Abstract
Previous genome-wide association studies have identified two independent variants in HNF1B as susceptibility loci for prostate cancer risk. To fine-map common genetic variation in this region, we genotyped 79 single nucleotide polymorphisms (SNPs) in the 17q12 region harboring HNF1B in 10 272 prostate cancer cases and 9123 controls of European ancestry from 10 case–control studies as part of the Cancer Genetic Markers of Susceptibility (CGEMS) initiative. Ten SNPs were significantly related to prostate cancer risk at a genome-wide significance level of P < 5 × 10−8 with the most significant association with rs4430796 (P = 1.62 × 10−24). However, risk within this first locus was not entirely explained by rs4430796. Although modestly correlated (r2= 0.64), rs7405696 was also associated with risk (P = 9.35 × 10−23) even after adjustment for rs4430769 (P = 0.007). As expected, rs11649743 was related to prostate cancer risk (P = 3.54 × 10−8); however, the association within this second locus was stronger for rs4794758 (P = 4.95 × 10−10), which explained all of the risk observed with rs11649743 when both SNPs were included in the same model (P = 0.32 for rs11649743; P = 0.002 for rs4794758). Sequential conditional analyses indicated that five SNPs (rs4430796, rs7405696, rs4794758, rs1016990 and rs3094509) together comprise the best model for risk in this region. This study demonstrates a complex relationship between variants in the HNF1B region and prostate cancer risk. Further studies are needed to investigate the biological basis of the association of variants in 17q12 with prostate cancer.
INTRODUCTION
Of all cancers, prostate cancer is one of the most heritable with genetic factors estimated to account for 42% of the risk (1). Genome-wide association studies (GWAS) have been highly successful in discovering susceptibility loci for prostate cancer and at least 30 loci have been identified to date (2–15). One of the earliest loci to be discovered for prostate cancer was a variant, rs4430796, in HNF1B at chromosome 17q12 in men of European background (6). In a subsequent GWAS in Japanese men, the same locus was identified (15). A second independent variant, rs11649743, located at chromosome 17q12 and separated by a recombination hotspot from the first variant, was subsequently found to be associated with risk (10). The HNF1B locus as well as two other prostate cancer susceptibility loci, chromosome 7p15.2 (JAZF1) and chromosome 2p21 (THADA), have also been shown to be associated with diabetes risk (6,16). Although epidemiologic studies have shown that diabetes is inversely associated with prostate cancer (17), variants in HNF1B and JAZF1 do not explain the association between diabetes and prostate cancer (18).
The strongest variants associated with prostate cancer risk at chromosome 17q12 localize to a region that harbors HNF1B, a gene that encodes a POU homeodomain-containing transcription factor. POU transcription factors help regulate the development of neuroendocrine organs, and HNF1B has been shown to play a regulatory role in nephron and pancreas development (19,20). Rare mutations in HNF1B have been associated with maturity-onset diabetes of the young type 5 (MODY5), kidney disorders, pancreatic atrophy, and genital malformations (21,22). Although the biological mechanism by which HNF1B may affect prostate cancer risk has not been elucidated, differential expression of HNF1B has been associated with prostate cancer recurrence (23).
To further characterize genetic variation in the HNF1B region and the risk of prostate cancer, we conducted a large-scale fine mapping study using tag single nucleotide polymorphisms (SNPs) based on HapMap data in 10 272 prostate cancer cases and 9123 controls of European ancestry from 10 case–control studies as part of the Cancer Genetic Markers of Susceptibility (CGEMS) initiative. A total of 79 SNPs in the HNF1B region that were genotyped and passed quality control criteria were analyzed in this study.
