Abstract
Motivation: Advances in high-throughput genotyping and next generation sequencing have generated a vast amount of human genetic variation data. Single nucleotide substitutions within protein coding regions are of particular importance owing to their potential to give rise to amino acid substitutions that affect protein structure and function which may ultimately lead to a disease state. Over the last decade, a number of computational methods have been developed to predict whether such amino acid substitutions result in an altered phenotype. Although these methods are useful in practice, and accurate for their intended purpose, they are not well suited for providing probabilistic estimates of the underlying disease mechanism.
Results: We have developed a new computational model, MutPred, that is based upon protein sequence, and which models changes of structural features and functional sites between wild-type and mutant sequences. These changes, expressed as probabilities of gain or loss of structure and function, can provide insight into the specific molecular mechanism responsible for the disease state. MutPred also builds on the established SIFT method but offers improved classification accuracy with respect to human disease mutations. Given conservative thresholds on the predicted disruption of molecular function, we propose that MutPred can generate accurate and reliable hypotheses on the molecular basis of disease for ∼11% of known inherited disease-causing mutations. We also note that the proportion of changes of functionally relevant residues in the sets of cancer-associated somatic mutations is higher than for the inherited lesions in the Human Gene Mutation Database which are instead predicted to be characterized by disruptions of protein structure.
Availability: http://mutdb.org/mutpred
Contact: predrag@indiana.edu; smooney@buckinstitute.org
1 INTRODUCTION
With rapid advances in high-throughput genotyping and next-generation sequencing technologies, a vast amount of genetic variation has been discovered and deposited in databases, with much more still to come (Mooney, 2005). Currently, there are over 50 000 coding region mutations, causing or associated with human genetic disease, listed in the Human Gene Mutation Database (HGMD) (http://www.hgmd.org; Stenson et al., 2009), of which ∼40 000 are amino acid substitutions. Amino acid substitutions have the potential to alter the function of their corresponding protein, either directly or via disruption of structure. Hence they are of particular interest as candidates for further experimental assessment (Cargill et al., 1999; Ng and Henikoff, 2006).
There is a need to effectively and efficiently identify functionally important variants which may be deleterious or disease-causing and to identify their molecular effects. For this purpose, a number of computational methods, based on amino acid sequence, structure and evolutionary information, have been proposed (Hon et al., 2008; Karchin, 2009; Mooney, 2005; Steward et al., 2003). One of the earliest tools developed in this area, Sorting Intolerant From Tolerant (SIFT), uses sequence homology to classify amino acid substitutions as tolerated or deleterious (Ng and Henikoff, 2001, 2003). Owing to its impressive predictive power and simplicity, SIFT continues to be used as a benchmark for other methods and approaches (Bao and Cui, 2005; Bromberg and Rost, 2007; Chan et al., 2007; Kulkarni et al., 2008). Protein structure information has been incorporated through the rule-based approach of Wang and Moult (2001) and supervised methods by Yue et al. (2005) and Yue and Moult (2006). PolyPhen also exploits sequence conservation, structure, and rich annotations from protein databases to classify damaging amino acid substitutions (Ramensky et al., 2002; Sunyaev et al., 2001). Scores utilizing more sophisticated alignments based on hidden Markov models from protein families were incorporated with the development of the Substitution Position-Specific Evolutionary Conservation (subPSEC) scores in the PANTHER database (Thomas et al., 2003). Features from comparative protein structural models have been incorporated via the LS-SNP approach (Karchin et al., 2005), while the usefulness of ab initio structural models was evaluated by Saunders and Baker (2002). Finally, servers such as SNAP (Bromberg and Rost, 2007), PMUT (Ferrer-Costa et al., 2005), or CanPredict (Kaminker et al., 2007a, b) use a number of sequence-based, structural and evolutionary features, combined with various classification approaches and training sets.
Currently, there are three general areas in need of improvement for the development of better classification approaches. First, experimentally validated and unbiased training sets of disease causing or deleterious mutations need to be developed. Ideally, these data sets would be quantitatively phenotyped at both the organismal and molecular level. Second, new mutational attributes (i.e. beyond sequence composition, structure and evolutionary conservation) that improve classification accuracy and yield hypotheses on molecular mechanisms of disease, need to be identified and characterized. Finally, new computational approaches need to be developed that improve classification accuracy when similar attributes and training sets are used. The second and third areas are the focus of this work.
Although current approaches are valuable in selecting and prioritizing mutations by yielding the most likely disease-causing polymorphisms, few of them provide clues as to the putative molecular mechanisms by which the mutations affect phenotypes (Wang and Moult, 2001; Yue et al., 2005; Yue and Moult, 2006), and even fewer when protein structure is not available. This is a severe limitation since it is unreasonable to expect that a high-resolution 3D structure will be available for all proteins and their mutants, either due to difficulties in structure determination and modeling, or because the proteins are intrinsically disordered (Dunker et al., 2001). Therefore, introducing novel sequence-based methods is important.
