Abstract
Under the impact of human activity, global extinction rates have risen a thousand times higher than shown in the fossil record. The resources available for conservation are insufficient to prevent the loss of much of the world's threatened biodiversity during this crisis. Conservation planners have been forced to prioritize their protective activities, in the context of great uncertainty. This has become known as ‘the agony of choice’. A range of methods have been proposed for prioritizing species for conservation attention; one of the most strongly supported is prioritizing those species that maximize phylogenetic distinctiveness (PD). We evaluate how a composite measure of extinction risk and phylogenetic isolation (EDGE) has been used to prioritize species according to their degree of unique evolutionary history (evolutionary distinctiveness, ED) weighted by conservation urgency (global endangerment, GE). We review PD-based approaches and provide an updated list of EDGE mammals using the 2010 IUCN Red List. We evaluate how robust this method is to changes in phylogenetic uncertainty, knowledge of taxonomy and extinction risk, and examine how mammalian species that rank highly in EDGE score are representative of the collective from which they are drawn.
Keywords: charismatic species, comparative methods, conservation prioritization, decline, phylogenetically distinct, phylogeny
1. Introduction
In the current era of unprecedented global change, where the rate of biodiversity loss continues unabated [1,2], decision-making about the focus of conservation investment has become a central part of both academic research and conservation action. It has been strongly argued that maximizing phylogenetic diversity should be one of the main goals of priority-setting for conservation [3–6]. This is owing to the fact that species represent different amounts of evolutionary history, reflecting different rates of divergence across any given phylogenetic tree. As such, limited conservation resources should be focused on those species that represent the greatest amounts of unique evolutionary history, whose loss would be felt most keenly.
There are two main arguments for choosing prioritization techniques that aim to conserve the maximum possible amount of evolutionary history. The first is a pragmatic perspective: phylogenetic distinctiveness (PD) is a compound metric of all forms of genotypic, phenotypic (‘feature’ or ‘character’ diversity) and functional diversity, both measurable and unmeasurable [7], so maximizing PD thereby provides biological systems with the most options to respond to a changing world, both at species level and community level. Moreover, PD could be used as a measure of ecosystem function, as phylogenies may reflect integrated phenotypic differences among taxa and so be a more encapsulating measure than sets of singular, discretely measured traits [8,9]. Prioritizing conservation by evolutionary history has been demonstrated to be an effective approach for capturing the range of morphological and ecological diversity that has evolved in a given phylogenetic group, reflecting the positive correlation between amount of evolutionary change and amount of time elapsed [10,11]. The second is from more of an ethical perspective, whereby maximizing the conservation of PD best preserves the immense history of the Earth [12].
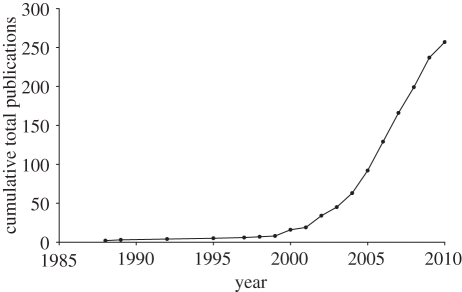
Evolution, in terms of both evolutionary history and evolutionary potential, is increasingly being recognized as the ‘missing component’ for conservation prioritization and planning (e.g. [13,14]). Now, more than at any other time, tools to both measure and incorporate evolutionary history into conservation priority-setting and planning are available. The expanding number of phylogenetic trees plotting the relationships among species (figure 1; [15]) and ever-increasing amount of information for conservation decision-making (e.g. species conservation status; [16]) are creating a wealth of knowledge that can be applied to address the current biodiversity crisis.
Figure 1.
Cumulative total number of publications containing the search term ‘supertree*’ from the ISI Web of Knowledge database.
Prioritizing conservation efforts on the basis of evolutionary history are of further importance because phylogenetic comparisons have revealed that the current human-caused loss of species is taxonomically selective rather than random, that extinction risk is clustered, and that mammals, birds, plants and other taxa with few close relatives are particularly likely to be at risk [17–19]. Evolutionarily distinctive species are known to have already experienced greater levels of extinction both during the recent historical era [20] and during recent millennia [21], leading to increasing imbalance in the mammalian phylogeny over the course of the Holocene Epoch. This pattern is associated in mammals and birds with the elevated level of extinctions of island species, including many ancient species-poor mammal lineages (e.g. Bibymalagasia, Solenodontidae, Thylacinidae; [22]).
The correlation between PD and species richness can be quite close, although there is substantial variation in this relationship [23,24]. However, PD is not often effectively captured by straightforward taxon-based conservation policies [25], because variance among grid cells in species richness is far greater than the variance in species' PD. Gains in taxon richness and PD can also be decoupled, particularly when the underlying phylogeny is unbalanced and species are not randomly distributed on the landscape [26]. Therefore, in general terms, conservation approaches that maximize species richness, such as endemic species hotspots [27], may not always protect PD, particularly at sub-global scales. This has led to the development of conservation programmes that aim to objectively assign PD values to species so that decisions can be made as to the most urgent focus of conservation action. In this study, we briefly review PD-based approaches to conservation priority-setting before examining how one measure, EDGE [28], has been implemented in mammals. We evaluate how robust this method is to changes in knowledge of taxonomy and extinction risk, examine how mammalian species that rank highly in EDGE score are representative of the collective from which they are drawn, assess the barriers to using PD, and report a set of conservation recommendations for new taxa.
(a). Review of phylogenetic diversity-based approaches for species conservation
The concept of using evolutionary history in conservation prioritization has been around for at least two decades [4,5]. These first approaches developed metrics concerning taxonomic distinctiveness (TD; relative to other species; [5]) and PD (sum of phylogenetic branch length of species in a given region; [4]). Many derivations have subsequently been developed from these approaches (see [12]), but these essentially all fall under one of these two categories.
Evolutionary history consists of two distinct components, the branching pattern of a phylogenetic tree and the length of its branches, and early attempts to integrate PD into conservation priority-setting were typically restricted to using information on branching pattern alone (i.e. they represented scores of TD; see [29]). However, the increasing availability of temporally calibrated branch lengths for phylogenies of large taxonomic groups has made it possible to calculate PD using both components [15]. This in turn has led to the development of a family of related measures of PD-based priority-setting approaches, which differ in their use of scoring methods for distributing among species the shared component of evolutionary history represented by deep phylogenetic branches, and in different methods for calculating and incorporating the extinction risk of different species across the phylogeny.
In addition to the original concept of PD [4], the two most widely followed scoring methods in the recent literature are equal splits (ES; [30,31]) and evolutionary distinctiveness (ED; [28]), also known as fair proportion (and which is very close to another, evolutionarily stable strategy game inspired measure, the Shapley index; [32], see [33] for wider review). ES hierarchically partitions branch lengths by the number of descendent edges, such that for a given branch, a descendent species receives credit equal to 0.5 to a power equal to the number of splits between the branch and the species. ED instead partitions branches by the total number of species descending from them, regardless of nested tree structure, such that the contribution of a given ancestral branch to the ED score is 1/number of descendants of that branch. Further modifications to the ED approach have been proposed, allowing it to also include abundance information to generate a metric of abundance-weighted ED that can be used to prioritize populations, species, habitats and biogeographical regions [12]. These alternative methods for scoring evolutionary history are then combined with a measure of threat to provide prioritization indices to inform conservation, as first carried out by Weitzman [34] and Avise [35]. The EDGE approach combines ED scores directly with a ranked measure of extinction risk (global endangerment; GE) based on the quantitative and objective framework provided by the International Union for Conservation of Nature (IUCN) Red List, to generate EDGE scores [28]. ES scores were similarly combined with a probability of extinction score (Pe) to generate a species-specific expected loss of evolutionary history (EL) metric [31]. EDGE is, in effect, a special case of EL in which each increase in the Red List category represents a doubling of extinction risk, an arbitrary approach that avoids the resultant list being dominated by species of only the highest threat category.
