Abstract
Breast cancers demonstrate substantial biological, clinical and etiological heterogeneity. We investigated breast cancer risk associations of eight susceptibility loci identified in GWAS and two putative susceptibility loci in candidate genes in relation to specific breast tumor subtypes. Subtypes were defined by five markers (ER, PR, HER2, CK5/6, EGFR) and other pathological and clinical features. Analyses included up to 30 040 invasive breast cancer cases and 53 692 controls from 31 studies within the Breast Cancer Association Consortium. We confirmed previous reports of stronger associations with ER+ than ER− tumors for six of the eight loci identified in GWAS: rs2981582 (10q26) (P-heterogeneity = 6.1 × 10−18), rs3803662 (16q12) (P = 3.7 × 10−5), rs13281615 (8q24) (P = 0.002), rs13387042 (2q35) (P = 0.006), rs4973768 (3p24) (P = 0.003) and rs6504950 (17q23) (P = 0.002). The two candidate loci, CASP8 (rs1045485, rs17468277) and TGFB1 (rs1982073), were most strongly related with the risk of PR negative tumors (P = 5.1 × 10−6 and P = 4.1 × 10−4, respectively), as previously suggested. Four of the eight loci identified in GWAS were associated with triple negative tumors (P ≤ 0.016): rs3803662 (16q12), rs889312 (5q11), rs3817198 (11p15) and rs13387042 (2q35); however, only two of them (16q12 and 2q35) were associated with tumors with the core basal phenotype (P ≤ 0.002). These analyses are consistent with different biological origins of breast cancers, and indicate that tumor stratification might help in the identification and characterization of novel risk factors for breast cancer subtypes. This may eventually result in further improvements in prevention, early detection and treatment.
INTRODUCTION
Breast tumors are biologically and clinically heterogeneous, and consist of several histo-pathological subtypes that are associated with different disease outcome and responses to treatment (1–4). Epidemiological studies have provided evidence that breast cancer risk factors vary by tumor characteristics (5–9). Therefore, detailed characterization of tumors may deepen our understanding of breast cancer etiology, facilitate the discovery of novel risk factors and potentially enable risk prediction for specific tumor types.
Recent genome-wide association studies (GWAS) have identified common variants associated with breast cancer risk at multiple genetic loci (10–15). In addition, large combined analyses of associations have provided evidence for association with coding variants in the caspase 8 (CASP8) and transforming growth factor beta 1 (TGFB1) genes (16). For several of these loci, we and others recently reported on the heterogeneity of genetic associations with risk of developing tumor subtypes defined by estrogen receptor (ER), progesterone receptor (PR) and other tumor characteristics, with only a few studies reporting on human epidermal growth factor receptor-2 (HER2) expression (12–14,17–19). The strongest evidence of heterogeneity to date is for the variant rs2981582 in intron 2 of the fibroblast growth factor 2 (FGFR2) gene that has been found to be associated primarily with increased risk of ER positive (ER+) disease (12–14,17,18). Further, the genes harboring these susceptibility SNPs were found differentially expressed in the breast cancer subtypes (20). Studies with larger sample sizes and further tumor characterization might be able to identify additional heterogeneity of breast cancer susceptibility loci.
In clinical practice, tumors are routinely classified according to protein expression of ER, PR and amplification of HER2 to guide the choice of treatment. More recently, gene expression profiling studies, primarily on relatively small sets of cases with frozen tumors, have identified at least four major breast cancer subtypes associated with distinctly different gene expression patterns and more importantly, a significant difference in clinical outcome (1,2). These molecular breast tumor subtypes include the luminal A and B tumors which are characterized by the expression of ER/PR and other luminal epithelial cell markers and are associated with the best clinical outcomes, particularly luminal A tumors that often lack HER2 overexpression. Additional subtypes include HER2 enriched tumors that tend to be hormone-receptor negative and overexpress HER2, and the basal-like tumors characterized by the expression of basal myoepithelial cell markers and are frequently triple negative tumors (ER−&PR−&HER2−) (1,2). Translation into large clinical or epidemiological studies has been challenging because of the limited availability of frozen tumors in these studies, coupled with the costs and technical difficulties in obtaining high-quality gene profiling data from paraffin embedded tumor material. As a result, immunohistochemistry (IHC) surrogate panels based on the expression of three markers used in routine clinical practice (ER, PR, HER2) and two basal markers, cytokeratin 5/6 (CK5/6) and epidermal growth factor receptor (EGFR), have been used to identify breast tumor subtypes in large studies, although the correspondence with subtypes defined by expression profiling is only approximate (21–26).
In this study, we investigated whether common breast cancer susceptibility loci were associated with specific tumor subtypes defined by five markers (ER, PR, HER2, CK5/6 and EGFR), as well as other important tumor characteristics (histology, grade of differentiation, tumor size, nodal involvement and stage at diagnosis). This report includes analysis of all known susceptibility loci identified through GWAS at the time of analyses, [rs2981582 (10q26), rs3803662 (16q12), rs889312 (5q11), rs13281615 (8q24), rs3817198 (11p15), rs13387042 (2q35), rs4973768 (3p24), rs6504950 (17q23)], as well as two putative susceptibility single-nucleotide polymorphisms (SNPs) in candidate genes [rs1045485/rs17468277 (CASP8) and rs1982073 (TGFB1)] (10–14,16). Analyses were based on data from 31 case–control or cohort studies in the Breast Cancer Association Consortium (BCAC) that included over 30 000 invasive breast tumors, mostly among women of European origin. This paper expands and refines our previous reports on analyses by ER (and/or PR) status (14,16,17,27) by including up to 11 additional studies (representing ∼20–60% of additional data, depending on the specific analysis), as well as three additional tumor markers (HER2, CK5/6 and EGFR). The addition of these three markers that were used to identify tumors with the triple negative and basal phenotypes is the most novel aspect of this paper. Moreover, the combined analysis and the increased sample size are allowing us to make more definite conclusions than previous reports.
RESULTS
Analyses by ER and PR status of tumors
Most studies (29 out of 31) were conducted in populations of European background; therefore, the main analyses were restricted to these women. As expected, all susceptibility loci identified in previously published GWAS showed highly significant associations with breast cancer risk among subjects included in this report, with per-allele ORs similar to those previously reported (Table 1). The two candidate loci in CASP8 and TGFB1 showed weaker evidence than in previous reports based on a smaller number of cases and controls (16). Small differences in risk estimates compared with previous publications are likely to be due to random variation or overestimates in original publications with stronger influence of data from discovery studies.
Table 1.
Association between susceptibility loci and breast cancer risk overall among a total of up to 29 studies in populations of European background with data on ER and/or PR status
| Locus | Neighborhood genes | rs number | n studiesa | Controls | mafb in controls | Cases | OR | 95% CI | P-value |
|---|---|---|---|---|---|---|---|---|---|
| Susceptibility loci identified in GWAS | |||||||||
| 10q26 | FGRF2 | rs2981582 (C/T)c | 27 | 33 908 | 0.38 | 25 182 | 1.22 | 1.19–1.25 | 1.5 × 10−59 |
| 16q12 | TOX3/LOC643714 | rs3803662 (C/T) | 29 | 34 857 | 0.27 | 26 671 | 1.24 | 1.20–1.27 | 3.0 × 10−59 |
| 5q11 | MAP3K1/MGC33648/MIER3 | rs889312 (A/C) | 28 | 34 325 | 0.28 | 25 830 | 1.11 | 1.08–1.14 | 7.1 × 10−16 |
| 8q24 | FAM84B/c-MYC | rs13281615 (A/G) | 26 | 29 849 | 0.41 | 23 172 | 1.11 | 1.08–1.13 | 3.5 × 10−15 |
| 11p15 | LSP1/H19 | rs3817198 ((T/C) | 24 | 31 891 | 0.31 | 23 879 | 1.06 | 1.03–1.09 | 1.0 × 10−05 |
| 2q35 | TNP1/IGFBP5/IGFBP2/TNS1 | rs13387042 (G/A) | 25 | 38 120 | 0.52 | 26 334 | 1.14 | 1.12–1.17 | 1.7 × 10−29 |
| 3p24 | SLC4A7/NEK10 | rs4973768 (C/T) | 21 | 34 386 | 0.46 | 22 506 | 1.11 | 1.09–1.14 | 1.1 × 10−17 |
| 17q23 | COX11/STXBP4/TOM1L1 | rs6504950 (G/A) | 26 | 34 236 | 0.28 | 26 204 | 0.94 | 0.92–0.97 | 3.2 × 10−05 |
| Putative susceptibility loci in candidate genes | |||||||||
| 2q33-q34 | CASP8 | rs1045485; rs17468277 (G/C;C/T) | 23 | 36 976 | 0.13 | 24 406 | 0.95 | 0.92–0.98 | 0.004 |
| 19q13 | TGFB1 | rs1982073 (G/C) | 17 | 27 745 | 0.38 | 16 123 | 1.04 | 1.02–1.08 | 0.003 |
aFor none of the 10 SNPs, genotype data were available from all 31 studies. Studies in Asian populations are not included in this table (MEC-Japanese, TBCS, TWBCS; see Supplementary Material, Table S7 for estimates in studies of Asian populations). Analyses included only cases with tumor marker information (defined as having at least information on ER and/or PR status).
bMinor allele frequency.
cMajor/minor allele.
