Abstract
Chloride binding in photosystem II (PSII) is essential for photosynthetic water oxidation. However, the functional roles of chloride and possible binding sites, during oxygen evolution, remain controversial. This paper examines the functions of chloride based on its binding site revealed in the X-ray crystal structure of PSII at 1.9 Å resolution. We find that chloride depletion induces formation of a salt-bridge between D2-K317 and D1-D61 that could suppress proton transfer to the lumen.
Keywords: Photosystem II, water splitting, Oxygen-Evolving Complex, allosteric regulation, chloride, MD simulations, MCCE method BRIEFS Effect of chloride binding on O2 evolution in photosystem II
Light-driven oxygen evolution in photosynthetic organisms is catalyzed by photosystem II (PSII), a ~350 kDa complex of 20 proteins embedded in the thylakoid membranes of green plant chloroplasts, or internal membranes of cyanobacteria (2). Dioxygen evolves from water oxidation catalyzed by the oxygen-evolving complex (OEC) of PSII, a Mn4CaO5 cluster embedded in protein subunit D1. This catalytic cluster is of great biological and technological interest since it is composed of inexpensive earth-abundant metals (e.g., Mn and Ca) and has an efficient turnover number as high as 50 dioxygen molecules per second for direct solar-driven water oxidation that is yet to be matched by artificial systems (Fig. 1) (3). Recent breakthroughs in X-ray crystallography (2) provide evidence of a cuboidal cluster MnCaMn3, similar to previous proposals (4–6), with a ‘dangling’ Mn bound to the CaMn3 cube, as well as information on the position of Cl− cofactor ions (2, 7). These breakthroughs open an opportunity for studies of structure/functions relations that might provide fundamental insight on the potential role of Cl− in the reaction mechanism. Cl− concentration is regulated very effectively under a variety of conditions in chloroplasts (8, 9) and it is well known that Cl− depletion suppresses O2 evolution (10–12), although the specific roles played by Cl− in the underlying catalytic process remains to be established at the molecular level (13, 14). This paper addresses the potential influence of Cl− binding on the hydrogen bonds of amino acid residues in close contact with the OEC that might facilitate fundamental processes that are essential for oxygen evolution, including water gating and proton transfer to the lumen.
Figure 1.
Top left: Position of Cl− relative to the OEC in the X-ray structure of PSII at 1.9 A (colored ball and sticks) and the 2006 DFT-QM/MM model (blue ball and sticks), aligned to minimize the RMSD of Mn1, Mn2, Mn3 and Ca2+. Top right: Waters modeled in the 1.9 Å structure (gray spheres) next to the Cl−, the OEC and residues D1-D61 and D2-K317. Bottom: Ligation of the OEC defined for MD and MCCE simulations, according to our new S1 state DFT-QM/MM model (colored ball and sticks), see supporting information (SI) (1). X-ray structure is shown in purple for comparison.
The catalytic mechanism of O2 evolution by water oxidation in PSII is driven by solar light absorption. The absorbed photon energy is transferred to the reaction center where it is used to oxidize a chlorophyll a species called P680, forming the radical-pair state P680+• – QA−• by reducing the protein-bound quinone cofactor, QA. The oxidizing power accumulated in the radical P680+• is used to oxidize YZ, which in turn oxidizes the OEC through a series of S states, with S0 and S4 the most reduced and oxidized states, respectively. The S4 state has enough oxidizing potential to oxidize water, regenerating the S0 state for the next catalytic turnover cycle. Experimental evidence shows that oxidation state transitions beyond the S2 state are suppressed upon Cl− depletion, or competitive binding of acetate in place of Cl− (15, 16). However, the specific functional/structural roles of Cl− during the catalytic cycle have yet to be established. An early hypothesis of Cl− ions bridging Mn atoms in the OEC core (9) was ruled out by X-ray absorption spectroscopy experiments (17, 18) in the early ‘90s, but the functional role of Cl− remained obscure. Fundamental information on structural characterization of the PSII protein complex (19) and the OEC (6) was achieved with the beginning of the “solid structural era” of PSII (20). In spite of that, the characterization of Cl− binding sites in PSII by X-ray crystallography has remained elusive until very recently (2, 13, 21). The absence of crystallographic information on Cl− positions around the OEC, triggered a series of studies aimed to reveal the binding sites of chloride in the proximity of the OEC and its functional role in PSII (10, 11, 18, 22–29). Some of the proposed binding sites involved ligation of chloride to manganese (10, 11, 22–26). Other models suggested association with amino acid residues in the Mn coordination shell (18, 27), and its potential functional role in the regulation of the redox potentials of the MnCaMn3 cluster (28), participation in hydrogen bond networks (4, 29, 30), and activation of the substrate water (29). Recent X-ray crystallographic studies of bromide-substituted and iodine-substituted PSII revealed the presence of two binding sites for halide anions in the proximity of the OEC (13). This experimental evidence suggested that bromide (and by analogy chloride) can facilitate the access of substrate water to removal of protons from the OEC and supported the idea of a proton-relay network constituted by chloride and charged amino acid side chains (30). The position of chloride determined in the crystal structure of cyanobacterial photosystem II (21) was found to match one of the two binding sites determined for bromide. In the most recent X-ray structural structure of T. vulcanus PSII at 1.9 Å resolution (2) two chloride binding sites near the OEC were finally resolved.
