biol 7: 1111–1116 [DOI] [PubMed] [Google Scholar]
- Sigel RKO, Sashital DG, Abramovitz DL, Palmer AG III, Butcher SE, Pyle AM 2004. Solution structure of domain 5 of a group II intron ribozyme reveals a new RNA motif. Nature Struct Mol Biol 11: 187–192 [DOI] [PubMed] [Google Scholar]
- Simon DM, Kelchner SA, Zimmerly S 2009. A broad-scale phylogenetic analysis of group II intron RNAs and intron-encoded reverse transcriptases. Mol Biol Evol 26: 2795–2808 [DOI] [PubMed] [Google Scholar]
- Simon DM, Clarke NA, McNeil BA, Johnson I, Pantuso D, Dai L, Chai D, Zimmerly S 2008. Group II introns in Eubacteria and Archaea: ORF-less introns and new varieties. RNA 14: 1704–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Lambowitz AM 2001. Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J Mol Biol 309: 361–386 [DOI] [PubMed] [Google Scholar]
- Singh RN, Saldanha RJ, D'Souza LM, Lambowitz AM 2002. Binding of a group II intron-encoded reverse transcriptase/maturase to its high affinity intron RNA binding site involves sequence-specific recognition and autoregulates translation. J Mol Biol 318: 287–303 [DOI] [PubMed] [Google Scholar]
- Smith D, Zhong J, Matsuura M, Lambowitz AM, Belfort M 2005. Recruitment of host functions suggests a repair pathway for late steps in group II intron retrohoming. Genes Dev 19: 2477–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solem A, Zingler N, Pyle AM 2006. A DEAD protein that activates intron self-splicing without unwinding RNA. Mol Cell 24: 611–617 [DOI] [PubMed] [Google Scholar]
- Stabell FB, Tourasse NJ, Kolstø AB 2009. A conserved 3′ extension in unusual group II introns is important for efficient second-step splicing. Nucleic Acids Res 37: 3202–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA 1993. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci 90: 6498–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR 2010. Chloroplast RNA metabolism. Ann Rev Plant Biol (in press) [DOI] [PubMed] [Google Scholar]
- Suchy M, Schmelzer C 1991. Restoration of the self-splicing activity of a defective group II intron by a small trans-acting RNA. J Mol Biol 222: 179–187 [DOI] [PubMed] [Google Scholar]
- Till B, Schmitz-Linneweber C, Williams-Carrier R, Barkan A 2001. CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7: 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Zimmerly S 2002. Identification of a family of group II introns encoding LAGLIDADG ORFs typical of group I introns. RNA 8: 1373–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Hausner G, Zimmerly S 2001. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA 7: 1142–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Fedorova O, Rajashankar K, Wang J, Pyle AM 2010. Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA 16: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM 2008a. Crystal structure of a self-spliced group II intron. Science 320: 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Rajashankar K, Keating KS, Pyle AM 2008b. Structural basis for exon recognition by a group II intron. Nat Struct Mol Biol 15: 1221–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Robart AR, Christianson J, Zimmerly S 2006. Self-splicing of a group IIC intron: 5′ exon recognition and alternative 5′ splicing events implicate the stem-loop motif of a transcriptional terminator. Nucleic Acids Res 34: 6461–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro N, Jiménez-Zurdo JI, García-Rodríguez FM 2007. Bacterial group II introns: Not just splicing. FEMS Microbiol Rev 31: 342–358 [DOI] [PubMed] [Google Scholar]
- Toro N, Molina-Sánchez M, Fernández-López M 2002. Identification and characterization of bacterial class E group II introns. Gene 299: 245–250 [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Mohammadi A, Jaladat Y, Geisler S 2009. Protein-free small nuclear RNAs catalyze a two-step splicing reaction. Proc Natl Acad Sci 106: 11901–11906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallès Y, Halanych KM, Boore JL 2008. Group II introns break new boundaries: Presence in a bilaterian's genome. PLoS ONE 3: e1488 10.1371/journal.pone.0001488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen R, Kwakman JH, Grivell LA 1987. Mutations at the lariat acceptor site allow self-splicing of a group II intron without lariat formation. EMBO J 6: 3827–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Börner T, Hess WR 1999. Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res 27: 3866–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wank H, San Filippo J, Singh RN, Matsuura M, Lambowitz AM 1999. A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol Cell 4: 239–250 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Lambowitz AM 2004. High-affinity binding site for a group II intron-encoded reverse transcriptase/maturase within a stem-loop structure in the intron RNA. RNA 10: 1433–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Eickbush T 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9: 3353–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mohr G, Perlman PS, Lambowitz AM 1998. Group II intron mobility in yeast mitochondria: Target DNA-primed reverse transcription activity of aI1 and reverse splicing into DNA transposition sites in vitro. J Mol Biol 282: 505–523 [DOI] [PubMed] [Google Scholar]
- Zhang L, Doudna JA 2002. Structural insights into group II intron catalysis and branch-site selection. Science 295: 2084–2088 [DOI] [PubMed] [Google Scholar]
- Zhao J, Niu W, Yao J, Mohr S, Marcotte EM, Lambowitz AM 2008. Group II intron protein localization and insertion sites are affected by polyphosphate. PLoS Biol 6: e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Lambowitz AM 2003. Group II intron mobility using nascent strands at DNA replication forks to prime reverse transcription. EMBO J 22: 4555–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang F, Karberg M, Perutka J, Lambowitz AM 2009a. EcI5, a group IIB intron with high retrohoming frequency: DNA target site recognition and use in gene targeting. RNA 15: 432–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang F, Mastroianni M, White TB, Lambowitz AM 2009b. Linear group II intron RNAs can retrohome in eukaryotes and may use nonhomologous end-joining for cDNA ligation. Proc Natl Acad Sci 106: 18189–18194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerly S, Hausner G, Wu X 2001. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res 29: 1238–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerly S, Guo H, Perlman PS, Lambowitz AM 1995. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell 82: 545–554 [DOI] [PubMed] [Google Scholar]
- Zoschke R, Nakamura M, Liere K, Sugiura M, Börner T, Linneweber-Schmitz C 2010. An organellar maturase associated with multiple group II introns. Proc Natl Acad Sci 107: 3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]