Small differences in point estimates and significance levels compared with previous publications are due to the different inclusion/exclusion criteria for cases and controls. P-value <0.05 was used to select findings mentioned in the results and discussion sections, and indicated in bold-face. Findings were interpreted in light of the strength of evidence based on the estimated OR's, 95% CI, P-values and prior knowledge of the hypothesis being tested.
Six (10q26, 16q12, 8q24, 2q35, 3p24, 17q23) of the eight loci identified in GWAS exhibited stronger associations with ER+ than ER− tumors (Table 2). Evidence for differences by ER status was strongest for rs2981582 (10q26) [per-allele OR = 1.28 (95% CI = 1.24–1.31) for ER+ versus OR = 1.05 (1.01–1.09) for ER−; case only P-heterogeneity = 6.1 × 10−18] and rs3803662 (16q12) [per-allele OR = 1.26 (1.23–1.30) for ER+ versus OR = 1.15 (1.10–1.20) for ER−, P-heterogeneity = 3.7 × 10−5]. Although associations were stronger for ER+ than ER− tumors, all six loci were also associated with the risk of ER− tumors (P ≤ 0.021), except for rs6504950 (17q23) [OR for ER− tumors = 1.00 (0.95–1.05) P = 0.938]. The strongest evidence of association with ER− disease was for SNP rs3803662 (16q12) [per-allele OR = 1.15 (1.10–1.20) P = 2.1 × 10−10]. The other two GWAS SNPs, rs889312 (5q11) (P-heterogeneity = 0.531) and rs3817198 (11p15) (P-heterogeneity = 0.426), showed no evidence of an association with ER− status. The associations with the two putative susceptibility loci in the candidate genes, CASP8 and TGFB1, did not appear to differ strongly by ER status, although the data for CASP8 suggested a stronger protective effect against ER− than ER+ disease (Table 2, P-heterogeneity = 0.038).
Table 2.
Odds ratios for breast cancer by ER expression in tumors (up to 29 studies in populations of European backgrounda)
| Case–control analyses |
Case only | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ER+ tumors versus controls |
ER− tumors versus controls |
|||||||||||
| Locus/gene | SNP | Controls | maf | ER+ cases | ORb | 95% CI | P-value | ER− cases | ORb | 95% CI | P-value | P-valuec |
| Susceptibility loci identified in GWAS | ||||||||||||
| 10q26 | rs2981582 | 33 908 | 0.38 | 18 310 | 1.28 | 1.24–1.31 | 4.7 × 10−73 | 5613 | 1.05 | 1.01–1.09 | 0.020 | 6.1 × 10−18 |
| 16q12 | rs3803662 | 34 857 | 0.27 | 19 420 | 1.26 | 1.23–1.30 | 9.6 × 10−60 | 5968 | 1.15 | 1.10–1.20 | 2.1 × 10−10 | 3.7 × 10−05 |
| 5q11 | rs889312 | 34 325 | 0.28 | 18 835 | 1.11 | 1.08–1.15 | 9.3 × 10−14 | 5735 | 1.09 | 1.05–1.14 | 6.0 × 10−05 | 0.531 |
| 8q24 | rs13281615 | 29 849 | 0.41 | 16 888 | 1.13 | 1.10–1.16 | 2.7 × 10−18 | 5098 | 1.06 | 1.01–1.10 | 0.012 | 0.002 |
| 11p15 | rs3817198 | 31 891 | 0.31 | 17 427 | 1.07 | 1.04–1.10 | 1.4 × 10−05 | 5223 | 1.05 | 1.00–1.09 | 0.056 | 0.426 |
| 2q35 | rs13387042 | 38 120 | 0.52 | 19 310 | 1.16 | 1.13–1.19 | 8.5 × 10−30 | 5770 | 1.09 | 1.05–1.13 | 2.9 × 10−05 | 0.006 |
| 3p24 | rs4973768 | 35 104 | 0.46 | 17 067 | 1.13 | 1.10–1.16 | 2.8 × 10−18 | 4854 | 1.05 | 1.01–1.10 | 0.021 | 0.003 |
| 17q23 | rs6504950 | 34 386 | 0.28 | 16 455 | 0.93 | 0.90–0.95 | 7.2 × 10−07 | 4774 | 1.00 | 0.95–1.05 | 0.938 | 0.002 |
| Putative susceptibility loci in candidate genes | ||||||||||||
| CASP8 | rs1045485; rs17468277 | 36 976 | 0.13 | 17 805 | 0.96 | 0.93–1.00 | 0.058 | 5347 | 0.90 | 0.84–0.96 | 0.001 | 0.038 |
| TGFB1 | rs1982073 | 27 745 | 0.38 | 11 495 | 1.04 | 1.01–1.08 | 0.011 | 3503 | 1.06 | 1.00–1.11 | 0.033 | 0.540 |
aDifferences in total number of ER+ (22 866) and ER− cases (7174), (Supplementary Material, Table S3) is due to missing genotype data.
bOR are adjusted by study.
cP-value from logistic regression models including only cases, with ER status as the outcome adjusted by study. P-values <0.05 are indicated in bold-face.
To evaluate the combined effects of the ten SNPs on ER+ and ER− disease, we calculated relative risks at the 10th, 50th and 90th centiles of the polygenic risk distribution by ER status under a log-additive (multiplicative) risk model. Calculations were based on estimates of per-allele ORs and allele frequencies for the nine loci with P < 0.05 for ER+ and eight loci for ER− shown in Table 2. The estimated risk distribution on a log relative risk scale was approximately normal with mean close to zero and variances of 0.085 and 0.022 for ER+ and ER− disease, respectively. The relative risk of breast cancer for women at the 10th, 50th and 90th centiles of the risk distribution, when compared with the population average, was 0.66, 0.96 and 1.39, respectively, for ER+ tumors. The corresponding relative risks for ER− tumors were 0.82, 0.99 and 1.20. According to the risk distribution by ER status, the AUC was slightly higher for ER+ (AUC = 58.2%) than for ER− disease (AUC = 54.3%).
Analyses by PR status for the eight SNPs identified in GWAS generally showed a similar pattern to that observed by ER status, as would be expected given the positive correlation between these two markers (data not shown). On the other hand, the SNPs in CASP8 and TGFB1 showed stronger evidence of differential in associations by PR than ER status. In both cases, the strongest statistical evidence for an association was with PR− tumors [CASP8: per-allele OR = 0.88 (0.83–0.93) P = 5.1 × 10−6; TGFB1: OR = 1.09 (1.04–1.14) P = 4.1 × 10−4; Table 3].
Table 3.
Odds ratios for breast cancer by PR expression in tumors for two putative susceptibility loci in CASP8 and TGFB1 (up to 21 studies in populations of European backgrounda)
| Gene/SNP | Genotype | Controls | Case–control analyses |
Case only | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PR+ tumors versus controls |
PR− tumors versus controls |
||||||||||
| PR+ cases | ORb | 95% CI | P-value | PR− cases | ORb | 95% CI | P-value | P-valuec | |||
| CASP8 | GG/CC | 28 109 | 10 105 | 1.00 | 5558 | 1.00 | |||||
| rs1045485/ | GC/CT | 8206 | 2833 | 0.97 | 0.92–1.02 | 0.181 | 1393 | 0.87 | 0.81–0.93 | 2.0 × 10−05 | |
| rs17468277d | CC /TT | 661 | 202 | 0.83 | 0.70–0.98 | 0.026 | 107 | 0.81 | 0.65–1.00 | 0.047 | |
| per allele | 36 976 | 13 140 | 0.95 | 0.91–0.99 | 0.027 | 7058 | 0.88 | 0.83–0.93 | 5.1 × 10−06 | 0.009 | |
| TGFB1 | GG | 10 824 | 2984 | 1.00 | 1649 | 1.00 | |||||
| rs1982073 | CG | 12 885 | 3745 | 1.06 | 1.00–1.12 | 0.050 | 2065 | 1.04 | 0.97–1.12 | 0.278 | |
| CC | 4036 | 1164 | 1.05 | 0.97–1.14 | 0.215 | 752 | 1.21 | 1.10–1.33 | 1.1 × 10−04 | ||
| per allele | 27 745 | 7893 | 1.03 | 0.99–1.07 | 0.091 | 4466 | 1.09 | 1.04–1.14 | 4.1 × 10−04 | 0.045 | |
aDifferences in total number of PR+ (16 997) and PR− (9392) cases (Supplementary Material, Table S3) is due to missing genotype data.
bOR are adjusted by study.
cP-value from logistic regression models including only cases, with PR status as the outcome adjusted by study.
dBecause of some technical difficulties (for some technologies) in genotyping the originally reported SNP [rs1045485 (G/C)] in CASP8, another SNP in strong LD [rs17468277 (C/T)] was used as a surrogate (r2 = 1 in HapMap CEU) for a subset of studies. Thirteen studies provided genotype data on rs1045485 and 19 studies on rs17468277. Five of those studies reported genotype data on both SNPs. For individuals that had the two SNPs genotyped, rs17468277 was used. P-values <0.05 are indicated in bold-face.
