Abstract
The loss of heterologous protein expression is one of the major problems faced by industrial cell line developers and has been reported by several authors. Therefore, the understanding of the mechanisms involved in the generation of stable and high producer cell lines is a critical issue, especially for those processes based on long term continuous cultures. We characterized two recombinant NS0 myeloma cell lines expressing Nimotuzumab, a humanized anti-human epidermal growth factor receptor (EGFR) antibody. The hR3/H7 clone is a stable producer obtained from the unstable hR3/t16 clone. The unstable clone was characterized by a bimodal distribution of intracellular immunoglobulin staining using flow cytometry. Loss of antibody production was due to the emergence of a non-producer cell subpopulation that increased with cell generation number. Immunoglobulin heavy chain (HC) and light chain (LC) ratio (HC/LC) was lower for the unstable phenotype. Proteomic maps using two dimensional gel electrophoresis (2DE) were obtained for both clones, at initial cell culture time and after 40 generations. Fifteen proteins potentially associated with the phenomenon of production stability were identified. The hR3/H7 stable clone showed an up-regulated expression pattern for most of these proteins. The regulation of recombinant antibody production by the host NS0 myeloma cell line most likely involves simultaneously cellular processes such as DNA transcription, mRNA processing, protein synthesis and folding, vesicular transport, glycolysis and energy production, according to the proteins identified in the present proteomic study.
Electronic supplementary material
The online version of this article (doi:10.1007/s10616-011-9348-7) contains supplementary material, which is available to authorized users.
Keywords: Recombinant antibody, Protein expression stability, NS0 myeloma cell line, Proteomics
Introduction
Mammalian cells are commonly used for the production of recombinant proteins in biotechnological industry. One of the major reasons for this is their ability to correctly fold the target protein and carry out a range of posttranslational modifications (Wurm 2004). Among the mammalian cells used for the production of recombinant antibodies, non-secreting myeloma cells are of particular commercial importance (Barnes et al. 2003). In particular, the NS0 cell line was derived from a mineral-oil-induced plasmacytoma (MOPC-21) in a female BALB/c mouse. Although it contains the necessary machinery (Köhler and Milstein 1976; Barnes et al. 2000), this cell line was selected in vitro for the non-immunoglobulin secretion phenotype (Whitford 2003), most likely caused by an epigenetic regulation mechanism.
One of the most important criteria for the successful production at large scale of a therapeutic protein is to obtain a recombinant cell line with a high and stable production rate. However, the production of heterologous proteins by a recombinant cell line is modulated by molecular events at different cellular processes, ranging from transcription, posttranscriptional processing, translation, posttranslational processing, to secretion (Barnes et al. 2003, 2006, 2007). Transgene can often undergo the so-called ‘‘position effect’’, whereby the surrounding genomic regions at the site of incorporation can decrease, or even silence, the recombinant DNA expression (Wallrath 1998; Wilson et al. 1990). In general, the causes at the molecular level of protein production instability are in many cases unknown. Therefore, in most cases the identification of stable cell lines is performed via empirical procedures based on analysis of the growth and productivity of potential cell lines over extended periods of time in culture.
A number of investigators have studied the unstable recombinant protein production phenotype from GS-NS0 cell lines, through the comparison of different producer cell lines at the mRNA level (Barnes and Dickson 2006). These studies were performed with the aim of better understanding the molecular mechanisms responsible for this phenomenon and to find predictor factors for stability. Proteomic approaches have also been used to define the “molecular fingerprint” determining the translational or secretory capacity of the host cell lines, looking for operational criteria of an “industrial” cell line (Smales et al. 2004; Seth et al. 2007).
In the present study we addressed the question of whether the stability of recombinant antibody production by NS0 myeloma cells could be achieved by a “gain of function” phenomenon through an epigenetic regulation mechanism, in opposition to the “loss of function” mechanism that has been largely documented as the cause of production instability.
In our experience, recombinant NS0 clones loss immunoglobulin expression along the culture time course. However, we have previously shown that stable producer clones can be selected from long-term cell cultures even of unstable clones (Rojas et al. 2005). The main goal of the present work was to characterize at the protein level changes explaining the emergence of a stable producer clone from an unstable one. For this purpose, we chose as a model two different recombinant NS0 clones expressing a humanized anti-human epidermal growth factor receptor (EGFR) antibody, the hR3/h7 stable producer clone (Castillo et al. 2005) and its parental hR3/t16 unstable producer clone (Mateo et al. 1997). A detailed kinetic study was performed to evaluate the growth and production rates of both clones. Using two-dimensional gel electrophoresis (2DE) and mass spectrometry, we found some up-regulated proteins potentially associated with antibody production stability. Our experimental data indicate that the stability in the production of the recombinant antibody by NS0 cells is a result of the upregulation of several cellular processes.
Materials and methods
Cell lines and culture conditions
Two cell lines were used in this work, the hR3/t16 clone, which was obtained by limiting dilution cloning of the original humanized anti-human EGFR hR3 antibody-producing NS0 cells (Mateo et al. 1997), and the hR3/H7 cell line, which was obtained by limiting dilution cloning after batch culture of the hR3/t16 cells. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F12 (GIBCO-BRL, Paisley, UK) supplemented with 5% (v/v) fetal bovine serum (FBS) (GIBCO BRL).