Several systematic studies have shown that predicting the molecular underpinnings of disease is feasible in practical terms: for example, by comparing the crystal structures or 3D models between wild-type and mutant proteins, Wang and Moult (2001) were able to catalog amino acid substitutions into five classes according to their effects on molecular function. These classes included changes in protein stability, but also direct changes in protein function via disrupted ligand binding, catalysis, allosteric regulation, and post-translational modifications, without necessarily affecting the stability of the molecule. Gains of N-linked glycosylation sites causatively implicated in disease were studied by Vogt et al. (Vogt et al., 2005, 2007). In our previous work, mutations predicted to cause a gain or loss of phosphorylation target sites were found to be significantly more prevalent among somatic cancer mutations than in control data sets, suggesting that phosphorylation target site mutation is an active general mechanism of dysregulation in cancer (Radivojac et al., 2008).
In this study, we extend our approach to include a number of additional structural and functional properties and show that predictions of the gain and loss of such properties can provide good classification accuracy, but also have the potential to estimate, in probabilistic terms, the underlying biochemical cause of disease.
2 METHODS
2.1 Data sets
A collection of five data sets of human amino acid substitutions were constructed from online databases and the literature (Table 1). Four of these data sets are composed of disease-associated mutations (Cancer, Kinase, Hgmd and SPd), whereas the remaining data set (SPp) contains inherited, putatively neutral polymorphisms. The Cancer data set comprises somatic mutations in genes re-sequenced from 22 cell lines from breast and colorectal cancer tissues (Sjoblom et al., 2006). The Kinase data set contains somatic mutations from sequencing kinase genes from 210 individual tumors (Greenman et al., 2007). Both of these sets are expected to contain mutations that lead to neoplastic progression (drivers) as well as neutral mutations that do not influence tumorigenesis (passengers). Mutations annotated with evidence for being disease-causing in HGMD constitute inherited disease-causing mutations in the HGMD data set. Finally, the Swiss-Prot database (Boeckmann et al., 2003) contains amino acid substitutions that are either disease-causing (SPd) or polymorphic (SPp). The full sequences of proteins harboring these mutations were downloaded and used for attribute generation. Table 1 describes the data sets, excluding the substitutions for which at least one of the attributes could not be computed.
Table 1.
Summary of data sets
| Data set | Substitutions | Proteins | Type |
|---|---|---|---|
| Cancer | 653 | 519 | Disease, somatic |
| Kinase | 422 | 231 | Disease, somatic |
| Hgmd | 26 655 | 1305 | Disease, inherited |
| SPd | 6606 | 680 | Disease, inherited |
| SPp | 23 426 | 8728 | Neutral, inherited |
2.2 Attribute construction
We generated a broad array of attributes from protein sequences and utilized them in classification. These attributes can be grouped into three classes: (i) attributes based on predicted protein structure and dynamics, (ii) attributes based on predicted functional properties and (iii) attributes based on amino acid sequence and evolutionary information. Sequence-based predictions of structural features or protein dynamics included secondary structure (Rost, 1996), solvent accessibility (Rost, 1996), transmembrane helices (Krogh et al., 2001), coiled-coil structure (Delorenzi and Speed, 2002), stability (Capriotti et al., 2005), B-factor (Radivojac et al., 2004), and intrinsic disorder (Peng et al., 2006). Predictions of functional sites included DNA-binding residues (Ahmad et al., 2004), catalytic residues (Xin and Radivojac, unpublished data), calmodulin-binding targets (Radivojac et al., 2006), as well as the phosphorylation (Iakoucheva et al., 2004), methylation (Daily et al., 2005), ubiquitination (Radivojac et al., 2009) and glycosylation (Radivojac, unpublished data) sites. Note that the calmodulin-binding target classifier predicts short structured or loosely structured helical segments within otherwise disordered regions and was used as a substitute for predicting Molecular Recognition Fragments (MoRFs), as proposed by Mohan et al. (2006). Each predictor, with the exception of N-linked glycosylation, was constructed in a supervised learning scenario, whereas the changes of N-linked glycosylation were modeled by simply counting whether a binding motif NX[ST] had been introduced or removed by the amino acid substitution (all structural and functional properties are listed in Table 2). Evolutionary attributes were generated from PSI-BLAST and also included a SIFT score, Pfam profile score (Finn et al., 2008), and transition frequencies. The transition frequencies, as proposed by Bromberg and Rost (2007), measure the likelihood of observing a given mutation in the UniRef80 database and Protein Data Bank.
Table 2.
