Abstract
Spatial data on species distributions are available in two main forms, point locations and distribution maps (polygon ranges and grids). The first are often temporally and spatially biased, and too discontinuous, to be useful (untransformed) in spatial analyses. A variety of modelling approaches are used to transform point locations into maps. We discuss the attributes that point location data and distribution maps must satisfy in order to be useful in conservation planning. We recommend that before point location data are used to produce and/or evaluate distribution models, the dataset should be assessed under a set of criteria, including sample size, age of data, environmental/geographical coverage, independence, accuracy, time relevance and (often forgotten) representation of areas of permanent and natural presence of the species. Distribution maps must satisfy additional attributes if used for conservation analyses and strategies, including minimizing commission and omission errors, credibility of the source/assessors and availability for public screening. We review currently available databases for mammals globally and show that they are highly variable in complying with these attributes. The heterogeneity and weakness of spatial data seriously constrain their utility to global and also sub-global scale conservation analyses.
Keywords: species distribution, species records, point locations, habitat suitability model, species distribution model, geographical range
1. Introduction
The mammalian clade receives a disproportionate share of conservation attention, as it includes many focal and flagship species [1]. Yet comprehensive data, including spatial distributions, only became available for all mammals very recently. The latest advances in the compilation of large datasets on mammals offer new opportunities for global analyses, particularly on conservation prioritization and on evaluating the effectiveness of conservation actions. Currently available data at a global scale only partially cover the entire mammalian class and are of five types: taxonomy [2], phylogeny [3], distribution [4,5], life history [6] and conservation status with supplementary information on threats [5]. These datasets have been used in a variety of analyses ranging from large-scale biogeographic and macro-ecological studies (e.g. [7,8]) to site scale conservation planning [9] and attempts to predict the impacts of climatic change on species extinction risk [10,11].
Accurate maps of current species distributions are a key component for the assessment of species conservation status [12–14] and for the identification of conservation priorities [15–18]. Spatial conservation prioritization analyses in particular, including gap analysis and systematic identification of conservation sites, are sensitive to commission (false presence in the map) and omission (false absence in the map) errors [19], with the former type of error being particularly problematic as it induces underestimation of the number of gap species (species not well protected by the existing protected areas) [9] and the further amount of area that needs to be protected to fill the gaps [20].
Spatial data on species distribution is generally available in two main forms: point locations and distribution maps [19,21]. Point locations represent (explicitly or not) the mainstay of most spatially explicit databases and consist mainly (considering mammals) in museum records and lists of locations of species sightings and captures. However, to be usable in conservation planning, points need to be transformed into areas [19]. Most collections of point locations are often temporally and spatially biased and discontinuous [22,23], and require extensive treatment before being used for conservation planning. Gridded atlases are the most basic form of transformation of points into areas and are essentially a type of distribution map [24]. They are generally obtained through organizing species records on a map with a superimposed grid of varying cell size, most commonly in the range of 100–10 000 km2 (e.g. [25]), but uneven sampling is often the most serious limitation to represent real species distributions. However, points can be used to help build predictive distribution models which can in turn be used to generate maps of species' predicted distributions and reduce the amount of omission error inherent to any point dataset.
The simplest modelling process that can be applied to a collection of points is based on interpolation of known locations and expert knowledge [14] to produce polygon range maps (e.g. minimum convex polygons and similar [26]). Polygon range maps, however, commonly overestimate species presence within the range [21], often introducing an unknown amount of commission error that reduces the reliability of maps for conservation applications [19]. In the last few decades, following the increased availability of geographical information systems (GISs) and of detailed digital maps of several environmental variables (e.g. climate, land use, altitude, human densities, vegetation and geology), a number of increasingly sophisticated statistical and computational techniques have been used to generate species distribution maps from point locations and polygon ranges [21,27,28]. A modelling approach uses point locations integrated with environmental data to calibrate and evaluate species distribution models (SDMs) based on the relationship between the presence of a species at known locations and a set of environmental variables [29]. This approach essentially aims at eliminating the omission errors of the point data within the range boundaries defined by polygon data. Another approach uses polygon data integrated with environmental data to generate habitat suitability models (HSMs [30]), aiming at eliminating the commission errors in polygon data by refining the polygons to remove parts where habitats are unsuitable for the species. This last approach needs point locations to evaluate the models.
In this paper, we review the specific attributes of spatial data (point locations and distribution maps) for the development of mammal conservation plans. We particularly focus on point locations that represent the baseline of any knowledge on species distribution and on maps produced by different techniques of interpolation and extrapolation of point locations. We then examine the attributes of the different global datasets currently available on mammals. We restrict our discussion to databases that are global in coverage, but focus our attention on the possibility for data to allow high-quality studies on a scale that is effectively useful for real-life conservation. Although we focus on mammal spatial datasets, we suggest that the same attributes should be met in conservation planning of any other taxonomic group.
2. Attributes point location data must meet for generating distribution maps
It is not possible to make a unique and prescriptive list of the main attributes that a set of point data should comply with in order to be used in the development of distribution maps, as this depends on variables such as the natural history of the species considered [31], the ecological context of the study area [32], the modelling techniques employed [32] and final purpose of the distribution map. Here, we outline the main factors to take into consideration when assessing the usefulness of point locations for conservation analyses (Box 1), building from a discussion initiated, among others, by Elith & Leathwick [27], Franklin [28] and Kremen et al. [33]. In general, the treatment of this issue in the literature is associated with the development of SDMs, because their inference is based on analytical, quantitative methods. Yet, many of the considerations we make also apply to the point locations used for other modelling procedures, including interpolations and extrapolations based on expert judgement and model evaluations.
Box 1. Attributes of species spatial data for global conservation applications.