Classification of tumors according to the co-expression of ER and PR status suggested a weaker association of rs2981582 (10q26) with ER−&PR+ (per-allele OR = 1.14 (1.04–1.26) than ER+&PR+ tumors (OR = 1.29 (1.25–1.33) P-heterogeneity = 0.019; data not shown); and a weaker association of rs3803662 (16q12) with ER+&PR− (per-allele OR = 1.16 (1.09–1.23) than ER+&PR+ tumors (OR = 1.28 (1.24–1.33) P-heterogeneity = 0.001; data not shown). These analyses suggest that the variant in 10q26 is primarily associated with ER status, whereas rs3803662 (16q12) might be primarily associated with PR status. No other differences (P > 0.10) were found in ORs for ER+&PR− and ER−&PR+ tumors compared with ER+&PR+ tumors (data not shown). In spite of these relatively small differences, ER+&PR+, ER+&PR− and ER−&PR+ tumors were combined as ER+ and/or PR+ (luminal) tumors before further stratification by HER2 status. This decision was made a priori based on the co-expression of ER and PR in defining tumor subtypes in studies using expression arrays (21–24).
Analyses by ER, PR and HER2 status of tumors
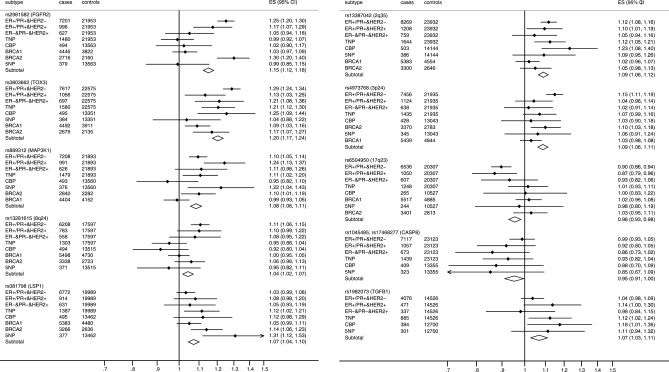
Figure 1 summarizes the findings from analyses of tumor subtypes defined by ER, PR and HER2 (tumor subtype nomenclature explanation: / = ‘and/or’, & = and). Within luminal tumors expressing ER and/or PR, three loci showed differences by HER2 expression: rs3803662 (16q12) and rs4973768 (3p24) showed a stronger association with ER+/PR+&HER2− than ER+/PR+&HER2+ tumors (P-heterogeneity 0.013 and 0.02, respectively), whereas rs889312 (5q11) showed a stronger association with ER+/PR+&HER2+ than ER+/PR+&HER2− tumors (P-heterogeneity = 0.014; Table 4 upper panel).
Figure 1.
Association of susceptibility loci with different tumor subtypes: a summary of the findings. The per-allele OR's (ES) are shown. Tumor subtype nomenclature explanation: / = ‘and/or’, & = and. Tumors were classified into four subtypes, two receptor positive or ‘luminal' subtypes: ER+/PR+&HER2− and ER+/PR+&HER2+, and two receptor negative subtypes or ‘non-luminal' ER−&PR−&HER2+ and triple negative tumors (TN: ER−&PR−&HER2−). Cases with TN tumors were further stratified according to the expression of basal markers into the core basal phenotype (CBP) (CK5/6, CK5 or EGFR positive) and the five-negative phenotype (5NP) (CK5/6 or CK5, and EGFR negative).
Table 4.
Odds ratios for breast cancer risk by ER, PR and HER2 expression in tumors (18 studies in populations of European background)
| Case–control analyses |
Case only | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls | ER+/PR+&HER2− |
ER+/PR+&HER2+ |
|||||||||
| Locus/gene | SNP | n | n | ORa | 95% CI | P-value | n | ORa | 95% CI | P-value | P-valueb |
| 10q26 | rs2981582 | 21 953 | 7201 | 1.25 | 1.20–1.30 | 2.2 × 10−29 | 996 | 1.18 | 1.07–1.29 | 5.5 × 10−04 | 0.233 |
| 16q12 | rs3803662 | 22 575 | 7617 | 1.29 | 1.24–1.34 | 6.8 × 10−34 | 1056 | 1.13 | 1.03–1.25 | 0.011 | 0.013 |
| 5q11 | rs889312 | 21 893 | 7208 | 1.10 | 1.05–1.14 | 2.6 × 10−05 | 991 | 1.24 | 1.13–1.37 | 1.2 × 10−05 | 0.014 |
| 8q24 | rs13281615 | 17 597 | 6208 | 1.11 | 1.06–1.15 | 3.2 × 10−06 | 783 | 1.10 | 0.99–1.22 | 0.069 | 0.968 |
| 11p15 | rs3817198 | 19 989 | 6772 | 1.03 | 0.99–1.08 | 0.147 | 914 | 1.08 | 0.98–1.20 | 0.134 | 0.349 |
| 2q35 | rs13387042 | 23 932 | 8269 | 1.12 | 1.08–1.16 | 1.7 × 10−09 | 1208 | 1.10 | 1.01–1.19 | 0.029 | 0.684 |
| 3p24 | rs4973768 | 21 935 | 7456 | 1.15 | 1.11–1.19 | 6.5 × 10−13 | 1124 | 1.04 | 0.96–1.14 | 0.335 | 0.020 |
| 17q23 | rs6504950 | 20 307 | 6536 | 0.90 | 0.86–0.94 | 2.8 × 10−06 | 1050 | 0.87 | 0.79–0.96 | 0.007 | 0.577 |
| CASP8 | rs1045485; rs17468277 | 23 323 | 7607 | 0.99 | 0.93–1.04 | 0.633 | 1117 | 0.93 | 0.81–1.06 | 0.258 | 0.303 |
| TGFB1 | rs1982073 | 14 526 | 4076 | 1.03 | 0.97–1.08 | 0.327 | 471 | 1.15 | 1.01–1.32 | 0.035 | 0.081 |
| Triple negative phenotype | ER−&PR−&HER2+ | P-valuec | |||||||||
| 10q26 | rs2981582 | 21 953 | 1480 | 0.99 | 0.92–1.07 | 0.841 | 627 | 1.05 | 0.94–1.18 | 0.396 | 0.150 |
| 16q12 | rs3803662 | 22 575 | 1580 | 1.21 | 1.11–1.30 | 3.1 × 10−06 | 697 | 1.21 | 1.08–1.36 | 0.001 | 0.827 |
| 5q11 | rs889312 | 21 893 | 1479 | 1.11 | 1.02–1.20 | 0.016 | 626 | 1.11 | 0.98–1.26 | 0.094 | 0.363 |
| 8q24 | rs13281615 | 17597 | 1303 | 0.95 | 0.88–1.04 | 0.266 | 558 | 1.08 | 0.95–1.22 | 0.223 | 0.095 |
| 11p15 | rs3817198 | 19 989 | 1387 | 1.11 | 1.03–1.21 | 0.011 | 631 | 1.05 | 0.93–1.19 | 0.407 | 0.479 |
| 2q35 | rs13387042 | 23 932 | 1644 | 1.12 | 1.05–1.21 | 0.001 | 759 | 1.05 | 0.94–1.16 | 0.387 | 0.243 |
| 3p24 | rs4973768 | 21935 | 1435 | 1.07 | 0.99–1.16 | 0.076 | 638 | 1.02 | 0.91–1.14 | 0.759 | 0.392 |
| 17q23 | rs6504950 | 20 307 | 1248 | 1.01 | 0.93–1.11 | 0.748 | 607 | 0.93 | 0.82–1.06 | 0.290 | 0.207 |
| CASP8 | rs1045485; rs17468277 | 23 323 | 1531 | 0.92 | 0.82–1.03 | 0.151 | 706 | 0.86 | 0.73–1.02 | 0.086 | 0.267 |
| TGFB1 | rs1982073 | 14 526 | 885 | 1.11 | 1.01–1.23 | 0.038 | 337 | 0.98 | 0.84–1.15 | 0.809 | 0.270 |
aOR are adjusted by study.
bP-value from logistic regression models including only cases comparing ER+/PR+&HER2+ versus ER+/PR+& HER2− tumors.
cP-value from logistic regression models including only cases comparing non-luminal HER2+ to triple negative tumors. P-values <0.05 are indicated in bold-face.
We found no differences (P ≥ 0.095) in the per-allele ORs for TN tumors and ER−&PR−&HER2+ tumors for any of the SNPs (Table 4, lower panel), although theses analyses were limited by small numbers of cases in each category. Of note, we did not find evidence for an association with TN tumors for the rs2981582 (10q26) [per-allele OR = 0.99 (0.92–1.07) P = 0.841] that showed one of the strongest associations overall with the luminal tumors. However, an effect size similar to that for all ER− disease could not be excluded. Five of the 10 SNPs showed associations (P ≤ 0.02) with TN disease [rs3803662 (16q12), rs889312 (5q11), rs3817198 (11p15), rs13387042 (2q35) and rs1982073 (TGFB1); Table 4]. No additional differences were found when we classified tumors as HER2− and HER2+, independently of ER and PR status (data not shown).
