Abstract
CHARGE is a multiple congenital anomaly disorder and a common cause of pubertal defects, olfactory dysfunction, growth delays, deaf-blindness, balance disorders and congenital heart malformations. Mutations in CHD7, the gene encoding chromodomain helicase DNA binding protein 7, are present in 60–80% of individuals with the CHARGE syndrome. Mutations in CHD7 have also been reported in the Kallmann syndrome (olfactory dysfunction, delayed puberty and hypogonadotropic hypogonadism). CHD7 is a positive regulator of neural stem cell proliferation and olfactory sensory neuron formation in the olfactory epithelium, suggesting that the loss of CHD7 might also disrupt development of other neural populations. Here we report that female Chd7Gt/+ mice have delays in vaginal opening and estrus onset, and erratic estrus cycles. Chd7Gt/+ mice also have decreased circulating levels of luteinizing hormone and follicle-stimulating hormone but apparently normal responsiveness to gonadotropin-releasing hormone (GnRH) agonist and antagonist treatment. GnRH neurons in the adult Chd7Gt/+ hypothalamus and embryonic nasal region are diminished, and there is decreased cellular proliferation in the embryonic olfactory placode. Expression levels of GnRH1 and Otx2 in the hypothalamus and GnRHR in the pituitary are significantly reduced in adult Chd7Gt/+ mice. Additionally, Chd7 mutant embryos have CHD7 dosage-dependent reductions in expression levels of Fgfr1, Bmp4 and Otx2 in the olfactory placode. Together, these data suggest that CHD7 has critical roles in the development and maintenance of GnRH neurons for regulating puberty and reproduction.
INTRODUCTION
In humans, heterozygous mutations in chromodomain helicase DNA binding protein 7 (CHD7) cause CHARGE syndrome, a clinically variable, multiple congenital anomaly condition with an estimated incidence of 1:8500–1:12 000 in newborns (1–3). CHARGE is characterized by ocular coloboma, heart defects, atresia of the choanae, retarded growth and development, genital hypoplasia and ear abnormalities including deafness and vestibular disorders (4). Genital hypoplasia occurs in ∼62% of CHARGE individuals with confirmed CHD7 mutations, and gonadotropins [luteinizing hormone (LH) and follicle-stimulating hormone (FSH)] are deficient in 81% of males and 93% of females (5–12).
The gonadotropic hormones LH and FSH are generated and secreted from the pituitary in response to gonadotropin-releasing hormone (GnRH) from the median eminence (13–16). GnRH production is dependent upon multiple signaling mechanisms, including sex-steroid feed-back regulation, kisspeptin-GPR54 signaling and leptin signaling (14,15,17). Transcription of GnRH1 requires the paired-like homeodomain transcription factor OTX2 (18–20). OTX2 is also required for neurogenesis in multiple tissues (21,22). GnRH neurogenesis is partially regulated by fibroblast growth factor (FGF) signaling, and mutations in FGF8 and FGFR1 cause hypogonadotropic hypogonadism and olfactory dysfunction in humans and mice (23–28).
Mice with the heterozygous loss of Chd7 (Chd7Gt/+) have olfactory defects (29) similar to those reported in CHARGE individuals, and are an excellent model for the CHARGE syndrome. Chd7 is highly expressed in the developing mouse and human olfactory epithelium (29–31), where GnRH neurons are born (32). In mice and humans, Chd7 is expressed in the developing (30,31) and mature hypothalamus, and in GnRH neuronal cell lines (8). CHD7 is hypothesized to influence gene expression by regulating access to chromatin through binding and unwinding chromatin (33–37), and CHD7 is likely to participate in multiple protein complexes that regulate transcription. Based on prior studies showing that CHD7 is critical for olfactory neural stem cell proliferation and regeneration of olfactory sensory neurons (29), we hypothesized that CHD7 may also regulate one or more aspects of GnRH neurogenesis. To test this, we analyzed Chd7Gt/+ mice for pubertal development and underlying cellular mechanisms involved in hypogonadotropic hypogonadism.
RESULTS
Analysis of puberty and estrus cycles
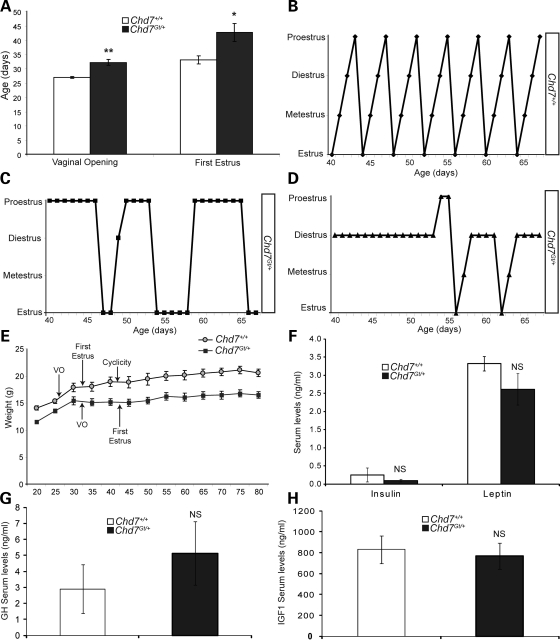
Endocrine defects including delayed puberty, hypogonadotropic hypogonadism and genital hypoplasia have been reported in CHARGE individuals (5–11). Prior studies of the reproductive system in Chd7Whi/+ mice, a model of the CHARGE syndrome, reported an increased time to first litter and hypoplasia of the testes, clitoris and uterus compared with wild-type mice (38,39). Based on these reproductive defects, we hypothesized that mice deficient for Chd7 might have underlying endocrine abnormalities. To test this, we analyzed female Chd7Gt/+ mice which are heterozygous for a gene trapped loss of function lacZ reporter allele (30). Homozygous Chd7Gt/Gt embryos die by E11, presumably from cardiac or other internal organ defects (30). Recently weaned female wild-type (n =5) and Chd7Gt/+ (n =6) littermate mice were examined for vaginal opening and first estrus, two well-established markers of rodent sexual maturity (Fig. 1A) (40). Vaginal opening was significantly delayed and occurs an average of 5 days later in Chd7Gt/+ female mice (postnatal day 32) compared with wild-type littermates (postnatal day 27) (Fig. 1A). Vaginal smears taken from both wild-type and Chd7Gt/+ littermate females showed that Chd7Gt/+ female mice achieve first estrus on postnatal day 43, 10 days later than their wild-type littermates (postnatal day 33) (Fig. 1A).
Figure 1.
Chd7Gt/+ females have delayed puberty and erratic estrus cycles. (A) Wild-type and Chd7Gt/+ female littermates were examined for vaginal opening and first estrus. (B) In wild-type females, estrus cyclicity is obtained at postnatal day 39, 9 days after first estrus. (C and D) Chd7Gt/+ female littermates never achieve cyclicity and have erratic estrus cycles. (E) Pubertal markers [vaginal opening (VO), first estrus and cyclicity] in relation to body size are depicted for wild-type and Chd7Gt/+ females. (F) Circulating levels of insulin and leptin are similar in wild-type and Chd7Gt/+ female littermate mice. (G and H) Circulating levels of GH or IGF1 are similar in wild-type and Chd7Gt/+ female littermate mice. *P < 0.05, **P < 0.01 by unpaired Student's t-test. NS, not significant.
Establishment of estrus cyclicity is another measure of reproductive health and development in mice, and the timing of cyclicity onset can be variable between inbred strains (40). Cyclicity in mice is defined by the sequential attainment of each stage of estrus during a 4–7-day period of time (40). To determine the onset of estrus cyclicity, vaginal smears were analyzed daily in female wild-type (n =5) and Chd7Gt/+ (n =6) mice for 3 months (Fig. 1B–D). In contrast to wild-type littermates, female Chd7Gt/+ mice have highly erratic estrus cycles, with no apparent pattern of timing or duration of cycle stages (Fig. 1B–D).
Body size can influence pubertal development in both humans and mice (40–45). For example, vaginal opening in wild-type female mice is dependent upon attainment of a threshold body size (40). Both male and female Chd7-deficient mice and humans with CHARGE syndrome have postnatally acquired growth delays and reduced body size (12,30,39). We examined female wild-type (n =5) and Chd7Gt/+ (n =6) littermate mice for pubertal markers with respect to body size. Vaginal opening corresponds to attainment of a body size of ∼15gm in both wild-type and Chd7Gt/+ female littermates (Fig. 1E). However, wild-type females have first estrus at ∼18 gm, while Chd7Gt/+ females have first estrus at ∼15 gm.
Metabolic cues and nutritional state are also key elements involved in proper pubertal development (42–48). Defects in the leptin, insulin, growth hormone (GH) or insulin-like growth factor 1 (IGF1) signaling pathway have been reported to cause delayed puberty, hypogonadotropic hypogonadism and/or infertility (49–55). Levels of leptin, insulin, GH and IGF1 are also sensitive to nutritional state, with circulating levels that vary between 2- and 5-fold after fasting (56). To further explore factors that may influence pubertal development, female wild-type (n =4) and Chd7Gt/+ (n =4) littermate mice were analyzed for circulating levels of insulin and leptin (Fig. 1F). Insulin and leptin levels were not altered in Chd7Gt/+ females compared with wild-type (Fig. 1F). Additionally, circulating levels of GH and IGF1 were similar in wild-type (n =4) and Chd7Gt/+ (n =4) littermate females (Fig. 1G and H) and males (data not shown). Although we could detect no significant differences in insulin, leptin, GH or IGF1, Chd7Gt/+ mice (male and female) are significantly smaller than their wild-type littermates (30). Our data cannot completely rule out a more subtle defect in metabolism or nutrition that may influence reproduction in Chd7Gt/+ mice.