Structural and functional properties used by MutPred
| Structural properties | Functional properties |
|---|---|
| Secondary structure | DNA-binding residues |
| Solvent accessibility | Catalytic residues |
| Stability | MoRFs |
| Intrinsic disorder | Phosphorylation sites |
| B-factor | Methylation sites |
| Transmembrane helix | Glycosylation sites |
| Coiled-coil structure | Ubiquitination sites |
Most of the computational models used in this study estimate the posterior probability that a residue has a given structural or functional property p, for example that a residue has high helical propensity or that it is post-translationally modified. Then, we can calculate the probability of loss or gain of such a property, i.e. mutant proteins can be predicted to either introduce or eliminate structural or functional properties. More specifically, given a protein sequence s, the probability of the loss of a particular property p at residue si can be expressed as:
| (1) |
where P(p|siw) is the probability of the presence of property p at residue si in the wild-type protein and (1−P(p|sim)) is the probability of absence of p at residue si in the mutant. The two events, corresponding to two physically separate molecules, are considered independent. Similarly, the gain of structural or functional property p can be expressed as:
| (2) |
It is clear from Equations (1) and (2) that the greater the difference between P(p|siw) and P(p|sim), the greater the probability of gain or loss of the property. However, note that a reduction of score from 1.0 to 0.9 corresponds to the probability of loss of 0.1, whilst the reduction of score from 0.5 to 0.4 corresponds to the probability of the loss of property of 0.3.
The gain and loss of a property at residue si does not necessarily suggest that the amino acid substitution occurred at position i. This situation is particularly interesting for single-residue functional sites such as post-translational modifications, because impacts of substitutions of neighboring residues cannot be easily detected even if the functional site is known. We refer to such cases as functional neighborhood mutations, whereas the direct changes of functional sites are referred to as functional site mutations. In general, we expect that the probability of gain or loss of function at si will be inversely correlated with the distance of the substitution site from si (here we consider residues between positions −5 and +5 from the substitution site). Thus, the largest impact on protein function is likely to be for the functional site mutations. In the case of a loss of property, this results in P(p|sim)=0 and therefore the probability that the wild-type sequence is functional at si equals the probability that the function will be lost. An example of such a situation is when a phosphorylatable serine residue is substituted by a non-phosphorylatable residue such as alanine. Similarly, in the case of a gain of function, the probability that si is non-functional in the wild-type equals 1, i.e. P(p|siw)=0. Hence, for functional site mutations, the prediction of protein property in the mutated and wild-type protein also predicts the gain or loss of that property, respectively. In total, seven structural and seven functional properties were used in this study (Table 2).
2.3 Classification models
To discriminate between disease-associated mutations and neutral polymorphisms, we applied and compared support vector machine (SVM) and random forest (RF) classifiers. SVM is a machine learning model that maximizes the margin of separation between examples of two classes projected into high-dimensional space (Vapnik, 1998), and have previously been applied successfully to mutation data (Karchin et al., 2005; Krishnan and Westhead, 2003; Yue and Moult, 2006). In the current study, we used SVMperf v2.50 (Joachims, 2005) with linear and non-linear kernels and kept the capacity parameter at its default value. SVMperf was trained to maximize the area under the ROC curve. Another machine learning technique used here was random forests (Breiman, 2001), which became popular and has been extensively used in bioinformatics applications in part due to its simplicity and interpretability (Bao and Cui, 2005; Kaminker et al., 2007a). In the training stage, an RF builds a committee of decision trees and in the test stage it averages the results from all trees as the final output. In the tree-growing procedure, a random subset of attributes is selected at each node and the best one is used for splitting. The R package randomForest v4.5-30 was used for this purpose.
2.4 Performance evaluation
To measure the ability of classifiers to discriminate between disease and neutral substitutions, we plotted receiver operating characteristic (ROC) curves and calculated the area under the ROC curve (AUC). ROC curve shows the true positive rate (or sensitivity, sn) as a function of the false positive rate. The false positive rate is typically denoted as 1−sp, where specificity (sp) is the accuracy on the negative data points. In this study, disease-associated mutations were considered to be positive examples, whereas the neutral polymorphisms were negative. For the classification using Cancer data set, the set of neutral polymorphisms was constructed by retaining only those substitutions found in cancer-associated proteins, as provided by the Cancer Gene Census (Futreal et al., 2004). Similarly, only kinases from the BRENDA database (Chang et al., 2009) were used to select a subset of neutral polymorphisms in evaluating the performance on Kinase. In total, the number of neutral polymorphisms from the Cancer and Kinase data sets was 1625 (480 proteins) and 1803 (394 proteins), respectively. Classification models were evaluated using per-protein 10-fold cross-validation.
2.5 Predicting molecular mechanism of disease
Owing to unknown class priors, it is not possible to estimate the precision of most structural and functional predictors accurately. Thus, our predictions of molecular mechanisms of disease are based upon the assumption that the majority of phenotypically neutral polymorphisms are unlikely to affect protein structure or function significantly. In such a situation, a distribution of scores can be created for each gain and loss of property using the data set SPp (or its filtered versions in the cases of Cancer and Kinase data). Then, each gain or loss score sc from Equations (1) and (2) of a disease mutation can be assigned a P-value P, i.e. the probability that a randomly selected neutral polymorphism will have the same score sc or higher. We refer to such P-values as property scores.
3 RESULTS
3.1 Discrimination between disease and neutral mutations using machine learning
The performance of individual classifiers is summarized in Table 3. For each classifier, we report sensitivity, specificity, accuracy and the area under the ROC curve. To alleviate the potential negative influence of unbalanced data between disease and neutral polymorphisms on classification performance, we also trained classifiers on equal-sized disease and neutral sets. We found that balancing the training data had almost no effect on the area under the ROC curve, but that it slightly impacted the classification accuracy. Since SIFT is an established method for distinguishing deleterious from putatively neutral polymorphisms and is easily portable, we used it for benchmarking. Thus, the AUC of SIFT scores provide our baseline measure.