Analyses by ER, PR, HER2 and basal markers (CK5/6 or CK5 and EGFR2) status of tumors
Figure 1 summarizes the findings from analyses of tumor subtypes defined by ER, PR, HER2 and basal markers. Of the five SNPs that showed associations with TN disease, three were also associated with the risk of core basal phenotype (CBP): rs3803662 (16q12) [per-allele OR = 1.25 (1.09–1.44) P = 0.001], rs13387042 (2q35) [OR = 1.23 (1.08–1.40) P = 0.002] and rs1982073 (TGFB1) [OR = 1.18 (1.01–1.36) P = 0.032]. In each case, however, the per-allele OR did not differ for TN tumors that did not exhibit basal markers (5NP) when compared with those that did (CBP), P > 0.132). The SNPs in 5q11 and 11p15 were associated with 5NP but not CBP tumors.
Analyses by other tumor characteristics
We evaluated associations between genotypes and tumor subtypes defined by grade of differentiation (Table 5), histopathology (Supplementary Material, Table S5), tumor size (Supplementary Material, Table S6), nodal involvement (Supplementary Material, Table S7) and stage at diagnosis (Supplementary Material, Table S8). Generally, we observed expected results based on the known correlations between these tumor features and the expression of tumor markers evaluated earlier. Four of the SNPs [rs2981582 (10q26), rs3803662 (16q12), rs13387042 (2q35) and rs973768 (3p24)] showed stronger associations with tumors of lower grade Table 5. However, only two of these remained significant (P < 0.05) after adjustment for ER status: adjusted case-only P = 0.0003 for rs2981582 (10q26) and 5.7 × 10−8 for rs13387042 (2q35). The association between 10q26 and ER status remained strongly significant after adjusting for grade (adjusted case-only P = 5.9 × 10−9); however, 2q35 was no longer associated with ER after adjustment for grade (adjusted case-only P = 0.80). Stratification of tumors by ER and grade among 24 studies with data on these two features (19 997 cases and 31 540 controls) suggested that the strongest association of rs2981582 (10q26) is with ER+/low grade tumors and that there is no association for ER−/high grade tumors: per-allele ORs for ER+/grade 1 [19% of tumors; OR = 1.29 (1.23–1.35)], for ER+/grade 2 [42% of tumors; 1.29 (1.25–1.34)], for ER+/grade 3 [17% of tumors OR = 1.19 (1.13–1.25)] and for ER−/grade 1 [1% of tumors; OR = 1.09 (0.92–1.29)], ER−/grade 2 [7.0% of tumors; OR = 1.13 (1.04, 1.22)], ER−/grade3 [15% of tumors; OR = 1.00 (0.95–1.06)]. Association with the histopathological types suggested a higher risk for lobular compared with other tumors, for rs3803662 (16q12), rs13281615 (8q24) and rs13387042 (2q35) (P = 0.032, 0.039 and 0.017, respectively). Conversely, CASP8 (rs1045485/rs17468277) appeared to be specifically associated with the risk of ductal tumors (Supplementary Material, Table S5). Only weak associations were observed with tumor size, and these were not significant (P < 0.05) after adjustment for ER status.
Table 5.
Odds ratios for breast cancer by grade of differentiation of the tumors (up to 27 studies in populations of European background)
| Cases (grade) |
Grade 1 |
Grade 2 |
Grade 3 |
Case only | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus/gene | SNP | Controls | 1 | 2 | 3 | ORa | 95% CI | ORa | 95% CI | ORa | 95% CI | P-valueb |
| Susceptibility loci identified in GWAS | ||||||||||||
| 10q26 | rs2981582 | 31 540 | 4886 | 11 380 | 7475 | 1.29 | 1.23–1.34 | 1.27 | 1.23–1.31 | 1.10 | 1.06–1.14 | 1.49 × 10−10 |
| 16q12 | rs3803662 | 32 486 | 5107 | 12 034 | 8042 | 1.29 | 1.23–1.35 | 1.23 | 1.19–1.27 | 1.18 | 1.14–1.23 | 8.34 × 10−04 |
| 5q11 | rs889312 | 31 941 | 4985 | 11 685 | 7711 | 1.09 | 1.04–1.15 | 1.11 | 1.07–1.15 | 1.10 | 1.06–1.14 | 0.854 |
| 8q24 | rs13281615 | 27 550 | 4676 | 10 534 | 7230 | 1.13 | 1.08–1.18 | 1.14 | 1.10–1.18 | 1.07 | 1.03–1.11 | 0.022 |
| 11p15 | rs3817198 | 29 544 | 4726 | 10 610 | 7222 | 1.09 | 1.04–1.14 | 1.07 | 1.03–1.11 | 1.08 | 1.03–1.12 | 0.910 |
| 2q35 | rs13387042 | 39 890 | 5417 | 12 771 | 8434 | 1.24 | 1.19–1.29 | 1.16 | 1.13–1.20 | 1.07 | 1.04–1.11 | 1.06 × 10−08 |
| 3p24 | rs4973768 | 36 997 | 5016 | 11 689 | 7603 | 1.17 | 1.12–1.22 | 1.11 | 1.07–1.14 | 1.08 | 1.04–1.12 | 8.96 × 10−04 |
| 17q23 | rs6504950 | 35 794 | 4487 | 10 940 | 7353 | 0.94 | 0.89–0.99 | 0.91 | 0.88–0.95 | 0.97 | 0.93–1.01 | 0.083 |
| Putative susceptibility loci in candidate genes | ||||||||||||
| CASP8 | rs1045485; rs17468277 | 37 571 | 4925 | 11 632 | 7876 | 0.93 | 0.87–0.99 | 0.97 | 0.92–1.01 | 0.95 | 0.90–1.00 | 0.764 |
| TGFB1 | rs1982073 | 27 361 | 3479 | 7778 | 5088 | 1.03 | 0.98–1.09 | 1.03 | 1.00–1.07 | 1.09 | 1.04–1.13 | 0.154 |
aOR are adjusted by study.
bP-value from case-only analyses using a polytomous logistic regression model constraining the effect size to increase linearly across levels. P-values <0.05 are indicated in bold-face.
Analysis of Asian populations
On the basis of data from three studies including Asian women, only the SNPs rs2981582 (10q26) and rs3803662 (16q12) showed associations with breast cancer risk overall (P = 5.2 × 10−8 and 0.027, respectively; Supplementary Material, Table S9). Because of the relatively small number of cases and controls, we only carried out analyses by ER status since this was the marker that was most strongly related to SNPs in Caucasian populations. None of the SNPs appeared to be differently associated with ER-defined disease [P-heterogeneity = 0.962 and 0.816, for rs rs2981582 (10q26) and rs3803662 (16q12), respectively, Supplementary Material, Table S10]. There was some evidence for differences in case-only ORs by ethnic group (Caucasians versus Asians) for rs2981582 (P = 0.042) but not for rs3803662 (P = 0.661). Notably, rs2981582 (10q26) was more strongly associated with ER− disease [OR = 1.34 (1.14–1.57)] in Asian compared with European populations.
DISCUSSION
Our data confirm previous reports and provide convincing evidence for heterogeneity in the strength of the associations of ten breast cancer susceptibility loci with respect to the risk of tumor subtypes defined by ER status and grade of differentiation of the tumors. In addition, stratification of tumors by the additional markers provided further insights into etiological heterogeneity (Fig. 1). These results suggest that low-risk susceptibility loci predict the pathological subtype of breast cancer and provide support for the hypothesis that breast tumor subtypes arise through distinct etiological pathways.
Six out of the eight susceptibility loci previously identified through GWAS, rs2981582 (10q26), rs3803662 (16q12), rs13281615 (8q24), rs13387042 (2q35), rs4973768 (3p24) and rs6504950 (17q23), showed stronger associations with ER+ than ER− disease. Findings for SNPs in 10q26, 16q12 and the 8q24 region have been previously indentified in a subset of this data (16,17), and are confirmed in this report after including data from additional studies not available in the previous publications. Analysis of combined SNP effects by ER status according to a log-additive polygenetic risk model, showed higher relative risks and a slightly higher discrimination power for ER+ (AUC = 58.2%) than ER− (AUC = 54.3%) disease, consistent with previous reports (19).
The predominance of loci identified to date that are associated with ER+ disease might reflect that the majority of invasive tumors express ER, and thus current GWAS including cases unselected for ER status had greater power to detect SNPs associated with ER+ disease than those associated with a similar relative risk for ER− disease. Therefore, subtype stratification in GWAS analyses and well-powered GWAS of more homogenous tumor types could help the identification of additional breast cancer risk loci. This is exemplified by the discovery of a variant on chromosome 19p13 discovered in a GWAS of BRCA1 mutation carriers who tend to develop TN tumors, and that showed an increased risk of ER− (particularly TN) but not ER+ disease in the general population (28). Studying different ethnic groups can also lead to the identification of additional loci, such as the loci reported in chromosome 6q25.1, located upstream of the gene encoding ER alpha, which was identified in a GWAS in Asian populations, and which was more strongly associated with ER− compared with ER+ disease (15).