EDGE and EL scores assigned to species are independent of the conservation status of other taxa. However, other approaches for combining extinction risk also take the conservation status of related species into account. This is based on the consideration that some future ‘sets’ of species are more likely to persist than others as a result of interspecific variation in extinction risk; at-risk species with close relatives that are also threatened with extinction should represent higher conservation priorities, because such species are predicted to represent a higher amount of unique evolutionary history in the future [36]. The corollary of this is that systems like EDGE might overestimate the importance of species with safe relatives. For example, extinction of a 1 Myr-old species lineage would result in the loss of one million years of evolution, but the future extinction of its currently threatened sister species results in the loss of another one million years of evolution as well as the deeper branch connecting the now-extinct species pair to the rest of the phylogeny. The probability of losing an internal branch in a phylogeny is, therefore, related to the number of descendent species and is the product of their probabilities of extinction, which is not accounted for by the EDGE or EL approaches.
In order to account for this issue, the basic PD approach [4] was modified by Witting & Loeschcke [6], Witting et al. [37] and Faith [36] to provide a measure of the expected or probabilistic PD for a given species that will result from different extinction scenarios affecting other species of varying relatedness. A similar method has also been developed by Steel et al. [38], where a heightened ED score (HED) is used to generate a HEDGE score. There has been criticism of EDGE and EL approaches owing to their dependency on a static apportioning of credit for branches and their failure to incorporate extinction probabilities of related species [36]. In particular, Faith argued that PD-based conservation initiatives should instead adopt probabilistic PD to properly take complementarity into account [36]. However, in reality this modification is unlikely to make much of a difference in conservation prioritization, because it has been demonstrated that most species derive the majority of their ED from terminal branches [28], and comparisons of HED and ED scores show very strong correlations (e.g. 0.94 for prosimians; [38]).
A second debate has also addressed appropriate methods for quantifying conservation status. IUCN Red List categories are ranks representing probabilities of extinction [39,40]. However, extinction risk ranks need to be assigned numerical values (an ‘urgency score’) when they are combined with other criteria, such as when integrated with phylogenetic trees to develop EDGE-style priority rankings. This raises the question, does movement between ranks represent a constant change in probability or is it nonlinear [41]? The EDGE approach treats IUCN Red List categories as equivalent intervals of risk [28,41,42]; however, alternative approaches can also be adopted, for example, by using empirical estimates from population viability analyses [43] for data-rich taxa. The greatest variation between PD-based priority rankings is caused by assuming latent risk (the ‘pessimistic’ approach of Mooers et al. [43]), which gives higher weight to PD because all taxa are considered to be at some risk of extinction, and includes species that are less threatened (see [44,45]). Some authors have also included Data Deficient (DD) species in PD-based prioritization approaches, for example, by arbitrarily (though probably conservatively) estimating their extinction risk as being between the Least Concern and Near Threatened categories [45]. While this may be a legitimate assumption with birds [46], evidence suggests that the probable status of DD species in many taxonomic groups might be more likely to be threatened [47]; at the very least, some unknown proportion of DD species are threatened, so treating DD species as a single value is not informative.
2. Methods
(a). EDGE scores
We collated mammal conservation status data from the IUCN Red List [48,49], and included genuine change in status from Hoffmann et al. [50]. We used a composite ‘supertree’ phylogeny [51–53] to calculate ED scores for mammals, following the procedure reported in Isaac et al. [28]. Briefly, we divided the total phylogenetic diversity of each clade among its members by applying a value to each branch equal to its length divided by the number of descendent species. The ED of a species is simply the sum of these values for all branches from which the species is descended, to the root of the phylogeny. The new mammal EDGE list presented here is constructed using an updated mammal taxonomy and the most recent Red List assessments, but also differs in several other ways, detailed below.
The new list uses the third edition of Mammal Species of the World (MSW3; [54]), whereas the original list of Isaac et al. [28] used the second edition taxonomy (MSW2; [55]). A phylogenetic tree in the MSW3 taxonomy was provided by Fritz et al. [53], who converted it from the MSW2 format tree of Bininda-Emonds et al. [51]. MSW3 contains 5416 species, when compared with 4629 species in MSW2. Only 291 of the additional species have been newly described since MSW2, so taxonomic changes (splitting and lumping) have accounted for a net gain of nearly 500 species (i.e. more than 10% growth). Such instability in taxonomic status presents wide-scale technical and philosophical challenges for research applications that use species lists, especially in evolutionary and conservation biology [56–58]. However, phylogenetic metrics such as EDGE are somewhat less sensitive to taxonomic change than alternative biodiversity measures such as counts of endemic or threatened species [28].
Species values of ED were calculated as the geometric mean of scores under the three sets of branch lengths. The algorithm for calculating ED scores [28] was applied with a modification to the way in which scores were corrected for polytomies (nodes with more than two descendants) and uncertainty in the estimated divergence times. Polytomies in supertrees result from poor or conflicting data rather than a true representation of the speciation process, so the distinctiveness of branches subtending them is overestimated, thus leading to biased ED scores. Isaac et al. [28] used a statistical fit to simulated data in order to correct the ED scores of nodes descended from polytomies. Their correction factor decreased to zero for nodes with large numbers (more than 20) of descendants, which leads to an underestimate of the ED score of many species in poorly resolved areas of the phylogeny (in this study, mainly bats and rodents). To deal with uncertainty in the branch length estimates, Isaac et al. [28] reported the geometric mean ED scores based on three sets of node ages (best, upper and lower) from Bininda-Emonds et al. [51].
For the new list, we calculated ED scores for each of 1000 supertrees, each of which was resolved using Bayesian methods described in Kuhn et al. [59]. These fully resolved supertrees represent the pseudo-posterior distribution of the underlying mammalian phylogeny. We modified the PolytomyResolver R script [59] in order to incorporate uncertainty in the estimates of individual node ages by placing a normally distributed prior constraint onto each resolved node of the starting tree [53]. These priors each had a mean equal to the best age estimate reported in Fritz et al. [53], and a standard deviation of (best–worst estimate)/1.96, where the worst estimate is defined as the estimate (upper or lower) that was furthest from the best. We created 1000 resolved trees using BEAST (v. 1.6.1) [60] to analyse five independent runs of approximately 2 000 000 iterations and a sampling interval of 1000. We assessed the burnin, convergence and mixing manually for each run using Tracer v. 1.5 [61] and produced the final distribution by combining all independent runs and subsampling to every 9000 iterations.
The MSW3 format phylogeny [53] contains 5020 species, i.e. 396 valid names were missing. Of these, 75 are known to be extinct [22]. We estimated ED scores for 250 of the extant missing species as the mean ED of congeneric species, such that only 71 extant species still lacked ED scores. We also estimated ED for two recently described species, Laonastes aenigmamus and Pseudoryx nghetinhensis, which were likely to represent EDGE priorities on the basis of their high taxonomic distinctiveness. ‘Surrogate’ ED scores for these species were crudely estimated as the likely time of divergence based on available molecular data (see electronic supplementary material, table S1, references S2 and S4). Finally, IUCN categories were matched for 5123 species, of which 692 are DD, producing a list of 4431 EDGE scores.
Changes in EDGE score between [28] and the results reported in this study are due to a number of reasons that are not mutually exclusive. EDGE score may change owing to reassessment of the conservation status of the species (i.e. updated Red List status, which may or may not be owing to a genuine change in species status; [42]), or a change in taxonomic status between MSW2 and MSW3. The latter is further complicated by new species discoveries and by the splitting and lumping of existing species, resulting in a changed phylogeny for both a given species and any sister taxa it may have. We tracked changes in taxonomy and Red List status between the old and new EDGE lists, recording changes in taxonomic status as new species described, species split, species lumped, or non-nested [56], in which there is no simple relationship between the species taxonomy in MSW2 and MSW3.
(b). Trait analysis
We followed the method of Redding et al. [10] to evaluate how mammal species that rank highly in EDGE score are representative of the collective from which they are drawn. We used six mammalian trait measures drawn from Jones et al. [62] of reproductive, behavioural, geographical and morphological species traits: body mass (grams), gestation length (days), home range size (square kilometres), litter size, geographical range size (square kilometres) and latitudinal midpoint of range (decimal degrees). Each trait was log10 transformed to lessen the effect of outliers and equalize variance. For each species value, we calculated absolute mean distance from the median value of the trait for the order; the greater the distance from the median value, the more unusual that species is in a given trait for its order.