Point locations
1. Spatial coverage: to include all regions where a species is present
2. Spatial accuracy: precise and unbiased
3. Currency: reflecting current distribution and habitat
4. Biological significance: to reflect areas where the species is found permanently and naturally
5. Credibility: scientific credibility of the source (including correct taxonomic identification)
6. Availability: point locations made available to the public for scrutiny and use
Distribution maps (further to the attributes above)
7. Spatial accuracy: reduce commission and omission errors
8. Spatial scale: to match the intended conservation application
9. Credibility: evaluated with independent data or reviewed by the best expert for the taxon/geographical area
10. Availability: maps (and point locations which they are based upon) made available to the public for scrutiny and use
(a). Sample size and spatial coverage
There is no set rule for calculating the minimum number of records required for developing a sufficiently accurate species distribution map. Nonetheless, the basic rule is that the point location dataset should be representative of the entire geographical distribution as well as of the full variation in environmental conditions where the species can be found. Whereas the geographical coverage is important especially when the points are used to interpolate a range map [26], a good representation of environmental conditions is particularly important for SDMs [27]. This has three immediate implications: (i) the number of point locations should increase along with the heterogeneity of the predictors used for the model (i.e. with habitat heterogeneity), (ii) the number of points should increase along with the size of the species distribution, and (iii) a specific sampling design is often necessary to collect an adequate sample of locations (or to integrate an existing but insufficient dataset) that represents the full parameter space available for the species. For example, in most ecological contexts across all continents, land cover, one of the environmental variables commonly used for SDMs, has been deeply affected by human impacts that have caused increased fragmentation [34]. The resulting highly heterogeneous mosaics would require adequate sampling in order to capture their full (multi-scalar) variation.
The number of records should account also for the measurement error of the variables (e.g. the accuracy of a land use map), admittedly difficult to quantify but almost certain to occur in GIS layers of environmental variables. To account for errors that vary across the layers there is little solution but to increase the number of species records as much as possible.
For SDMs, several authors have proposed minimum numbers of data points to ensure good maps. Elith & Leathwick [27] and Kremen et al. [33] suggested that as few as 30 records might be sufficient in certain situations. However, such a small number would be acceptable only in highly homogeneous environments for all parameters of the model. From a purely statistical point of view, the simple rule of thumb for regression models suggests at least 10 (but, more cautiously, up to 20–40) records for each predictor used in the regression.
There have been claims of good model predictive power also with numbers of records smaller than 50 but it is likely that the outcome is just a description of broad trends, probably good for large-scale (grain and extent) biogeographic studies but of limited use for conservation applications. Coudon & Gégout [35] suggested that 50 records are a good compromise between a small sample and good model performance and other authors raised the minimum to 50–100 records [36,37]. As a general guideline, 50–100 records appear to be the minimum to calibrate a model and safely account for all types of variation, adequately cover large extents and reduce the impact of the limitations described in the following paragraphs. However, 100–500 appear to be very often a more secure quantity to rely on, as it could help solving other potential issues of the sample (e.g. effect size [28]) and include the independent dataset needed for model evaluation. Note that considering interpolation techniques (e.g. convex polygons) instead of SDMs would require a much higher number of points, providing at the same time a much more homogeneous coverage of the study area. The challenge, however, is that a dataset with as few as 50–100 records will then have to also meet all the attributes discussed below.
(b). Autocorrelation
All point data, especially if there are a small number of records, and specifically for developing SDMs, need to be independent from each other [38]. Practically, we should consider that the presence of a species in one location is independent (i.e. not influenced) by the presence/absence in the neighbouring location. At least two main types of independence are relevant when considering species records. The first is spatial autocorrelation where the position of one data point in space depends on the position of the previous data point collected. It occurs, for example, when animals are sampled at congregatory locations (e.g. waterholes, breeding areas) or when sampling the same animal within short time periods (e.g. radio- or Global Positioning System (GPS)-tracking records). The second type of autocorrelation is biological, when the sampled animals are related within a social structure (e.g. herds, packs and any animal association) and the movements of one is dependent on those of the other member of the group. This type of autocorrelation is most common, for example, when modelling at local scale using observations from radio-telemetry, sightings or sampling animals from migrating herds. There are several techniques and rules of thumb to ensure full data independence [39], and there are also instances in which autocorrelation can help improving prediction success [40]. The main point here is that we cannot simply ignore the issue and use whatever data are available.
(c). Bias
Most species records are the result of opportunistic collections without a coherent sampling strategy to cover the full variation of environmental variables across the entire range. Biases are therefore very common: records can be spatially biased towards the most popular (e.g. the urban and protected areas) or easily accessible sites (e.g. along road networks), and temporally biased towards certain times of the day or of the year when the species can be easily observed [22,41–44]. This is especially true when occasional data are collected using methods that sample only one aspect of the biology of the species, such as sightings at preferred sites, road kills, hunting bags, etc. Biases are likely to occur also when the species live at various densities across the range and presence data selectively reflects only areas of higher density and easier detection. Temporal biases are also common, owing to irregular recordings over time, causing biased coverage of the environmental variation.
Plotting on two- or three-dimensional graphs the values of environmental variables in stratified locations across the entire species range and overlapping those with the values of the locations available for modelling (both presence and absence ideally) can help in understanding to what extent the database available covers well the parameter space.
The problem of biased dataset can be partially solved by weighting the presence data according to their distribution patterns [27] but at the cost of introducing substantial subjectivity in the model.
(d). Accuracy
Point data collected in the last 10–20 years are likely to have been georeferenced using a GPS, which ensures high accuracy. However, most of the species records stored by museums and research centres have been collected when locations were defined simply by toponyms (i.e. the name of a local geographical feature) and they are normally associated with a moderate to high level of inaccuracy [45]. The centroid of the toponym is often used to transform the toponym into precise geographical coordinates. However, this procedure merely masks the inaccuracy of the original data and does not solve their limitations [46]. Ideally, species records with unknown or unacceptable level of accuracy should be eliminated from the set of data points used to develop maps. For many poorly known restricted-range mammals these might be the only available records, but this introduces a further problem as for these species in particular, accuracy is of paramount importance. So, effort should be made to try to correct inaccurate locations for restricted-range species a posteriori on the basis of corollary information (e.g. for species known only from the type locality and restricted to high elevation, a relatively common situation for tropical mammals, move the record according to a digital elevation model of the area).