Consistent with our previous report, this larger analysis confirmed that rs2981582 (at 10q26 within FGFR2) is most strongly associated with ER+/low grade tumors, with no association observed for ER−, high-grade tumors. Further stratification by the other tumor markers, not included in our previous publication, showed the strongest association with ER+/PR+&HER2− tumors, and no evidence for an association with the risk of triple negative tumors or tumors with the CBP (Fig. 1). FGFR2 is a receptor tyrosine kinase and its strong association with luminal-like tumors is consistent with the involvement of FGFR2 in estrogen-related breast carcinogenesis (29). This finding is also consistent with the evidence provided by the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) consortium; they showed an association of the FGFR2 locus with breast cancer risk in BRCA2 mutation carriers but not in BRCA1 mutation carriers (30,31). CIMBA has evaluated the associations between common susceptibility loci and risk of breast cancer in carriers of BRCA1 and BRCA2 mutations. Although analyses were not stratified by tumor subtypes, BRCA1 carriers tend to develop hormone receptor negative tumors, whereas tumors in BRCA2 carriers show similar subtype distributions as tumors developed in non-carriers (i.e. a predominance of ER+ tumors) (32). CIMBA analyses indicated that common susceptibility loci identified in populations not selected for carrier status have similar associations with risk among BRCA2 mutation carriers; however, the evidence for an association is weaker for BRCA1 carriers (30,31,33).
The rs3803662 (16q12) locus was associated with the increased risk of all tumor subtypes, with the strongest association being for ER+/PR+&HER2− tumors (Fig. 1). This locus also showed the strongest association with TN tumors and those with the CBP among the 10 loci examined. Interestingly, rs3803662 in 16q12 shows the strongest evidence for associations with the risk in BRCA1 mutation carriers (30,31,33), and in our data, the SNPs also showed the strongest evidence for association with the risk of developing tumors with the CBP (Fig. 1). Although this report only includes a relatively small number of tumors with the CBP, our data suggest that they might be a distinct subtype within TN tumors. The locus at 2q35 was also differentially associated with breast tumor subtypes defined by grade, with the strongest association being for tumors of low grade.
Although estimates of relative risk for TN tumors (particularly the subset of tumors identified as having the CBP), were relatively imprecise, our data suggest that two SNPs, rs13281615 in the 8q24 region and rs6504950 in the 17q23 region, might be associated with an increased risk for all tumor subtypes, except for TN and CBP, On the other hand, rs3817198 (11p15), rs13387042 (2q35) and rs4973768 (3p24) seemed to be associated with increased risk of all tumor subtypes. The different patterns of association with specific tumor subtypes observed in this study suggest that tumor subtypes have some common as well as distinct pathogenic pathways (34).
Previous studies provide strong evidence for a reduced risk association between the D302H variant in CASP8 that encodes caspase 8, an important initiator of apoptosis (16). Our data support the previous observations that this association might be stronger for tumors that do not express PR. The evidence for a risk association with rs1982073 (L10P) in TGFB1, encoding for the transforming growth factor B1, is weaker (16). Our data suggest that rs1982073 at TGFB1 is associated with increased risk of PR negative tumors with aggressive characteristics, particularly tumors with the CBP, and tumors diagnosed at advanced stages. However, Rebbeck et al. (35) observed that there is no association between TGFB1 rs1982073 and breast cancer risk in either BRCA1 or BRCA2 mutation carriers. Therefore, additional evidence is needed to confirm a potential association between TGFB1 rs1982073 and risk of breast cancer subtypes, particularly sporadic basal-like cancers through mechanisms that might be independent of the BRCA1 pathway.
It is unclear whether PR status has an effect on breast carcinogenesis independent of ER status. About 65% of ER+ breast cancers are also PR+, and there is a high correlation between ER and PR expression (21,22). Actually, ER−/PR+ tumors (4% in this study) are suspected to be misclassified ER+ tumors. Our data suggest that risk for SNPs in 16q12, CASP8 and TGFB1 might be associated with PR expression, independent of ER expression, indicating that PR might possibly have a role on tumor etiology beyond its roles as a co-expressor with ER.
A major strength of our study is the very large sample size and consistency of findings across studies, in spite of heterogeneity of study designs. The majority of subjects included in this study were of white European origin, with only three studies including women from Asian populations. Because the sample size was considerably smaller for Asian studies, the main conclusions from this manuscript are based on analyses among white European women. Of note, some of the strongest findings from this study, i.e. the modification of the breast cancer risk conferred by loci rs2981582 (10q26) and rs3803662 (16q12) by ER status, were not supported by analyses in Asian populations. Future studies including larger numbers of Asians and subjects of other ethnicities are necessary to clarify the consistency of findings across ethnic groups.
A limitation of our study is the use of non-standardized data on tumor markers, since data were derived from studies using different tissue collection and processing protocols, IHC assays and criteria for pathology review. In addition, although most cases from studies in this report had data on ER and PR status of the tumors, only a subset of studies had data on other tumor markers and the percentage of missing data within these studies was higher than that for ER and PR status. This resulted in relatively small number of tumors classified by a combination of three or more markers, and thus findings from these analyses need to be interpreted with caution. For instance, while we were able to identify 1865 TN tumors, only 509 tumors were classified as having the CBP. As a result, the power to evaluate associations with these tumor subtypes, which are known to be characterized by a poor response to available treatments, is limited.
Missing data and misclassification probabilities are likely to be independent of susceptibly loci, and thus would tend to underestimate associations rather than create spurious associations. Misclassification is likely to be particularly strong for HER2, especially for studies that inferred HER2 status based on IHC scoring (36). Recently, de Ronde et al. (24) showed a high discordance between HER expression based on IHC and mRNA, 60% of the tumors classified HER2+ by IHC did not display elevated levels by mRNA expression. In spite of these limitations, we observed consistent associations across studies, indicating that our findings are robust and that reduction of misclassification and missing data might improve our ability to identify association between risk factors and tumor subtypes (37). To address these limitations, we are currently conducting a study aimed at standardization of scoring of tumor markers in TMA using automated image analysis technologies, in situ hybridization assays for HER2 amplification and web based systems for pathological scoring. This work will also provide quantitative or semi-quantitative measures of tumor maker expression to evaluate dose–response relationships.
In summary, our data provides strong evidence for etiological heterogeneity of breast cancer subtypes, particularly those defined by the expression of ER status and grade of differentiation of the tumors. Future etiological studies should consider ER positive and negative tumors as distinct breast cancer subtypes and evaluate the value of additional classifications to expand our understanding of the etiology of this heterogeneous tumor. Further characterization and understanding of the underlying etiological heterogeneity of breast cancer can provide biological insights on the mechanisms of carcinogenesis, and eventually result in improvement in prevention (population risk stratification in screening programs), early detection and treatment.
MATERIALS AND METHODS
Study populations
Thirty-one breast cancer studies participating in BCAC provided tumor marker data on at least ER and/or PR tumor status and genotype data for at least one of the 10 susceptibility loci evaluated; rs2981582 (at 10q26 within FGFR2), rs3803662 (at 16q12 near TOX3, previously indicated as TNRC9), rs889312 (at 5q11 near MAP3K1), rs13281615 (at 8q24), rs3817198 (at 11p15 near LSP1), rs13387042 (at 2q35), rs4973768 (at 3p24), rs6504950 (at 17q23), rs1045485/rs17468277 (at 2q33-q34 within/near CASP8) and rs1982073 (at 19q13 within TGFB1). Additionally, data on age, gender and ethnicity were provided. Twenty-eight studies included women of European background in Europe, North America and Australia, one study (MEC) included similar numbers of women in the US of European and Japanese backgrounds and two studies, from Taiwan (TWBCS) and Thailand (TBCS), included East Asians (see Supplementary Material, Table S1 for a more detailed description of participating studies).
All studies were approved by their institutional review committees and written informed consent was obtained from all participants, where applicable.
For studies including mostly women of European background, we excluded from the analyses the small number of women of other ethnic backgrounds to reduce heterogeneity within studies. Analyses were restricted to women with invasive breast cancer and female controls. This resulted in a total of 38 360 invasive breast cancer cases and 53 692 controls from 31 case–control or prospective cohort studies eligible for study. Twenty of these studies (with a total of 23 839 invasive breast cancer cases and 26 928 controls) were included in our previous BCAC report on five breast cancer susceptibility loci in relation to tumor subtypes defined by ER and PR status and some pathological characteristics (17). Due to missing tumor marker data, analyses in the present study were based on a maximum of 30 040 cases with ER data and smaller number of cases for analyses including other tumor markers. The final numbers of cases and controls with genotype and pathology data included in each analysis are shown in the tables of results.
Pathology and tumor markers
Most studies provided information, regarding the tumors of the cases, on histopathological subtype (27 studies: 75% ductal, 14% lobular, 1% medullary and 10% other histologies), grade of differentiation (27 studies: 20% grade 1, 49% grade 2 and 31% grade 3+), tumor size (21 studies: 20% 1 cm or less, 43% >1–2 cm and 36% >2 cm), nodal involvement (30 studies with 38% node positive) and stage at diagnosis (23 studies: 51% stage I, 40% stage II, 8% stage III and 2% stage IV). The percentages in brackets represent the distribution for each characteristic.