RESULTS
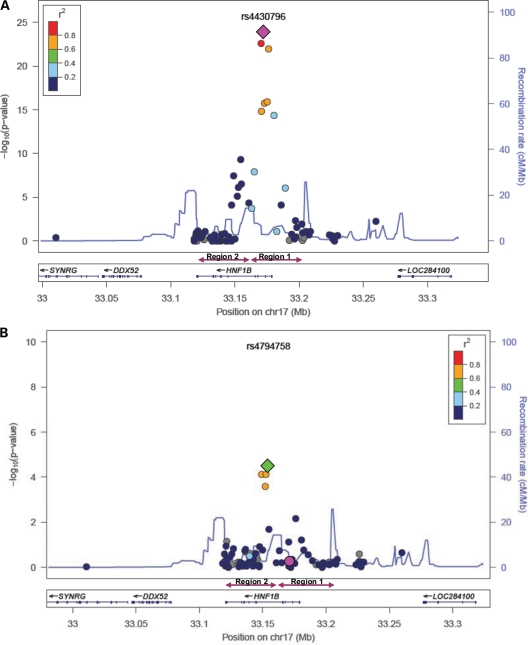
We analyzed 79 SNPs located in a 249 kb region surrounding HNF1B (chromosome 17: 33,010,707–33,259,778) in 10 272 prostate cancer cases and 9123 controls from 10 studies (Supplementary Material, Table S1). Ten SNPs were significantly associated with prostate cancer risk below the threshold of genome-wide significance (P < 5 × 10−8); the most significant association observed was the previously identified SNP rs4430796 (P = 1.62 × 10−24) (Table 1, Fig. 1A). Eight of the 10 significant SNPs were located in the first HNF1B region associated with prostate cancer, and all eight were highly correlated in controls with D′ ≥ 0.6 and pairwise r2 values between 0.13 and 0.94. However, risk within this first locus was not entirely explained by rs4430796. Although modestly correlated (r2= 0.64), rs7405696 was also associated with prostate cancer risk (P = 9.35 × 10−23). After conditioning on rs4430796, rs7405696 retained an association with risk (P = 0.007). None of the other SNPs in region 1 remained associated with risk after adjustment for rs4430796 (Supplementary Material, Table S2).
Table 1.
SNPs in the HNF1B region that were associated with prostate cancer at genome-wide significance levels (P < 5.0 × 10−8)
| SNP | Base pair position | Region | Minor allele | Other allele | Minor allele frequency: cases | Minor allele frequency: controls | No. of cases | No. of controls | OR (95% CI)a | P-value | Heterogeneity P | I2 (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs4430796 | 33172153 | 1 | G | A | 0.420 | 0.473 | 10220 | 9073 | 0.81 (0.77–0.84) | 1.62 × 10−24 | 0.64 | 0 |
| rs2005705 | 33170413 | 1 | T | C | 0.394 | 0.445 | 10268 | 9120 | 0.81 (0.78–0.85) | 2.54 × 10−23 | 0.96 | 0 |
| rs7405696 | 33176148 | 1 | C | G | 0.485 | 0.433 | 10258 | 9117 | 1.23 (1.18–1.28) | 9.35 × 10−23 | 0.16 | 31.5 |
| rs7501939 | 33175269 | 1 | T | C | 0.351 | 0.394 | 10247 | 9100 | 0.84 (0.80–0.87) | 1.22 × 10−16 | 0.56 | 0 |
| rs4239217 | 33173100 | 1 | G | A | 0.352 | 0.395 | 10266 | 9120 | 0.84 (0.80–0.87) | 1.57 × 10−16 | 0.77 | 0 |
| rs757210 | 33170628 | 1 | A | G | 0.326 | 0.366 | 10134 | 8974 | 0.84 (0.80–0.88) | 1.39 × 10−15 | 0.95 | 0 |
| rs3760511 | 33180426 | 1 | C | A | 0.379 | 0.339 | 10261 | 9116 | 1.19 (1.14–1.24) | 4.45 × 10−15 | 0.33 | 12.1 |
| rs4794758 | 33154541 | 2 | T | C | 0.247 | 0.275 | 10045 | 8893 | 0.86 (0.82–0.90) | 4.95 × 10−10 | 0.79 | 0 |
| rs3744763 | 33164998 | 1 | C | T | 0.383 | 0.411 | 10263 | 9117 | 0.89 (0.85–0.92) | 1.21 × 10−08 | 0.79 | 0 |
| rs11649743 | 33149092 | 2 | A | G | 0.176 | 0.197 | 10261 | 9103 | 0.86 (0.82–0.91) | 3.54 × 10−08 | 0.44 | 0.1 |
aOdds ratio per minor allele.
Figure 1.
Prostate cancer risk associated with SNPs in the HNF1B region: (A) results from the unconditional analysis; (B) results after conditioning on rs4430796. The diamond indicates the most statistically significant SNP in the region.