(e). Time relevance
Species distributions are not static, they evolve and shift dynamically in time and space as a consequence of changed land use and other climatic and environmental conditions. Given the high land cover conversion rate in most regions (e.g. [47]), as a general guideline only records referring to a time period of about 10–20 years around the date of the predictor layers should be used for current distribution maps. As most species records are collected opportunistically over long time periods, the attribute of time relevance vastly reduces the amount of point data available.
The attribute of data currency is even tighter for SDMs based on land cover maps. An SDM is a model of the relationship between the species presence and its environment, although simplified by the selection of few predictors. It is therefore mandatory that the data points used to calibrate/evaluate the model and the environmental layers refer to the same time period. The assumption that the value of the predictors at each location reflects the condition at the time when the species was actually recorded [29] is a key and often forgotten condition to build SDMs. As most SDMs aim at modelling current distribution, recent data points are necessary to match current layers of the predictors. Assuming no habitat conversion even over a few years is not realistic, especially in highly human-dominated landscapes. For example, in Italy almost 52 per cent of the land cover has changed in the last 30 years in spite of rules on maintaining traditional landscapes [47], and this is true also for the existing protected areas [48].
(f). Reliability
Data points have normally been collected by a variety of people, not all equally reliable. The species may have been misidentified, the location and time may have been wrongly recorded and many other sources of errors may have been introduced in the datasets [49,50]. The credibility of the source should be carefully evaluated before accepting the dataset [51].
(g). Biological significance
The least recognized source of error in a point location dataset is the biological information linked to a record of presence. Location points are generally assumed to represent where the species is found permanently and naturally, not as a temporary transient or as a consequence of any human pressure and, in modelling habitat suitability, they are often assumed to indicate the optimal habitat of the species. However, reality is far from these assumptions as species may have been displaced or restricted to a portion of their range as a consequence of various factors, such as human impacts, interspecific competition and historical reasons, all common processes for most of the world where landscapes have been deeply changed by human activities [52]. These range shifts may have the consequence of pushing the species to poorly suitable habitat, and sampling it will result in misleading distribution models. Also, many species change habitat seasonally and point locations may reflect only some of a species' life stages. Mammals are often highly mobile and it is natural that during migrations, dispersal, occasional wanderings or accidents often occur outside their normal range or in unsuitable or poorly suitable habitat. A large dataset is expected to dilute the potential impact of these occurrences, which could otherwise have a significant weight if the dataset was small. Species live often also in sink areas (where λ < 0) as part of spatially structured metapopulations, and presence data from these areas would need to be balanced with data from the whole species range to ensure correct representation of all environmental variations. Again, a large dataset is expected to reduce the probability of biased representation, at least for species that are not spatially structured as a consequence of territorial behaviour.
Neglecting these potential sources of error is very risky as the resulting models could be biologically wrong even though their statistical performance is good [53]. Unfortunately, this biological information is lacking from most of the species records and it is not possible to filter the data according to them. In the absence of good quality data with associated ecological information, the only (partial) solution is to increase the number of data points as far as possible. In view of these considerations, we suggest that a minimum of 50–100 data points could be a reasonable rule of thumb for the greatest majority of SDMs, whatever the modelling technique used.
(h). Sources of species records
The attributes of a good dataset of species presence are so demanding that the only way to obtain it is often through a specific survey. A properly designed survey can ensure the completeness of coverage for the study area, required sample size, correct stratification of the environmental variables, prevalence of the species, resolution and precision of the records, and many other characteristics of the sample that are important for SDMs [28]. A specific survey could be designed to obtain estimates of the detectability of the species and provide an extremely useful parameter for more refined modelling techniques [54,55]. However, it is actually rare that a species modelling exercise also includes the collection of species point data in the field. If it can be done for small extents or in conjunction with surveys aimed at other goals than species modelling, it would certainly be impractical for studies at continental or global scale. Moreover, investing limited conservation resources in data collection should be always considered in the context of other options such as implementation of conservation action [56].
Therefore, the only alternative sources of data are the existing datasets derived by the opportunistic collections organized by museums and research centres or extracted from data of national biological inventories and conservation programmes. At the global scale, for most species these data repositories are the only sources of species records.
3. Further attributes of distribution maps
Any distribution map interpolated and/or extrapolated from point locations that do not reach minimum quality attributes, as discussed above, has a chance to be of diminished value. Even if it is based on point locations of reasonably good quality, a distribution map should be assessed against further attributes before being used in conservation applications (Box 1).
(a). Accuracy and scale
While accuracy for point locations is best summarized by the precision of locations and collective biases of points, for polygon ranges it is more appropriately measured by the rate of errors of commission and omission [57]. The way interpolations are applied to complete insufficient geographical coverage of the data determines the amount of the two types of error. Their impact is not always explicitly assessed, but is known to exist and depends on the analysis carried out [37]. Global studies of conservation priorities have often used coarse grain data represented in large grid cells or hexagons (e.g. [14,16,58]), which inevitably include a large amount of unoccupied area [52]. This is because environmental variables that are used to determine the distribution of species [29], particularly those linked to human pressures, are expected to have highly heterogeneous values across space and to be best represented at small resolution.
Ideally, spatial conservation prioritization analyses would need maps on a resolution that is fine-scaled enough to capture the fragmented and discontinuous distribution of species, and discriminate between sites on the scale of conservation action. Gridded data on a resolution lower than 100 km2 (and often 1–25 km2) may be necessary if the output is expected to have really useful conservation applications [59]. In practice, such data are far from being available for the vast majority of mammal species. To approximate this fine scale, inferences on which parts of a species' geographical range may be used by the species have been done for mammals on a global scale on the basis of expert opinion [14] or deterministic HSMs [30]. Whatever method is used to try to downscale geographical ranges to a fine grain, commission errors are reduced at the expense of an increase in omission errors [19]. Therefore, the result of the downscaling must be evaluated against sample point location data to check at least whether the reduction of commission error outweighs the increase in omission error [9,30].