Following Redding et al. [10], we used Pearson correlations to test for a relationship between EDGE score (and its components, ED and GE) and absolute distance from the median value for each trait. Owing to the repeated tests, we used a correction factor to account for false discoveries and the possibility of elevated type I errors. This procedure accounts for the number of false-positive hypotheses that would be accepted with raw p-values, given a predefined significance value of α = 0.05 [63,64]. All analyses were conducted in R v. 2.12.1 [65].
3. Results
Our new analysis of EDGE scores has generated a new priority list of mammals requiring urgent conservation attention on the basis of a combination of high ED and high threat status (table 1). Our data show that there has been some change in the ranks of species between EDGE lists, but that the overall priority set appears robust to these changes. These rank changes can be attributed to both changes in taxonomy (table 2 and figure 2) and changes in Red List status (figure 3). The taxonomy of the majority of species remains unchanged (approx. 70%; table 2). Of the changes to species taxonomic status, approximately 20 per cent have been split, 5 per cent are new species descriptions, and about 2.5 per cent have been either lumped or represent non-nested taxonomic changes. The relatively minor impact of these taxonomic changes on ED score is apparent from figure 2, which shows a strong correlation between ED scores derived from MSW2 [55] and those derived from MSW3 [54].
Table 1.
Top 100 mammal EDGE scores, representing the highest priority mammal species requiring urgent conservation attention on the basis of a combination of high ED and high threat status. Conservation attention was assessed following the methods used by Sitas et al. [66]. CR, Critically Endangered; EN, Endangered VU, Vulnerable.
| rank | species | order | family | status | ED | EDGE | conservation attention |
|---|---|---|---|---|---|---|---|
| 1= | Zaglossus attenboroughi | Monotremata | Tachyglossidae | CR | 55.21737845 | 6.801814656 | none |
| 1= | Zaglossus bartoni | Monotremata | Tachyglossidae | CR | 55.21737845 | 6.801814656 | limited |
| 1= | Zaglossus bruijnii | Monotremata | Tachyglossidae | CR | 55.21737845 | 6.801814656 | none |
| 4 | Mystacina robusta | Chiroptera | Mystacinidae | CR | 54.10322232 | 6.781796918 | none |
| 5 | Lipotes vexillifer | Cetacea | Lipotidae | CR | 38.67180179 | 6.453229375 | none |
| 6 | Burramys parvus | Diprotodontia | Burramyidae | CR | 32.75582928 | 6.291741844 | active |
| 7= | Solenodon cubanus | Soricomorpha | Solenodontidae | EN | 61.69215212 | 6.217677816 | none |
| 7= | Solenodon paradoxus | Soricomorpha | Solenodontidae | EN | 61.69215212 | 6.217677816 | limited |
| 9 | Dicerorhinus sumatrensis | Perissodactyla | Rhinocerotidae | CR | 29.44751148 | 6.188592988 | active |
| 10 | Bunolagus monticularis | Lagomorpha | Leporidae | CR | 27.88392179 | 6.135873823 | active |
| 11 | Diceros bicornis | Perissodactyla | Rhinocerotidae | CR | 26.63195412 | 6.091561583 | active |
| 12 | Lasiorhinus krefftii | Diprotodontia | Vombatidae | CR | 25.98457399 | 6.067854091 | active |
| 13 | Camelus ferus | Artiodactyla | Camelidae | CR | 25.29566761 | 6.041992918 | limited |
| 14 | Rhinoceros sondaicus | Perissodactyla | Rhinocerotidae | CR | 24.64177112 | 6.016811428 | active |
| 15 | Laonastes aenigmamus | Rodentia | Diatomyidae | EN | 44.3 | 5.892748574 | none |
| 16 | Bradypus pygmaeus | Pilosa | Bradypodidae | CR | 20.88097152 | 5.858206101 | none |
| 17 | Elephas maximus | Proboscidea | Elephantidae | EN | 39.76418423 | 5.7872454 | active |
| 18 | Octodon pacificus | Rodentia | Octodontidae | CR | 18.43970169 | 5.739906176 | none |
| 19 | Ailuropoda melanoleuca | Carnivora | Ursidae | EN | 36.77014331 | 5.710960473 | active |
| 20 | Tapirus indicus | Perissodactyla | Tapiridae | EN | 36.03587836 | 5.69132867 | active |
| 21 | Abrocoma boliviensis | Rodentia | Abrocomidae | CR | 17.4621309 | 5.688310378 | none |
| 22= | Monachus monachus | Carnivora | Phocidae | CR | 16.79398976 | 5.651449469 | active |
| 22= | Monachus schauinslandi | Carnivora | Phocidae | CR | 16.79398976 | 5.651449469 | active |
| 24 | Ailurops melanotis | Diprotodontia | Phalangeridae | CR | 16.65441885 | 5.643574834 | none |
| 25 | Natalus jamaicensis | Chiroptera | Natalidae | CR | 16.59446732 | 5.640173218 | none |
| 26 | Coleura seychellensis | Chiroptera | Emballonuridae | CR | 16.5694351 | 5.638749473 | limited |
| 27 | Natalus primus | Chiroptera | Natalidae | CR | 16.40073806 | 5.629101345 | none |
| 28 | Choeropsis liberiensis | Artiodactyla | Hippopotamidae | EN | 33.17906211 | 5.611054779 | limited |
| 29 | Indri indri | Primates | Indridae | EN | 33.00886339 | 5.60606272 | active |
| 30 | Galagoides rondoensis | Primates | Galagidae | CR | 15.61252133 | 5.58274543 | none |
| 31 | Myrmecobius fasciatus | Dasyuromorphia | Myrmecobiidae | EN | 32.0385503 | 5.577116612 | active |
| 32 | Pharotis imogene | Chiroptera | Vespertilionidae | CR | 15.302246 | 5.563891612 | none |
| 33 | Aproteles bulmerae | Chiroptera | Pteropodidae | CR | 15.29611383 | 5.563515386 | none |
| 34 | Phalanger matanim | Diprotodontia | Phalangeridae | CR | 15.26573074 | 5.561649208 | none |
| 35 | Potorous gilbertii | Diprotodontia | Potoroidae | CR | 15.14476359 | 5.554184483 | active |
| 36 | Marmosops handleyi | Didelphimorphia | Didelphidae | CR | 14.89316215 | 5.538477686 | none |
| 37 | Varecia variegata | Primates | Lemuridae | CR | 14.71875348 | 5.527443211 | active |
| 38 | Amorphochilus schnablii | Chiroptera | Furipteridae | EN | 30.2569337 | 5.521682772 | none |
| 39 | Tapirus bairdii | Perissodactyla | Tapiridae | EN | 30.00565773 | 5.513611237 | active |
| 40 | Romerolagus diazi | Lagomorpha | Leporidae | EN | 29.85224334 | 5.508651007 | none |
| 41 | Prolemur simus | Primates | Lemuridae | CR | 14.3982973 | 5.506845661 | active |
| 42 | Pentalagus furnessi | Lagomorpha | Leporidae | EN | 29.4589476 | 5.495821338 | limited |
| 43 | Beatragus hunteri | Artiodactyla | Bovidae | CR | 14.12584734 | 5.488993747 | limited |
| 44 | Pseudoryx nghetinhensis | Artiodactyla | Bovidae | CR | 13.68 | 5.459074745 | limited |
| 45 | Pongo abelii | Primates | Hominidae | CR | 13.66284712 | 5.45790561 | active |
| 46 | Rhynchocyon chrysopygus | Macroscelidea | Macroscelididae | EN | 28.12701704 | 5.451107706 | limited |
| 47 | Hapalemur alaotrensis | Primates | Lemuridae | CR | 13.49001975 | 5.446048842 | active |