Two SNPs located in the second identified region by Sun et al. (10) were also associated with the prostate cancer risk at a significance level of 5 × 10−8. As expected, the previously identified SNP in the second region, rs11649743, was associated with prostate cancer risk (P = 3.54 × 10−8); however, the association within this second locus was stronger for rs4794758 (P = 4.95 × 10−10). The two SNPs were correlated in controls (r2 = 0.61), but when rs11649743 and rs4794758 were included in the same model, only rs4794758 remained associated with risk (P = 0.002 for rs4794758; P = 0.32 for rs11649743). Interestingly, it accounted for the risk associated with rs11649743.
To examine the interdependence of the signals observed on chromosome 17q12, we first conducted a set of sequential conditional analyses, conditioning on the most significant SNP from the unconditional analysis and each conditional analysis sequentially until no SNPs remain nominally associated with risk (P < 0.05) (Supplementary Material, Table S2). After conditioning on the most significant SNP (rs4430796 in region 1), six SNPs remained nominally associated with risk (P < 0.05) with the most significant SNP being rs4794758 in region 2 (Fig. 1B, Supplementary Material, Table S2). Although rs4430796 and rs4794758 were separated by a modest recombination hotspot, there was some correlation between them (r2 = 0.04) and consequently the P-value for rs4794758 was attenuated after conditioning on rs4430796 (P = 3.45 × 10−5). After conditioning on both rs4430796 and rs4794758, four SNPs were nominally associated with risk (P < 0.05) with the most significant SNP being rs1016990 (P = 0.009). This SNP was only nominally associated with risk in the unconditional model (P = 0.0002) and not significantly associated with risk after conditioning on rs4430796 only. Further sequential conditional analyses yielded rs7405696 (P = 0.01) as the most significant SNP after conditioning on rs4430796, rs4794758 and rs1016990, followed by rs3094509 (P = 0.02) after conditioning on rs4430796, rs4794758, rs1016990 and rs74056969. No other SNPs remained nominally associated with prostate cancer risk after conditioning on these five SNPs, suggesting that these SNPs (i.e. rs4430796, rs4794758, rs1016990, rs74056969 and rs3094509) capture the risk in this region.
To ascertain whether the same SNPs were identified using other statistical approaches, we performed forward stepwise regression adding SNPs below a significance level of 0.05. This method resulted in the inclusion of four SNPs: rs4430796, rs4794758, rs1016990 and rs74056969 in the model. Using lasso, five SNPs (rs4430796, rs2005705, rs7405696, rs4794758 and rs11649743) entered the model at a lambda >0.01. A comparison of the models selected by these three methods using Akaike information criterion (AIC) indicated that the SNPs using the sequential conditional analysis method yielded the best model (Table 2). The sequential conditional model was also found to be a better model than the model containing the most significant SNP from region 1 and region 2 (AIC: 25351.114 versus 25364.42 for the sequential and two SNP models, respectively). We imputed the SNPs from this region available from the 1000 Genomes Project and conducted a sequential conditional analysis with the imputed data. The results were quite similar with at least four of the five SNPs either being the same SNP or a highly correlated SNP (r2> 0.95) as observed in our sequential conditional analysis with directly genotyped SNPs (Supplementary Material, Table S3).
Table 2.