Chd7Gt/+ mice have decreased levels of circulating gonadotropic hormones
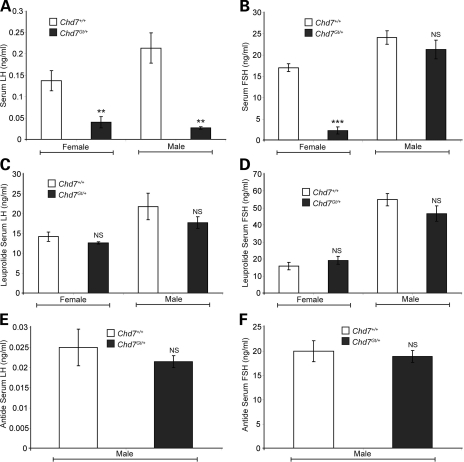
CHARGE individuals are reported to have hypogonadotropic hypogonadism and variable response to GnRH stimulation (5–11). To test the integrity of the hypothalamic-pituitary axis, we assayed circulating levels of LH and FSH in 3–4-month-old wild-type (males n =4, females n =5) and Chd7Gt/+ (males n =5, females n =6) sex-matched littermate mice (Fig. 2A and B). Circulating levels of female gonadotropic hormones fluctuate dependent upon stage of estrus (57); therefore, we collected serum from all females at late estrus. We found that Chd7Gt/+ female mice have reduced levels of circulating LH (74% reduction) and FSH (83% reduction) in comparison to wild-type littermates (Fig. 2A and B). Chd7Gt/+ male mice also have reduced levels of circulating LH (86% reduction), whereas levels of FSH are normal (Fig. 2A and B). Interestingly, studies of Fshβ−/− mice reported that FSH is not required for male fertility but is required for female fertility (58,59). In contrast, Fshβ mutations in human males cause azoospermia and infertility (60,61). Fshβ is also expressed independent of GnRH signaling in an activin-dependent pathway (62,63), and LH and FSH are differentially regulated by GnRH during development (64).
Figure 2.
Circulating LH and FSH are decreased in Chd7Gt/+ mice. (A) Chd7Gt/+ mice analyzed for circulating LH have decreased serum levels compared with wild-type littermates. The LH reportable range was 0.04–37.4 ng/ml. (B) Chd7Gt/+ females have decreased levels of circulating FSH but Chd7Gt/+ males have normal serum levels. The FSH reportable range was 2.3–20.0 ng/ml. (C and D) GnRH agonist, leuprolide, administered to wild-type and Chd7Gt/+ mice caused similar responses in the production and circulation of LH and FSH. The reportable range for LH was 0.04–37.4 ng/ml and for FSH was 6.7–75.0 ng/ml. (E and F) Wild-type and Chd7Gt/+ males were administered GnRH antagonist, antide. Wild-type and Chd7Gt/+ males have similar decreases in circulating levels of LH and FSH. The reportable range for LH was 0.04–37.4 ng/ml and for FSH was 6.7–75.0 ng/ml. **P < 0.01, ***P < 0.001 by unpaired Student's t-test. NS, not significant.
Chd7Gt/+ mice respond normally to GnRH agonist stimulation
Hypogonadotropic hypogonadism can be caused by defects in the hypothalamus, the pituitary or both. Mutations in the GnRH receptor gene, GnRHR, cause pituitary gonadotrope insensitivity to GnRH stimulation and subsequent hypogonadotropic hypogonadism (65,66), whereas mutations in genes such as GPR54 or GnRH1 affect GnRH neuronal function at the level of the hypothalamus (67,68). Mutations in genes affecting GnRH neuronal function could therefore mask a normal ability of pituitary gonadotropes to respond to GnRH. To test this, we administered the GnRH agonist, leuprolide, to 3–4-month-old wild-type (males n =4, females n =3) and Chd7Gt/+ (males n =4, females n =4) sex-matched, late estrus-matched (females) littermate mice. In mice, leuprolide causes a rapid (between 70 and 180 min) increase in production and circulation of LH and FSH (69,70). Wild-type and Chd7Gt/+ mice had similar circulating levels of LH and FSH measured 2 h following leuprolide administration (Fig. 2C and D). These observations are consistent with normal pituitary gonadotrope responsiveness to GnRH agonist in Chd7Gt/+ mice.
Male Chd7Gt/+ mice have normal baseline circulating levels of FSH (Fig. 2B). To determine whether pituitary gonadotropes in Chd7Gt/+ males constitutively produce FSH independent of GnRH signal, we administered the GnRH antagonist antide to 3-month-old male wild-type (n = 4) and Chd7Gt/+ (n = 4) littermate mice. In mice, antide causes a rapid (2 h) decrease in the production and circulation of LH and FSH (71,72). Wild-type and Chd7Gt/+ mice had no difference in circulating levels of FSH or LH 2 h following antide administration (Fig. 2E and F). Additionally, no abnormalities in pituitary histology were found by hematoxylin and eosin staining of sections from E10.5, E14.5, E18.5 and adult Chd7Gt/+ mice (data not shown; 30).
GnRH neurons are decreased in Chd7-deficient mice
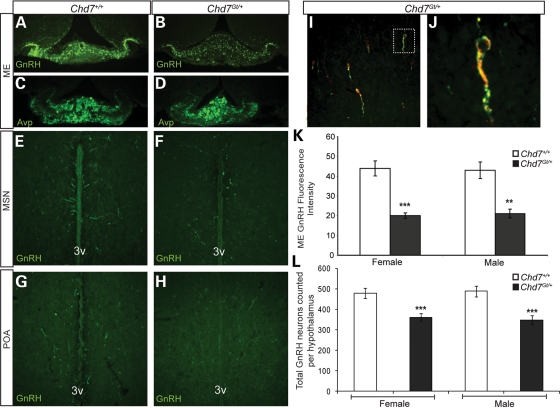
Reduced levels of circulating LH and FSH could reflect defects in GnRH neurons in the hypothalamus, as a result of reductions in GnRH neuronal number, defective migration to the hypothalamus and/or aberrant hormone production and/or release (14). To measure GnRH neurons in Chd7Gt/+ mice, immunofluorescence was performed with anti-GnRH antibody on hypothalamic sections from 3–4-month-old wild-type (males n =4, females n =4) and Chd7Gt/+ (males n =4, females n =4) sex-matched, late estrus-matched (females) littermates. A visually apparent decrease in GnRH immunofluorescence was found in the median eminence in both male and female Chd7Gt/+ mice compared with wild-type littermates (Fig. 3A and B). In contrast, there was no difference between wild-type and Chd7Gt/+ mice in immunofluorescence of arginine vasopressin (AVP)-positive fibers in the median eminence (Fig. 3C and D), providing evidence against a general defect in neural development or maintenance. Quantitation of anti-GnRH immunofluorescence in the median eminence using ImageJ software showed a 54% reduction in anti-GnRH immunofluorescence in Chd7Gt/+ females and a 51% reduction in Chd7Gt/+ males compared with wild-type littermates (Fig. 3K).
Figure 3.
Chd7Gt/+ mice have decreased GnRH neurons in the hypothalamus. (A and B) Immunofluorescence using anti-GnRH (green) revealed decreased GnRH immunofluorescence in the median eminence in Chd7Gt/+ mice compared with wild-type littermates. (C and D) Immunofluorescence using anti-AVP showed no difference in the median eminence in Chd7Gt/+ mice compared with wild-type littermates. (E–H) Immunofluorescence using anti-GnRH (green) showed decreased numbers of GnRH neurons in the MSN and POA in Chd7Gt/+ mice compared with wild-type littermates. (I and J) Chd7 is expressed in GnRH neurons in vivo. Immunofluorescence of Chd7Gt/+ hypothalamic sections using anti-β-galactosidase (β-gal) and anti-GnRH showed that most GnRH-positive neurons in the hypothalamus are also positive for the β-galactosidase reporter for Chd7 expression. White box in (I) indicates the co-labeled neuron magnified in (J). (K) Quantitation of anti-GnRH immunofluorescence in the median eminence using ImageJ software showed significantly decreased GnRH in Chd7Gt/+ mice compared with the wild-type. (L) Quantitation of GnRH neurons per hypothalamus showed decreased numbers of GnRH neurons in both male and female Chd7Gt/+ mice compared with sex-matched wild-type littermates. Sections are in the coronal plane. **P < 0.01, ***P < 0.001 by unpaired Student's t-test. 3v, third ventricle.
Reduced GnRH in the median eminence could be a result of fewer GnRH neurons in the hypothalamus or defects in GnRH neuronal function. Adult GnRH neurons have a rostral–caudal distribution that spans from the nasal region to the hypothalamus, but the vast majority of GnRH neurons reside in specific regions of the hypothalamus (73). To quantify GnRH neurons in the hypothalamus, immunofluorescence was performed with anti-GnRH antibody on hypothalamic sections from 3–4-month-old wild-type (males n =4, females n =4) and Chd7Gt/+ sex-matched (males n =4, females n =4), late estrus-matched (for females) littermates. GnRH neuronal cell counts taken from the hypothalamus including the medial septal nucleus (MSN) (Fig. 3E and F) and preoptic area (POA) (Fig. 3G and H) showed a significant (35%) reduction in the number of GnRH-positive cell bodies in the hypothalamus of Chd7Gt/+ females and Chd7Gt/+ males compared with wild-type littermates (Fig. 3L). The average total number of GnRH-positive neurons counted in the hypothalamus is: (for females) wild-type = 487 (standard error =26), Chd7Gt/+= 348 (standard error =21; P <0.001); (for males) wild-type= 473 (standard error =24), Chd7Gt/+= 361 (standard error =18; P <0.001). To determine whether GnRH neurons exhibit migration abnormalities in Chd7Gt/+ mice, immunofluorescence was performed with anti-GnRH antibody on both coronal and sagittal sections from 3–4-month-old wild-type (males n =4, females n =4) and Chd7Gt/+ (males n =4, females n =4) sex-matched, late estrus-matched (females) littermates. GnRH neurons were distributed similarly throughout the rostral–caudal axis from the nasal region to the cerebellum in wild-type and Chd7Gt/+ mice. Thus, we did not identify a major defect in GnRH neuronal migration in Chd7Gt/+ mice.
Chd7 is expressed in immortalized GnRH neuronal cell lines (8), but Chd7 expression in GnRH neurons in vivo had not been previously examined. We analyzed adult Chd7Gt/+ hypothalamic sections using anti-β-galactosidase and anti-GnRH and found that most GnRH-positive neurons were also positive for the β-galactosidase reporter (Fig. 3I and J). These data suggest that CHD7 may also regulate GnRH neuronal development or function.
Chd7Gt/+ embryos have fewer GnRH neurons in the nasal region
The timing of GnRH neuron formation, their migratory paths and molecules regulating this migration have been extensively studied (74). In mice, GnRH neurons arise from the E9.5–11.5 olfactory placode and then migrate along olfactory tracts leading to the forebrain (32). GnRH neurons reside in the nasal region until E13.5, at which time most GnRH neurons have migrated into the forebrain; GnRH neurons continue to migrate until E16.5 when they reach the hypothalamus and have assumed their adult-like distribution (32).