| 48 | Tokudaia muenninki | Rodentia | Muridae | CR | 13.48531689 | 5.44572423 | none |
| 49 | Gymnobelideus leadbeateri | Diprotodontia | Petauridae | EN | 27.62231266 | 5.433628118 | active |
| 50 | Dugong dugon | Sirenia | Dugongidae | VU | 56.07711486 | 5.430697607 | active |
| 51 | Neohylomys hainanensis | Erinaceomorpha | Erinaceidae | EN | 27.36334182 | 5.424539072 | none |
| 52 | Podogymnura aureospinula | Erinaceomorpha | Erinaceidae | EN | 27.22727204 | 5.419730146 | none |
| 53= | Chinchilla chinchilla | Rodentia | Chinchillidae | CR | 12.97919099 | 5.410158588 | none |
| 53= | Chinchilla lanigera | Rodentia | Chinchillidae | CR | 12.97919099 | 5.410158588 | none |
| 55 | Spilocuscus rufoniger | Diprotodontia | Phalangeridae | CR | 12.78305286 | 5.396028506 | none |
| 56 | Mystacina tuberculata | Chiroptera | Mystacinidae | VU | 54.10322232 | 5.395502557 | active |
| 57 | Sminthopsis aitkeni | Dasyuromorphia | Dasyuridae | CR | 12.69165084 | 5.389374942 | active |
| 58 | Lepilemur septentrionalis | Primates | Lepilemuridae | CR | 12.65667849 | 5.386817391 | limited |
| 59 | Micropotamogale lamottei | Afrosoricida | Tenrecidae | EN | 26.20431682 | 5.382817209 | none |
| 60 | Platanista gangetica | Cetacea | Platanistidae | EN | 26.19122582 | 5.382335883 | limited |
| 61 | Bradypus torquatus | Pilosa | Bradypodidae | EN | 25.33866497 | 5.350479552 | limited |
| 62 | Hipposideros lamottei | Chiroptera | Hipposideridae | CR | 11.8752596 | 5.327896332 | none |
| 63 | Phocoena sinus | Cetacea | Phocoenidae | CR | 11.82671575 | 5.324118886 | limited |
| 64 | Oreonax flavicauda | Primates | Atelidae | CR | 11.61379513 | 5.307379789 | limited |
| 65 | Propithecus perrieri | Primates | Indridae | CR | 11.59144115 | 5.305606032 | limited |
| 66 | Loris tardigradus | Primates | Lorisidae | EN | 23.67408268 | 5.28519495 | limited |
| 67 | Cavia intermedia | Rodentia | Caviidae | CR | 11.25459124 | 5.278489384 | none |
| 68 | Gorilla gorilla | Primates | Hominidae | CR | 11.21914344 | 5.275592579 | active |
| 69 | Trichechus inunguis | Sirenia | Trichechidae | VU | 47.24796806 | 5.262648075 | limited |
| 70 | Nilopegamys plumbeus | Rodentia | Muridae | CR | 11.02530903 | 5.259602237 | none |
| 71 | Catagonus wagneri | Artiodactyla | Tayassuidae | EN | 22.66555597 | 5.243462198 | active |
| 72 | Neamblysomus gunningi | Afrosoricida | Chrysochloridae | EN | 22.35386779 | 5.230204156 | none |
| 73 | Balaenoptera physalus | Cetacea | Balaenopteridae | EN | 22.24687411 | 5.225612218 | active |
| 74 | Tapirus pinchaque | Perissodactyla | Tapiridae | EN | 22.14517275 | 5.22122778 | limited |
| 75 | Balaenoptera musculus | Cetacea | Balaenopteridae | EN | 22.06062052 | 5.217567965 | active |
| 76 | Dendromus kahuziensis | Rodentia | Nesomyidae | CR | 10.47272767 | 5.212561434 | none |
| 77 | Chrysospalax trevelyani | Afrosoricida | Chrysochloridae | EN | 21.90862527 | 5.210955031 | none |
| 78 | Leporillus apicalis | Rodentia | Muridae | CR | 10.02143241 | 5.1724305 | none |
| 79 | Hypogeomys antimena | Rodentia | Nesomyidae | EN | 20.86613092 | 5.164380448 | active |
| 80 | Tylomys bullaris | Rodentia | Cricetidae | CR | 9.894517263 | 5.160848382 | none |
| 81 | Callicebus barbarabrownae | Primates | Pitheciidae | CR | 9.869378529 | 5.158538249 | none |
| 82= | Sorex sclateri | Soricomorpha | Soricidae | CR | 9.853968549 | 5.1571195 | none |
| 82= | Sorex stizodon | Soricomorpha | Soricidae | CR | 9.853968549 | 5.1571195 | none |
| 84 | Tylomys tumbalensis | Rodentia | Cricetidae | CR | 9.789790788 | 5.151189112 | none |
| 85 | Bettongia penicillata | Diprotodontia | Potoroidae | CR | 9.741552342 | 5.146708339 | active |
| 86 | Cryptotis nelsoni | Soricomorpha | Soricidae | CR | 9.73005188 | 5.145637114 | none |
| 87 | Mesocapromys sanfelipensis | Rodentia | Capromyidae | CR | 9.715985838 | 5.144325352 | none |
| 88 | Mesocapromys nanus | Rodentia | Capromyidae | CR | 9.707420414 | 5.14352572 | none |
| 89 | Manis pentadactyla | Pholidota | Manidae | EN | 20.35572093 | 5.140761205 | limited |
| 90 | Manis javanica | Pholidota | Manidae | EN | 20.30695965 | 5.138475305 | limited |
| 91 | Brachyteles hypoxanthus | Primates | Atelidae | CR | 9.65114119 | 5.138255763 | limited |
| 92= | Trichechus manatus | Sirenia | Trichechidae | VU | 41.57146606 | 5.137478579 | active |
| 92= | Trichechus senegalensis | Sirenia | Trichechidae | VU | 41.57146606 | 5.137478579 | active |
| 94 | Potorous longipes | Diprotodontia | Potoroidae | EN | 20.22170929 | 5.134466223 | active |
| 95 | Cremnomys elvira | Rodentia | Muridae | CR | 9.532109427 | 5.127017354 | none |
| 96 | Millardia kondana | Rodentia | Muridae | CR | 9.299543578 | 5.104688304 | none |
| 97 | Crateromys australis | Rodentia | Muridae | CR | 9.257893072 | 5.100636187 | none |
| 98 | Viverra civettina | Carnivora | Viverridae | CR | 9.189373539 | 5.09393409 | none |
| 99 | Habromys chinanteco | Rodentia | Cricetidae | CR | 9.169551079 | 5.09198679 | none |
| 100 | Amblysomus marleyi | Afrosoricida | Chrysochloridae | EN | 19.23849603 | 5.087028077 | none |
Table 2.
Change in taxonomic status of mammal species 1993–2005. Numbers refer to species in the new (third) edition of Mammal Species of the World [55] and expressed relative to species status in the second edition [54].
| taxonomic status | no. of species | proportion of species |
|---|---|---|
| new species described | 291 | 0.054 |
| split | 1099 | 0.203 |
| lumped | 142 | 0.026 |
| non-nested | 128 | 0.024 |
| unchanged | 3756 | 0.693 |
| total | 5416 |
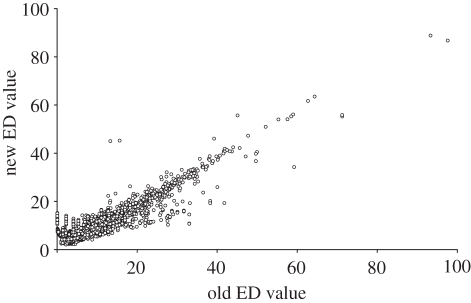
Figure 2.
Comparison of ED values between EDGE lists. Old ED status is the value for each species as reported in Isaac et al. [28]; new ED status is the value for each species calculated in this study.
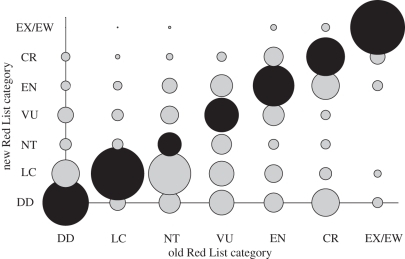
Figure 3.
Comparison of Red List status between EDGE lists. Old Red List status is the categories of species reported in [28]; new Red List status is the category of species calculated in this study, following Schipper et al. [48]. Black shading indicates where no category change has taken place. Bubble size is scaled to the number of cases of a given category, as a proportion of the species that used to be in that category in the previous version of the Red List. n = 4708 mammal species for which direct comparison could be made. DD, Data Deficient, LC, Least Concern, NT, Near Threatened, VU, Vulnerable, EN, Endangered, CR, Critically Endangered, EX/EW, Extinct/Extinct in the Wild.