In one binding site (BS1), Cl− is bound to the NH groups of D1-N338, D1-F339 and CP43-E354 backbones, distant 7.40 Å from the OEC (i.e. from Mn(1)). Here, Cl− is not directly interacting with a counterion and the closest protonable residue is D1-H337 (at 6.20 Å, see SI and Figure S2), indicating that Cl− located in this binding pocket should not affect the protonation state of amino acid residues close to the OEC. In the second binding pocket (BS2), Cl− is located in proximity to the positively charged D2-K317 side chain (at 6.67 Å from Mn(4), see Fig. 1) (2, 13, 14), interacting with the backbone of D1-E333 and the polar sidechain of D1-N181 (see Figure S2). The most recent X-ray structure revealed that the D1-D61 side chain occupies a critical position in between the MnCaMn3 cluster of OEC and this chloride-binding pocket. Therefore, we focus on the potential influence of chloride binding on the protonation state of D1-D61, which can be modified by hydrogen bonding and electrostatic interactions with nearby amino acid residues, including D2-K317 and D1-E333. These fundamental interactions might play important roles in water gating and proton transfer mechanisms since D1-D61 is the first residue leading from the OEC to the lumenal side of the membrane (see Figure 1b) (6, 31, 32). Morevoer, oxygen-release in D61N and D61A point-mutants from Synechocystis sp. PCC 6803 is slowed by a factor of 8–10 (compared to WT), with flash-spectrophotometric measurements indicating that residue D1-D61 interacts directly with the MnCaMn3 unit and selectively affects the redox potentials of the OEC cluster (33). Interestingly, the D61E mutation in the cyanobacterium does not change the oxygen-release of PSII with respect to WT, suggesting that charged (and protonable) residue at position 61 is crucial for the efficiency of photosynthetic function.
In this work, we analyze the effect of chloride binding on specific hydrogen bonding interactions by using Molecular Dynamics (MD) (34) and Monte Carlo (MC) within Multi-Conformer Continuum Electrostatics (MCCE) (35). MCCE binding constants show the strong affinity of Cl− (36) to both sites with BS2 estimated to bind ≈10 kcal/mol tighter than BS1. The pKa of D1-D61 is estimated to be lower than 4.9 when both Cl− are bound. In BS1, the interactions between Cl− and the backbones of the D1 C-terminal loop and CP43 polypeptide appear to stabilize the structural conformation by which D1-D342, D1-A344 and CP43-E354 are bound to the OEC, stabilizing the Mn cofactor in PSII. The higher binding affinity for Cl− in BS2 with respect to BS1 is due to its interaction with the positively charged D2-K317 side chain. Moreover, Cl− binding in BS2 could affect the protonation state of amino acid residues close to the OEC, such as D1-D61, playing a role in the proton transfer mechanism.