By definition, all studies included in these analyses provided data on ER or PR status of the tumors, and a subset of studies also provided data on HER2 (n = 18 studies), CK5/6 or CK5 (n = 8 studies) and EGFR (n = 5 studies). Supplementary Material, Table S2 shows the source of information and methods used by each study to determine tumor marker status. The most common source of data for ER, PR and HER2 status was from medical records, followed by IHC performed on tumor tissue microarrays (TMAs) or whole section tumor slides. Data on CK5/6 (or CK5) and EGFR tumor status were derived from IHC performed on TMAs or whole sections. For the majority of studies, data submitted were dichotomous, i.e. a marker was reported as being positive or negative for each tumor markers. The cut-offs used in each study are shown in Supplementary Material, Table S2. Most commonly, ER, PR, CK5/6 and EGFR negative were defined as <10% of the tumor cells stained, HER2 negative was typically defined as either as zero cells stained or a score of 1+.
Figure 2 shows the number of cases with tumor marker data available and the classification scheme we used based on combinations of markers (Fig. 2; nomenclature explanation, / indicates ‘and/or’, & indicates ‘and’). Of the 38 368 cases eligible for analysis, 30 040 cases had data on ER status [22 866 ER+ and 7174 (24%) ER−] and 26 389 cases had data on PR status [16 997 PR+ and 9392 (36%) PR−] (Supplementary Material, Table S3). For cases with ER and PR data, 15 797 (60%) were ER+&PR+, 3755 (14%) were ER+&PR−, 1120 (4%) were ER−&PR+ and 5550 (21%) were ER−&PR−. A subset of 18 studies provided data on HER2 expression, in addition to ER and PR, for a total of 13 385 cases (Supplementary Material, Table S3). Given the strong co-expression between ER, PR and HER2, tumors were classified into four subtypes, two receptor positive or ‘luminal' subtypes: ER+/PR+&HER2− (n = 8952; 68%) and ER+/PR+&HER2+ (n = 1386; 11%) and two receptor negative subtypes or ‘non-luminal' ER−&PR−&HER2+ (n = 890; 7%) and triple negative tumors (TN: ER−&PR−&HER2−; n = 1865; 14%).
Figure 2.
Classification of breast cancer tumors according to the expression of ER, PR, HER2, CK5/6 and EGFR in tumor tissues. Tumor subtype nomenclature explanation: / = ‘and/or’, & = and.
Cases with TN tumors were further stratified according to the expression of basal markers in eight studies with data on CK5/6 [513 negative and 377 (42%) positive] and five studies with EGFR data [326 negative and 309 (49%) positive; Supplementary Material, Table S4]. Data on these markers were used to stratify TN tumors into the CBP (n = 509: CK5/6, CK5 or EGFR positive) and the five-negative phenotype (5NP) (n = 390: CK5/6 or CK5, and EGFR negative).
Genotyping
This report includes all known susceptibility loci for breast cancer identified in GWAS at the time of analyses, as well as two putative susceptibility loci from candidate gene studies. Genotyping was performed in the framework of BCAC as described previously (10,14,16,38). Twenty-nine studies genotyped cases and controls for rs2981582, 30 studies for rs3803662, 30 for rs889312, 28 for rs13281615, 26 for rs3817198, 26 for rs13387042, 22 for rs4973768 and 25 for rs6504950. Because of some technical difficulties (for some technologies) in genotyping the originally reported SNP rs1045485 in CASP8, another SNP in strong LD (rs17468277) was used as a surrogate (r2 = 1 in HapMap CEU) for a subset of studies. Thirteen studies provided genotype data on rs1045485 and 19 studies on rs17468277. Five of those studies reported genotype data on both SNPs. For individuals that had the two SNPs genotyped, rs17468277 was used. Two studies in Asian populations (TBCS and TWCS) provided genotype data on rs1045485 and rs17468277, respectively, but genotype data were not included in the analyses due to the low MAF of these SNPs in these populations. Genotype data on rs1982073 in TGFB1 were available from 17 studies. Twelve of the 31 studies had genotype data available for all 10 SNPs. Genotype data were excluded from the analysis on a study-by-study basis according to BCAC quality control (QC) guidelines: (i) any sample that consistently failed genotyping for >20% of the SNPs; (ii) all samples on any one plate that had an SNP call rate <90%; (iii) all genotype data for any SNP where overall call rate was <95%; (iv) all genotype data for any SNP where duplicate concordance was <98%. In addition, for any SNP, where the P-value for departures from Hardy–Weinberg equilibrium among controls was <0.005, clustering of the intensity plots was reviewed manually and the data excluded if clustering was judged to be poor.
Statistical analyses
Polytomous logistic regression was used to estimate odds ratios (OR) adjusted by study and associated 95% confidence intervals (CI), as measures of association between genotypes and risk of breast cancer subtypes (comparing case subtypes to all controls). All models included terms for study [dummy variables for each study, with two terms for each ethnicity in one study (MEC) that included women of both East Asian and Caucasian origin]. Further adjustment by age at diagnosis/enrolment did not substantially influence OR estimates (data not shown) and so this variable was not included in final models to avoid dropping women with missing age information from the analyses. We assessed the association for each SNP in terms of genotype-specific ORs and per-allele ORs (assuming a log-additive model). Heterogeneity between genotype ORs for different tumor subtypes was assessed using logistic regression analyses restricted to cases (case-only analyses) with the tumor characteristic as the outcome variable. For tumor subtypes with more than two levels (i.e. grade, size, stage), we used a polytomous logistic regression model constraining the effect size to increase linearly across levels (e.g. the parameter for grade 3 versus grade1 was constrained to be twice that for grade2 versus grade1). To evaluate which of the several correlated tumor features was most important in determining genotype associations, we performed stratified and adjusted analyses. For adjusted analyses, we fitted logistic regression models with one of the tumor features as the outcome and the genotype and other tumor features as explanatory variables. A P-value <0.05 was used to select findings mentioned in the results and discussion sections. Findings were interpreted in light of the strength of evidence for an association, based on the estimated ORs, 95% CI, P-values and prior knowledge of the hypothesis being tested (38).
To evaluate the combined effects of the 10 SNPs on ER+ and ER− disease, we calculated relative risks at the 10th, 50th and 90th centiles of the polygenic risk distribution by ER status (39). Calculations assumed a log-additive (multiplicative) risk model, which is consistent with analyses of the combined effects of the 10 SNPs reported in this manuscript (data not shown), and were based on estimated allele frequencies and per-allele ORs. The estimated risk distribution on a log-relative risk scale is approximately normal with mean close to zero. The area under the curve (AUC) of the receiver operating characteristic curve was calculated as a measure of discrimination of the estimated risk distribution. An AUC of 50% indicates random classification of cases and controls and an AUC of 100% indicates perfect classification.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by funding from the European Community's Seventh Framework Programme under grant agreement no. 223175 (HEALTH-F2-2009-223175). The BCAC is funded by CR-UK (C1287/A10118, C1287/ A7497, C1287/A12014). Meetings of the BCAC have been funded by the European Union COST program (BM0606). D.F.E. is a Principal Research Fellow of CR-UK. The ABCFS study was supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the National Cancer Institute, National Institutes of Health under RFA-CA-06-503 and through cooperative agreements with members of the Breast Cancer Family Registry (CFR) and P.I.s. The ABCS study was supported by the Dutch Cancer Society (grants NKI 2001-2423; 2007-3839) and the Dutch National Genomics Initiative. The BBCC study was partly funded by a Grand of ELAN Funding of the University of Erlangen. P.A.F. is partly funded by Dr Mildred Scheel Stiftung of the Deutsche Krebshilfe e.V. The BSUCH study was supported by the Dietmar-Hopp Foundation and the Helmholtz Society. The CGPS study was supported by the Chief Physician Johan Boserup and Lise Boserup Fund, the Danish Medical Research Council and Copenhagen University Hospital, Herlev Hospital. The CNIO-BCS study was supported by the Red Temática de Investigación Cooperativa en Cáncer and grants from the Asociación Española Contra Cáncer and the Fondo de Investigación Sanitario (PI081120 to JB and PI081583 to RLM). The GENICA study was supported by the German Human Genome Project and funded by the Federal Ministry of Education and Research (BMBF) Germany (grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114). The Robert Bosch Foundation of Medical Research, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Institute for Prevention and Occupational Medicine of the German Social Accident Insurance (IPA), Bochum as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany. The GESBC study was supported by the Deutsche Krebshilfe e. V. (70492) and GESBC genotyping in part by the state of Baden-Württemberg through the Medical Faculty of the University of Ulm (P.685). The HABCS study was supported by an intramural grant from Hannover