Final prostate cancer risk models for HNF1B SNPs from the sequential conditional, stepwise regression and lasso analysesa
| Sequential conditional model |
Forward stepwise regression model |
Lasso model | Two SNP model | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Base pair position | Region | Minor allele | Other allele | ORb (95% CI) | P-value | ORb (95% CI) | P-value | ORb (95% CI) | P-value | ORb (95% CI) | P-value |
| rs4430796 | 33172153 | 1 | G | A | 0.87 (0.80–0.93) | 9.23E−05 | 0.86 (0.80–0.92) | 3.79E−05 | 0.93 (0.83–1.04) | 0.21 | 0.82 (0.79–0.86) | 5.10E−19 |
| rs2005705 | 33170413 | 1 | T | C | – | – | – | – | 0.95 (0.86–1.04) | 0.26 | – | – |
| rs7405696 | 33176148 | 1 | C | G | 1.11 (1.03–1.18) | 0.005 | 1.09 (1.02–1.17) | 0.01 | 1.10 (1.02–1.18) | 0.009 | – | – |
| rs4794758 | 33154541 | 2 | T | C | 0.88 (0.84–0.93) | 5.73E−07 | 0.89 (0.85–0.94) | 4.22E−06 | 0.94 (0.87–1.02) | 0.13 | 0.91 (0.86–0.95) | 4.50E−05 |
| rs11649743 | 33149092 | 2 | A | G | – | – | – | – | 0.94 (0.87–1.03) | 0.19 | – | – |
| rs1016990 | 33163028 | 1 | G | C | 1.07 (1.01–1.12) | 0.02 | 1.07 (1.02–1.13) | 0.007 | – | – | – | – |
| rs3094509 | 33136412 | 2 | T | C | 1.06 (1.01–1.10) | 0.02 | – | – | – | – | – | – |
| AIC | 25351.114 | 25354.881 | 25361.176 | 25364.42 | ||||||||
aFor comparison, all analyses were restricted to subjects with complete genotyping for these SNPs, so that the total number of subjects included in each model was the same.
bOdds ratio per minor allele.
Our sequential conditional model included three SNPs from region 1 (rs4430796, rs74056969, rs1016990) and two SNPs from region 2 (rs4794758, rs3094509). A haplotype analysis of these five SNPs revealed that the most significant haplotype associated with risk carried the protective allele at rs4430796, rs74056969 and rs4794758 (P = 2.78 × 10−8) (Supplementary Material, Table S4). When the combined risk of all five SNPs was examined, a dose–response was observed with increasing number of risk variants (Ptrend= 1.94 × 10−26, Table 3). Men with eight or more risk alleles had a 1.88-fold increased risk of prostate cancer compared with men with zero to two risk alleles (95% CI: 1.52–2.33, P = 4.29 × 10−9). In comparison, when only the most significant SNP from region 1 (rs4430796) and region 2 (rs4794758) were examined, men with four or more risk alleles had a 1.69-fold risk compared with men with no risk alleles (95% CI: 1.40–2.04, P = 6.66 × 10−8) with Ptrend = 1.80 × 10−25. The P-value for the number of risk alleles for the three other SNPs from the sequential model modeled as a continuous variable was 1.63 × 10−5. The variance explained by the five SNPs was 0.9% compared with 0.7% for the two best SNPs from region 1 and region 2.
Table 3.
Risk of prostate cancer associated with the count of HNF1B risk allelesa
| No. of risk alleles | No. of cases | No. of controls | OR (95% CI) | P-value |
|---|---|---|---|---|
| 0–2 | 335 | 413 | 1.0 | |
| 3 | 1030 | 1167 | 1.10 (0.93–1.31) | 0.26 |
| 4 | 2158 | 2053 | 1.29 (1.10–1.52) | 0.001 |
| 5 | 2596 | 2408 | 1.33 (1.13–1.55) | 0.0004 |
| 6 | 2320 | 1721 | 1.66 (1.42–1.95) | 3.61E−10 |
| 7 | 1110 | 803 | 1.71 (1.44–2.03) | 1.13E−09 |
| 8–10 | 426 | 276 | 1.88 (1.52–2.33) | 4.92E−09 |
aIncludes the five SNPs identified in the sequential conditional analysis (rs4430796, rs7405696, rs4794758, rs1016990 and rs3094509).
Finally, we conducted stratified analyses by family history of prostate cancer and did observe a qualitative interaction for rs2107131 (Pinteraction = 4.29 × 10−5) that remained statistically significant after adjustment for multiple testing (Padj = 0.003) (Supplementary Material, Table S5). Among men with a family history of prostate cancer, the T allele at rs2107131 was associated with a reduced risk of prostate cancer (OR = 0.85; 95% CI: 0.74–0.97), whereas among men without a family history, it was associated with an increased risk of prostate cancer (OR = 1.13; 95% CI: 1.08–1.19). Stratified analyses were also performed for prostate cancer aggressiveness (Gleason < 7 and Stage A/B versus Gleason ≥ 7 or Stage C/D); however, there were no significant differences beyond what would be expected by chance (Supplementary Material, Table S6). We did not observe any significant heterogeneity between studies beyond what would be expected by chance (Supplementary Material, Table S1), except for rs1058166 (Pheterogeneity= 0.0002).