Table 3.
Performance accuracy of different classification models on four data sets containing disease-associated amino acid substitutions versus the data set of inherited polymorphisms
| Data set | Sensitivity (%) |
Specificity (%) |
Accuracy (%) |
AUC (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SIFT | SVM | RF | SIFT | SVM | RF | SIFT | SVM | RF | SIFT | SVM | RF | |
| Cancer | 50.8 | 43.1 | 57.5 | 67.1 | 77.1 | 70.1 | 62.4 | 67.4 | 63.8 | 61.4 | 62.0 | 68.6 |
| Kinase | 51.7 | 37.6 | 55.1 | 61.8 | 80.1 | 69.8 | 59.9 | 72.1 | 62.4 | 58.3 | 59.3 | 66.1 |
| Hgmd | 70.8 | 75.6 | 72.6 | 73.9 | 76.2 | 80.9 | 72.3 | 75.9 | 76.5 | 77.6 | 82.4 | 83.5 |
| SPd | 74.1 | 77.6 | 76.5 | 73.9 | 78.7 | 81.4 | 74.0 | 78.4 | 80.3 | 79.1 | 84.6 | 85.3 |
Compared are SIFT, a linear SVM and a RF approach using 1000 trees. Bold fonts indicate the best performing models (P<0.001 in each pairwise comparison with SIFT; t-test based on 10 repeats).
Unsurprisingly, SIFT scores alone worked well on the Hgmd, and SPd data sets (Table 3). The relatively inferior performance of SIFT on Cancer and Kinase data suggests large differences in evolutionary conservation between the amino acid residues harboring inherited and somatic mutations. The performance of other classifiers also resulted in a relatively low accuracy on the Cancer and Kinase data sets; this may be due to the fact that somatic mutations in cancer are likely to contain a large number of so-called passenger mutations (Futreal et al., 2005).
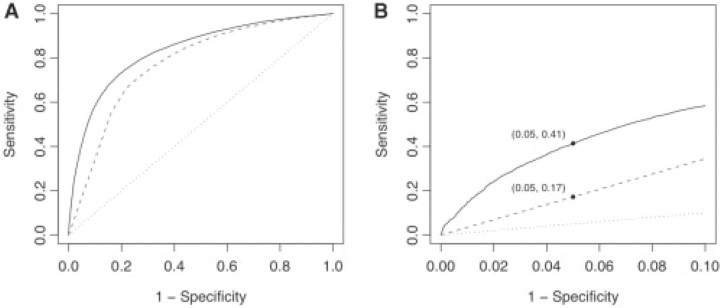
The AUCs of the SVM across all data sets, were 3.0 percentage points greater than those of SIFT, suggesting that a linear SVM had a limited ability to extract useful information from additional features. With limited parameter variation, non-linear SVMs were even less successful (data not shown). RFs outperformed SIFT on all data sets by 6.8 percentage points and SVMs by 3.8 percentage points. The full ROC curves for data set Hgmd are shown in Figure 1A. Figure 1B presents the same curve in the range of false positive rates from 0 to 0.1. Note that for the specificity level of 0.95, the sensitivity of MutPred and SIFT were 0.414 and 0.172, respectively. Similarly, sensitivities on Cancer were 0.193 versus 0.087 and on Kinase were 0.160 versus 0.076. Thus, MutPred appears to be well suited to prioritization of those amino acid substitutions which are most likely to be involved in disease.
Fig. 1.
ROC curves for Hgmd data set. (A) full curves and (B) curves in the false positive rate range of [0, 0.1]. The solid black curve represents the MutPred general score, the dashed gray curve represents SIFT, and the dotted line is the random model.
It is noteworthy that the classification performance using RF models with more than 1000 trees was rather stable. Thus, 1000 trees were sufficient for the purpose of the current study. Since RFs performed better than the SVMs, further analyses and our final predictive model, MutPred, are based on these classifiers. We refer to the output of the RF classifier as the MutPred general score.
3.2 Assessing the molecular mechanism of disease
Since there are relatively few amino acid substitutions for which the molecular mechanism of disease is known, it is not currently possible to directly evaluate the accuracy of approaches designed to predict such mechanisms. Thus, our approach relies on the postulate that inherited polymorphic sites have little or no influence on the structure and function of proteins. Under this assumption, one can generate a distribution of scores for each gain or loss of property over neutral polymorphisms and output the P-value as a quantitative score for each property prediction.
We consider a prediction of the underlying mechanism of disease to be an actionable hypothesis if the MutPred general score is >0.5 and the property score P < 0.05. The prediction is considered a confident hypothesis if the MutPred general score corresponds to a specificity of 0.95 (false positive rate of 0.05) as estimated during the performance evaluation on the Hgmd data and the individual property score P < 0.05. Finally, a prediction is considered to be very confident if the general score corresponds to a specificity of 0.95 and the gain/loss property score P < 0.01. Note that a separate null hypothesis distribution was created for each individual gain/loss of property attribute. Ideally, one should use a false discovery rate to control for the number of false predictions [fdr = n0 · (1 − sp)/(n0 · (1 − sp)+n1 · sn), where n0 is the number of negative data points and n1 is the number of positive data points]. However, since the number of truly positive sites (n1) in the human proteome is unknown, an accurate estimate of the false discovery rate is not currently possible.