Medical School. The HEBCS study has been financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (110663), the Finnish Cancer Society and the Sigrid Juselius Foundation. The KARBAC study was supported by The Swedish Cancer Society, The Gustav V Jubilee Foundation And The Bert von Kantzow Foundation. The KBCP was supported by the Kuopio University Central EVO Research Fund, Academy of Finland, the Finnish Cancer Society, the University of Kuopio and EVO research funding of Vaasa Hospital District. The kConFab and its Clinical Follow-Up study were funded by grants from the National Breast Cancer Foundation, the National Health and Medical Research Council (NHMRC) and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia and the Cancer Foundation of Western Australia, as well as by NHMRC (grants 145684, 288704 and 454508). The AOCS study was supported by the US Army Medical Research and Material Command (DAMD17-01-1-0729), the Cancer Council Tasmania and Cancer Foundation of Western Australia and The National Health and Medical Research Council of Australia (199600). Amanda B. Spurdle is supported by an NHMRC Senior Research Fellowship, and Georgia Chenevix-Trench by an NHMRC Senior Principal Research Fellowship The LMBC stduy was supported by European Union Framework Programme 6 (Project LSHC-CT-2003-503297) and by the ‘Stichting tegen Kanker’ (232–2008). The MARIE study was supported by the Deutsche Krebshilfe e.V., (grant70-2892-BR I), the Hamburg Cancer Society, the German Cancer Research Center and the German Federal Ministry of Education and Research (01KH0402). The MCBCS was supported by National Institutes of Health (grant R01 CA122340), and an NCI Specialized Program of Research Excellence (SPORE) in breast cancer (P50 CA116201). The MCCS study was supported by Cancer Council Victoria and by NHMRC (grants 209057, 251533, 396414,504711, 504715). The MEC study was supported by National Institutes of Health (grants R01-CA63464, R37-CA54281). The NBCS study was supported by grants from the Norwegian Research council, (155218/V40, 175240/S10) to ALBD, FUGE-NFR (181600/V11) to VNK and a Swizz Bridge Award to ALBD. The OFBCR was supported by the National Cancer Institute, National Institutes of Health under (grant CA- 06-503) and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and Principal Investigators, including Cancer Care Ontario (U01 CA69467), Northern California Cancer Center (U01 CA69417), and University of Melbourne (U01 CA69638) and by Cancer Care Ontario. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR nor does the mention of trade names, commercial products or organizations imply endorsement by the US Government or the BCFR. The ORIGO study was supported by the Dutch Cancer Society. The PBCS was supported by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. The RBCS study was supported by the Dutch Cancer Society (grant DDHK 2004-3124). The SASBAC study was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US National Institute of Health (NIH) and the Susan G. Komen Breast Cancer Foundation. The SBCS study was funded by the Breast Cancer Campaign (grant 2004Nov 49) and Yorkshire Cancer Research core funding. SEARCH study was supported by Cancer Research UK grants (C490/A1102, C8197/A10123, C490/A10119, C490/A11020, C1287/A10118) and AMD was funded by CR-UK grant (C8197/A10865). The pathology work in Cambridge was supported by the NIHR Cambridge Biomedical Research Centre and by the Cambridge Experimental Cancer Medicine Centre. The SZBCS was supported by Grant (PBZ_KBN_122/P05/2004). The TBCS was funded by The National Cancer Institute Thailand. The TWBCS study was supported by the Institute of Biomedical Sciences, Academia Sinica, National Sciences Counciland Taiwan Biobank. The UCIBCS study was supported by the National Institutes of Health, National Cancer Institute (grants CA-58860) and the Lon V Smith Foundation (grant LVS-39420).
Supplementary Material
ACKNOWLEDGEMENTS
The ABCFS would like to acknowledge The University of Melbourne (U01 CA69638) who contributed data to this study. The content of this manuscript does not necessarily reflect the views or the policies of the National Cancer Institute or any of the collaborating centres in the CFR nor does the mention of trade names, commercial products or organizations imply endorsement by the US Government or the CFR. We extend our thanks to the many women and their families that generously participated in the Australian Breast Cancer Family Study and consented to us accessing their pathology material. J.L.H. is a National Health and Medical Research Council Australia Fellow. M.C.S. is a National Health and Medical Research Council Senior Research Fellow. J.L.H. and M.C.S. are both group leaders of the Victoria Breast Cancer Research Consortium. The ABCS would like to acknowledge Hans Peterse, Flora van Leeuwen, Rob Tollenaar, Renate Udo and other contributors to the ‘BOSOM’ study. The BSUCH thanks all participants and colleagues of the University Womeńs Clinic Heidelberg who supported this study. Especially, we would like to thank Anne Langheinz for genotyping. The CNIO-BCS thanks Anna González-Neira, Charo Alonso, Tais Moreno and Guillermo Pita. GENICA acknowledges contributions from Christian Baisch, Volker Harth and Sylvia Rabstein. GESBC thanks Ursula Eilber and Tanya Koehler for competent technical assistance. HEBCS thanks R.N. Hanna Jäntti for the help with the patient data and Drs Ari Ristimäki, Mira Heinonen and Laura Hautala for their help with the TMA studies, and gratefully acknowledges the Finnish Cancer Registry for the cancer data. KBCP is grateful to Mrs Eija Myöhänen and Mrs Aija Parkkinen for their skilful assistance. The kConFab study wishes to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics and the Clinical Follow-Up Study for their contributions to this resource and the many families who contributed to kConFab. The AOCS Management Group (D. Bowtell, G. Chenevix-Trench, A. deFazio, D. Gertig, A. Green, P. Webb) gratefully acknowledges the contribution of all the clinical and scientific collaborators (see http:// www.aocstudy.org/). LMBC thanks Natacha Lays and Gilian Peuteman for technical assistance. MARIE would like to thank the pathology institutes and R. Salazar and W. Höppner from BioGlobe GmBH, Hamburg for their valuable contributions, and S. Behrens, R. Birr, W. Busch, U. Eilber, B. Kaspereit, N. Knese, K. Smit, for their excellent technical assistance. MCBCS thanks Vicki Cafourek for abstraction of medical records and other contributors to the MCBCS study. OFBCR wishes to thank the participants in the Ontario Familial Breast Cancer Registry. We wish to thank Gord Glendon, Teresa Selander, Elaine Maloney and Nayana Weerasooriya from Cancer Care Ontario and members of the Ontario Cancer Genetics Network for their contributions to the study. ORIGO wishes to thank E. Krol-Warmerdam and J. Blom for patient accrual, administering questionnaires and managing clinical information. The PBCS thanks Dr Louise Brinton from the Division of Cancer Epidemiology and Genetics of the National Cancer Institute, USA, Drs Neonila Szeszenia-Dabrowska and Beata Peplonska of the Nofer Institute of Occupational Medicine (Lodz, Poland), Witold Zatonski of the Department of Cancer Epidemiology and Prevention, The M. Sklodowska-Curie Cancer Center and Institute of Oncology (Warsaw, Poland) and Pei Chao and Michael Stagner from Information Management Services (Silver Spring, MD, USA), for their valuable contributions to the study. The RBCS would like to acknowledge Petra Bos, Jannet Blom, Ellen Crepin, Elisabeth Huijskens and Annette Heemskerk for their contribution in data managing. The SBCS would like to thank Sue Higham, Gordon MacPherson and Ian Brock from the University of Sheffield for their contributions to this study. We also thank all the participants in all the participating studies.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Sorlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.B., van de Rijn M., Jeffrey S.S., et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. doi:10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perou C.M., Sorlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. doi:10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Korde L.A., Lusa L., McShane L., Lebowitz P.F., Lukes L., Camphausen K., Parker J.S., Swain S.M., Hunter K., Zujewski J.A. Gene expression pathway analysis to predict response to neoadjuvant docetaxel and capecitabine for breast cancer. Breast Cancer Res. Treat. 2010;119:685–699. doi: 10.1007/s10549-009-0651-3. doi:10.1007/s10549-009-0651-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams P.D., Cheon S., Havaleshko D.M., Jeong H., Cheng F., Theodorescu D., Lee J.K. Concordant gene expression signatures predict clinical outcomes of cancer patients undergoing systemic therapy. Cancer Res. 2009;69:8302–8309. doi: 10.1158/0008-5472.CAN-09-0798. doi:10.1158/0008-5472.CAN-09-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Althuis M.D., Fergenbaum J.H., Garcia-Closas M., Brinton L.A., Madigan M.P., Sherman M.E. Etiology of hormone receptor-defined breast cancer: a systematic review of the literature. Cancer Epidemiol. Biomarkers Prev. 2004;13:1558–1568. [PubMed] [Google Scholar]
- 6.Anderson W.F., Jatoi I., Devesa S.S. Distinct breast cancer incidence and prognostic patterns in the NCI's SEER program: suggesting a possible link between etiology and outcome. Breast Cancer Res. Treat. 2005;90:127–137. doi: 10.1007/s10549-004-3777-3. doi:10.1007/s10549-004-3777-3. [DOI] [PubMed] [Google Scholar]