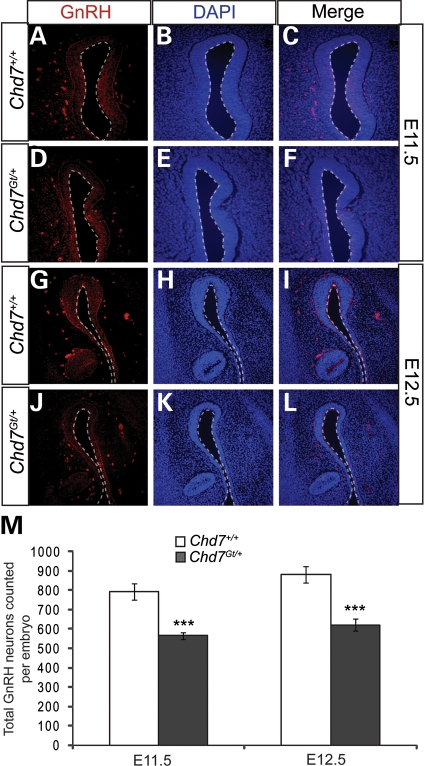
Hypogonadotropic hypogonadism is often associated with abnormalities in GnRH neurons, and can be caused by defects in neurogenesis, migration or survival (75). Mouse models with mutations affecting genes such as Pkr2, Epha5 and Nhlh2 are reported to have defects in GnRH neuronal migration and/or survival (76–78), while mutations in Fgf8 and Fgfr1 cause reduced neurogenesis (26–28). To determine whether GnRH neuronal numbers are altered in Chd7Gt/+ embryos, GnRH-positive cells were counted in wild-type (n = 4 each time point) and Chd7Gt/+ (n = 4 each time point) littermate embryos at E11.5 and E12.5. We found a 30% reduction in the number of GnRH neurons in the nasal region of Chd7Gt/+ embryos at both E11.5 and E12.5 (Fig. 4). The average total number of GnRH-positive neurons counted in the embryonic nasal region is: (for E11.5) wild-type = 793 (standard error =42), Chd7Gt/+= 565 (standard error =19; P <0.001); (for E12.5) wild-type = 882 (standard error =40), Chd7Gt/+= 622 (standard error =32; P <0.001). Decreased GnRH neurons in Chd7Gt/+ embryos are consistent with fewer GnRH neurons in the hypothalamus of adult Chd7Gt/+ mice.
Figure 4.
GnRH neurons are reduced in Chd7Gt/+ embryos. (A–L) Immunofluorescence using antibody against GnRH (red) and counterstained with DAPI (blue) showed a decrease in anti-GnRH staining at E11.5 (A–D) and E12.5. (M) The total number of GnRH neurons per embryo was significantly decreased at each time point in Chd7Gt/+ embryos compared with wild-type littermates. Sections are in the transverse plane. ***P < 0.001 by unpaired Student's t-test.
Chd7Gt/+ embryos have defects in cellular proliferation in the olfactory placode
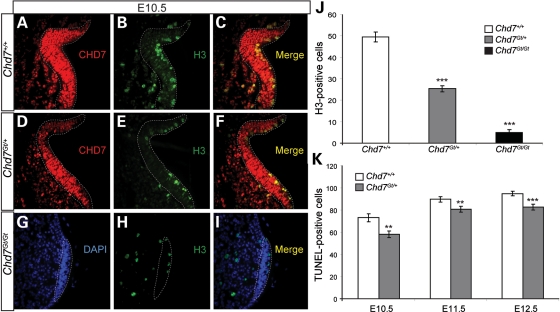
Chd7 is highly expressed in the olfactory placode by E10.5 (30), and continues to be expressed in olfactory tissues throughout development and into adulthood (29). To test cellular proliferation in the olfactory placode, immunofluorescence was performed with anti-CHD7 and the proliferation marker anti-phospho-histone H3 in wild-type (n = 4), Chd7Gt/+ (n = 4) and Chd7Gt/Gt (n = 3) littermate embryos at E10.5 (Fig. 5A–I). A 49% reduction in phospho-histone H3-positive cells was observed in E10.5 Chd7Gt/+ embryos compared with the wild-type (Fig. 5A–F and J). The vast majority (97%) of phospho-histone H3-positive cells in the olfactory placode were CHD7 positive, similar to previously published results showing CHD7 in 98% of proliferating cells in the adult olfactory epithelium (29). In Chd7Gt/Gt embryos, we found a 90% reduction in H3-positive cells compared with the wild-type, and an 80% reduction in H3-positive cells compared with Chd7Gt/+ embryos (Fig. 5A–J). The olfactory placode was also reduced in size in Chd7Gt/Gt embryos but appeared unchanged in Chd7Gt/+ compared with the wild-type. Together, these data suggest that Chd7 deficiency negatively impacts cellular proliferation in the developing olfactory placode, consistent with fewer GnRH neurons available for migration into the hypothalamus and with our earlier observation that there are fewer olfactory sensory neurons in the adult Chd7Gt/+ olfactory epithelium (29).
Figure 5.
Chd7Gt/+ embryos have reduced cellular proliferation in the olfactory placode. (A–F) Immunofluorescence of the wild-type and Chd7Gt/+ at E10.5 using antibodies against CHD7 (red) and phospho-histone H3 (H3) (green). (G–I) Immunofluorescence of Chd7Gt/Gt embryos at E10.5 using antibodies against CHD7 (red) and phospho-histone H3 (H3) (green) counterstained with DAPI (blue). (J) Cell counts showed decreased cellular proliferation in the olfactory placode in Chd7Gt/+ and Chd7Gt/Gt embryos compared with wild-type littermates. (K) Cell counts showed a decrease in TUNEL-positive cells at E10.5, E11.5 and E12.5 in Chd7Gt/+ embryos compared with the wild-type. Sections are in the transverse plane. **P < 0.01, ***P < 0.001 by ANOVA (proliferation) and unpaired Student's t-test (TUNEL).
Decreased numbers of adult GnRH neurons could also result from defects in cell survival (78). To test this, wild-type (n = 4 each time point) and Chd7Gt/+ (n = 4 each time point) littermate embryos at E10.5, E11.5 and E12.5 were analyzed for apoptosis by the TUNEL assay. Chd7Gt/+ embryos have significantly fewer TUNEL-positive cells in the nasal region compared with wild-type embryos at all time points analyzed [E10.5 (21% reduction), E11.5 (11% reduction) and E12.5 (13% reduction)] (Fig. 5K). The reduction in TUNEL-positive cells at all three time points may reflect fewer cells in the nasal region of Chd7Gt/+ embryos, or decreased susceptibility to apoptosis. In E10.5 Chd7Gt/+ embryos, the 49% reduction in H3-positive cells in the olfactory placode is greater than the 21% reduction in E10.5 TUNEL-positive cells, consistent with a net loss of GnRH precursors. Wild-type and Chd7Gt/+ embryos at E10.5 appear to have normal invagination of the olfactory placode to form the olfactory pit (Fig. 5A–F). However, Chd7Gt/Gt embryos appear to lack olfactory pits, suggesting that invagination of the olfactory placode is disrupted (Fig. 5G–I). Altogether, our data suggest that defects in GnRH neurogenesis (rather than increased cell death) lead to reduced GnRH neurons in embryonic and adult Chd7Gt/+ mice.
Reduced CHD7 dosage alters gene expression in olfactory placode, pituitary and hypothalamus
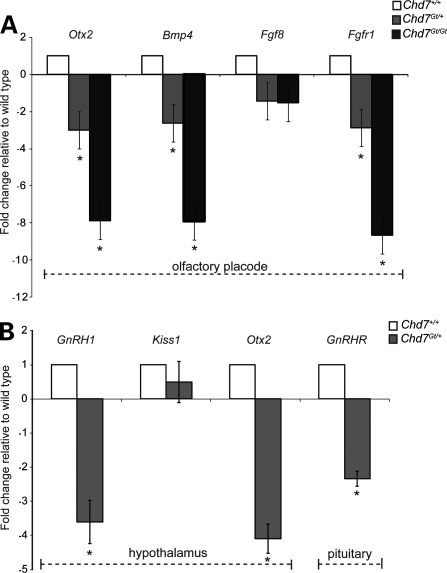
CHD7 may regulate neural progenitors by directly influencing the expression of morphogens such as the bone morphogenetic proteins (BMPs) and FGFs. CHD7 may also regulate the expression and/or function of transcription factors involved in neurogenesis. Chd7 is highly expressed throughout the entire olfactory placode, whereas the expression of morphogens is often regionally restricted within the olfactory placode (23,79). To determine whether CHD7 regulates expression of Fgfr1, Fgf8, Bmp4 or Otx2, we analyzed microdissected olfactory placode from E10.5 wild-type (n = 3), Chd7Gt/+ (n = 3) and Chd7Gt/Gt (n = 4) littermate embryos using TaqMan gene expression assays. We found that Chd7Gt/+ embryos have decreased expression of Otx2 (fold change: −3.01 ± 0.32), Bmp4 (fold change: −2.61 ± 0.15) and Fgfr1 (fold change: −2.88 ± 0.23) compared with wild-type littermates (Fig. 6A). Chd7Gt/Gt embryos also have decreased expression of Otx2 (fold change: −7.87 ± 0.46), Bmp4 (fold change: −7.91 ± 0.37) and Fgfr1 (fold change: −8.65 ± 0.29) compared with wild-type littermates (Fig. 6A). There was no significant difference in Fgf8 expression in Chd7Gt/+ or Chd7Gt/Gt embryos compared with wild-type littermates (Fig. 6A). Potential changes in spatial distribution of gene expression within the Chd7 mutant placode could not be determined from these studies.
Figure 6.
Reduced CHD7 dosage alters gene expression in olfactory placode, adult hypothalamus and adult pituitary. TaqMan gene expression assays were done on microdissected tissues. (A) Expression of Otx2, Bmp4 and Fgfr1 is decreased in E10.5 Chd7Gt/+ and Chd7Gt/Gt olfactory placode compared with wild-type littermates, whereas Fgf8 expression is unchanged. (B) Expression of Otx2 and GnRH1 is decreased in Chd7Gt/+ hypothalamus compared with wild-type littermates, whereas Kiss1 expression is unchanged. Expression of GnRHR is decreased in Chd7Gt/+ pituitary compared with wild-type littermates. *P< 0.05 by DataAssist Software.