(b). Credibility and availability
To ensure that the information is the best available and conservation is planned on the most credible foundation, the species distribution maps should be (i) evaluated against independent data, if derived from statistical SDM and/or expert based HSMs, or (ii) compiled or reviewed and approved by species experts if based on expert judgement. In addition, we advocate that maps based on expert judgement should be accompanied by a rationale explaining how the distribution was drawn. Different species experts may have different attitudes towards, for example, the risk of interpolating distribution maps over large areas of unsampled but potentially suitable habitat between locations where a species has been found. As managing the number and diversity of mammals exceeds the knowledge of any individual expert, it is inevitable that global datasets for mammal distribution need the support of a large network of experts. The different perspectives of species experts are then likely to be reflected in sometime large differences in species distribution maps. An explicit rationale for the maps would facilitate further re-evaluation of the maps, and over time increase the consistency of inferences among experts, thus among species. Moreover, if species distribution maps are expected to have a real impact on conservation, it is of paramount importance that they are of public domain and transparently open to scrutiny and critique (Box 1).
4. Global spatial datasets on mammals
(a). Point locations
The single largest repository of point locations for mammals is the Global Biodiversity Information Facility (GBIF) [4], an international institution dedicated to facilitating free and open access to biodiversity data through a web portal where data are submitted by voluntary providers and downloadable by users. GBIF currently (February 2011) lists data from 11 708 datasets by 322 institutes worldwide, accounting for more than 260 millions records in total and almost 3 400 000 geo-referenced records belonging to more than 3700 mammal species. More than 60 per cent of the data are composed from observations and less than 40 per cent from digitized natural history collections data, and list among the others, information such as coordinates, coordinate precision, collection date and identification date.
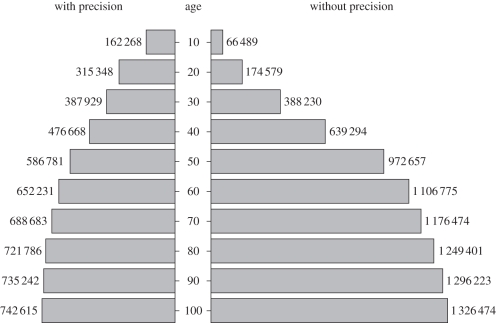
The GBIF database is in constant expansion, and between 2008 and 2010 alone the records with coordinates pertaining to mammals have more than doubled, bringing the current amount of points collected in the last 30, 20 and 10 years, respectively, to 776 159, 489 927 and 228 757, accounting for 36.3, 22.9 and 10.7 per cent of all points collected after 1950. This, along with the fact that points are unevenly distributed among species and that for most records information on precision is not available (figure 1), brings the number of mammal species with more than 100 records with coordinates, an indication of coordinate precision, and less than 20 years of age (i.e. potentially, reasonably accurate and current) to be only 174.
Figure 1.
Numbers of point locations on mammals available in GBIF ordered by age (years) with/without information on coordinates precision (February 2011).
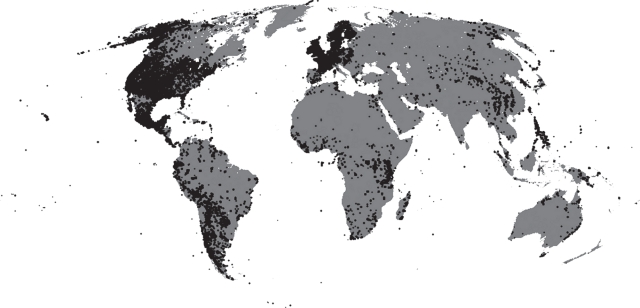
The spatial coverage of mammal point locations in GBIF (figure 2) does not reflect the known global patterns of mammal richness [14], as the maximum density of point locations in GBIF is found in North America and Europe, while the maximum richness in mammal species is in the tropics. This is strong evidence of an overall spatial bias in data collection, which is expected in a meta-database like GBIF because there is no systematic plan and means of acquiring data. The natural history collections that form the basis of the dataset have in general displayed a high incidence of species, geographical and temporal bias [22,42,45,46,60,61]. Also field data collection, the other source of point locations, is more likely to be carried out in developed, species poor but funds rich countries. Another limitation of the GBIF dataset is that without access to the original data, it is impossible to assess the independence of single data points. Multiple points could in fact refer to the same individual subject to a monitoring campaign, introducing further bias in the data. While this may be negligible with large amounts of data, it may become relevant for species with scarce data.
Figure 2.
The distribution of point locations for all mammal species available in GBIF (February 2011).
However unsatisfying the quantity and quality of point locations may be, they are by definition the only primary source of knowledge on species distribution (although not all point locations used to derive the existing global mammal distribution maps are from GBIF), and as such are used for expert interpolation/extrapolation, model calibration and evaluation. When metadata on point locations are available (spatial error, age, identification date) the amount of error they contain can be quantified directly and weighted in modelling, an advantage over distribution maps where the error can be estimated indirectly through point locations (HSMs and SDMs) or not quantified at all (range maps drawn by experts).
(b). Distribution maps
Several datasets of global mammal distribution maps have been assembled in the last decade. The first two global sets of range maps to cover all mammal species were compiled by Sechrest [62] and Ceballos et al. [58] for 4735 and 4795 species, respectively. The first set was built from consulting over 2000 publications and gathering all types of available spatial data filtered for currency, accuracy and source reliability. Maps were built with subjective but conservative interpretation whenever point locations or different maps had to be reconciled into a final map. This database has been used, among other studies, for the first global gap analysis aimed at evaluating the effectiveness of the existing network of protected areas [16]. The second dataset was assembled using literature sources. Ranges were superimposed on a 10 000 km2 grid for subsequent analyses of conservation priorities [8]. Several studies have been based on this dataset (e.g. [63]).