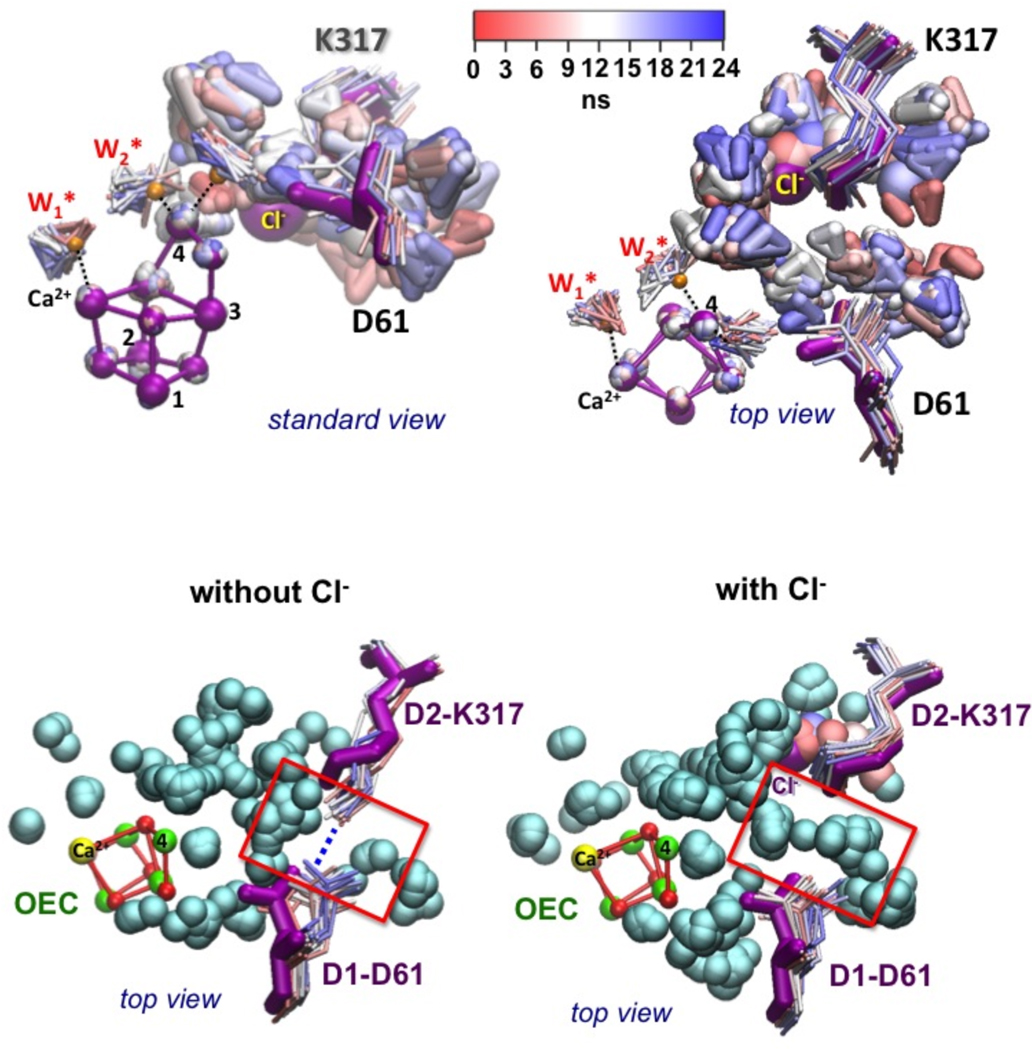
We find evidence of significant changes in the interactions of the side chain of D1-D61 induced by chloride depletion (Fig. 2). Specifically, MC and MD simulations predict that Cl− depletion induces formation of a salt-bridge between the negatively charged carboxylate group of D1-D61 and the positively charged side chain of D2-K317 (Figure 2, left) As a consequence, Cl− depletion would stabilize an inactive configuration of charged amino acid side chains in the hydrogen-bonding network close to the MnCaMn3 cluster, where D1-D61 is unable to extract protons from the OEC. We observed that the formation of the D1-D61/D2-K317 salt-bridge is independent of the protonation state of the µ-oxo linking Mn(3) and Mn(4). The absence of Cl− could, thus, affect the deprotonation of the different S-states in the Kok cycle.
Figure 2.
Superposition of instantaneous configurations along MD simulations, including H2O molecules (W1* and W2*), Cl−, residues D1-D61 and D2-K317, and bound waters (triangles colored from red–blue for 0–24 ns). Salt-bridge formed between K317 and D61 (left, without Cl−), and interrupted by water (right, with Cl−). X-ray structure is shown in magenta.