To determine whether CHD7 regulates the expression of genes required for normal adult hypothalamic-pituitary signaling, we analyzed the expression of GnRH1, Otx2, Kiss1 and GnRHR using TaqMan gene expression assays. We analyzed RNA isolated from the hypothalamus and pituitary of 3–4-month-old wild-type (n =4) and Chd7Gt/+ (n =4) littermate mice. We found decreased expression of GnRHR (fold change: −2.34 ± 0.22) in Chd7Gt/+ pituitary (Fig. 6B), consistent with decreased GnRH signal (80–82) rather than a direct effect of reduced Chd7 dosage. We also found decreased expression of GnRH1 (fold change: −3.61 ± 0.63) and Otx2 (fold change: −4.09 ± 0.43) in Chd7Gt/+ hypothalamus compared with wild-type littermates (Fig. 6B). There was no significant difference in expression of Kiss1 in the Chd7Gt/+ hypothalamus compared with wild-type littermates (Fig. 6B).
DISCUSSION
We report here that Chd7Gt/+ mice have delayed puberty, erratic estrus cycles, decreased levels of circulating gonadotropins and reduced GnRH neurons in the hypothalamus. Chd7Gt/+ embryos also have fewer GnRH neurons and reduced cellular proliferation in the olfactory placode. Additionally, reduced CHD7 dosage impacts expression of genes involved in proliferation and/or neurogenesis (Fgfr1, Bmp4 and Otx2) in the olfactory placode and GnRH signaling (Otx2 and GnRH1) in the adult hypothalamus. Together, our data suggest that defects in neural progenitor proliferation in the developing olfactory epithelium may underlie the Kallmann-like features found in Chd7Gt/+ mice and in CHD7 mutation-positive humans with the CHARGE syndrome. These observations also provide evidence that CHD7 positively regulates cellular proliferation potentially by either directly or indirectly regulating transcription of Fgfr1, Bmp4 and Otx2. Combined with previous published results in the olfactory system (29) and the developing inner ear (83), our data imply a broad role for CHD7 in promoting neurogenesis in multiple tissues.
Although no binding site consensus sequences have been identified for CHD7, we compared our gene expression results to CHD7 CHiP-seq data (derived from embryonic stem cells) deposited in GEO by Schnetz et al. (84). We found CHD7-binding sites at the medium threshold for Otx2 (one site inside the gene and one upstream close to the transcriptional start site), Bmp4 (three sites downstream of the gene) and Fgfr1 (five sites in the second intron and several sites both upstream and downstream of the gene). We also found a potential CHD7-binding site for Fgf8 in the adjacent gene Mgea5, which could contain an enhancer region for Fgf8. There were no CHD7-binding sites at the low, medium or high thresholds for GnRH1, GnRHR or Kiss1. It is important to note that comparison of our data to that of prior ChIP-seq experiments is limited due to differences in cell types analyzed. Moreover, our results do not provide information on whether changes in gene expression result from direct or indirect effects of CHD7.
Roles for CHD7 in regulation of gene expression during tissue development and maintenance are emerging for multiple organs. In a study of neural crest formation in Xenopus, CHD7 was implicated to regulate multipotent neural crest-like cells by binding to PBAF components, including BRG1, BAF170, BAF155, BAF57, PB1, ARID2 and BRD7 (85). In mesenchymal stem cells, CHD7 regulates cell fate specification during osteoblast and adipocyte differentiation (86). CHD7 forms a complex with NLK, SETDB1 and PPAR-γ, and binds to methylated lysine 4 and lysine 9 residues on histone H3 at PPAR-γ target promoters, which suppresses ligand-induced transactivation of PPAR-γ target genes (86). Additionally, the CHD7 Drosophila orthologue, Kismet, is involved in transcriptional elongation by RNA polymerase II through recruitment of ASH1 and TRX, and may help maintain stem cell pluripotency by regulating methylation of histone H3 lysine 27 (87). These data provide evidence that the role of CHD7 in tissue development and maintenance is dependent upon coordination of multiple components. Our results showing that CHD7 affects GnRH neurogenesis and signaling, potentially by influencing transcriptional regulation of key target genes, add to this emerging evidence.
Previous reports of reproductive phenotypes in Chd7 haploinsufficient mice using the Chd7Whi allele (which contains a nonsense mutation) (39) showed that Chd7Whi/+ mice have genital hypoplasia, hypogonadism and increased time to first litter (38). Similar to Chd7Gt/+ mice, Chd7Whi/+ mice were also reported to have reduced GnRH neurons in the adult hypothalamus (38). However, the mechanism underlying decreased GnRH neurons in Chd7Whi/+ mice was not evaluated. Our data provide a potential cellular mechanism for the reduced gonadal size and decreased fertility associated with CHD7 haploinsufficiency. Compared with wild-type littermates, Chd7Gt/+ mice have reduced capacity for GnRH signal to pituitary gonadotropes, leading to less LH and FSH available to act on the gonads for sex-steroid production. Reductions in circulating LH and FSH negatively affect both gonad size and fertility (88). Although our data do not exclude the possibility of a subtle defect in GnRH migration, reduced GnRH neurons in Chd7Gt/+ mice are explained at least in part by defects in GnRH neurogenesis during development. Together, these data support our model for an underlying intrinsic cellular mechanism for hypogonadotropic hypogonadism in Chd7 haploinsufficient mice.
Neural progenitors must be tightly regulated during development by factors that are likely to be temporally and spatially restricted (89). The neurogenic potential of progenitors is influenced by both BMP and FGF signaling in the central and peripheral nervous systems (90–94). The opposing effects of FGF and BMP signaling must be tightly regulated during olfactory development to obtain the proper proportions of olfactory sensory and olfactory respiratory cell types; notably, reductions in either FGF8 or BMP4 can cause decreased cellular proliferation in the olfactory placode (95). In addition, BMP4 and BMP7 have dosage- and threshold-dependent effects on neurogenesis in the olfactory epithelium (90–92,96,97). Although BMP activity is generally thought to inhibit neurogenesis, low doses of BMP4 are required for promoting neurogenesis in multiple tissues including the olfactory placode, hippocampus and subventricular zone (95–100). Neurogenesis in the olfactory epithelium is also regulated by FGF8 and FGFR1, and mutations in Fgf8 or Fgfr1 in humans and mice cause reduced numbers of olfactory sensory neurons and GnRH neurons (23–28). FGF2 induces neurogenesis and increases cellular proliferation of progenitors in olfactory tissues (96), and FGF2 signals via FGFR1 to induce proliferation of olfactory bulb progenitors in the subventricular zone (101). Our data provide the first evidence that reduced CHD7 dosage results in decreased expression of Fgfr1 and Bmp4, and that CHD7 is required for proper cellular proliferation and genesis of GnRH neurons during development.
Proper CHD7 dosage is critical for development in humans and mice (30,31,39,102). However, CHD7 function in adult tissues and post-mitotic cells has not been fully explored. Although adult Chd7 expression in the brain is mostly restricted to proliferative regions, cells expressing Chd7 are scattered throughout the adult mouse hypothalamus [data available on GENSAT website (http://www.gensat.org/index.html)]. CHD7 is also expressed in GnRH neurons in vivo (Fig. 3I and J) and in GnRH neuronal cell lines (8), but it is unknown whether CHD7 has a functional role in post-mitotic GnRH neurons. Our data provide the first evidence that CHD7 haploinsufficiency in mice results in decreased GnRH1 and Otx2 expression in the adult hypothalamus. These data are consistent with a recent report showing that the loss of CHD7 in the developing mouse inner ear causes reduced expression of Otx2 (83). Interestingly, humans with OTX2 mutations have some clinical features similar to those seen in CHARGE, including pituitary hormone deficiency, short stature and ocular colobomata (103,104). Thus, fewer GnRH neurons in Chd7Gt/+ mice and defects in GnRH signaling may be exacerbated by decreased expression of GnRH1. Together, these results provide evidence that CHD7 controls multiple facets of normal hypothalamic-pituitary signaling, by regulating GnRH neuronal cell number as well as expression of transcription factors and morphogens required for proper GnRH signaling.
MATERIALS AND METHODS
Mice
Chd7Gt/+ mice were generated and characterized as previously described (30). Mice were maintained by backcrossing with 129S1/Sv1mJ (Jackson Laboratory) mice to generation N6–N8. Mice are housed with a 10/14 h dark/light cycle and fed ad libitum. Serum was collected in the afternoon between 2:00 and 3:00 pm by cardiac exsanguinations after anesthesia with 250 mg/kg body weight tribromoethanol. Timed pregnancies were established, and the morning of plug identification designated as E0.5. Embryos were collected after cervical dislocation and hysterectomy, and washed briefly in phosphate buffered saline (PBS). Amniotic sacs were collected and DNA isolated for PCR genotyping as described (30). All procedures were approved by The University of Michigan University Committee on Use and Care of Animals (UCUCA).
Vaginal smears
Vaginal smears were taken at the same time daily over a 3-month period. One drop of water from a Pasteur pipette was gently expelled into the vagina and aspirated four to six times, and then transferred to a microscope slide. The smear was dried at room temperature and then fixed with 100% methanol. Fixed slides were stained for 30 min in 2% Giemsa (GibcoBRL, Grand Island, NY, USA). Stained slides were then analyzed for stage of estrus cycle.