DISCUSSION
Our fine mapping study of a region on 17q12 associated with prostate cancer confirmed the previously established signals (6,7,10) and found evidence that additional variants contribute to the risk of prostate cancer. Although rs4430796 was the most significant SNP associated with risk, rs7405696 was also significant at a genome-wide significance level and explained part of the risk associated with the first HNF1B locus, suggesting a more complex genetic architecture for common variants in this region. Since this study used SNP markers, further work is needed to investigate the biological basis of the association with common variants in 17q12, which may regulate HNF1B, or perhaps another gene in prostate cancer. It is plausible that multiple variants are directly associated with prostate cancer susceptibility.
In the second HNF1B locus, we found that rs4794758 was more strongly associated with risk than the previously identified variant, rs11649743. When both variants were included in the same model, rs4794758 explained all of the risk associated with rs11649743, indicating that this variant more aptly captured the risk attributable to this locus. Although the first and second HNF1B loci are separated by recombination hotspot (10), the two loci are not completely independent and the risk associated with rs4794758 was attenuated after conditioning on the most significant SNP in the first locus, rs4430796.
In our study, the best model for prostate cancer risk in the HNF1B region included five SNPs (rs4430796, rs7405696, rs4794758, rs1016990 and rs3094509). Although three of these SNPs reached genome-wide significance in unconditional models, rs1016990 was only nominally associated with risk (P = 0.0002) and rs3094509 was not associated with risk (P = 0.80) in unconditional models. Together, these five variants provided a better model for the data than other combinations, suggesting that these SNPs together capture more of the risk associated with this region. Furthermore, haplotype analysis revealed that risk was not explained by a single haplotype, suggesting a role for multiple variants or combinations of variants. Imputation using data from the 1000 Genomes Project yielded similar results; however, additional variants discovered in future next generation sequencing may also contribute to risk.
Interestingly, we observed a significant interaction between a SNP located in the second HNF1B locus, rs2107131, and family history of prostate cancer. Family history was only captured at baseline for participants and could have changed over time, and the number of subjects with a positive family history of prostate cancer was limited (N = 2182 men), so this finding should be interpreted with caution. However, it is possible that the interaction reflects haplotype segregation with uncommon disease-causing alleles associated with familial cases and common disease-causing alleles associated with sporadic cases.
The biological mechanism by which 17q12 variants may alter the regulation or splicing of a plausible candidate gene, such as HNF1B, is not clear. Down-regulation of HNF1B expression has been associated with renal cell cancer progression (24), and differential expression of HNF1B has been associated with prostate cancer recurrence (23). HNF1B is a transcription factor that encodes three isoforms in humans. Isoforms HNF1B(A) and HNF1B(B) appear to be transcriptional activators, whereas isoform HNF1B(C) is a transcriptional repressor (25). Differences in isoform-specific HNF1B expression have also been observed between normal and malignant prostate tissue, with prostate tumors displaying greater isoform HNF1B(B) expression but less isoform HNF1B(C) expression than normal prostate tissue (26). It is possible that variants in HNF1B contribute to the altered the expression of HNF1B isoforms in prostate cancer. It is also plausible that variants in this region of 17q12 could also influence the regulation or expression of other genes at a distance.
In conclusion, this large-scale fine mapping study revealed that the association between variants in the 17q12 region and prostate cancer risk is more complex than earlier studies have indicated. Additional sequencing and functional studies are needed to pinpoint the variants in this region that are directly associated with prostate cancer risk and the biological mechanisms involved.