In 2008, the International Union for Conservation of Nature (IUCN) completed the Global Mammal Assessment (GMA; including the distribution maps for 5489 species or ca 95% of the mammals) as part of the larger Red List programme [14]. The GMA database is freely available [5] and open to scrutiny and includes, for each species, information on taxonomy, spatially explicit distribution, population trends, habitats, threats, human use and conservation measures in place and needed. This dataset has been used to study the patterns of mammalian threat [14] and is currently used for a variety of other geography-based studies, including two in this issue (the modelling of species distribution through HSMs at 0.09 km2 resolution for 5027 of 5330 known terrestrial mammals [30]; and a study of the effect of using phylogenetic data for the identification of spatial priorities for mammals [64]).
It is unfortunate that none of the existing global datasets of mammal distribution maps has retained the information about which point locations each map is based on (or this information has not been made available). Points were degraded into the grid scheme or lost in drawing the polygons. Therefore, it is impossible to assess the quality of the original point locations used. Of the three global datasets on mammal distributions, the GMA is the only one that has been validated through expert review (the Species Survival Commission of IUCN is composed of more than 8000 experts), is subject to periodic revision to include updates, and has been made available to the public through a website (see above). The flexibility and openness of the GMA can make its distribution maps a community standard for the development of strategies on a global scale. To achieve this, two main issues need to be resolved. One is to improve the link between the maps and the underlying point locations, by making transparent which point locations have been used, and what expert inference has been made to transform these points into a continuous map. The other is to increase map resolution to match that of conservation action, without losing accuracy to an unacceptable level. At the same time, it is necessary to improve models' accuracy and evaluation for all species.
(c). Sub-global datasets
Whereas this discussion focuses on datasets that are global in coverage, it is relevant also for analyses on sub-global scales. Much of the work on conservation prioritization and action happens on sub-global scales, where strategies are more easily implemented and resources mobilized. Several spatial datasets have been compiled for mammals of large geographical regions, such as sub-Saharan Africa ([65], 942 species mapped on a 1° grid), all Africa ([66], 328 large mammal species mapped at the resolution of 1 km2), Southeast Asia ([9], 1086 species at 1 km2 resolution), Europe ([25], all species at the resolution of 2500 km2). All these datasets have been successfully used for many large-scale conservation studies (e.g. [67–69]), although some of the maps were based on very coarse data and on scales and resolution of little significance for site-level conservation. Finally, complete datasets of distribution maps (gridded atlases and polygon ranges) are available for many countries, assembled on a great diversity of scales and resolutions [28].
5. Conclusion
Species data are but one piece of the information needed in conservation planning, and other datasets (e.g. habitats, threats) need to also be considered in developing global and local strategies [30,70,71]. However, species distribution maps remain the foundation of any conservation planning and managers must build on whatever is available. The heterogeneity of mammal spatial data appears to be a serious constraint for global strategies. A combination of paucity of data and lack of standardized criteria to accept minimum attributes of data quantity and quality for spatial analyses and conservation strategies hinders the potential of the most advanced methods of spatial prioritization and reduces the application on scales useful for conservation action. The negative impact of inaccurate maps on conservation strategies is difficult to quantify but can be potentially very dangerous for conserving species. We advocate the need for more rigorous screening of the datasets being used for map production, the explicit discussion of the data according to their attributes and the adoption of the most conservative modelling approach [19].
Current datasets are limited and with different types of problems. Datasets of point locations are quantitatively and qualitatively poor for global analyses and appear largely insufficient to generate SDMs for all mammals. Polygon ranges are now available through the GMA and allow the production of HSMs for almost all species [30] and obtaining distribution maps for conservation analyses. Finally, we advocate an increased openness of the datasets, and the possibility for users to contribute with information and critique will help in enhancing the quality of the datasets and gradually building a dynamic database to become the world reference for the distribution of all mammal species.
Acknowledgements
We are grateful for long discussions on these issues to Bob Pressey and for the insightful comments of two anonymous reviewers and Ana Rodrigues.
References
- 1.Leader-Williams N., Dublin H. T. 2000. Charismatic megafauna as ‘flagship species’. In Priorities for the conservation of mammalian diversity (eds Entwistle A., Dunstone N.), pp. 53–81 Cambridge, UK: Cambridge University Press [Google Scholar]