Immunofluorescence
Embryos were fixed in 4% paraformaldehyde for 30 min (E10.5) to 1.5h (E12.5), and then washed in PBS and dehydrated in an ethanol gradient. Embryos were subsequently embedded in paraffin and sectioned at 7 μm. Three- to 4-month-old Chd7+/+ and Chd7Gt/+ sex-matched littermate mice were anesthetized with 250 mg/kg body weight tribromoethanol and perfusion fixed with 4% paraformaldehyde. Mice were then decapitated and heads placed in 4% paraformaldehyde overnight at 4°C. Heads were incubated in RDO Rapid Decalcifier (Apex Engineering, Aurora, IL, USA) for 4–6 h followed by 30% sucrose protection overnight at 4°C. Tissue was flash frozen in O.C.T. embedding medium (Tissue Tek, Torrance, CA, USA) for sectioning at 14 μm. Following paraffin or cryosectioning, sections were processed for immunofluorescence with antibodies against CHD7 (1:1000; Abcam, Cambridge, MA, USA), GnRH (1:150; Abcam (ab5617)), β-galactosidase (1:200; Vector Laboratories, Burlington, CA, USA), AVP (1:500; Abcam) or phospho-histone H3 (1:200; Millipore, Temecula, CA, USA). Secondary antibodies were used at 1:200 and were conjugated with Alexa 488, Alexa 555 or biotin with streptavidin-HRP (Vector Laboratories) and or biotinylated secondary antibodies conjugated with streptavidin-Alexa488 or streptavidin-Alexa555 (Molecular Probes, Eugene, OR, USA and Invitrogen, Carlsbad, CA, USA). Images were captured by single channel fluorescence microscopy on a Leica upright DMRB microscope and processed in Photoshop v.9.0 (San Jose, CA, USA). NIH ImageJ software (Bethesda, MD, USA) was used to analyze anti-GnRH fluorescence intensity over at least 30 images of the median eminence per animal. Fluorescence intensity was measured in restricted areas of the median eminence and background was subtracted to obtain the final measurements. GnRH neuronal cell counts were performed by counting cell bodies only on at least four adults (4 pairs male and 4 pairs female) and four embryos of each genotype at each time point. A minimum of 30 sections per animal were used for all cell counts. Cell counts were analyzed for significance by Student's t-test using two-tailed unequal variance.
Mouse insulin and leptin ELISA assay
Circulating serum levels of insulin and leptin were assayed by ELISA from 3–4-month-old Chd7+/+ and Chd7Gt/+ stage of estrus cycle-matched littermate mice. Insulin was measured using the Ultra Sensitive Mouse Insulin ELISA Kit (Crystal Chem, Downers Grove, IL, USA). Leptin was measured using the Mouse Leptin ELISA Kit (Crystal Chem). Absorbance (A450) for insulin and leptin was measured and analyzed using a standard curve. Differences in absorbance were analyzed for significance by Student's t-test using two-tailed unequal variance.
Mouse GH and IGF1 ELISA assay
Circulating levels of GH and IGF1 were assayed by ELISA. GH was measured in serum collected from 3–4-month-old Chd7+/+ and Chd7Gt/+ stage of estrus cycle-matched littermate mice using the ELISA Kit Cat# EZRMGH-45K (LINCO Research, St Charles, MO, USA). IGF1 was measured in serum using the ELISA Kit Cat# DSL-10-29200 (Diagnostic Systems Laboratories, Webster, TX, USA). Absorbance (A450) for GH and IGF1 was measured and analyzed using a standard curve. Differences in absorbance were analyzed for significance by Student's t-test using two-tailed unequal variance.
Mouse LH sandwich assay (MLHS)
Circulating levels of LH were assayed at the University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core using the following methods: LH was measured in serum by a sensitive two-site sandwich immunoassay (105,106) using monoclonal antibodies against bovine LH (no. 581B7) and against the human LH-beta subunit (no. 5303: Medix Kauniainen, Finland) as previously described (106). The tracer antibody (no. 518B7) was kindly provided by Dr Janet Roser (107) (Department of Animal Science, University of California, Davis) and iodinated by the chloramine T method and purified on Sephadex G-50 columns. The capture antibody (no. 5303) was biotinylated and immobilized on avidin-coated polystyrene beads (7 mm; Epitope Diagnostics, Inc., San Diego, CA, USA). Mouse LH reference prep (AFP5306A; provided by Dr A.F. Parlow and the National Hormone and Peptide program) was used as standard. The mouse LH sandwich assay (MLHS) has a sensitivity of 0.07 ng/ml.
Mouse FSH radioimmunoassay (RIA)
Circulating levels of FSH were assayed at the University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core using the following methods: FSH was assayed by radioimmunoassay (RIA) using reagents provided by Dr A.F. Parlow and the National Hormone and Peptide Program, as previously described (108). Mouse FSH reference prep AFP5308D was used for assay standards and mouse FSH antiserum (guinea pig; AFP-1760191) diluted to a final concentration of 1:400 000 was used as primary antibody. The secondary antibody was purchased from Equitech-Bio, Inc. and diluted to a final concentration of 1:25. The assay has a sensitivity of 2.0 ng/ml and <0.5% cross-reactivity with other pituitary hormones.
GnRH agonist and antagonist assays
Three- to 4-month-old Chd7+/+ and Chd7Gt/+ sex-matched littermate mice were given a subcutaneous injection of either the GnRH agonist leuprolide (Sigma, St Louis, MO, USA) or the GnRH antagonist antide (Sigma). Leuprolide was dissolved in 0.9% saline and administered at a dose of 1 mg/kg body weight 2h prior to cardiac exsanguination (69,70). Antide was dissolved in 20% propylene glycol and 0.9% saline and administered at a dose of 3.0 mg/kg body weight 2h prior to cardiac exsanguination (71,72). Serum was collected and analyzed by MLHS and mouse FSH RIA.
Cellular proliferation assays
Wild-type, Chd7Gt/+ and Chd7Gt/Gt E10.5 embryos were processed as above for immunofluorescence. Serial adjacent sections were labeled with rabbit anti-H3 (1:200, Millipore) followed by incubation with anti-rabbit AlexaFluor488 conjugated secondary antibody (Invitrogen). A minimum of 30 sections per embryo were photographed by single channel fluorescence microscopy on a Leica upright DMRB microscope and processed for dual channel imaging with Adobe Photoshop software v9.0 (San Jose, CA, USA). Cell counts were performed on at least four embryos of each genotype. Statistical significance was determined using one-way analysis of variance (ANOVA) analysis with GraphPad Prism 5 software.
TUNEL assays
Wild-type and Chd7Gt/+ E10.5, E11.5 and E12.5 embryos were processed as above for immunofluorescence. Serial adjacent sections were assayed for apoptosis using Fluorescein-FragEL DNA Fragmentation Detection Kit (Calbiochem, Darmstadt, Germany). A minimum of 30 sections per embryo were photographed by single channel fluorescence microscopy on a Leica upright DMRB microscope and processed for dual channel imaging with Adobe Photoshop software v9.0. Cell counts were performed on at least four embryos of each genotype at each time point. Cell counts were analyzed for significance by Student's t-test using two-tailed unequal variance.
RNA isolation and real-time PCR
Three- to 4-month-old Chd7+/+ and Chd7Gt/+ sex-matched littermate mice were euthanized by cervical dislocation and decapitated. The brain and pituitary were removed from the head. The hypothalamus was microdissected from whole brain and placed in ice-cold TRIzol (Invitrogen) and mechanically homogenized prior to RNA isolation. Pituitary RNA was isolated using the RNAqueous-Micro RNA Isolation Kit (Ambion, Austin, TX, USA). Wild-type, Chd7Gt/+ and Chd7Gt/Gt E10.5 embryos were harvested as described above. The olfactory placode was microdissected and RNA isolated using the RNAqueous-Micro RNA Isolation Kit (Ambion). Isolated RNA from adult and embryonic tissues was treated with DNase I prior to cDNA synthesis. cDNA was generated using Superscript First-Strand cDNA Synthesis system for reverse transcriptase-polymerase chain reaction (RT-PCR) (Invitrogen) with random primers.
Relative expression levels were assayed utilizing TaqMan Gene Expression Master Mix and TaqMan probes (Applied Biosystems, Foster City, CA, USA) for GnRH1, GnRHR, Kiss1, Otx2, Bmp4, Fgfr1, Fgf8 and Gapdh. Reactions were run in triplicate in an Applied Biosystems StepOne-Plus Real-Time PCR System. The level of Gapdh was used as an internal control. The difference in CT between the assayed gene and Gapdh for any given sample was defined as ΔCT(X). The difference in ΔCT(x) between two samples was defined as ΔΔCT(X), which represents a relative difference in expression of the assayed gene. The fold change of the assayed gene relative to Gapdh was defined as 2−ΔΔCT (109). DataAssist software (Applied Biosystems) was used for statistical analysis and to confirm ΔCT(X) calculation.
FUNDING
This work was supported by the National Institutes of Health (F31DC010955-01 to W.S.L., R01DC009410 and an ARRA supplement to D.M.M.).