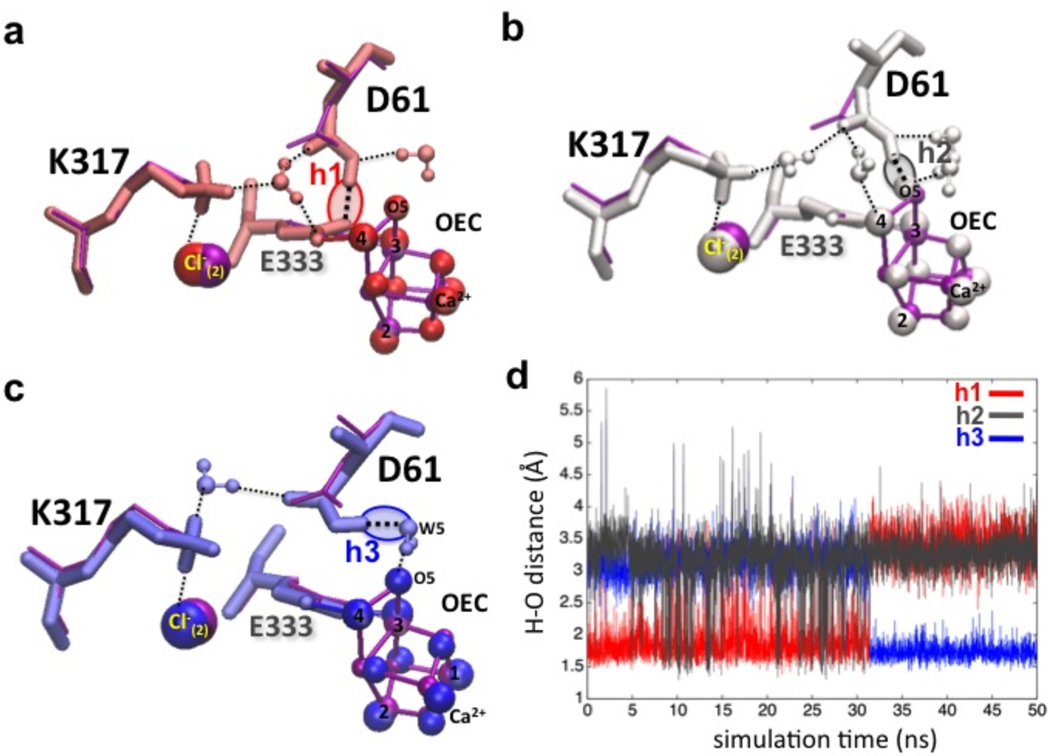
In contrast, in the presence of Cl−, the carboxylate group of the anionic D1-D61 moves and is often hydrogen bonded to a water ligand of the ‘dangling’ Mn as well as to a water in contact with the (D2-K317)-NH3+/Cl− ion-pair (Figure 2, right). MD simulations show thermal fluctuations in the position of Cl−, correlated to fluctuations in the configurations of surrounding water moieties and side-chains of D2-K317 and D1-D61 (see Figure 2). MC analysis also shows multiple positions of the D61 side chain (see Figure S3). However, partial solvation screens the electrostatic interactions between the negatively charged carboxylate group of D1-D61 and the proximal (D2-K317)- NH3+/Cl− ion pair, preventing D1-D61 from displacing Cl− and interacting directly with D2-K317. Both MCCE electrostatic analysis and MD simulations show that the protein could stabilize the protonated form of D1-D61. This is due to desolvation of D1-D61 by burial in the protein and its proximity to nearby anionic species, including the carboxylate group of D1-E333 and the µ-oxo bridge linking Mn(3) and Mn(4). MD simulations show vidence of specific H-bonding interactions for the neutral form of D1-D61 (Figure 3). Figure 3 (panel d) shows that thermal fluctuations allow the hydroxyl group of D1-D61 to switch hydrogen bonds between these polar species in the ns timescale. The relative stability of these three types of hydrogen bonds is also (14) manifested by their corresponding lifetimes. H-bonds with D1-E333 (h1), or with a water moiety H-bonded to the µ-oxo linking Mn(3) and Mn(4) (h3) are very stable, while the H-bond with the µ-oxo bridge (h2) is observed only transiently (see Figure 3). These relative life-times suggest that a protonated state of the µ-oxo bridge would be destabilized by thermal fluctuations of the carboxylate group of D1-D61, with proton transfer from the OEC to D1-D61 leading to formation of h1 or h3. Therefore, different conformations, protonation states and hydrogen bonding of D1-D61 might be observed during the S-state cycle, supporting the idea that conformational changes of D1-D61 coupled with Cl− might be an essential aspect of the OEC deprotonation mechanism (37). MCCE analysis shows that a hydrogen bond can be made between the anionic D1-D61 and terminal water bound to Mn(4) or between the protonated D1-D61 and a terminal hydroxide bound to Mn(4), supporting the notion that this could be a site for proton transfer. Interestingly, MD simulations of Cl− depleted PSII indicated that, in the presence of a neutral D1-D61, the positively charged D2-K317 is attracted to the negatively charged side chain, D1-D65, which is, however, too distant to form an alternative salt-bridge. This supports the idea that the main effect of Cl− depletion is to alter the functional role of D1-D61 as “molecular gate” for proton transfer from the OEC core to the lumen by allowing D1-D61 to form a stable (and catalytically inactive) salt-bridge with D2-K317. We propose that a primary role of Cl− as allosteric regulator of PSII is to stabilize a configuration of charged side chains close to the OEC that favors a flexible conformation of the basic center (D1-D61), assisting the proton abstraction mechanism from the OEC core. We considered the possibility of having the protonated form of D1-D61 hydrogen bonded to Cl−. However, MD simulations initialized by placing Cl− in between D2-K317 and D1-D61 show fast diffusion of Cl− back into the Cl− binding pocket found in the X-ray structure in < 3 ns, at 6.7 Å from Mn(4). Here the backbone of D1-E333, polar side chain of N181 and two water molecules provide more favorable H-bonding interactions (see Figure S1b).