Although there is an overall strong correlation between the ED scores reported in this study and previous estimates (figure 2), several anomalies do exist. For example, Ochotona nubrica has a large increase in ED score between the MSW2 (and 3) and the current estimate, resulting in an EDGE rank of 739 (a climb of 106 in the ranking). This difference in ED scores results from an error in the node age estimates of the original supertree, where the upper and lower age estimates appear to be reversed. This type of node age estimate issue is only relevant to eight of the 2503 nodes of the supertree, and does not dramatically alter ED scores for species other than O. nubrica.
Variation in Red List ranking has a much greater impact on the composite measure making up the EDGE score (figures 3 and 4). This variation reveals two clear patterns. Firstly, there is considerable movement of species between threatened categories (Critically Endangered, Endangered, Vulnerable) and non-threatened categories (Near Threatened, Least Concern), which probably reflects the gathering of new data and/or reassessments of the quality of old data on species threat status. Secondly, movement within the threatened categories is usually only by one category, and changes are rare for species that were already listed as threatened. It should be noted that of these changes, only 195 represent a change in Red List status brought about by a genuine deterioration or improvement in the status of the species [50], rather than a change in knowledge about the species (1246 species); and only 71 of these represent ‘EDGE species’, i.e. threatened species with above-average ED score.
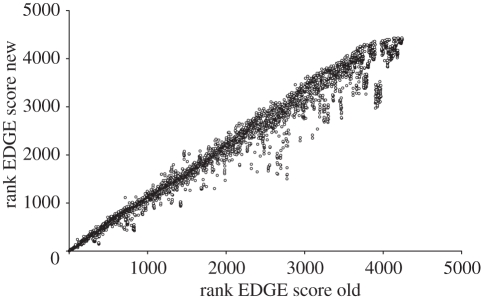
Figure 4.
Comparison of EDGE ranks between EDGE lists. Rank EDGE score old is the value for each species as reported in Isaac et al. [28]; rank EDGE score new is the value for each species calculated in this study.
In the EDGE species trait analysis, we evaluated how species that rank highly in EDGE score are representative of the collective from which they are drawn. Overall there was strong support for the positive correlation of mammal trait oddness and high EDGE score (table 3). This was also true of the component parts of the EDGE score, ED and GE (see electronic supplementary material, tables S2 and S3). The greatest support across orders was for geographical range (eight orders showed significant correlation), followed by body mass and gestation length (table 4). Tests on the component parts of species EDGE score revealed that the relationship with geographical range is driven by the GE component (i.e. correlates of extinction risk), whereas morphological and reproductive traits (body mass, gestation length and litter size) showed strong correlation with ED score (electronic supplementary material, tables S2 and S3).
Table 3.
Pearson correlations of EDGE scores against distance of species-specific traits from the median value for each trait for 11 mammalian orders. ρ denotes correlation coefficient; all statistical values adjusted as per Benjamini & Yekutieli [64].
| Afrosoricida |
Artiodactyla |
Carnivora |
Chiroptera |
Dasyuromorphia |
Didelphimorphia |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| trait | d.f. | ρ | d.f. | ρ | d.f. | ρ | d.f. | ρ | d.f. | ρ | d.f. | ρ |
| body mass | 35 | 0.012 | 175 | 0.007 | 218 | 0.153 | 615 | 0.158** | 55 | 0.161 | 51 | −0.088 |
| gestation length | 7 | 0.920* | 150 | 0.18 | 154 | 0.141 | 148 | 0.078 | 33 | 0.036 | 6 | −0.035 |
| home range | — | — | 60 | −0.045 | 96 | 0.111 | — | — | 6 | −0.565 | 8 | 0.393 |
| litter size | 20 | 0.035 | 154 | 0.207* | 187 | −0.061 | 429 | −0.122* | 45 | 0.167 | 24 | 0.382 |
| geographical range | 42 | 0.400* | 185 | 0.338*** | 198 | 0.393*** | 841 | 0.406*** | 62 | 0.265 | 43 | 0.434 |
| latitudinal midpoint | 42 | 0.460** | 185 | −0.203* | 198 | −0.105 | 841 | −0.022 | 62 | 0.007 | 43 | 0.101 |
| Diprotodontia | Lagomorpha | Primates | Rodentia | Soricomorpha | ||||||||
| body mass | 110 | 0.082 | 58 | 0.039 | 236 | 0.129 | 1199 | 0.059* | 164 | 0.401* | ||
| gestation length | 41 | 0.317 | 33 | 0.01 | 132 | 0.171 | 402 | 0.297*** | 37 | 0.198 | ||
| home range | 29 | −0.075 | 22 | −0.281 | 141 | 0.099 | 225 | 0.057 | 24 | −0.072 | ||
| litter size | 102 | 0.072 | 54 | 0.316 | 184 | −0.05 | 835 | 0.039 | 107 | 0.104 | ||
| geographical range | 129 | 0.350*** | 80 | 0.409** | 291 | 0.082 | 1565 | 0.361*** | 291 | 0.344*** | ||
| latitudinal midpoint | 129 | 0.202 | 80 | 0.086 | 291 | 0.127 | 1565 | −0.119*** | 291 | −0.178** | ||
*p < 0.05, **p < 0.01, ***p < 0.001.
Table 4.
Proportion of mammal orders showing significant positive support for relationship between trait oddity and EDGE score, ED score and GE score.
| trait | EDGE | ED | GE |
|---|---|---|---|
| body mass | 0.27 | 0.55 | 0.09 |
| geographical range | 0.73 | 0.09 | 0.73 |
| gestation length | 0.18 | 0.40 | 0.10 |
| home range | 0.11 | 0 | 0 |
| latitudinal midpoint | 0.09 | 0 | 0.09 |
| litter size | 0.09 | 0.27 | 0 |
4. Discussion
It is important that approaches to conservation priority-setting are able to satisfy two conditions: they must capture biodiversity, a complex and multi-faceted concept, and must be robust to uncertainty. As knowledge continues to develop about the relationships among species and the extinction risk that these species face, techniques such as the one presented here must allow for the prospect that lists of priority species may change. This is a necessary part of incorporating new knowledge to the best effect into prioritization initiatives. Nevertheless, the most appropriate approaches will often be those that are least subject to the vagaries of these inevitable changes in our knowledge of extinction risk and taxonomy. The EDGE method appears on the evidence presented here to represent a robust approach to incorporating evolutionary history into priority-setting in mammals.
The majority of the changes in species ranks between this and the previous version of the EDGE list [28] are due to changes in Red List status. The 195 changes in mammal conservation status, which principally represent changes to more threatened categories of Red List status, are of serious concern; they are leading to a net deterioration in conservation status across the group and an erosion of biodiversity [50]. This study also reveals the number of reassessments of mammalian species extinction risk category owing to non-genuine impacts on status, i.e. changing taxonomy, new information and reassessment of the quality of existing data used for assessments in the light of new understanding. While such changes are potentially problematic for priority-setting schemes, such as EDGE, the expanding knowledge of species conservation status is undoubtedly a positive advance for conservation. Nevertheless, such changes are quite numerous, even in a well-studied group like mammals. These changes appear to be greatest for species in non-threatened or DD categories, whereas threatened species tend only to move a single category. The result of such patterns will be that large jumps in ranking will be experienced by species undergoing the greatest steps of change between threatened and non-threatened categories, and vice versa. The level to which this matters depends on the scale of conservation we wish to achieve. Lower down the ranking, even a small change in score can lead to a large change in rank. However, species actually classed as EDGE species reside towards the top of the ranking, where such changes are far smaller.
By avoiding priority lists that are dominated by highly threatened species, an approach that uses a roughly equal weighting of extinction risk and PD buffers high-ranking species from change, in cases such as mammals where the range of the two component scores is similar (roughly two orders of magnitude). The EDGE programme, which implements practical on-the-ground conservation actions for focal species from the top 100 list of EDGE species (table 1), makes use of this benefit. Although the EDGE programme is important for identifying species as foci for conservation at the global scale, the approach is not yet suitable for application to regional conservation planning (e.g. reserve design). As discussed above, this is because EDGE does not incorporate the principle of complementarity [36]. Modification of the EDGE algorithm is possible (a ‘HEDGE’ list, [38]) but this has yet to be implemented.