ACKNOWLEDGEMENTS
The authors would like to thank Dr Sally Camper for insightful discussions and critical reading of the manuscript. We also thank Gina Leinninger for help with ELISA assays and Joseph Micucci for help analyzing CHiP-seq data sets.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Harris J., Robert E., Kallen B. Epidemiology of choanal atresia with special reference to the CHARGE association. Pediatrics. 1997;99:363–367. doi: 10.1542/peds.99.3.363. [DOI] [PubMed] [Google Scholar]
- 2.Issekutz K.A., Graham J.M., Jr, Prasad C., Smith I.M., Blake K.D. An epidemiological analysis of CHARGE syndrome: preliminary results from a Canadian study. Am. J. Med. Genet. A. 2005;133:309–317. doi: 10.1002/ajmg.a.30560. [DOI] [PubMed] [Google Scholar]
- 3.Kallen K., Robert E., Mastroiacovo P., Castilla E.E., Kallen B. CHARGE Association in newborns: a registry-based study. Teratology. 1999;60:334–343. doi: 10.1002/(SICI)1096-9926(199912)60:6<334::AID-TERA5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 4.Hall B.D. Choanal atresia and associated multiple anomalies. J. Pediatr. 1979;95:395–398. doi: 10.1016/s0022-3476(79)80513-2. [DOI] [PubMed] [Google Scholar]
- 5.Asakura Y., Toyota Y., Muroya K., Kurosawa K., Fujita K., Aida N., Kawame H., Kosaki K., Adachi M. Endocrine and radiological studies in patients with molecularly confirmed CHARGE syndrome. J. Clin. Endocrinol. Metab. 2008;93:920–924. doi: 10.1210/jc.2007-1419. [DOI] [PubMed] [Google Scholar]
- 6.Jongmans M.C., Admiraal R.J., van der Donk K.P., Vissers L.E., Baas A.F., Kapusta L., van Hagen J.M., Donnai D., de Ravel T.J., Veltman J.A., et al. CHARGE syndrome: the phenotypic spectrum of mutations in the CHD7 gene. J. Med. Genet. 2006;43:306–314. doi: 10.1136/jmg.2005.036061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jongmans M.C., van Ravenswaaij-Arts C.M., Pitteloud N., Ogata T., Sato N., Claahsen-van der Grinten H.L., van der Donk K., Seminara S., Bergman J.E., Brunner H.G., et al. CHD7 mutations in patients initially diagnosed with Kallmann syndrome—the clinical overlap with CHARGE syndrome. Clin. Genet. 2009;75:65–71. doi: 10.1111/j.1399-0004.2008.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H.G., Kurth I., Lan F., Meliciani I., Wenzel W., Eom S.H., Kang G.B., Rosenberger G., Tekin M., Ozata M., et al. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am. J. Hum. Genet. 2008;83:511–519. doi: 10.1016/j.ajhg.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lalani S.R., Safiullah A.M., Fernbach S.D., Harutyunyan K.G., Thaller C., Peterson L.E., McPherson J.D., Gibbs R.A., White L.D., Hefner M., et al. Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am. J. Hum. Genet. 2006;78:303–314. doi: 10.1086/500273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogata T., Fujiwara I., Ogawa E., Sato N., Udaka T., Kosaki K. Kallmann syndrome phenotype in a female patient with CHARGE syndrome and CHD7 mutation. Endocr. J. 2006;53:741–743. doi: 10.1507/endocrj.k06-099. [DOI] [PubMed] [Google Scholar]
- 11.Pinto G., Abadie V., Mesnage R., Blustajn J., Cabrol S., Amiel J., Hertz-Pannier L., Bertrand A.M., Lyonnet S., Rappaport R., et al. CHARGE syndrome includes hypogonadotropic hypogonadism and abnormal olfactory bulb development. J. Clin. Endocrinol. Metab. 2005;90:5621–5626. doi: 10.1210/jc.2004-2474. [DOI] [PubMed] [Google Scholar]
- 12.Zentner G.E., Layman W.S., Martin D.M., Scacheri P.C. Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am. J. Med. Genet. A. 2010;152A:674–686. doi: 10.1002/ajmg.a.33323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camper S., Suh H., Raetzman L., Douglas K., Cushman L., Nasonkin I., Burrows H., Gage P., Martin D. In: Mouse Development Patterning, Morphogenesis, and Organogenesis. Rossant J., Tam P., editors. San Diego: Academic Press; 2002. pp. 499–518. [Google Scholar]
- 14.Krsmanovic L.Z., Hu L., Leung P.K., Feng H., Catt K.J. The hypothalamic GnRH pulse generator: multiple regulatory mechanisms. Trends Endocrinol. Metab. 2009;20:402–408. doi: 10.1016/j.tem.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maffucci J.A., Gore A.C. Chapter 2: hypothalamic neural systems controlling the female reproductive life cycle gonadotropin-releasing hormone, glutamate, and GABA. Int. Rev. Cell Mol. Biol. 2009;274:69–127. doi: 10.1016/S1937-6448(08)02002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wray S. Development of gonadotropin-releasing hormone-1 neurons. Front. Neuroendocrinol. 2002;23:292–316. doi: 10.1016/s0091-3022(02)00001-8. [DOI] [PubMed] [Google Scholar]
- 17.Chu Z., Andrade J., Shupnik M.A., Moenter S.M. Differential regulation of gonadotropin-releasing hormone neuron activity and membrane properties by acutely applied estradiol: dependence on dose and estrogen receptor subtype. J. Neurosci. 2009;29:5616–5627. doi: 10.1523/JNEUROSCI.0352-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelley C.G., Lavorgna G., Clark M.E., Boncinelli E., Mellon P.L. The Otx2 homeoprotein regulates expression from the gonadotropin-releasing hormone proximal promoter. Mol. Endocrinol. 2000;14:1246–1256. doi: 10.1210/mend.14.8.0509. [DOI] [PubMed] [Google Scholar]
- 19.Larder R., Mellon P.L. Otx2 induction of the gonadotropin-releasing hormone promoter is modulated by direct interactions with Grg co-repressors. J. Biol. Chem. 2009;284:16966–16978. doi: 10.1074/jbc.M109.002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dateki S., Kosaka K., Hasegawa K., Tanaka H., Azuma N., Yokoya S., Muroya K., Adachi M., Tajima T., Motomura K., et al. Heterozygous orthodenticle homeobox 2 mutations are associated with variable pituitary phenotype. J. Clin. Endocrinol. Metab. 2010;95:756–764. doi: 10.1210/jc.2009-1334. [DOI] [PubMed] [Google Scholar]
- 21.Omodei D., Acampora D., Mancuso P., Prakash N., Di Giovannantonio L.G., Wurst W., Simeone A. Anterior-posterior graded response to Otx2 controls proliferation and differentiation of dopaminergic progenitors in the ventral mesencephalon. Development. 2008;135:3459–3470. doi: 10.1242/dev.027003. [DOI] [PubMed] [Google Scholar]
- 22.Vernay B., Koch M., Vaccarino F., Briscoe J., Simeone A., Kageyama R., Ang S.L. Otx2 regulates subtype specification and neurogenesis in the midbrain. J. Neurosci. 2005;25:4856–4867. doi: 10.1523/JNEUROSCI.5158-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawauchi S., Shou J., Santos R., Hebert J.M., McConnell S.K., Mason I., Calof A.L. Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development. 2005;132:5211–5223. doi: 10.1242/dev.02143. [DOI] [PubMed] [Google Scholar]
- 24.Hebert J.M., Lin M., Partanen J., Rossant J., McConnell S.K. FGF signaling through FGFR1 is required for olfactory bulb morphogenesis. Development. 2003;130:1101–1111. doi: 10.1242/dev.00334. [DOI] [PubMed] [Google Scholar]
- 25.Hsu P., Yu F., Feron F., Pickles J.O., Sneesby K., Mackay-Sim A. Basic fibroblast growth factor and fibroblast growth factor receptors in adult olfactory epithelium. Brain Res. 2001;896:188–197. doi: 10.1016/s0006-8993(01)02173-4. [DOI] [PubMed] [Google Scholar]
- 26.Chung W.C., Moyle S.S., Tsai P.S. Fibroblast growth factor 8 signaling through fibroblast growth factor receptor 1 is required for the emergence of gonadotropin-releasing hormone neurons. Endocrinology. 2008;149:4997–5003. doi: 10.1210/en.2007-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falardeau J., Chung W.C., Beenken A., Raivio T., Plummer L., Sidis Y., Jacobson-Dickman E.E., Eliseenkova A.V., Ma J., Dwyer A., et al. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J. Clin. Invest. 2008;118:2822–2831. doi: 10.1172/JCI34538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai P.S., Moenter S.M., Postigo H.R., El Majdoubi M., Pak T.R., Gill J.C., Paruthiyil S., Werner S., Weiner R.I. Targeted expression of a dominant-negative fibroblast growth factor (FGF) receptor in gonadotropin-releasing hormone (GnRH) neurons reduces FGF responsiveness and the size of GnRH neuronal population. Mol. Endocrinol. 2005;19:225–236. doi: 10.1210/me.2004-0330. [DOI] [PubMed] [Google Scholar]
- 29.Layman W.S., McEwen D.P., Beyer L.A., Lalani S.R., Fernbach S.D., Oh E., Swaroop A., Hegg C.C., Raphael Y., Martens J.R., et al. Defects in neural stem cell proliferation and olfaction in Chd7 deficient mice indicate a mechanism for hyposmia in human CHARGE syndrome. Hum. Mol. Genet. 2009;18:1909–1923. doi: 10.1093/hmg/ddp112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurd E.A., Capers P.L., Blauwkamp M.N., Adams M.E., Raphael Y., Poucher H.K., Martin D.M. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm. Genome. 2007;18:94–104. doi: 10.1007/s00335-006-0107-6. [DOI] [PubMed] [Google Scholar]
- 31.Sanlaville D., Etchevers H.C., Gonzales M., Martinovic J., Clement-Ziza M., Delezoide A.L., Aubry M.C., Pelet A., Chemouny S., Cruaud C., et al. Phenotypic spectrum of CHARGE syndrome in fetuses with CHD7 truncating mutations correlates with expression during human development. J. Med. Genet. 2006;43:211–217. doi: 10.1136/jmg.2005.036160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wray S., Grant P., Gainer H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc. Natl Acad. Sci. USA. 1989;86:8132–8136. doi: 10.1073/pnas.86.20.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Becker P.B., Horz W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 34.Eberharter A., Becker P.B. ATP-dependent nucleosome remodelling: factors and functions. J. Cell Sci. 2004;117:3707–3711. doi: 10.1242/jcs.01175. [DOI] [PubMed] [Google Scholar]