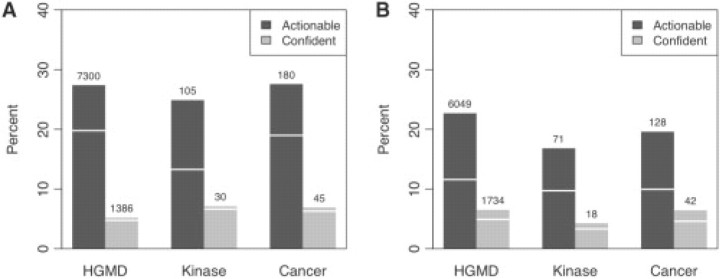
In Figure 2, we show the percentage and number of human amino acid substitutions with a predicted mechanism of disease as a consequence of gain or loss of functional (Fig. 2A) and structural (Fig. 2B) properties. We show the number of mutations for which actionable and confident hypotheses can be generated for all data sets, except SPd which contained 79% of mutations already available in Hgmd. Our results indicate that some explanation of the mechanism of disease may be created in as many as 41.1% of mutations in HGMD, while confident hypotheses may be generated in 11.2% of cases (111 confident predictions overlapped between gain/loss of structure and function). Confident hypotheses can be generated for 11.6% of disease-causing mutations in Swiss-Prot, as well as 10.2 and 12.1% of somatic mutations in Kinase and Cancer data sets. Finally, we note that very confident hypotheses can be generated for 5.5% of mutations in both HGMD and Swiss-Prot, and for 1.2% of substitutions in somatic mutation data sets.
Fig. 2.
The percentage and number of amino acid substitutions for (A) functional properties and (B) structural properties, that represent actionable (dark gray) and confident (light gray) hypotheses on the molecular cause of disease on three data sets (SPd is omitted due to large overlap with Hgmd). White line indicates the number of mutations predicted to be influencing more than one functional or structural property.
3.3 Importance of gain/loss of properties on inherited and somatic amino acid substitutions
To investigate the influence of individual attributes on the sets of inherited and somatic mutations, we compared the most common actionable hypotheses of the molecular mechanism of disease in the Hgmd, Cancer, and Kinase data sets. We selected Hgmd as a representative of the set of inherited mutations owing to its large size and the fact that most of the mutations from SPd are already listed in Hgmd (79%).
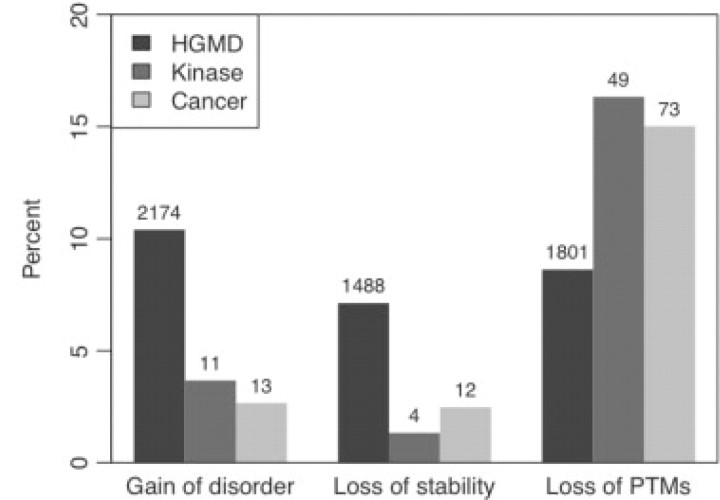
The most common actionable hypotheses that characterized inherited, but not somatic mutations, were order-to-disorder transition (10.2% of all actionable hypotheses in Hgmd, 2.7% in Cancer, 3.7% in Kinase) and loss of stability (7.1% in Hgmd, 2.5% in Cancer, 1.3% in Kinase), thereby emphasizing the disruption of structure in monogenic disease. By contrast, Cancer and Kinase data were enriched in the disruption of post-translational modifications (25.0% in Hgmd, 31.1% in Cancer, 39.0% in Kinase). Using a complete set of 30 categories, we determined that the differences between the Hgmd and Cancer (P=5.0 × 10−5; χ2 test) and Kinase (P=5.0 × 10−5; χ2 test) data sets were statistically significant. Figure 3 shows the most significant differences among the three sets.
Fig. 3.
The percentage of actionable hypotheses on Hgmd, Kinase, and Cancer data sets. P-values are calculated between Hgmd versus Kinase and Hgmd versus Cancer: gain of disorder (3.4×10−9; 2.7 ×10−22), loss of stability (1.0×10−15; 3.4×10−10), loss of post-translationally modified (PTM) target sites (3.8×10−4; 1.0×10−4).