We also find evidence across numerous mammalian orders to support findings previously identified in small-scale studies [10,11] that species with high EDGE scores are biologically and/or ecologically atypical of the groups from which they are drawn [10]. However, correlations of trait values with ED or EDGE are not consistent across mammalian orders, providing further evidence that threatened and evolutionarily distinct species represent a truly unique set of taxa, comprising varied traits that contribute disproportionately to biodiversity. One common criticism of phylogeny-based conservation prioritization is that it may preferentially select relictual species that might be less likely to contribute to future evolutionary radiations. The only study to our knowledge that has explicitly evaluated this question [10] found no strong tendency for primate species with high EDGE scores to have ancestral characteristics, suggesting that such species instead possess both rare and derived characters. While such an analysis was beyond the scope of this study, it would be an obvious avenue for further research. Furthermore, while we followed Redding et al.'s [10] method to calculate biological oddness, this method probably works better for species traits with distributions that have a strong central tendency (e.g. life-history traits; [67]). One could potentially scale trait deviations by branch length to identify species that deviate more or less than expected under a Brownian null model of trait evolution. An alternative avenue for future research would be to restrict such correlations to a ‘biologically interesting’ subset, perhaps the upper quartile or even just the top 100. This is because the majority of species have extremely low ED and EDGE scores such that they contribute more ‘noise’ than ‘signal’ to any correlation.
EDGE-style approaches are increasingly being adopted to diagnose conservation priorities within an evolutionary framework. For example, Agnarsson et al. [45] used both EDGE and HEDGE approaches to assess conservation priorities for mammalian carnivore species. Although these authors recognized that priority rankings were strongly dependent on the particular chosen parameters, a consistent series of species were high-ranking in most analyses. Similar analyses have also used the EDGE approach to prioritize conservation of evolutionarily significant units within species [68]. Other recent studies, while not formally quantifying ED, have also adopted EDGE's conceptual framework to make conservation recommendations on the basis of relative ages of different clades (e.g. [69]) or TD [70,71], or at least to acknowledge ED as a key component of conservation prioritization (e.g. [72]). Other approaches have recently incorporated EDGE into other contexts, e.g. biogeographic/ecoregion analyses of conservation priorities or evolutionary history [12,44]. However, despite this growing body of literature citing evidence for the importance of evolutionary history and its incorporation into conservation priority-setting [13], species PD levels remain completely uncorrelated with levels of conservation attention [66], and many of the species identified as conservation priorities in these recent EDGE-style approaches have been acknowledged to be receiving little or no conservation attention [45]. Indeed, 64 per cent of the top 100 ranked species in our new EDGE mammal list (table 1) are currently receiving little or no conservation attention.
We have conducted our EDGE priority-setting approach on mammals because they are one of the best-studied groups, with near-complete data now available on both phylogenetic relationships and extinction risk for component species. Unfortunately, most higher = order taxonomic groups still lack sufficient phylogenetic data to permit calculation of ED scores, and also lack any formal IUCN Red List assessment (although see [73,74]). Given that large numbers of evolutionarily distinct species are inadequately served by existing conservation strategies, the priority must be to fast-track the necessary Red Listing [75] and phylogeny-building exercises to ensure that an imminent loss of large quantities of our global evolutionary heritage does not occur.
Acknowledgements
We thank Arne Mooers, Dave Redding and Gavin Thomas for discussions, Luigi Boitani and Carlo Rondinini for inviting us to contribute to this issue, and one anonymous reviewer for comments on the manuscript. We are grateful to the Rufford Foundation (B.C.), the Royal Society (S.T.T.), and Synchronicity Earth for funding. The EDGE programme is supported by public donations through www.edgeofexistence.org. While the EDGE scores reported in this paper may undergo some change in future due to Red List changes or an updated mammal supertree, this paper should be considered the appropriate reference for the EDGE mammal list.
References
- 1.Butchart S. H. M., et al. 2010. Global biodiversity decline continues. Science 328, 1164–1168 10.1126/science.1187512 (doi:10.1126/science.1187512) [DOI] [PubMed] [Google Scholar]
- 2.Collen B., Loh J., Holbrook S., McRae L., Amin R., Baillie J. E. M. 2009. Monitoring change in vertebrate abundance: the Living Planet Index. Conserv. Biol. 23, 317–327 10.1111/j.1523-1739.2008.01117.x (doi:10.1111/j.1523-1739.2008.01117.x) [DOI] [PubMed] [Google Scholar]
- 3.Crozier R. H. 1997. Preserving the information content of species: genetic diversity, phylogeny, and conservation worth. Annu. Rev. Ecol. Syst. 28, 243–268 10.1146/annurev.ecolsys.28.1.243 (doi:10.1146/annurev.ecolsys.28.1.243) [DOI] [Google Scholar]
- 4.Faith D. P. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10 10.1016/0006-3207(92)91201-3 (doi:10.1016/0006-3207(92)91201-3) [DOI] [Google Scholar]
- 5.Vane-Wright R. I., Humphries C. J., Williams P. H. 1991. What to protect—systematics and the agony of choice. Biol. Conserv. 55, 235–254 10.1016/0006-3207(91)90030-D (doi:10.1016/0006-3207(91)90030-D) [DOI] [Google Scholar]
- 6.Witting L., Loeschcke V. 1995. The optimization of biodiversity conservation. Biol. Conserv. 71, 205–207 10.1016/0006-3207(94)00041-N (doi:10.1016/0006-3207(94)00041-N) [DOI] [Google Scholar]
- 7.Faith D. P. 2002. Quantifying biodiversity: a phylogenetic perspective. Conserv. Biol. 16, 248–252 10.1046/j.1523-1739.2002.00503.x (doi:10.1046/j.1523-1739.2002.00503.x) [DOI] [PubMed] [Google Scholar]
- 8.Cadotte M. W., Cardinale B. J., Oakley T. H. 2008. Evolutionary history and the effect of biodiversity on plant productivity. Proc. Natl Acad. Sci. USA 105, 17 012–17 017 10.1073/pnas.0805962105 (doi:10.1073/pnas.0805962105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cadotte M. W., Cavender-Bares J., Tilman D., Oakley T. H. 2009. Using phylogenetic, functional and trait diversity to understand patterns of plant community productivity. PLoS ONE 4, e5695. 10.1371/journal.pone.0005695 (doi:10.1371/journal.pone.0005695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redding D. W., DeWolff C. V., Mooers A. O. 2010. Evolutionary distinctiveness, threat status and ecological oddity in primates. Conserv. Biol. 24, 1052–1058 10.1111/j.1523-1739.2010.01532.x (doi:10.1111/j.1523-1739.2010.01532.x) [DOI] [PubMed] [Google Scholar]