- 35.Lusser A., Kadonaga J.T. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25:1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- 36.Narlikar G.J., Fan H.Y., Kingston R.E. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 37.Smith C.L., Peterson C.L. ATP-dependent chromatin remodeling. Curr. Top. Dev. Biol. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- 38.Bergman J.E., Bosman E.A., van Ravenswaaij-Arts C.M., Steel K.P. Study of smell and reproductive organs in a mouse model for CHARGE syndrome. Eur. J. Hum. Genet. 2009;18:171–177. doi: 10.1038/ejhg.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bosman E.A., Penn A.C., Ambrose J.C., Kettleborough R., Stemple D.L., Steel K.P. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum. Mol. Genet. 2005;14:3463–3476. doi: 10.1093/hmg/ddi375. [DOI] [PubMed] [Google Scholar]
- 40.Nelson J.F., Karelus K., Felicio L.S., Johnson T.E. Genetic influences on the timing of puberty in mice. Biol. Reprod. 1990;42:649–655. doi: 10.1095/biolreprod42.4.649. [DOI] [PubMed] [Google Scholar]
- 41.Lee J.M., Appugliese D., Kaciroti N., Corwyn R.F., Bradley R.H., Lumeng J.C. Weight status in young girls and the onset of puberty. Pediatrics. 2007;119:e624–e630. doi: 10.1542/peds.2006-2188. [DOI] [PubMed] [Google Scholar]
- 42.Ahima R.S., Dushay J., Flier S.N., Prabakaran D., Flier J.S. Leptin accelerates the onset of puberty in normal female mice. J. Clin. Invest. 1997;99:391–395. doi: 10.1172/JCI119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foster D.L., Ebling F.J., Micka A.F., Vannerson L.A., Bucholtz D.C., Wood R.I., Suttie J.M., Fenner D.E. Metabolic interfaces between growth and reproduction. I. Nutritional modulation of gonadotropin, prolactin, and growth hormone secretion in the growth-limited female lamb. Endocrinology. 1989;125:342–350. doi: 10.1210/endo-125-1-342. [DOI] [PubMed] [Google Scholar]
- 44.Frisch R.E., McArthur J.W. Menstrual cycles: fatness as a determinant of minimum weight for height necessary for their maintenance or onset. Science. 1974;185:949–951. doi: 10.1126/science.185.4155.949. [DOI] [PubMed] [Google Scholar]
- 45.Kennedy G.C. Interactions between feeding behavior and hormones during growth. Ann. N. Y. Acad. Sci. 1969;157:1049–1061. doi: 10.1111/j.1749-6632.1969.tb12936.x. [DOI] [PubMed] [Google Scholar]
- 46.Budak E., Fernandez Sanchez M., Bellver J., Cervero A., Simon C., Pellicer A. Interactions of the hormones leptin, ghrelin, adiponectin, resistin, and PYY3–36 with the reproductive system. Fertil. Steril. 2006;85:1563–1581. doi: 10.1016/j.fertnstert.2005.09.065. [DOI] [PubMed] [Google Scholar]
- 47.Plum L., Schubert M., Bruning J.C. The role of insulin receptor signaling in the brain. Trends Endocrinol. Metab. 2005;16:59–65. doi: 10.1016/j.tem.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Schneider J.E. Energy balance and reproduction. Physiol. Behav. 2004;81:289–317. doi: 10.1016/j.physbeh.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 49.Bruning J.C., Gautam D., Burks D.J., Gillette J., Schubert M., Orban P.C., Klein R., Krone W., Muller-Wieland D., Kahn C.R. Role of brain insulin receptor in control of body weight and reproduction. Science. 2000;289:2122–2125. doi: 10.1126/science.289.5487.2122. [DOI] [PubMed] [Google Scholar]
- 50.Chehab F.F., Lim M.E., Lu R. Correction of the sterility defect in homozygous obese female mice by treatment with the human recombinant leptin. Nat. Genet. 1996;12:318–320. doi: 10.1038/ng0396-318. [DOI] [PubMed] [Google Scholar]
- 51.He D., Funabashi T., Sano A., Uemura T., Minaguchi H., Kimura F. Effects of glucose and related substrates on the recovery of the electrical activity of gonadotropin-releasing hormone pulse generator which is decreased by insulin-induced hypoglycemia in the estrogen-primed ovariectomized rat. Brain Res. 1999;820:71–76. doi: 10.1016/s0006-8993(98)01358-4. [DOI] [PubMed] [Google Scholar]
- 52.Hiney J.K., Srivastava V., Nyberg C.L., Ojeda S.R., Dees W.L. Insulin-like growth factor I of peripheral origin acts centrally to accelerate the initiation of female puberty. Endocrinology. 1996;137:3717–3728. doi: 10.1210/endo.137.9.8756538. [DOI] [PubMed] [Google Scholar]
- 53.Longo K.M., Sun Y., Gore A.C. Insulin-like growth factor-I effects on gonadotropin-releasing hormone biosynthesis in GT1–7 cells. Endocrinology. 1998;139:1125–1132. doi: 10.1210/endo.139.3.5852. [DOI] [PubMed] [Google Scholar]
- 54.Mounzih K., Lu R., Chehab F.F. Leptin treatment rescues the sterility of genetically obese ob/ob males. Endocrinology. 1997;138:1190–1193. doi: 10.1210/endo.138.3.5024. [DOI] [PubMed] [Google Scholar]
- 55.Schneider J.E., Wade G.N. Availability of metabolic fuels controls estrous cyclicity of Syrian hamsters. Science. 1989;244:1326–1328. doi: 10.1126/science.2734610. [DOI] [PubMed] [Google Scholar]
- 56.Luque R.M., Park S., Kineman R.D. Severity of the catabolic condition differentially modulates hypothalamic expression of growth hormone-releasing hormone in the fasted mouse: potential role of neuropeptide Y and corticotropin-releasing hormone. Endocrinology. 2007;148:300–309. doi: 10.1210/en.2006-0592. [DOI] [PubMed] [Google Scholar]
- 57.Staley K., Scharfman H. A woman's prerogative. Nat. Neurosci. 2005;8:697–699. doi: 10.1038/nn0605-697. [DOI] [PubMed] [Google Scholar]
- 58.Kumar T.R., Wang Y., Lu N., Matzuk M.M. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat. Genet. 1997;15:201–204. doi: 10.1038/ng0297-201. [DOI] [PubMed] [Google Scholar]
- 59.Layman L.C., McDonough P.G. Mutations of follicle stimulating hormone-beta and its receptor in human and mouse: genotype/phenotype. Mol. Cell Endocrinol. 2000;161:9–17. doi: 10.1016/s0303-7207(99)00217-8. [DOI] [PubMed] [Google Scholar]
- 60.Layman L.C., Porto A.L., Xie J., da Motta L.A., da Motta L.D., Weiser W., Sluss P.M. FSH beta gene mutations in a female with partial breast development and a male sibling with normal puberty and azoospermia. J. Clin. Endocrinol. Metab. 2002;87:3702–3707. doi: 10.1210/jcem.87.8.8724. [DOI] [PubMed] [Google Scholar]
- 61.Phillip M., Arbelle J.E., Segev Y., Parvari R. Male hypogonadism due to a mutation in the gene for the beta-subunit of follicle-stimulating hormone. N. Engl. J. Med. 1998;338:1729–1732. doi: 10.1056/NEJM199806113382404. [DOI] [PubMed] [Google Scholar]
- 62.Lamba P., Santos M.M., Philips D.P., Bernard D.J. Acute regulation of murine follicle-stimulating hormone beta subunit transcription by activin A. J. Mol. Endocrinol. 2006;36:201–220. doi: 10.1677/jme.1.01961. [DOI] [PubMed] [Google Scholar]
- 63.Weiss J., Guendner M.J., Halvorson L.M., Jameson J.L. Transcriptional activation of the follicle-stimulating hormone beta-subunit gene by activin. Endocrinology. 1995;136:1885–1891. doi: 10.1210/endo.136.5.7720634. [DOI] [PubMed] [Google Scholar]
- 64.Wen S., Ai W., Alim Z., Boehm U. Embryonic gonadotropin-releasing hormone signaling is necessary for maturation of the male reproductive axis. Proc. Natl Acad. Sci. USA. 2010;152:1515–1526. doi: 10.1073/pnas.1000423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pask A.J., Kanasaki H., Kaiser U.B., Conn P.M., Janovick J.A., Stockton D.W., Hess D.L., Justice M.J., Behringer R.R. A novel mouse model of hypogonadotrophic hypogonadism: N-ethyl-N-nitrosourea-induced gonadotropin-releasing hormone receptor gene mutation. Mol. Endocrinol. 2005;19:972–981. doi: 10.1210/me.2004-0192. [DOI] [PubMed] [Google Scholar]
- 66.Wu S., Wilson M.D., Busby E.R., Isaac E.R., Sherwood N.M. Disruption of the single copy gonadotropin-releasing hormone receptor in mice by gene trap: severe reduction of reproductive organs and functions in developing and adult mice. Endocrinology. 2010;151:1142–1152. doi: 10.1210/en.2009-0598. [DOI] [PubMed] [Google Scholar]
- 67.Messager S., Chatzidaki E.E., Ma D., Hendrick A.G., Zahn D., Dixon J., Thresher R.R., Malinge I., Lomet D., Carlton M.B., et al. Kisspeptin directly stimulates gonadotropin-releasing hormone release via G protein-coupled receptor 54. Proc. Natl Acad. Sci. USA. 2005;102:1761–1766. doi: 10.1073/pnas.0409330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gill J.C., Wadas B., Chen P., Portillo W., Reyna A., Jorgensen E., Mani S., Schwarting G.A., Moenter S.M., Tobet S., et al. The gonadotropin-releasing hormone (GnRH) neuronal population is normal in size and distribution in GnRH-deficient and GnRH receptor-mutant hypogonadal mice. Endocrinology. 2008;149:4596–4604. doi: 10.1210/en.2008-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sofianos Z.D., Katsila T., Kostomitsopoulos N., Balafas V., Matsoukas J., Tselios T., Tamvakopoulos C. In vivo evaluation and in vitro metabolism of leuprolide in mice—mass spectrometry-based biomarker measurement for efficacy and toxicity. J. Mass Spectrom. 2008;43:1381–1392. doi: 10.1002/jms.1416. [DOI] [PubMed] [Google Scholar]
- 70.Yamazaki I., Okada H. A radioimmunoassay for a highly active luteinizing hormone-releasing hormone analogue and relation between the serum level of the analogue and that of gonadotropin. Endocrinol. Jpn. 1980;27:593–605. doi: 10.1507/endocrj1954.27.593. [DOI] [PubMed] [Google Scholar]
- 71.Danforth D.R., Arbogast L.K., Friedman C.I. Acute depletion of murine primordial follicle reserve by gonadotropin-releasing hormone antagonists. Fertil. Steril. 2005;83:1333–1338. doi: 10.1016/j.fertnstert.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 72.Ljungqvist A., Feng D.M., Hook W., Shen Z.X., Bowers C., Folkers K. Antide and related antagonists of luteinizing hormone release with long action and oral activity. Proc. Natl Acad. Sci. USA. 1988;85:8236–8240. doi: 10.1073/pnas.85.21.8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bennett-Clarke C., Joseph S.A. Immunocytochemical distribution of LHRH neurons and processes in the rat: hypothalamic and extrahypothalamic locations. Cell Tissue Res. 1982;221:493–504. doi: 10.1007/BF00215698. [DOI] [PubMed] [Google Scholar]