To additionally validate the strength of gain and loss of structural/functional properties, we trained a prediction model based only on these attributes with the goal of discriminating between amino acid substitutions on which the overall model had score >0.5 and neutral substitutions. The area under the ROC curve using these attributes ranged between 71.0% (SPd) and 79.1% (Hgmd), indicating, somewhat surprisingly, that the gain and loss of structural/functional properties alone have good discriminatory power and are suitable for the purpose of assessing the mechanism of disease.
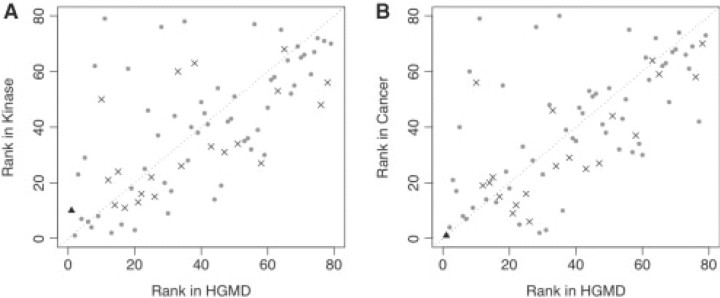
In Figure 4, we show the relative ranking of various attributes on different training data, using the gini index. Loss and gain of structural and functional properties are indicated by ×'s, whereas SIFT is represented by a black triangle. As expected, the SIFT score was the single highest-ranking attribute in Hgmd. However, there were significant differences between the inherited and somatic amino acid substitutions. In the somatic mutations data sets, the gain/loss of most structural/functional properties were higher ranked in Cancer and Kinase than in Hgmd (presented as ×'s below the diagonal line).
Fig. 4.
Relative ranking of attributes across the Hgmd and Kinase (A) and Hgmd and Cancer (B) data sets. Gain and loss of structural and functional properties are represented by ×'s. SIFT is represented by a black triangle.
3.4 Case studies
We searched the literature and found direct experimental evidence for several disease-causing mutations with a high score for a gain or loss of structural or functional properties. The first example is phosphatase and tensin homolog (gene name: PTEN), a tumor suppressor gene that negatively regulates the AKT/PKB signaling pathway. PTEN acts as both dual-specificity protein phosphatase and lipid phosphatase that is considered to be critical for its suppressor function. Many PTEN gene mutations were identified and associated with classical Cowden syndrome (CS), Bannayan-Riley-Ruvalcaba syndrome, Proteus syndrome and Proteus-like syndrome (Eng, 2003). PTEN mutations, especially those located in exons 5, 7 and 8, have been found in 80% of CS patients (Marsh et al., 1998, 1999). Exon 5, which encodes the phosphatase core motif, accounts for 40% all PTEN mutations (Eng, 2003) and includes catalytic residues C124 and D92 (Lee et al., 1999). Substitutions C124R and D92E, listed in HGMD, are associated with Cowden syndrome and are known to affect PTEN's phosphatase function (Eng, 2003). The MutPred general score for C124R was 0.61 (SIFT score was 0; SIFT predictions below 0.05 are considered positive) and the catalytic residue predictor yielded a loss of property prediction of 0.57, which resulted in the property score P=0.048 for the loss of catalytic residue propensity. Substitution D92E, on the other hand, was attributed a MutPred score of 0.98 (SIFT score 0) and a loss of catalytic residue score of 0.42 (P=0.18). Although D92E is not considered an actionable hypothesis by our criteria, its strong score may serve as an indicator that the thresholds selected in Section 3.3 are stringent.
The second example involves human PTP synthase (gene name: PTS), a carbon-oxygen lyase that catalyzes triphosphate elimination yielding 6-pyruvoyltetrahydrobiopterin. PTP synthase has 19 documented amino acid substitutions in Swiss-Prot release 56.6 and defects in PTP synthase are known to be associated with hyperphenylalaninemia (HPA). HPA is an autosomal recessive disorder with serious neurological symptoms that represents a mild form of phenylketonuria. The mutation R16C is documented to diminish phosphorylation of S19 by PKG and also to cause HPA (Oppliger et al., 1995; Scherer-Oppliger et al., 1999; Thony et al., 1994). The MutPred general score for this mutation is 0.87 (SIFT score 0.01) whereas the phosphorylation score for S19 decreased from 0.85 to 0.49 (loss of phosphorylation score of 0.43; P=0.006 over all functional neighborhood mutations; P=0.058 over all functional site and neighborhood mutations) upon introducing mutation R16C, as predicted by DisPhos (Iakoucheva et al., 2004). Probabilistically quantifying such a reduction in phosphorylation likelihood is difficult even if it is known that S19 is a phosphorylation site. Therefore, the use of computational models is highly important.