MATERIALS AND METHODS
Study subjects
As described previously (11), prostate cancer cases and controls of European ancestry were drawn from 10 studies in the USA and Europe: Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial (972 cases/927 controls); Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC) (906 cases/868 controls); American Cancer Society Cancer Prevention Study II (CPSII) (1643 cases/1640 controls); Health Professionals Follow-up Study (HPFS) (595 cases/589 controls); CeRePP French Prostate Case-Control Study (FPCC) (998 cases/952 controls); Multiethnic Cohort Study (MEC) (676 cases/682 controls); European Prospective Investigation into Cancer and Nutrition (EPIC) (682 cases/990 controls); Cohort of Norway (CONOR) (606 cases/662 controls); Cancer of the Prostate in Sweden (CAPS) (2213 cases/1362 controls); and a hospital-based case–control from the Johns Hopkins Hospital (JHH) (990 cases/451 controls). A total of 10 272 prostate cancer cases and 9123 controls were available for this study. Aggressive prostate cancer was defined at Gleason score ≥7 or stage C/D. Among the cases with information on disease aggressiveness, 4824 cases were defined as aggressive and 4337 were non-aggressive (Gleason score <7 and stage A/B). Family history of prostate cancer was obtained by self-report from the participants and available for 8 of the 10 studies. A positive family history of prostate cancer was defined as a first degree relative with prostate cancer. Each study obtained an informed consent from study participants and approval from their respective institutional review boards for this study.
SNP selection
A total of 88 SNPs within a 249 071 bp region encompassing HNF1B were selected for fine mapping. The SNPs were chosen based on the 0.2 cM HapMap recombination region flanking the most significant SNP (rs4430796) in the HNF1B locus from the second stage of the CGEMS GWAS (7). The entire region was tagged to capture SNPs with a minor allele frequency ≥5% at a D′ > 0.6 based on the HapMap CEU population (Build 26). All three SNPs with a P < 10−3 from the second stage of CGEMS in this region (i.e. rs4430796, rs7501939 and rs11649743) were selected as obligated SNPs to be included. The final tag SNPs were selected if they were found to be correlated with an r2 of ≥0.8 in the HapMap CEU, YRI or JPT + CHB populations with the obligated SNPs. The tag SNP selection was performed using the GLU software package (http://code.google.com/p/glu-genetics/). The chosen SNPs were primarily located in first (chromosome 17: 33.161–33.205Mb, NCBI Build 36) and second (chromosome 17: 33.116–33.161Mb, NCBI Build 36) HNF1B regions identified to be associated with prostate cancer risk; however, other SNPs outside these regions were also included.
Genotyping
All SNPs for this study were genotyped on a custom Illumina iSelect assay panel as part of the third stage of CGEMS as described previously (11). In brief, a total of 6652 SNPs, including 1400 SNPs to monitor population stratification, were attempted in 22 057 samples, including quality control duplicates. Duplicate samples yielded a 99.97% concordance rate. Subjects with <90% completion (n = 1350), missing covariate data or sparse group (n = 104), likely an intra-study duplicate based on genotype concordance ≥99% (n = 18), or non-European ancestry defined by <0.80 European admixture as estimated using STRUCTURE (27) (n = 372) were excluded, leaving 10 272 cases and 9123 controls for analysis. SNPs with <80% completion within a study were removed from analysis for that study. SNPs that failed to provide genotypes for more than six studies (n = 1), had a minor allele count ≤10 (n = 7) or had genotypes that were inconsistent with Hardy–Weinberg proportions among controls (P < 0.001) (n = 1) were excluded from analysis, leaving 79 SNPs for analysis.
Imputation
To explore the possibility that other variants in the region could be related to risk, we imputed the 249 071 bp region encompassing our genotyped SNPs using IMPUTE2 and the 1000 Genomes Project data (June 2010 release). A total of 653 SNPs were imputed from this region, but only SNPs with a quality score (i.e. info) >0.40 were considered for analysis (N = 353). All imputed SNPs were analyzed using SNPTEST v.2.2.0 accounting for imputation uncertainty.
Statistical analysis
Principal components analysis was conducted using EIGENSTRAT (28) with 1399 SNPs that were genotyped, passed quality control criteria and selected for population stratification; these SNPs were chosen because they had minimal correlation (r2< 0.004) (29). The Wilcoxon rank test was used to test the association between the top five eigenvectors and case–control status. Four eigenvectors displayed a significant or borderline significant association with prostate cancer (P < 0.08) and were included in the analysis. The association between each SNP and prostate cancer risk was estimated using logistic regression adjusting for age (<50, 50–59, 60–69, 70–79, ≥80), study (including country for EPIC) and significant principal components. Stratified analyses were conducted to examine differences by disease aggressiveness, family history and study. Heterogeneity between aggressive and non-aggressive disease was assessed in a case-only analysis using logistic regression. Heterogeneity between studies and risk modification by family history was assessed using a likelihood ratio test comparing the model with and without the cross-product term(s). P-values for interactions tests were adjusted for the false discovery rate (30).