- 2.Wilson D. E., Reeder D. M. 2005. Mammal species of the world. A taxonomic and geographic reference, 3rd edn. Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 3.Fritz S. A., Bininda-Emonds O. R. P., Purvis A. 2009. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol. Lett. 12, 538–549 10.1111/j.1461-0248.2009.01307.x (doi:10.1111/j.1461-0248.2009.01307.x) [DOI] [PubMed] [Google Scholar]
- 4.GBIF. 2011. Global Biodiversity Information Facility. See http://www.gbif.org (accessed 15 March 2011)
- 5.IUCN Red List. 2011. The International Union for the Conservation of Nature's Red List of Threatened Species. See http://www.iucnredlist.org/initiatives/mammals (accessed 15 March 2011)
- 6.Jones K. E., et al. 2009. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648. 10.1890/08-1494.1 (doi:10.1890/08-1494.1) [DOI] [Google Scholar]
- 7.Guralnick R. 2006. The legacy of past climate and landscape change on species' current experienced climate and elevation ranges across latitude: a multispecies study utilizing mammals in western North America. Global Ecol. Biogeogr. 15, 505–518 [Google Scholar]
- 8.Ceballos G., Ehrlich P. R. 2006. Global mammal distributions, biodiversity hotspots, and conservation. Proc. Natl Acad. Sci. USA 103, 19 374–19 379 10.1073/pnas.0609334103 (doi:10.1073/pnas.0609334103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catullo G., Masi M., Falcucci A., Maiorano L., Rondinini C., Boitani L. 2008. A gap analysis of Southeast Asian mammals based on habitat suitability models. Biol. Conserv. 141, 2730–2744 10.1016/j.biocon.2008.08.019 (doi:10.1016/j.biocon.2008.08.019) [DOI] [Google Scholar]
- 10.Cardillo M., Mace G. M., Gittleman J. L., Purvis A. 2006. Latent extinction risk and the future battlegrounds of mammal conservation. Proc. Natl Acad. Sci. USA 103, 4157–4161 10.1073/pnas.0510541103 (doi:10.1073/pnas.0510541103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maiorano L., Falcucci A., Zimmermann N. E., Psomas A., Pottier J., Baisero D., Rondinini C., Guisan A., Boitani L. 2011. The future of terrestrial mammals in the Mediterranean basin under climate change. Phil. Trans. R. Soc. B 366, 2681–2692 10.1098/rstb.2011.0121 (doi:10.1098/rstb.2011.0121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freitag S., Van Jaarsveld A. S. 1998. Sensitivity of selection procedures for priority conservation areas to survey extent, survey intensity and taxonomic knowledge. Proc. R. Soc. Lond. B 265, 1475–1482 10.1098/rspb.1998.0460 (doi:10.1098/rspb.1998.0460) [DOI] [Google Scholar]
- 13.Hoffmann M. M., et al. 2010. The impact of conservation on the status of the world's vertebrates. Science 329, 141–142 10.1126/science.1193522 (doi:10.1126/science.1193522) [DOI] [PubMed] [Google Scholar]
- 14.Schipper J., et al. 2008. The status of the world's land and marine mammals: diversity, threat, and knowledge. Science 322, 225–230 10.1126/science.1165115 (doi:10.1126/science.1165115) [DOI] [PubMed] [Google Scholar]
- 15.Eklund J., Arponen A., Visconti P., Cabeza M. 2011. Governance factors in the identification of global conservation priorities for mammals. Phil. Trans. R. Soc. B 366, 2661–2669 10.1098/rstb.2011.0114 (doi:10.1098/rstb.2011.0114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodrigues A. S. L., et al. 2004. Effectiveness of the global protected-area network in representing species diversity. Nature 428, 640–643 10.1038/nature02422 (doi:10.1038/nature02422) [DOI] [PubMed] [Google Scholar]
- 17.Visconti P., et al. 2011. Future hotspots of terrestrial mammal loss. Phil. Trans. R. Soc. B 366, 2693–2702 10.1098/rstb.2011.0105 (doi:10.1098/rstb.2011.0105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson K. A., Evans M. C., Di Marco M., Green D. C., Boitani L., Possingham H. P., Chiozza F., Rondinini C. 2011. Prioritizing conservation investments for mammal species globally. Phil. Trans. R. Soc. B 366, 2670–2680 10.1098/rstb.2011.0108 (doi:10.1098/rstb.2011.0108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rondinini C., Wilson K. A., Boitani L., Grantham H., Possingham H. P. 2006. Tradeoffs of different types of species occurrence data for use in systematic conservation planning. Ecol. Lett. 9, 1136–1145 10.1111/j.1461-0248.2006.00970.x (doi:10.1111/j.1461-0248.2006.00970.x) [DOI] [PubMed] [Google Scholar]
- 20.Rondinini C., Stuart S., Boitani L. 2005. Habitat suitability models and the shortfall in conservation planning for African vertebrates. Conserv. Biol. 19, 1488–1497 10.1111/j.1523-1739.2005.00204.x (doi:10.1111/j.1523-1739.2005.00204.x) [DOI] [Google Scholar]
- 21.Rondinini C., Boitani L. In press Mind the map: trips and pitfalls in making and reading maps of carnivore distribution. In Carnivore ecology and conservation. A handbook of techniques (eds Boitani L., Powell R. A.). Oxford, UK: Oxford University Press [Google Scholar]
- 22.Boakes E. H., McGowan P. J. K., Fuller R. A., Chang-qing D., Clark N. E., O'Connor K., Mace G. M. 2010. Distorted views of biodiversity: spatial and temporal bias in species occurrence data. PLoS Biol. 8, e1000385. 10.1371/journal.pbio.1000385 (doi:10.1371/journal.pbio.1000385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guisan A., Thuiller W. 2005. Predicting species distribution: offering more than simple habitat models. Ecol. Lett. 8, 993–1009 10.1111/j.1461-0248.2005.00792.x (doi:10.1111/j.1461-0248.2005.00792.x) [DOI] [PubMed] [Google Scholar]
- 24.Harrison J. A. 1989. Atlassing as a tool in conservation, with special reference to the Southern African Bird Atlas Project. In Biotic diversity in Southern Africa: concepts and conservation (ed. Huntley B. J.), pp. 157–169 Cape Town, South Africa: Oxford University Press [Google Scholar]