4 DISCUSSION
Here we introduced and evaluated a new computational model that builds upon SIFT by explicitly estimating probabilities of affecting various structural and functional properties, such as the loss of helical propensity, catalytic activity or post-translational modifications. Over the last decade, several methods have been proposed and tested to predict whether a particular amino acid substitution affects protein function leading to an altered phenotype. These approaches are reasonably accurate and useful. However, they do not generate hypotheses relating to the biochemical cause of disease. In our model, the loss and gain of each structural and functional property was directly modeled via posterior probabilities, thereby enabling us to directly estimate the contribution of a gain/loss of a given property in order to deduce the underlying mechanism of disease. In this way, our method indirectly exploits the structural and functional data available for functional prediction, effectively enlarging the training data sets beyond the characterized disease-causing events.
Attributes representing predictions of gain/loss of structural and functional properties also contributed to an improved classification performance over SIFT. The increase in classification performance ranged between 5.9 (Hgmd) and 7.8 (Kinase) percentage points. Hgmd was eventually used to train the final model, MutPred. The performance of random forest algorithms was better than that of support vector machines, probably due to the explicit modeling of the gain/loss of structural and functional properties which can be more easily exploited by decision trees.
The good prediction accuracy of MutPred on Cancer and Kinase data indicates that somatic sites can be predicted when compared to inherited polymorphisms, even when the final model was trained on Hgmd data alone (data not shown). This was not surprising since the amino acid residues which harbor somatic mutations are expected to be under a different set of evolutionary constraints than those harboring inherited polymorphisms. However, we believe that using molecular features to identify causative somatic mutations (drivers) may be more difficult than for inherited mutations because passenger mutations can introduce or disrupt functional sites even although they may not exert an effect within the context of a particular cell or tissue type. For example, amino acid substitutions in a kinase catalytic domain that is not expressed could be predicted to be damaging, but would in practice have no observable phenotypic effect since the protein product would not be present in the cell. To allow for tissue differences and particular predicted molecular events, future improvements in the classification models should incorporate more detailed information on the particular context of disease.
Based on sample data from the Protein Data Bank, Wang and Moult (2001) developed a rule-based approach to characterize molecular causes of disease. They also provided first assessments of the ways in which disease-causing mutations might affect protein function: >80% of inherited mutations were estimated to disrupt protein stability as a proxy to changed function, whereas <10% could be attributed to direct changes of functional residues. Interestingly, using a very different approach, we arrived at similar conclusions, with the advantage of being able to predict such events from sequence. Our results are also in agreement with the study of Torkamani and Schork (2007), who achieved improved prediction accuracy on a kinase-specific data set compared to SIFT. Furthermore, we find that somatic mutations, although predictable, may affect cellular functions in ways that are subtler and more diverse than for inherited disease mutations.
In conclusion, we used the most comprehensive data set of disease-associated mutations and incorporated new attributes for classification that directly model the gain/loss of structural and functional properties. We believe that this type of probabilistic evidence is informative and complements evidence that a conserved residue is disrupted.
ACKNOWLEDGEMENTS
The authors would like to thank Prateek Kumar and Pauline Ng from the J. Craig Venter Institute for their help with the SIFT tool. They also thank the anonymous reviewers on their comments and suggestions that improved the quality of this work.
Funding: National Institutes of Health R01LM009722 to S.D.M. and National Science Foundation DBI-0644017 to P.R.
Conflict of Interest: none declared.
REFERENCES
- Ahmad S, et al. Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. Bioinformatics. 2004;20:477–486. doi: 10.1093/bioinformatics/btg432. [DOI] [PubMed] [Google Scholar]
- Bao L, Cui Y. Prediction of the phenotypic effects of non-synonymous single nucleotide polymorphisms using structural and evolutionary information. Bioinformatics. 2005;21:2185–2190. doi: 10.1093/bioinformatics/bti365. [DOI] [PubMed] [Google Scholar]
- Boeckmann B, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E, et al. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill M, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Chan PA, et al. Interpreting missense variants: comparing computational methods in human disease genes CDKN2A, MLH1, MSH2, MECP2, and tyrosinase (TYR) Hum. Mutat. 2007;28:683–693. doi: 10.1002/humu.20492. [DOI] [PubMed] [Google Scholar]
- Chang A, et al. BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res. 2009;37:D588–D592. doi: 10.1093/nar/gkn820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily KM, et al. IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB). San Diego, California, USA: 2005. Intrinsic disorder and protein modifications: building an SVM predictor for methylation; pp. 475–481. [Google Scholar]
- Delorenzi M, Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18:617–625. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