To explore the interdependence of the associations observed, three separate approaches were taken: (i) sequential conditional analyses, (ii) stepwise regression, and (iii) lasso (31). Sequential conditional analyses were conducted by including the most significant SNP in the unconditional logistic regression model followed by sequential inclusion of the most significant SNP in each conditional model and examining the association with each of the remaining SNPs independently. Forward stepwise regression was performed using the SNPs that reached a nominal significance level in the unconditional model. SNPs were sequentially included in the model based on a minimal P < 0.05. Lasso was conducted in R using all SNPs without missing data and an unweighted penalty function.
Linkage disequilibrium measures (D′ and r2) were estimated in the controls using Haploview. Haplotypes were estimated using an expectation-maximization algorithm and analyzed using the generalized linear regression model implemented in HaploStats (32). Unless otherwise indicated, all analyses were conducted using PLINK or STATA.
SUPPLEMENTARY MATERIAL
FUNDING
This study was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health and in part by National Cancer Institute, National Institutes of Health (CA129684 to J.X.) and contract number HHSN261200800001E.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health (NIH). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products or organization indicate endorsement by the US Government. The authors thank Drs Christine Berg and Philip Prorok, Division of Cancer Prevention, NCI, the screening center investigators and staff of the PLCO Cancer Screening Trial, Mr Thomas Riley and staff at Information Management Services, Inc., and Ms Barbara O'Brien and staff at Westat, Inc. for their contributions to the PLCO. Finally, we are grateful to the study participants for donating their time and making this study possible.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Lichtenstein P., Holm N.V., Verkasalo P.K., Iliadou A., Kaprio J., Koskenvuo M., Pukkala E., Skytthe A., Hemminki K. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 2.Amundadottir L.T., Sulem P., Gudmundsson J., Helgason A., Baker A., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Cazier J.B., Sainz J., et al. A common variant associated with prostate cancer in European and African populations. Nat. Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 3.Yeager M., Orr N., Hayes R.B., Jacobs K.B., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 4.Gudmundsson J., Sulem P., Manolescu A., Amundadottir L.T., Gudbjartsson D., Helgason A., Rafnar T., Bergthorsson J.T., Agnarsson B.A., Baker A., et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 5.Haiman C.A., Patterson N., Freedman M.L., Myers S.R., Pike M.C., Waliszewska A., Neubauer J., Tandon A., Schirmer C., McDonald G.J., et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gudmundsson J., Sulem P., Steinthorsdottir V., Bergthorsson J.T., Thorleifsson G., Manolescu A., Rafnar T., Gudbjartsson D., Agnarsson B.A., Baker A., et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat. Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 7.Thomas G., Jacobs K.B., Yeager M., Kraft P., Wacholder S., Orr N., Yu K., Chatterjee N., Welch R., Hutchinson A., et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 8.Eeles R.A., Kote-Jarai Z., Giles G.G., Olama A.A., Guy M., Jugurnauth S.K., Mulholland S., Leongamornlert D.A., Edwards S.M., Morrison J., et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 9.Gudmundsson J., Sulem P., Rafnar T., Bergthorsson J.T., Manolescu A., Gudbjartsson D., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Blondal T., et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat. Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun J., Zheng S.L., Wiklund F., Isaacs S.D., Purcell L.D., Gao Z., Hsu F.C., Kim S.T., Liu W., Zhu Y., et al. Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat. Genet. 2008;40:1153–1155. doi: 10.1038/ng.