- 25.Mitchell-Jones A. J., et al. 1999. The atlas of European mammals. London: Academic Press [Google Scholar]
- 26.Burgman M. A., Fox J. C. 2003. Bias in species range estimates from minimum convex polygons: implications for conservation and options for improved planning. Anim. Conserv. 6, 19–28 10.1017/S1367943003003044 (doi:10.1017/S1367943003003044) [DOI] [Google Scholar]
- 27.Elith J., Leathwick J. R. 2009. Species distribution models: ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 40, 677–697 10.1146/annurev.ecolsys.110308.120159 (doi:10.1146/annurev.ecolsys.110308.120159) [DOI] [Google Scholar]
- 28.Franklin J. 2009. Mapping species distributions: spatial inference and prediction. Cambridge, UK: Cambridge University Press [Google Scholar]
- 29.Guisan A., Zimmermann N. E. 2000. Predictive habitat distribution models in ecology. Ecol. Model. 135, 147–186 10.1016/S0304-3800(00)00354-9 (doi:10.1016/S0304-3800(00)00354-9) [DOI] [Google Scholar]
- 30.Rondinini C., et al. 2011. Reconciling global mammal prioritization schemes into a strategy. Phil. Trans. R. Soc. B 366, 2722–2728 10.1098/rstb.2011.0112 (doi:10.1098/rstb.2011.0112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McPherson J. M., Jetz W. 2007. Effects of species' ecology on the accuracy of distribution models. Ecography 30, 135–151 10.1111/j.0906-7590.2007.04823.x (doi:10.1111/j.0906-7590.2007.04823.x) [DOI] [Google Scholar]
- 32.Elith J., et al. 2006. Novel methods improve prediction of species' distributions from occurrence data. Ecography 29, 129–151 10.1111/j.2006.0906-7590.04596.x (doi:10.1111/j.2006.0906-7590.04596.x) [DOI] [Google Scholar]
- 33.Kremen C., et al. 2008. Aligning conservation priorities across taxa in Madagascar with high-resolution planning tools. Science 320, 222–226 10.1126/science.1155193 (doi:10.1126/science.1155193) [DOI] [PubMed] [Google Scholar]
- 34.Fischer J., Lindenmayer D. B. 2007. Landscape modification and habitat fragmentation: a synthesis. Global Ecol. Biogeogr. 16, 265–280 10.1111/j.1466-8238.2007.00287.x (doi:10.1111/j.1466-8238.2007.00287.x) [DOI] [Google Scholar]
- 35.Coudon C., Gégout J. C. 2007. Quantitative prediction of the distribution and abundance of Vaccinium myrtillus with climatic and edaphic factors. J. Veg. Sci. 18, 517–524 10.1111/j.1654-1103.2007.tb02566.x (doi:10.1111/j.1654-1103.2007.tb02566.x) [DOI] [Google Scholar]
- 36.Kadmon R., Farber O., Danin A. 2003. A systematic analysis of factors affecting the performance of climatic envelope models. Ecol. Appl. 13, 853–867 10.1890/1051-0761(2003)013[0853:ASAOFA]2.0.CO;2 (doi:10.1890/1051-0761(2003)013[0853:ASAOFA]2.0.CO;2) [DOI] [Google Scholar]
- 37.Loiselle B. A., Howell C. A., Graham C. H., Goerck J. M., Brooks T. M., Smith K. G., Williams P. H. 2003. Avoiding pitfalls of using species distribution models in conservation planning. Conserv. Biol. 17, 1591–1600 10.1111/j.1523-1739.2003.00233.x (doi:10.1111/j.1523-1739.2003.00233.x) [DOI] [Google Scholar]
- 38.Dorman C. F. 2006. Effects of incorporating spatial autocorrelation into the analysis of species distribution data. Global Ecol. Biogeogr. 16, 129–138 10.1111/j.1466-8238.2006.00279.x (doi:10.1111/j.1466-8238.2006.00279.x) [DOI] [Google Scholar]
- 39.Dorman C. F., et al. 2007. Methods to account for spatial autocorrelation in the analysis of species distributional data: a review. Ecography 30, 609–628 10.1111/j.2007.0906-7590.05171.x (doi:10.1111/j.2007.0906-7590.05171.x) [DOI] [Google Scholar]
- 40.De Marco P., Diniz-Filho J. A. F., Mauricio B. L. 2008. Spatial analysis improves species distribution modelling during range expansion. Biol. Lett. 4, 577–580 10.1098/rsbl.2008.0210 (doi:10.1098/rsbl.2008.0210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keller C. M. E., Scallan J. T. 1999. Potential roadside biases due to habitat changes along breeding bird survey routes. Condor 101, 50–57 10.2307/1370445 (doi:10.2307/1370445) [DOI] [Google Scholar]
- 42.Funk V., Richardson K. 2002. Systematic data in biodiversity studies: use it or lose it. Syst. Biol. 51, 303–316 10.1080/10635150252899789 (doi:10.1080/10635150252899789) [DOI] [PubMed] [Google Scholar]
- 43.Reddy S., Davalos L. M. 2003. Geographic sampling bias and its implications for conservation priorities in Africa. J. Biogeogr. 30, 1719–1727 10.1046/j.1365-2699.2003.00946.x (doi:10.1046/j.1365-2699.2003.00946.x) [DOI] [Google Scholar]
- 44.Kadmon R., Farber O., Danin A. 2004. Effect of roadside bias on the accuracy of predictive maps produced by bioclimatic models. Ecol. Appl. 14, 401–413 10.1890/02-5364 (doi:10.1890/02-5364) [DOI] [Google Scholar]
- 45.Graham C., Ferrier S., Huettman F., Moritz C., Peterson A. 2004. New developments in museum-based informatics and applications in biodiversity analysis. Trends Ecol. Evol. 19, 497–503 10.1016/j.tree.2004.07.006 (doi:10.1016/j.tree.2004.07.006) [DOI] [PubMed] [Google Scholar]
- 46.Newbold T. 2010. Applications and limitations of museum data for conservation and ecology, with particular attention to species distribution models. Prog. Phys. Geogr. 34, 3–22 10.1177/0309133309355630 (doi:10.1177/0309133309355630) [DOI] [Google Scholar]
- 47.Falcucci A., Maiorano L., Boitani L. 2007. Changes in land-use/land-cover patterns in Italy and their implications for biodiversity conservation. Landscape Ecol. 22, 617–631 10.1007/s10980-006-9056-4 (doi:10.1007/s10980-006-9056-4) [DOI] [Google Scholar]