Figure 3.
Panel a-c: Representative configurations of H-bonds between protonated D1-D61 and polar moieties at 1 (red, a), 22 (gray, b) and 48 ns (blue, c), including interactions with D1-E333 (h1), the µ-oxo bridge linking Mn(3) and Mn(4) (h2), and a water moiety H-bonded to the µ-oxo bridge (h3). X-ray structure is shown in magenta. Panel d: H-bond distances for h1-h3, along a 50 ns MD simulation.
In summary, MD and MC simulations of PSII both show evidence of specific electrostatic interactions between Cl− and essential amino acid residues in close contact with the OEC. We conclude that Cl− depletion induces formation of a salt-bridge between D2-K317 and D1-D61 that could hinder proton transfer to the lumen, as previously proposed (37). Further work is required in order to establish the detailed mechanism of proton transfer in PSII.
Supplementary Material
ACKNOWLEDGMENT
We acknowledge financial support from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy (DE-SC 0001423). V.S.B acknowledges supercomputer time from NERSC and support from the NIH Grant 1R01-GM-084267-01 for the developments of methods and models implemented in this study. G.W.B acknowledges support from the NIH grant GM32715.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Description of computational methods and structural models. This material is available free of charge via the Internet http://pubs.acs.org.
REFERENCES
- 1.Luber S, Rivalta I, Umena Y, Kawakami K, Shen J-R, Kamiya N, Brudvig GW, Batista VS. Biochemstry. 2011 doi: 10.1021/bi200685w. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Umena Y, Kawakami K, Shen J-R, Kamiya N. Nature. 2011;473:55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- 3.Blankenship RE. Molecular Mechanisms of Photosynthesis. Oxford, UK: Blackwell Science; 2002. [Google Scholar]
- 4.Sproviero EM, Gascon JA, McEvoy JP, Brudvig GW, Batista VS. J Chem Theory Comput. 2006;2:1119–1134. doi: 10.1021/ct060018l. [DOI] [PubMed] [Google Scholar]
- 5.Sproviero EM, Shinopoulos K, Gascon JA, McEvoy JP, Brudvig GW, Batista VS. Philos T R Soc B. 2008;363:1149–1156. doi: 10.1098/rstb.2007.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Science. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- 7.Kawakami K, Umena Y, Kamiya N, Shen J-R. J. Photochem. Photobiol. B. 2011;104:9–18. doi: 10.1016/j.jphotobiol.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 8.Demmig B, Winter K. Planta. 1983;159:66–76. doi: 10.1007/BF00998816. [DOI] [PubMed] [Google Scholar]
- 9.Critchley C. Biochim. Biophys. Acta. 1985;811:33–46. [Google Scholar]
- 10.Sandusky PO, Yocum CF. Biochim. Biophys. Acta. 1984;766:603–611. [Google Scholar]
- 11.Brudvig GW, Beck WF, de Paula JC. Annu Rev Biophys Bio. 1989;18:25–46. doi: 10.1146/annurev.bb.18.060189.000325. [DOI] [PubMed] [Google Scholar]
- 12.Popelkova H, Yocum CF. Photosynth. Res. 2007;93:111–121. doi: 10.1007/s11120-006-9121-5. [DOI] [PubMed] [Google Scholar]
- 13.Murray JW, Maghlaoui K, Kargul J, Ishida N, Lai TL, Rutherford AW, Sugiura M, Boussac A, Barber J. Energ Environ Sci. 2008;1:161–166. [Google Scholar]