- 11.Magnuson-Ford K., Ingram R., Redding D. W., Mooers A. Ø. 2009. Rockfish (Sebastes) that are evolutionarily isolated are also large, morphologically distinctive and vulnerable to overfishing. Biol. Conserv. 142, 1787–1796 10.1016/j.biocon.2009.03.020 (doi:10.1016/j.biocon.2009.03.020) [DOI] [Google Scholar]
- 12.Cadotte M. W., Davies T. J. 2010. Rarest of the rare: advances in combining evolutionary distinctiveness and scarcity to inform conservation at biogeographical scales. Divers. Distrib. 16, 376–385 10.1111/j.1472-4642.2010.00650.x (doi:10.1111/j.1472-4642.2010.00650.x) [DOI] [Google Scholar]
- 13.Mace G. M., Gittleman J. L., Purvis A. 2003. Preserving the Tree of Life. Science 300, 1707–1709 10.1126/science.1085510 (doi:10.1126/science.1085510) [DOI] [PubMed] [Google Scholar]
- 14.Mace G. M., Purvis A. 2008. Evolutionary biology and practical conservation: bridging a widening gap. Mol. Ecol. 17, 9–19 10.1111/j.1365-294X.2007.03455.x (doi:10.1111/j.1365-294X.2007.03455.x) [DOI] [PubMed] [Google Scholar]
- 15.Bininda-Emonds O. R. P. 2004. The evolution of supertrees. Trends Ecol. Evol. 19, 315–322 10.1016/j.tree.2004.03.015 (doi:10.1016/j.tree.2004.03.015) [DOI] [PubMed] [Google Scholar]
- 16.Baillie J. E. M., Griffiths J., Turvey S. T., Loh J., Collen B. 2010. Evolution lost: status and trends of the world's vertebrates. UK: Zoological Society of London [Google Scholar]
- 17.Bennett P. M., Owens I. P. F. 1997. Variation in extinction risk among birds: chance or evolutionary predisposition. Proc. R. Soc. Lond. B 264, 401–408 10.1098/rspb.1997.0057 (doi:10.1098/rspb.1997.0057) [DOI] [Google Scholar]
- 18.Purvis A., Agapow P.-M., Gittleman J. L., Mace G. M. 2000. Nonrandom extinction and the loss of evolutionary history. Science 288, 328–330 10.1126/science.288.5464.328 (doi:10.1126/science.288.5464.328) [DOI] [PubMed] [Google Scholar]
- 19.Vamosi J. C., Wilson J. R. U. 2008. Nonrandom extinction leads to elevated loss of angiosperm evolutionary history. Ecol. Lett. 11, 1047–1053 10.1111/j.1461-0248.2008.01215.x (doi:10.1111/j.1461-0248.2008.01215.x) [DOI] [PubMed] [Google Scholar]
- 20.Russell G. J., Brooks T. M., McKinney M. M., Anderson C. G. 1998. Present and future taxonomic selectivity in bird and mammal extinctions. Conserv. Biol. 12, 1365–1376 10.1111/j.1523-1739.1998.96332.x (doi:10.1111/j.1523-1739.1998.96332.x) [DOI] [Google Scholar]
- 21.Mooers A. Ø, Goring S. J., Turvey S. T., Kuhn T. S. 2009. Holocene extinctions and the loss of feature diversity. In Holocene extinctions (ed. Turvey S. T.), pp. 263–277 Oxford, UK: Oxford University Press [Google Scholar]
- 22.Turvey S. T. 2009. Holocene Extinctions. Oxford, UK: Oxford University Press [Google Scholar]
- 23.Rodrigues A. S. L., Gaston K. J. 2002. Rarity and conservation planning across geopolitical units. Conserv. Biol. 16, 674–682 10.1046/j.1523-1739.2002.00455.x (doi:10.1046/j.1523-1739.2002.00455.x) [DOI] [Google Scholar]
- 24.Rodrigues A. S. L., et al. 2011. Complete, accurate, mammalian phylogenies aid conservation planning, but not much. Phil. Trans. R. Soc. B 366, 2652–2660 10.1098/rstb.2011.0104 (doi:10.1098/rstb.2011.0104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devictor V., Mouillot D., Meynard C., Jiguet F., Thuiller W., Mouquet N. 2010. Spatial mismatch and congruence between taxonomic, phylogenetic and functional diversity: the need for integrative conservation strategies in a changing world. Ecol. Lett. 13, 1030–1040 [DOI] [PubMed] [Google Scholar]
- 26.Forest F., et al. 2007. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 445, 757–760 10.1038/nature05587 (doi:10.1038/nature05587) [DOI] [PubMed] [Google Scholar]
- 27.Myers N., Mittermeier R. A., Mittermeier C. G., de Fonseca G. A. B., Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature 403, 853–858 10.1038/35002501 (doi:10.1038/35002501) [DOI] [PubMed] [Google Scholar]
- 28.Isaac N. J. B., Turvey S. T., Collen B., Waterman C., Baillie J. E. M. 2007. Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS ONE 2, e296. 10.1371/journal.pone.0000296 (doi:10.1371/journal.pone.0000296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavoine S., Ollier S., Dufour A. B. 2005. Is the originality of a species measurable? Ecol. Lett. 8, 579–586 10.1111/j.1461-0248.2005.00752.x (doi:10.1111/j.1461-0248.2005.00752.x) [DOI] [Google Scholar]
- 30.Redding D. W. 2003. Incorporating genetic distinctness and reserve occupancy into a conservation prioritisation approach. Masters Thesis, University of East Anglia, Norwich [Google Scholar]
- 31.Redding D. W., Mooers A. O. 2006. Incorporating evolutionary measures into conservation prioritization. Conserv. Biol. 20, 1670–1678 10.1111/j.1523-1739.2006.00555.x (doi:10.1111/j.1523-1739.2006.00555.x) [DOI] [PubMed] [Google Scholar]
- 32.Haake C.-J., Kashiwada A., Su F. E. 2008. The Shapley value of phylogenetic trees. J. Math. Biol. 56, 479–497 10.1007/s00285-007-0126-2 (doi:10.1007/s00285-007-0126-2) [DOI] [PubMed] [Google Scholar]
- 33.Redding D. W., Hartmann K., Mimoto A., Bokal D., DeVos M., Mooers A. Ø. 2008. Evolutionarily distinctive species often capture more phylogenetic diversity than expected. J. Theor. Biol. 51, 606–615 10.1016/j.jtbi.2007.12.006 (doi:10.1016/j.jtbi.2007.12.006) [DOI] [PubMed] [Google Scholar]
- 34.Weitzman M. L. 1993. What to preserve—an application of diversity theory to crane conservation. Q. J. Econ. 108, 157–183 10.2307/2118499 (doi:10.2307/2118499) [DOI] [Google Scholar]
- 35.Avise J. C. 2005. Phylogenetic units and currencies above and below the species level. In Phylogeny and conservation (eds Purvis A., Gittleman J. L., Brooks T.), pp. 76–101 Cambridge, UK: Cambridge University Press [Google Scholar]
- 36.Faith D. P. 2008. Threatened species and the potential loss of phylogenetic diversity: conservation scenarios based on estimated extinction probabilities and phylogenetic risk analysis. Conserv. Biol. 22, 1461–1470 10.1111/j.1523-1739.2008.01068.x (doi:10.1111/j.1523-1739.2008.01068.x) [DOI] [PubMed] [Google Scholar]
- 37.Witting L., Tomiuk J., Loeschcke V. 2000. Modelling the optimal conservation of interacting species. Ecol. Model. 125, 123–143 10.1016/S0304-3800(99)00177-5 (doi:10.1016/S0304-3800(99)00177-5) [DOI] [Google Scholar]
- 38.Steel M., Mimoto A., Mooers A. Ø. 2007. Hedging our bets: the expected contribution of species to future phylogenetic diversity. Evol. Bioinform. 3, 237–244 [PMC free article] [PubMed] [Google Scholar]
- 39.IUCN 2001. IUCN Red List categories and criteria version 3.1. Gland, Switzerland: IUCN [Google Scholar]
- 40.Mace G. M., Collar N. J., Gaston K. J., Hilton-Taylor C., Akcakaya H. R., Leader-Williams N., Milner-Gulland E. J., Stuart S. N. 2008. Quantification of extinction risk: IUCN's system for classifying threatened species. Conserv. Biol. 22, 1424–1442 10.1111/j.1523-1739.2008.01044.x (doi:10.1111/j.1523-1739.2008.01044.x) [DOI] [PubMed] [Google Scholar]
- 41.Purvis A., Cardillo M., Grenyer R., Collen B. 2005. Correlates of extinction risk: phylogeny, biology, threat and scale. In Phylogeny and conservation (eds Purvis A., Brooks T. M., Gittleman J. L.), pp. 295–316 Cambridge, UK: Cambridge University Press [Google Scholar]