- Dunker AK, et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Eng C. PTEN: one gene, many syndromes. Hum. Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- Ferrer-Costa C, et al. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, et al. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, et al. Somatic mutations in human cancer: insights from resequencing the protein kinase gene family. Cold Spring Harb. Symp. Quant. Biol. 2005;70:43–49. doi: 10.1101/sqb.2005.70.015. [DOI] [PubMed] [Google Scholar]
- Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon LS, et al. Computational approaches for predicting causal missense mutations in cancer genome projects. Curr. Bioinformatics. 2008;3:46–55. [Google Scholar]
- Iakoucheva LM, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachims T. International Conference on Machine Learning (ICML). Bonn, Germany, New York: ACM Press; 2005. A support vector method for multivariate performance measures; pp. 377–384. [Google Scholar]
- Kaminker JS, et al. CanPredict: a computational tool for predicting cancer-associated missense mutations. Nucleic Acids Res. 2007a;35:W595–W598. doi: 10.1093/nar/gkm405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JS, et al. Distinguishing cancer-associated missense mutations from common polymorphisms. Cancer Res. 2007b;67:465–473. doi: 10.1158/0008-5472.CAN-06-1736. [DOI] [PubMed] [Google Scholar]
- Karchin R. Next generation tools for the annotation of human SNPs. Brief Bioinformatics. 2009;10:35–52. doi: 10.1093/bib/bbn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchin R, et al. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- Krishnan VG, Westhead DR. A comparative study of machine-learning methods to predict the effects of single nucleotide polymorphisms on protein function. Bioinformatics. 2003;19:2199–2209. doi: 10.1093/bioinformatics/btg297. [DOI] [PubMed] [Google Scholar]
- Krogh A, et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kulkarni V, et al. Exhaustive prediction of disease susceptibility to coding base changes in the human genome. BMC Bioinformatics. 2008;9(Suppl. 9):S3. doi: 10.1186/1471-2105-9-S9-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- Marsh DJ, et al. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum. Mol. Genet. 1998;7:507–515. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- Marsh DJ, et al. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum. Mol. Genet. 1999;8:1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- Mohan A, et al. Analysis of molecular recognition features (MoRFs) J. Mol. Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- Mooney SD. Bioinformatics approaches and resources for single nucleotide polymorphism functional analysis. Brief Bioinformatics. 2005;6:44–56. doi: 10.1093/bib/6.1.44. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Oppliger T, et al. Structural and functional consequences of mutations in 6-pyruvoyltetrahydropterin synthase causing hyperphenylalaninemia in humans. Phosphorylation is a requirement for in vivo activity. J. Biol. Chem. 1995;270:29498–29506. doi: 10.1074/jbc.270.49.29498. [DOI] [PubMed] [Google Scholar]
- Peng K, et al. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, et al. Protein flexibility and intrinsic disorder. Protein Sci. 2004;13:71–80. doi: 10.1110/ps.03128904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, et al. Calmodulin signaling: analysis and prediction of a disorder-dependent molecular recognition. Proteins. 2006;63:398–410. doi: 10.1002/prot.20873. [DOI] [PubMed] [Google Scholar]
- Radivojac P, et al. Gain and loss of phosphorylation sites in human cancer. Bioinformatics. 2008;24:i241–i247. doi: 10.1093/bioinformatics/btn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, et al. Identification, analysis and prediction of protein ubiquitination sites. Proteins. 2009 doi: 10.1002/prot.22555. [Epub ahead of print, doi:10.1002/prot.22555, July 22, 2009] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V, et al. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- Saunders CT, Baker D. Evaluation of structural and evolutionary contributions to deleterious mutation prediction. J. Mol. Biol. 2002;322:891–901. doi: 10.1016/s0022-2836(02)00813-6. [DOI] [PubMed] [Google Scholar]
- Scherer-Oppliger T, et al. Serine 19 of human 6-pyruvoyltetrahydropterin synthase is phosphorylated by cGMP protein kinase II. J. Biol. Chem. 1999;274:31341–31348. doi: 10.1074/jbc.274.44.31341. [DOI] [PubMed] [Google Scholar]
- Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Stenson PD, et al. The human gene mutation database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward RE, et al. Molecular basis of inherited diseases: a structural perspective. Trends Genet. 2003;19:505–513. doi: 10.1016/S0168-9525(03)00195-1. [DOI] [PubMed] [Google Scholar]
- Sunyaev S, et al. Prediction of deleterious human alleles. Hum. Mol. Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thony B, et al. Hyperphenylalaninemia due to defects in tetrahydrobiopterin metabolism: molecular characterization of mutations in 6-pyruvoyl-tetrahydropterin synthase. Am. J. Hum. Genet. 1994;54:782–792. [PMC free article] [PubMed] [Google Scholar]
- Torkamani A, Schork NJ. Accurate prediction of deleterious protein kinase polymorphisms. Bioinformatics. 2007;23:2918–2925. doi: 10.1093/bioinformatics/btm437. [DOI] [PubMed] [Google Scholar]
- Vapnik V. Statistical Learning Theory. New York: John Wiley & Sons; 1998. [Google Scholar]
- Vogt G, et al. Gains of glycosylation comprise an unexpectedly large group of pathogenic mutations. Nat. Genet. 2005;37:692–700. doi: 10.1038/ng1581. [DOI] [PubMed] [Google Scholar]
- Vogt G, et al. Gain-of-glycosylation mutations. Curr. Opin. Genet. Dev. 2007;17:245–251. doi: 10.1016/j.gde.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Wang Z, Moult J. SNPs, protein structure, and disease. Hum. Mutat. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- Yue P, Moult J. Identification and analysis of deleterious human SNPs. J. Mol. Biol. 2006;356:1263–1274. doi: 10.1016/j.jmb.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Yue P, et al. Loss of protein structure stability as a major causative factor in monogenic disease. J. Mol. Biol. 2005;353:459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]