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeager M., Chatterjee N., Ciampa J., Jacobs K.B., Gonzalez-Bosquet J., Hayes R.B., Kraft P., Wacholder S., Orr N., Berndt S., et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al Olama A.A., Kote-Jarai Z., Giles G.G., Guy M., Morrison J., Severi G., Leongamornlert D.A., Tymrakiewicz M., Jhavar S., Saunders E., et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 13.Eeles R.A., Kote-Jarai Z., Al Olama A.A., Giles G.G., Guy M., Severi G., Muir K., Hopper J.L., Henderson B.E., Haiman C.A., et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat. Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudmundsson J., Sulem P., Gudbjartsson D.F., Blondal T., Gylfason A., Agnarsson B.A., Benediktsdottir K.R., Magnusdottir D.N., Orlygsdottir G., Jakobsdottir M., et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takata R., Akamatsu S., Kubo M., Takahashi A., Hosono N., Kawaguchi T., Tsunoda T., Inazawa J., Kamatani N., Ogawa O., et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat. Genet. 2010;42:751–754. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 16.Zeggini E., Scott L.J., Saxena R., Voight B.F., Marchini J.L., Hu T., de Bakker P.I., Abecasis G.R., Almgren P., Andersen G., et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasper J.S., Giovannucci E. A meta-analysis of diabetes mellitus and the risk of prostate cancer. Cancer Epidemiol. Biomarkers Prev. 2006;15:2056–2062. doi: 10.1158/1055-9965.EPI-06-0410. [DOI] [PubMed] [Google Scholar]
- 18.Stevens V.L., Ahn J., Sun J., Jacobs E.J., Moore S.C., Patel A.V., Berndt S.I., Albanes D., Hayes R.B. HNF1B and JAZF1 genes, diabetes, and prostate cancer risk. Prostate. 2010;70:601–607. doi: 10.1002/pros.21094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato N., Motoyama T. Hepatocyte nuclear factor-1beta(HNF-1beta) in human urogenital organs: its expression and role in embryogenesis and tumorigenesis. Histol. Histopathol. 2009;24:1479–1486. doi: 10.14670/HH-24.1479. [DOI] [PubMed] [Google Scholar]
- 20.Maestro M.A., Cardalda C., Boj S.F., Luco R.F., Servitja J.M., Ferrer J. Distinct roles of HNF1beta, HNF1alpha, and HNF4alpha in regulating pancreas development, beta-cell function and growth. Endocr. Dev. 2007;12:33–45. doi: 10.1159/000109603. [DOI] [PubMed] [Google Scholar]
- 21.Bellanne-Chantelot C., Chauveau D., Gautier J.F., Dubois-Laforgue D., Clauin S., Beaufils S., Wilhelm J.M., Boitard C., Noel L.H., Velho G., Timsit J. Clinical spectrum associated with hepatocyte nuclear factor-1beta mutations. Ann. Intern. Med. 2004;140:510–517. doi: 10.7326/0003-4819-140-7-200404060-00009. [DOI] [PubMed] [Google Scholar]
- 22.Edghill E.L., Bingham C., Ellard S., Hattersley A.T. Mutations in hepatocyte nuclear factor-1beta and their related phenotypes. J. Med. Genet. 2006;43:84–90. doi: 10.1136/jmg.2005.032854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glinsky G.V., Glinskii A.B., Stephenson A.J., Hoffman R.M., Gerald W.L. Gene expression profiling predicts clinical outcome of prostate cancer. J. Clin. Invest. 2004;113:913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchner A., Castro M., Hennig A., Popp T., Assmann G., Stief C.G., Zimmermann W. Downregulation of HNF-1B in renal cell carcinoma is associated with tumor progression and poor prognosis. Urology. 2010;76:507–511. doi: 10.1016/j.urology.2010.03.042. [DOI] [PubMed] [Google Scholar]
- 25.Bach I., Yaniv M. More potent transcriptional activators or a transdominant inhibitor of the HNF1 homeoprotein family are generated by alternative RNA processing. EMBO J. 1993;12:4229–4242. doi: 10.1002/j.1460-2075.1993.tb06107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harries L.W., Perry J.R., McCullagh P., Crundwell M. Alterations in LMTK2, MSMB and HNF1B gene expression are associated with the development of prostate cancer. BMC Cancer. 2010;10:315. doi: 10.1186/1471-2407-10-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 29.Yu K., Wang Z., Li Q., Wacholder S., Hunter D.J., Hoover R.N., Chanock S., Thomas G. Population substructure and control selection in genome-wide association studies. PLoS ONE. 2008;3:e2551. doi: 10.1371/journal.pone.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- 31.Tibshirani R. Regression shrinkage and selection via lasso. J. R. Stat. Soc. B. 1996;58:267–288. [Google Scholar]
- 32.Schaid D.J., Rowland C.M., Tines D.E., Jacobson R.M., Poland G.A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.