- 48.Maiorano L., Falcucci A., Boitani L. 2008. Size dependent resistance of protected areas to land-use change. Proc. R. Soc. B 275, 1297–1304 10.1098/rspb.2007.1756 (doi:10.1098/rspb.2007.1756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunn R., Harrison A. R., White R. C. 1990. Positional accuracy and measurement error in digital databases: an empirical study. Int. J. Geogr. Info. Sci. 4, 385–398 10.1080/02693799008941554 (doi:10.1080/02693799008941554) [DOI] [Google Scholar]
- 50.Meier R., Dikow T. 2004. Significance of specimen databases from taxonomic revisions for estimating and mapping the global species diversity of invertebrates and repatriating reliable specimen data. Conserv. Biol. 18, 478–488 10.1111/j.1523-1739.2004.00233.x (doi:10.1111/j.1523-1739.2004.00233.x) [DOI] [Google Scholar]
- 51.Williams P. H., Margules C. R., Hilbert D. W. 2002. Data requirements and data sources for biodiversity priority area selection. J. Biosci. 27, 327–338 10.1007/BF02704963 (doi:10.1007/BF02704963) [DOI] [PubMed] [Google Scholar]
- 52.Gaston K. J. 2003. The structure and dynamics of geographic ranges. Oxford, UK: Oxford University Press [Google Scholar]
- 53.Pulliam H. R. 2000. On the relationship between niche and distribution. Ecol. Lett. 3, 349–361 10.1046/j.1461-0248.2000.00143.x (doi:10.1046/j.1461-0248.2000.00143.x) [DOI] [Google Scholar]
- 54.Wintle B. A., McCarthy M. A., Parris K. M., Burgman M. A. 2004. Precision and bias of methods for estimating point survey detection probabilities. Ecol. Appl. 14, 703–712 10.1890/02-5166 (doi:10.1890/02-5166) [DOI] [Google Scholar]
- 55.MacKenzie D. I. 2006. Modeling the probability of resource use: the effect of, and dealing with, detecting a species imperfectly. J. Wildl. Manag. 70, 367–374 10.2193/0022-541X(2006)70[367:MTPORU]2.0.CO;2 (doi:10.2193/0022-541X(2006)70[367:MTPORU]2.0.CO;2) [DOI] [Google Scholar]
- 56.Grantham H. S., Moilanen A., Wilson K. A., Pressey R. L., Rebelo T. G., Possingham H. P. 2008. Diminishing return on investment for biodiversity data in conservation planning. Conserv. Lett. 1, 190–198 10.1111/j.1755-263X.2008.00029.x (doi:10.1111/j.1755-263X.2008.00029.x) [DOI] [Google Scholar]
- 57.Fielding A. H., Bell J. F. 1997. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 24, 38–49 10.1017/S0376892997000088 (doi:10.1017/S0376892997000088) [DOI] [Google Scholar]
- 58.Ceballos G., Ehrlich P. R., Soberon J., Salazar I., Fay J. P. 2005. Global mammal conservation: what must we manage? Science 309, 603–607 10.1126/science.1114015 (doi:10.1126/science.1114015) [DOI] [PubMed] [Google Scholar]
- 59.Seo C., Thorne J., Hannah L., Thuiller W. 2009. Scale effects in species distribution models: implications for conservation planning under climate change. Biol. Lett. 5, 39–43 10.1098/rsbl.2008.0476 (doi:10.1098/rsbl.2008.0476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Funk V., Zermoglio M., Nasir N. 1999. Testing the use of specimen collection data and GIS in biodiversity exploration and conservation decision making in Guyana. Biodivers. Conserv. 8, 727–751 10.1023/A:1008877222842 (doi:10.1023/A:1008877222842) [DOI] [Google Scholar]
- 61.Johnson K. G., et al. 2011. Climate change and biosphere response: unlocking the collections vault. Bioscience 61, 147–153 10.1525/bio.2011.61.2.10 (doi:10.1525/bio.2011.61.2.10) [DOI] [Google Scholar]
- 62.Sechrest W. W. 2003. Global diversity, endemism and conservation of mammals. Thesis, University of Virginia, Charlottesville, USA [Google Scholar]
- 63.Carwardine J., Wilson K. A., Ceballos G., Ehrlich P. R., Naidoo R., Iwamura T., Hajkowicz S. A., Possingham H. P. 2008. Cost-effective priorities for global mammal conservation. Proc. Natl Acad. Sci. USA 105, 11 446–11 450 10.1073/pnas.0707157105 (doi:10.1073/pnas.0707157105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodrigues A. S. L., et al. 2011. Complete, accurate, mammalian phylogenies aid conservation planning, but not much. Phil. Trans. R. Soc. B 366, 2652–2660 10.1098/rstb.2011.0104 (doi:10.1098/rstb.2011.0104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burgess N. D., de Klerk H., Fjeldsa J., Rahbek C. 2000. A preliminary assessment of congruence between biodiversity patterns in Afrotropical forest birds and forest mammals. Ostrich 71, 286–291 10.1080/00306525.2000.9639929 (doi:10.1080/00306525.2000.9639929) [DOI] [Google Scholar]
- 66.Boitani L., Corsi F., Reggiani G., Sinibaldi J., Trapanese P. 1999. A databank for the conservation and management of the African mammals. Roma, Italy: Istituto di Ecologia Applicata [Google Scholar]
- 67.Burgess N. D., Rahbek C., Wugt Larsen F., William P., Balmford A. 2002. How much of the vertebrate diversity of sub-Saharan Africa is catered for by recent conservation proposals? Biol. Conserv. 107, 327–339 10.1016/S0006-3207(02)00071-X (doi:10.1016/S0006-3207(02)00071-X) [DOI] [Google Scholar]
- 68.Balmford A., Moore J. L., Brooks T., Burgess N., Hansen L. A., Williams P., Rahbek C. 2001. Conservation conflicts across Africa. Science 291, 2616–2619 10.1126/science.291.5513.2616 (doi:10.1126/science.291.5513.2616) [DOI] [PubMed] [Google Scholar]
- 69.Wilson K., et al. 2010. Conserving biodiversity in production landscapes. Ecol. Appl. 20, 1721–1732 10.1890/09-1051.1 (doi:10.1890/09-1051.1) [DOI] [PubMed] [Google Scholar]
- 70.Cowling R. M., Knight A. T., Faith D. P., Ferrier S., Lombard A. T., Driver A., Rouget M., Maze K., Desmet P. G. 2004. Nature conservation requires more than a passion for species. Conserv. Biol. 18, 1674–1676 10.1111/j.1523-1739.2004.00296.x (doi:10.1111/j.1523-1739.2004.00296.x) [DOI] [Google Scholar]
- 71.Rondinini C., Rodrigues A. S. L., Boitani L. 2011. The key elements of a comprehensive global mammal conservation strategy. Phil. Trans. R. Soc. B 366, 2591–2597 10.1098/rstb.2011.0111 (doi:10.1098/rstb.2011.0111) [DOI] [PMC free article] [PubMed] [Google Scholar]