- 14.Kawakami K, Umena Y, Kamiya N, Shen JR. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8567–8572. doi: 10.1073/pnas.0812797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kühne H, Szalai VA, Brudvig GW. Biochemistry. 1999;38:6604–6613. doi: 10.1021/bi990341t. [DOI] [PubMed] [Google Scholar]
- 16.Wincencjusz H, van Gorkom HJ, Yocum CF. Biochemistry. 1997;36:3663–3670. doi: 10.1021/bi9626719. [DOI] [PubMed] [Google Scholar]
- 17.Yachandra VK, DeRose VJ, Latimer MJ, Mukerji I, Sauer K, Klein MP. Science. 1993;260:675–679. doi: 10.1126/science.8480177. [DOI] [PubMed] [Google Scholar]
- 18.Haumann M, Barra M, Loja P, Loscher S, Krivanek R, Grundmeier A, Andreasson LE, Dau H. Biochemistry. 2006;45:13101–13107. doi: 10.1021/bi061308r. [DOI] [PubMed] [Google Scholar]
- 19.Zouni A, Witt HT, Kern J, Fromme P, Krauss N, Saenger W, Orth P. Nature. 2001;409:739–743. doi: 10.1038/35055589. [DOI] [PubMed] [Google Scholar]
- 20.Rhee KH. Annu. Rev. Biophys. Biomol. Struct. 2001;30:307–328. doi: 10.1146/annurev.biophys.30.1.307. [DOI] [PubMed] [Google Scholar]
- 21.Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W. Nat. Struct. Mol. Biol. 2009;16:334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- 22.Hureau C, Blondin G, Charlot MF, Philouze C, Nierlich M, Cesario M, Anxolabehere-Mallart E. Inorg. Chem. 2005;44:3669–3683. doi: 10.1021/ic050243y. [DOI] [PubMed] [Google Scholar]
- 23.Yachandra VK, Sauer K, Klein MP. Chem. Rev. 1996;96:2927–2950. doi: 10.1021/cr950052k. [DOI] [PubMed] [Google Scholar]
- 24.Pecoraro VL, Baldwin MJ, Caudle MT, Hsieh WY, Law NA. Pure Appl. Chem. 1998;70:925–929. [Google Scholar]
- 25.Vrettos JS, Limburg J, Brudvig GW. Bba-Bioenergetics. 2001;1503:229–245. doi: 10.1016/s0005-2728(00)00214-0. [DOI] [PubMed] [Google Scholar]
- 26.Britt RD, Campbell KA, Peloquin JM, Gilchrist ML, Aznar CP, Dicus MM, Robblee J, Messinger J. Bba-Bioenergetics. 2004;1655:158–171. doi: 10.1016/j.bbabio.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Hasegawa K, Kimura Y, Ono TA. Biochemistry. 2002;41:13839–13850. doi: 10.1021/bi026595n. [DOI] [PubMed] [Google Scholar]
- 28.Boussac A, Rutherford AW. J. Biol. Chem. 1994;269:12462–12467. [PubMed] [Google Scholar]
- 29.McEvoy JP, Brudvig GW. Phys. Chem. Chem. Phys. 2004;6:4754–4763. [Google Scholar]
- 30.Olesen K, Andreasson LE. Biochemistry. 2003;42:2025–2035. doi: 10.1021/bi026175y. [DOI] [PubMed] [Google Scholar]
- 31.Ishikita H, Saenger W, Loll B, Biesiadka J, Knapp EW. Biochemistry. 2006;45:2063–2071. doi: 10.1021/bi051615h. [DOI] [PubMed] [Google Scholar]
- 32.Sproviero EM, Gascon JA, McEvoy JP, Brudvig GW, Batista VS. Coord. Chem. Rev. 2008;252:395–415. doi: 10.1016/j.ccr.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hundelt M, Hays AMA, Debus RJ, Junge W. Biochemistry. 1998;37:14450–14456. doi: 10.1021/bi981164j. [DOI] [PubMed] [Google Scholar]
- 34.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song YF, Mao JJ, Gunner MR. J. Comput. Chem. 2009;30:2231–2247. doi: 10.1002/jcc.21222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song YF, Gunner MR. J. Mol. Bio. 2009;387:840–856. doi: 10.1016/j.jmb.2009.01.038. [DOI] [PubMed] [Google Scholar]
- 37.Pokhrel R, McConnel I, Brudvig GW. Biochemistry. 2011;50:2725–2734. doi: 10.1021/bi2000388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.