- 42.Butchart S. H. M., Stattersfield A. J., Baillie J. E. M., Bennun L. A., Stuart S. N., Akçakaya H. R., Hilton-Taylor C., Mace G. M. 2005. Using Red List Indices to measure progress towards the 2010 target and beyond. Phil. Trans. R. Soc. B 360, 255–268 10.1098/rstb.2004.1583 (doi:10.1098/rstb.2004.1583) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mooers A. Ø., Faith D. P., Maddison W. P. 2008. Converting endangered species categories to probabilities of extinction for phylogenetic conservation prioritization. PLoS ONE 3, e3700. 10.1371/journal.pone.0003700 (doi:10.1371/journal.pone.0003700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies T. J., et al. 2008. Phylogenetic trees and the future of mammalian biodiversity. Proc. Natl Acad. Sci. USA 105, 11 556–11 563 10.1073/pnas.0801917105 (doi:10.1073/pnas.0801917105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agnarsson I., Kuntner M., May-Collado L. 2010. Dogs, cats, and kin: a molecular species-level phylogeny of Carnivora. Mol. Phylogenet. Evol. 54, 726–745 10.1016/j.ympev.2009.10.033 (doi:10.1016/j.ympev.2009.10.033) [DOI] [PubMed] [Google Scholar]
- 46.Butchart S. H. M., Bird J. P. 2010. Data Deficient birds on the IUCN Red List: what don't we know and why does it matter. Biol. Conserv. 143, 239–247 10.1016/j.biocon.2009.10.008 (doi:10.1016/j.biocon.2009.10.008) [DOI] [Google Scholar]
- 47.Collen B., Ram M., Dewhurst N., Clausnitzer V., Kalkman V. J., Cumberlidge N., Baillie J. E. M. 2009. Broadening the coverage of biodiversity assessments. In Wildlife in a changing world—an analysis of the 2008 IUCN Red List of threatened species (eds Vié J.-C., Hilton-Taylor C., Stuart S. N.), pp. 67–76 Gland, Switzerland: IUCN [Google Scholar]
- 48.Schipper J., et al. 2008. The status of the world's land and marine mammals: diversity, threat, and knowledge. Science 322, 225–230 10.1126/science.1165115 (doi:10.1126/science.1165115) [DOI] [PubMed] [Google Scholar]
- 49.IUCN 2010. IUCN Red List of threatened species. Version 2010.3. See www.iucnredlist.org
- 50.Hoffmann M., et al. 2010. The impact and shortfall of conservation on the status of the world's vertebrates. Science 330, 1503–1509 10.1126/science.1194442 (doi:10.1126/science.1194442) [DOI] [PubMed] [Google Scholar]
- 51.Bininda-Emonds O. R. P., et al. 2007. The delayed rise of present-day mammals. Nature 446, 507–512 10.1038/nature05634 (doi:10.1038/nature05634) [DOI] [PubMed] [Google Scholar]
- 52.Bininda-Emonds O. R. P., et al. 2008. The delayed rise of present-day mammals: corrigendum. Nature 456, 274. 10.1038/nature07347 (doi:10.1038/nature07347) [DOI] [PubMed] [Google Scholar]
- 53.Fritz S. A., Bininda-Emonds O. R. P., Purvis A. 2009. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol. Lett. 12, 538–549 10.1111/j.1461-0248.2009.01307.x (doi:10.1111/j.1461-0248.2009.01307.x) [DOI] [PubMed] [Google Scholar]
- 54.Wilson D. E., Reeder D. M. 2005. Mammal species of the world: a taxonomic and geographic reference, 3rd edn. Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 55.Wilson D. E., Reeder D. M. 1993. Mammal species of the world: a taxonomic and geographic reference, 2nd edn. Washington, DC: Smithsonian Institution Press [Google Scholar]
- 56.Agapow P.-M., Bininda-Emonds O. R. P., Crandall K. A., Gittleman J. L., Mace G. M., Marshall J. C., Purvis A. 2004. The impact of species concept on biodiversity studies. Q. Rev. Biol. 79, 161–179 10.1086/383542 (doi:10.1086/383542) [DOI] [PubMed] [Google Scholar]
- 57.Isaac N. J. B., Mallet J., Mace G. M. 2004. Taxonomic inflation: its influence on macroecology and conservation. Trends Ecol. Evol. 19, 464–469 10.1016/j.tree.2004.06.004 (doi:10.1016/j.tree.2004.06.004) [DOI] [PubMed] [Google Scholar]
- 58.Isaac N. J. B., Purvis A. 2004. The ‘species problem’ and testing macroevolutionary hypotheses. Divers. Distrib. 10, 275–281 10.1111/j.1366-9516.2004.00092.x (doi:10.1111/j.1366-9516.2004.00092.x) [DOI] [Google Scholar]
- 59.Kuhn T. S., Mooers A. O., Thomas G. H. 2011. A simple polytomy resolver for dated phylogenies. Methods Ecol. Evol. 2, 1–10 (early online edition.) (doi:10.1111/j.2041-210X.11.00103.x) [Google Scholar]
- 60.Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rambaut A., Drummond A. J. 2009. Tracer v1.5: an MCMC trace analysis tool. See http://beast.bio.ed.ac.uk/. (accessed 1 December 2009).
- 62.Jones K. E., et al. 2009. PanTHERIA: a species-level database of life history, ecology and geography of extant and recently extinct mammals. Ecology 90, 2648. 10.1890/08-1494.1 (doi:10.1890/08-1494.1) [DOI] [Google Scholar]
- 63.Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 [Google Scholar]
- 64.Benjamini Y., Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188 10.1214/aos/1013699998 (doi:10.1214/aos/1013699998) [DOI] [Google Scholar]
- 65.R Development Core Team 2010. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org [Google Scholar]
- 66.Sitas N., Baillie J. E. M., Isaac N. J. B. 2009. What are we saving? Developing a standardized approach for conservation action. Anim. Conserv. 12, 231–237 10.1111/j.1469-1795.2009.00244.x (doi:10.1111/j.1469-1795.2009.00244.x) [DOI] [Google Scholar]
- 67.Bielby J., Mace G. M., Bininda-Emonds O. R. P., Cardillo M., Gittleman J. L., Jones K. E., Orme C. D. L., Purvis A. 2007. The fast–slow continuum in mammalian life history: an empirical reevaluation. Am. Nat. 169, 748–757 10.1086/516847 (doi:10.1086/516847) [DOI] [PubMed] [Google Scholar]
- 68.Sato J. J., Yasuda S. P., Hosoda T. 2009. Genetic diversity of the Japanese marten (Martes melampus) and its implications for the conservation unit. Zool. Sci. 26, 457–466 10.2108/zsj.26.457 (doi:10.2108/zsj.26.457) [DOI] [PubMed] [Google Scholar]
- 69.Bell N. E., Hyvönen J. 2010. Phylogeny of the moss clade Polytrichopsida (BRYOPHYTA): generic-level structure and incongruent gene trees. Mol. Phylogenet. Evol. 55, 381–398 10.1016/j.ympev.2010.02.004 (doi:10.1016/j.ympev.2010.02.004) [DOI] [PubMed] [Google Scholar]
- 70.IUCN 2011. Amphibian Ark Conservation Planning. See http://www.amphibianark.org/about-us/aark-activities/planning-workshops/
- 71.Joseph L. N., Maloney R. F., Possingham H. P. 2009. Optimal allocation of resources among threatened species: a project prioritization protocol. Conserv. Biol. 23, 328–338 10.1111/j.1523-1739.2008.01124.x (doi:10.1111/j.1523-1739.2008.01124.x) [DOI] [PubMed] [Google Scholar]
- 72.Brandley M. C., Wang Y., Guo X., Nieto Montes de Oca A., Fería Ortíz M., Hikida T., Ota H., Rankin D. J. 2010. Bermuda as an evolutionary life raft for an ancient lineage of endangered lizards. PLoS ONE 5, e11375. 10.1371/journal.pone.0011375 (doi:10.1371/journal.pone.0011375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baillie J. E. M., et al. 2008. Towards monitoring global biodiversity. Conserv. Lett. 1, 18–26 10.1111/j.1755-263X.2008.00009.x (doi:10.1111/j.1755-263X.2008.00009.x) [DOI] [Google Scholar]
- 74.Collen B., Baillie J. E. M. 2010. The barometer of life: sampling. Science 329, 140. 10.1126/science.329.5988.140-a (doi:10.1126/science.329.5988.140-a) [DOI] [PubMed] [Google Scholar]
- 75.Stuart S. N., Wilson E. O., McNeely J. A., Mittermeier R. A., Rodriguez J. P. 2010. The barometer of life. Science 328, 117. 10.1126/science.1188606 (doi:10.1126/science.1188606) [DOI] [PubMed] [Google Scholar]