- 7.Anderson W.F., Chu K.C., Chang S., Sherman M.E. Comparison of age-specific incidence rate patterns for different histopathologic types of breast carcinoma. Cancer Epidemiol. Biomarkers Prev. 2004;13:1128–1135. [PubMed] [Google Scholar]
- 8.Ma H., Bernstein L., Pike M.C., Ursin G. Reproductive factors and breast cancer risk according to joint estrogen and progesterone receptor status: a meta-analysis of epidemiological studies. Breast Cancer Res. 2006;8:R43. doi: 10.1186/bcr1525. doi:10.1186/bcr1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X.R., Chang-Claude J., Goode E.L., Couch F.J., Nevanlinna H., Milne R.L., Gaudet M., Schmidt M.K., Broeks A., Cox A., et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. J. Natl Cancer Inst. 2011;103:250–263. doi: 10.1093/jnci/djq526. doi:10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. doi:10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. doi:10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stacey S.N., Manolescu A., Sulem P., Thorlacius S., Gudjonsson S.A., Jonsson G.F., Jakobsdottir M., Bergthorsson J.T., Gudmundsson J., Aben K.K., et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2008;40:703–706. doi: 10.1038/ng.131. doi:10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 13.Stacey S.N., Manolescu A., Sulem P., Rafnar T., Gudmundsson J., Gudjonsson S.A., Masson G., Jakobsdottir M., Thorlacius S., Helgason A., et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007;39:865–869. doi: 10.1038/ng2064. doi:10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 14.Ahmed S., Thomas G., Ghoussaini M., Healey C.S., Humphreys M.K., Platte R., Morrison J., Maranian M., Pooley K.A., Luben R., et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 2009;41:585–590. doi: 10.1038/ng.354. doi:10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng W., Long J., Gao Y.T., Li C., Zheng Y., Xiang Y.B., Wen W., Levy S., Deming S.L., Haines J.L., et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009;41:324–328. doi: 10.1038/ng.318. doi:10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox A., Dunning A.M., Garcia-Closas M., Balasubramanian S., Reed M.W., Pooley K.A., Scollen S., Baynes C., Ponder B.A., Chanock S., et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet. 2007;39:352–358. doi: 10.1038/ng1981. doi:10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Closas M., Hall P., Nevanlinna H., Pooley K., Morrison J., Richesson D.A., Bojesen S.E., Nordestgaard B.G., Axelsson C.K., Arias J.I., et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. doi:10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Closas M., Chanock S. Genetic susceptibility loci for breast cancer by estrogen receptor status. Clin. Cancer Res. 2008;14:8000–8009. doi: 10.1158/1078-0432.CCR-08-0975. doi:10.1158/1078-0432.CCR-08-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reeves G.K., Travis R.C., Green J., Bull D., Tipper S., Baker K., Beral V., Peto R., Bell J., Zelenika D., Lathrop M. Incidence of breast cancer and its subtypes in relation to individual and multiple low-penetrance genetic susceptibility loci. JAMA. 2010;304:426–434. doi: 10.1001/jama.2010.1042. doi:10.1001/jama.2010.1042. [DOI] [PubMed] [Google Scholar]
- 20.Nordgard S.H., Johansen F.E., Alnaes G.I., Naume B., Borresen-Dale A.L., Kristensen V.N. Genes harbouring susceptibility SNPs are differentially expressed in the breast cancer subtypes. Breast Cancer Res. 2007;9:113. doi: 10.1186/bcr1784. doi:10.1186/bcr1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen T.O., Hsu F.D., Jensen K., Cheang M., Karaca G., Hu Z., Hernandez-Boussard T., Livasy C., Cowan D., Dressler L., et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma 21. Clin. Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. doi:10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 22.Yang X.R., Pfeiffer R.M., Garcia-Closas M., Rimm D.L., Lissowska J., Brinton L.A., Peplonska B., Hewitt S.M., Cartun R.W., Mandich D., et al. Hormonal markers in breast cancer: coexpression, relationship with pathologic characteristics, and risk factor associations in a population-based study. Cancer Res. 2007;67:10608–10617. doi: 10.1158/0008-5472.CAN-07-2142. doi:10.1158/0008-5472.CAN-07-2142. [DOI] [PubMed] [Google Scholar]
- 23.Kreike B., van Kouwenhove M., Horlings H., Weigelt B., Peterse H., Bartelink H., van de Vijver M.J. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9:R65. doi: 10.1186/bcr1771. doi:10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Ronde J.J., Hannemann J., Halfwerk H., Mulder L., Straver M.E., Vrancken Peeters M.J., Wesseling J., van de Vijver M., Wessels L.F., Rodenhuis S. Concordance of clinical and molecular breast cancer subtyping in the context of preoperative chemotherapy response. Breast Cancer Res. Treat. 2010;119:119–126. doi: 10.1007/s10549-009-0499-6. doi:10.1007/s10549-009-0499-6. [DOI] [PubMed] [Google Scholar]
- 25.Kapp A.V., Jeffrey S.S., Langerod A., Borresen-Dale A.L., Han W., Noh D.Y., Bukholm I.R., Nicolau M., Brown P.O., Tibshirani R. Discovery and validation of breast cancer subtypes. BMC Genomics. 2006;7:231. doi: 10.1186/1471-2164-7-231. doi:10.1186/1471-2164-7-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blows F.M., Driver K.E., Schmidt M.K., Broeks A., van Leeuwen F.E., Wesseling J., Cheang M.C., Gelmon K., Nielsen T.O., Blomqvist C., et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10 159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. doi:10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milne R.L., Benitez J., Nevanlinna H., Heikkinen T., Aittomaki K., Blomqvist C., Arias J.I., Zamora M.P., Burwinkel B., Bartram C.R., et al. Risk of estrogen receptor-positive and -negative breast cancer and single-nucleotide polymorphism 2q35-rs13387042. J. Natl Cancer Inst. 2009;101:1012–1018. doi: 10.1093/jnci/djp167. doi:10.1093/jnci/djp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antoniou A.C., Wang X., Fredericksen Z.S., McGuffog L., Tarrell R., Sinilnikova O.M., Healey S., Morrison J., Kartsonaki C., Lesnick T., et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat. Genet. 2010;42:885–892. doi: 10.1038/ng.669. doi:10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luqmani Y.A., Graham M., Coombes R.C. Expression of basic fibroblast growth factor, FGFR1 and FGFR2 in normal and malignant human breast, and comparison with other normal tissues. Br. J. Cancer. 1992;66:273–280. doi: 10.1038/bjc.1992.256. doi:10.1038/bjc.1992.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antoniou A.C., Spurdle A.B., Sinilnikova O.M., Healey S., Pooley K.A., Schmutzler R.K., Versmold B., Engel C., Meindl A., Arnold N., et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am. J. Hum. Genet. 2008;82:937–948. doi: 10.1016/j.ajhg.2008.02.008. doi:10.1016/j.ajhg.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antoniou A.C., Beesley J., McGuffog L., Sinilnikova O.M., Healey S., Neuhausen S.L., Ding Y.C., Rebbeck T.R., Weitzel J.N., Lynch H.T., et al. Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: implications for risk prediction. Cancer Res. 2010;70:9742–9754. doi: 10.1158/0008-5472.CAN-10-1907. doi:10.1158/0008-5472.CAN-10-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lakhani S.R., van de Vijver M.J., Jacquemier J., Anderson T.J., Osin P.P., McGuffog L., Easton D.F. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, `, HER−2, and p53 in patients with mutations in BRCA1 and BRCA2. J. Clin. Oncol. 2002;20:2310–2318. doi: 10.1200/JCO.2002.09.023. doi:10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 33.Antoniou A.C., Sinilnikova O.M., McGuffog L., Healey S., Nevanlinna H., Heikkinen T., Simard J., Spurdle A.B., Beesley J., Chen X., et al. Common variants in LSP1, 2q35 and 8q24 and breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum. Mol. Genet. 2009;18:4442–4456. doi: 10.1093/hmg/ddp372. doi:10.1093/hmg/ddp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawson S.J., Provenzano E., Caldas C. Triple negative breast cancers: clinical and prognostic implications. Eur. J. Cancer. 2009;45((Suppl. 1)):27–40. doi: 10.1016/S0959-8049(09)70013-9. doi:10.1016/S0959-8049(09)70013-9. [DOI] [PubMed] [Google Scholar]
- 35.Rebbeck T.R., Antoniou A.C., Llopis T.C., Nevanlinna H., Aittomaki K., Simard J., Spurdle A.B., Couch F.J., Pereira L.H., Greene M.H., et al. No association of TGFB1 L10P genotypes and breast cancer risk in BRCA1 and BRCA2 mutation carriers: a multi-center cohort study. Breast Cancer Res. Treat. 2009;115:185–192. doi: 10.1007/s10549-008-0064-8. doi:10.1007/s10549-008-0064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gown A.M. Current issues in ER and HER2 testing by IHC in breast cancer. Mod. Pathol. 2008;21((Suppl. 2)):S8–S15. doi: 10.1038/modpathol.2008.34. doi:10.1038/modpathol.2008.34. [DOI] [PubMed] [Google Scholar]
- 37.Collins L.C., Marotti J.D., Baer H.J., Tamimi R.M. Comparison of estrogen receptor results from pathology reports with results from central laboratory testing. J. Natl Cancer Inst. 2008;100:218–221. doi: 10.1093/jnci/djm270. doi:10.1093/jnci/djm270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wacholder S., Chanock S., Garcia-Closas M., El Ghormli L., Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. doi:10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pharoah P.D., Antoniou A.C., Easton D.F., Ponder B.A. Polygenes, risk prediction, and targeted prevention of breast cancer. N. Engl. J. Med. 2008;358:2796–2803. doi: 10.1056/NEJMsa0708739. doi:10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.