- 74.Wierman M.E., Kiseljak-Vassiliades K., Tobet S. Gonadotropin-releasing hormone (GnRH) neuron migration: initiation, maintenance and cessation as critical steps to ensure normal reproductive function. Front. Neuroendocrinol. 2010;32:43–52. doi: 10.1016/j.yfrne.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Colledge W.H., Mei H., Tassigny X.D. Mouse models to study the central regulation of puberty. Mol. Cell Endocrinol. 2010;324:12–20. doi: 10.1016/j.mce.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 76.Matsumoto S., Yamazaki C., Masumoto K.H., Nagano M., Naito M., Soga T., Hiyama H., Matsumoto M., Takasaki J., Kamohara M., et al. Abnormal development of the olfactory bulb and reproductive system in mice lacking prokineticin receptor PKR2. Proc. Natl Acad. Sci. USA. 2006;103:4140–4145. doi: 10.1073/pnas.0508881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gamble J.A., Karunadasa D.K., Pape J.R., Skynner M.J., Todman M.G., Bicknell R.J., Allen J.P., Herbison A.E. Disruption of ephrin signaling associates with disordered axophilic migration of the gonadotropin-releasing hormone neurons. J. Neurosci. 2005;25:3142–3150. doi: 10.1523/JNEUROSCI.4759-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cogliati T., Delgado-Romero P., Norwitz E.R., Guduric-Fuchs J., Kaiser U.B., Wray S., Kirsch I.R. Pubertal impairment in Nhlh2 null mice is associated with hypothalamic and pituitary deficiencies. Mol. Endocrinol. 2007;21:3013–3027. doi: 10.1210/me.2005-0337. [DOI] [PubMed] [Google Scholar]
- 79.Compagnucci C., Fish J.L., Schwark M., Tarabykin V., Depew M.J. Pax6 regulates craniofacial form through its control of an essential cephalic ectodermal patterning center. Genesis. 2011;49:307–325. doi: 10.1002/dvg.20724. [DOI] [PubMed] [Google Scholar]
- 80.Kaiser U.B., Jakubowiak A., Steinberger A., Chin W.W. Regulation of rat pituitary gonadotropin-releasing hormone receptor mRNA levels in vivo and in vitro. Endocrinology. 1993;133:931–934. doi: 10.1210/endo.133.2.8393779. [DOI] [PubMed] [Google Scholar]
- 81.White B.R., Duval D.L., Mulvaney J.M., Roberson M.S., Clay C.M. Homologous regulation of the gonadotropin-releasing hormone receptor gene is partially mediated by protein kinase C activation of an activator protein-1 element. Mol. Endocrinol. 1999;13:566–577. doi: 10.1210/mend.13.4.0262. [DOI] [PubMed] [Google Scholar]
- 82.Yasin M., Dalkin A.C., Haisenleder D.J., Kerrigan J.R., Marshall J.C. Gonadotropin-releasing hormone (GnRH) pulse pattern regulates GnRH receptor gene expression: augmentation by estradiol. Endocrinology. 1995;136:1559–1564. doi: 10.1210/endo.136.4.7895666. [DOI] [PubMed] [Google Scholar]
- 83.Hurd E.A., Poucher H.K., Cheng K., Raphael Y., Martin D.M. The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development. 2010;137:3139–3150. doi: 10.1242/dev.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schnetz M.P., Handoko L., Akhtar-Zaidi B., Bartels C.F., Pereira C.F., Fisher A.G., Adams D.J., Flicek P., Crawford G.E., Laframboise T., et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010;6:e1001023. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bajpai R., Chen D.A., Rada-Iglesias A., Zhang J., Xiong Y., Helms J., Chang C.P., Zhao Y., Swigut T., Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takada I., Mihara M., Suzawa M., Ohtake F., Kobayashi S., Igarashi M., Youn M.Y., Takeyama K., Nakamura T., Mezaki Y., et al. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat. Cell Biol. 2007;9:1273–1285. doi: 10.1038/ncb1647. [DOI] [PubMed] [Google Scholar]
- 87.Srinivasan S., Dorighi K.M., Tamkun J.W. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 2008;4:e1000217. doi: 10.1371/journal.pgen.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cattanach B.M., Iddon C.A., Charlton H.M., Chiappa S.A., Fink G. Gonadotrophin-releasing hormone deficiency in a mutant mouse with hypogonadism. Nature. 1977;269:338–340. doi: 10.1038/269338a0. [DOI] [PubMed] [Google Scholar]
- 89.Ho L., Crabtree G.R. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peretto P., Cummings D., Modena C., Behrens M., Venkatraman G., Fasolo A., Margolis F.L. BMP mRNA and protein expression in the developing mouse olfactory system. J. Comp. Neurol. 2002;451:267–278. doi: 10.1002/cne.10343. [DOI] [PubMed] [Google Scholar]
- 91.Peretto P., Dati C., De Marchis S., Kim H.H., Ukhanova M., Fasolo A., Margolis F.L. Expression of the secreted factors noggin and bone morphogenetic proteins in the subependymal layer and olfactory bulb of the adult mouse brain. Neuroscience. 2004;128:685–696. doi: 10.1016/j.neuroscience.2004.06.053. [DOI] [PubMed] [Google Scholar]
- 92.Cate H.S., Sabo J.K., Merlo D., Kemper D., Aumann T.D., Robinson J., Merson T.D., Emery B., Perreau V.M., Kilpatrick T.J. Modulation of bone morphogenic protein signalling alters numbers of astrocytes and oligodendroglia in the subventricular zone during cuprizone-induced demyelination. J. Neurochem. 2010;115:11–22. doi: 10.1111/j.1471-4159.2010.06660.x. [DOI] [PubMed] [Google Scholar]
- 93.Kang K., Song M.R. Diverse FGF receptor signaling controls astrocyte specification and proliferation. Biochem. Biophys. Res. Commun. 2010;395:324–329. doi: 10.1016/j.bbrc.2010.03.174. [DOI] [PubMed] [Google Scholar]
- 94.Gonzalez-Quevedo R., Lee Y., Poss K.D., Wilkinson D.G. Neuronal regulation of the spatial patterning of neurogenesis. Dev. Cell. 2010;18:136–147. doi: 10.1016/j.devcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maier E., von Hofsten J., Nord H., Fernandes M., Paek H., Hebert J.M., Gunhaga L. Opposing Fgf and Bmp activities regulate the specification of olfactory sensory and respiratory epithelial cell fates. Development. 2010;137:1601–1611. doi: 10.1242/dev.051219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shou J., Murray R.C., Rim P.C., Calof A.L. Opposing effects of bone morphogenetic proteins on neuron production and survival in the olfactory receptor neuron lineage. Development. 2000;127:5403–5413. doi: 10.1242/dev.127.24.5403. [DOI] [PubMed] [Google Scholar]
- 97.Shou J., Rim P.C., Calof A.L. BMPs inhibit neurogenesis by a mechanism involving degradation of a transcription factor. Nat. Neurosci. 1999;2:339–345. doi: 10.1038/7251. [DOI] [PubMed] [Google Scholar]
- 98.Bonaguidi M.A., Peng C.Y., McGuire T., Falciglia G., Gobeske K.T., Czeisler C., Kessler J.A. Noggin expands neural stem cells in the adult hippocampus. J. Neurosci. 2008;28:9194–9204. doi: 10.1523/JNEUROSCI.3314-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Calof A.L., Bonnin A., Crocker C., Kawauchi S., Murray R.C., Shou J., Wu H.H. Progenitor cells of the olfactory receptor neuron lineage. Microsc. Res. Tech. 2002;58:176–188. doi: 10.1002/jemt.10147. [DOI] [PubMed] [Google Scholar]
- 100.Colak D., Mori T., Brill M.S., Pfeifer A., Falk S., Deng C., Monteiro R., Mummery C., Sommer L., Gotz M. Adult neurogenesis requires Smad4-mediated bone morphogenic protein signaling in stem cells. J. Neurosci. 2008;28:434–446. doi: 10.1523/JNEUROSCI.4374-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Garcia-Gonzalez D., Clemente D., Coelho M., Esteban P.F., Soussi-Yanicostas N., de Castro F. Dynamic roles of FGF-2 and Anosmin-1 in the migration of neuronal precursors from the subventricular zone during pre- and postnatal development. Exp. Neurol. 2010;222:285–295. doi: 10.1016/j.expneurol.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 102.Vissers L.E., van Ravenswaaij C.M., Admiraal R., Hurst J.A., de Vries B.B., Janssen I.M., van der Vliet W.A., Huys E.H., de Jong P.J., Hamel B.C., et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 103.Wyatt A., Bakrania P., Bunyan D.J., Osborne R.J., Crolla J.A., Salt A., Ayuso C., Newbury-Ecob R., Abou-Rayyah Y., Collin J.R., et al. Novel heterozygous OTX2 mutations and whole gene deletions in anophthalmia, microphthalmia and coloboma. Hum. Mutat. 2008;29:E278–E283. doi: 10.1002/humu.20869. [DOI] [PubMed] [Google Scholar]
- 104.Dateki S., Fukami M., Sato N., Muroya K., Adachi M., Ogata T. OTX2 mutation in a patient with anophthalmia, short stature, and partial growth hormone deficiency: functional studies using the IRBP, HESX1, and POU1F1 promoters. J. Clin. Endocrinol. Metab. 2008;93:3697–3702. doi: 10.1210/jc.2008-0720. [DOI] [PubMed] [Google Scholar]
- 105.Fallest P.C., Trader G.L., Darrow J.M., Shupnik M.A. Regulation of rat luteinizing hormone beta gene expression in transgenic mice by steroids and a gonadotropin-releasing hormone antagonist. Biol. Reprod. 1995;53:103–109. doi: 10.1095/biolreprod53.1.103. [DOI] [PubMed] [Google Scholar]
- 106.Haavisto A.M., Pettersson K., Bergendahl M., Perheentupa A., Roser J.F., Huhtaniemi I. A supersensitive immunofluorometric assay for rat luteinizing hormone. Endocrinology. 1993;132:1687–1691. doi: 10.1210/endo.132.4.8462469. [DOI] [PubMed] [Google Scholar]
- 107.Matteri R.L., Roser J.F., Baldwin D.M., Lipovetsky V., Papkoff H. Characterization of a monoclonal antibody which detects luteinizing hormone from diverse mammalian species. Domest. Anim. Endocrinol. 1987;4:157–165. doi: 10.1016/0739-7240(87)90011-7. [DOI] [PubMed] [Google Scholar]
- 108.Gay V.L., Midgley A.R., Jr, Niswender G.D. Patterns of gonadotrophin secretion associated with ovulation. Fed. Proc. 1970;29:1880–1887. [PubMed] [Google Scholar]
- 109.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]