For the study of the antibody expression stability the cell lines were cultured in T-75 flasks (Greiner Bio-One, Switzerland) over 40 days in repeated batch culture conditions, keeping them in an exponential growth phase. This was achieved by changing the fresh medium every 48 h and removing the excess of cells for maintaining the same cell concentration. We chose this phase of the culture trying to simulate the growth conditions of the industrial scale for the production of the recombinant antibody, in which the perfusion technology is used. Bioreactors are periodically “bleed”, in order to remove the excess of cells and add fresh medium, thus maintaining the culture in the exponential growth phase. Every 10 days, samples were seeded onto 24-well plates (Greiner Bio-One, Switzerland) at a density of 3 × 105 viable cells per mL. The supernatants were collected from each well 10 days later and stored at −70 °C before ELISA testing. 2.5 × 105 cells were taken both from day 0 and 40 of the expression stability study, washed twice with phosphate buffered saline (PBS), fixed with 70% ethanol for 16 h at 4 °C and then analyzed by flow cytometry.
Batch culture growth analyses were performed in duplicates in 250 mL spinner flasks. Initial seeding density was 3 × 105 viable cells per mL and cell concentration and viability were assessed at 24 h intervals by cell counting using the trypan blue exclusion method in Neubauer chambers. Cultures were stopped when viability was below 50%. Sample supernatants were then centrifuged and stored at −70 °C for IgG concentration measurement by ELISA. For the proteomic experiments cells were harvested at the mid-exponential growth phase. All cell cultivation was performed at 37 °C in an atmosphere with 5% CO2.
FACS analysis
For assessment of intracellular recombinant IgG, cells fixed with 70% ethanol were centrifuged for 5 min at 500×g at 4 °C in order to remove ethanol. After washing with PBS, cells were stained with a FITC-conjugated anti-human γ-chain antibody (Sigma, St. Louis, MO) for 30 min at 4 °C, followed by a second washing. Up to 10,000 events were acquired using a FACScan flow cytometer (Beckton Dickinson, Mansfield, MA) and analyzed using the CellQuest software.
Supernatant IgG quantification by ELISA
Supernatant samples were tested by ELISA as detailed by Faife et al. 2008.
Total protein extraction from cell lines
Samples from both cell lines corresponding to days 0 and 40 were harvested at the mid-exponential phase and total proteins were extracted as previously described by de la Luz et al. 2008.
Immunoblotting analysis of heavy and light chain expression
For analysis of intracellular heavy chain (HC) and light chain (LC) expression, 1 × 108 cells were prepared as described above. Supernatant samples and whole cell extracts were analyzed by electrophoresis on 10 and 12.5% SDS–polyacrylamide gels (Laemmli 1970), respectively. Samples were dissolved in non-reducing SDS-sample buffer and 15 μL per lane were applied. Three replicates for each condition were analyzed.
The electrophoresed proteins were transferred onto nitrocellulose membranes by means of a semi-dry electrophoretic transfer cell using transfer buffer (25 mM Tris, 192 mM glycine and 20% methanol) at 100 mA, for 1 h. Membranes were blocked for 1 h at 37 °C with 5% (w/v) non-fatty milk in PBS/0.1% Tween-20 and then washed three times for 5 min with PBS/0.1% Tween-20. Membranes were subsequently incubated with either horseradish peroxidase-conjugated goat anti-human κ-light chain antibody (Sigma) (1/1,000 dilution) or an alkaline phosphatase-conjugated anti-human γ-chain antibody (Sigma) (1/10,000 dilution). Proteins were detected by adding the respective chromogenic solutions: 500 mg diaminobenzidine dissolved in 500 μL of N, N-dimethylformamide and 20 μL of 30% hydrogen peroxide; or a mixture of Fast Red TR/Napthol AS MX dissolved in 0.1 M Tris, pH 8.0. Membrane images were captured with a calibrated Powerlook III prepress colour scanner (Amersham Pharmacia, UK) and the MagicScan software. Band densitometry analysis was performed using TotalLab 120 software (Nonlinear Dynamics, Newcastle, UK). Band intensity of extracellular HC and LC polypeptides (AHC, ALC) per lane was calculated as follows: AHC = AHC2-LC2 + AHC-LC and ALC = AHC2-LC2 + AHC-LC + ALC2.
Two-dimensional electrophoresis
For Two-dimensional electrophoresis (2-DE), 3 × 108 cells were prepared as described above. For each condition, three IPG strips (pI range 3–10, 17 cm) were focused in parallel. The strips were rehydrated passively overnight in 300 μL of lysis solution containing approximately 200 μg of proteins for analytical gels and 600 μg for preparative gels. The isoelectric focusing and SDS–PAGE were performed as previously described (de la Luz et al. 2008). For analytical gels, silver staining was used for protein detection as described by Gharahdaghi et al. 1999, while preparative gels were silver-stained with a mass spectrometry (MS) compatible procedure as described by Shevchenko et al. (1996).
2-DE image analysis
Stained gels were captured with a calibrated Powerlook III prepress colour scanner (Amersham Pharmacia, UK) and the images were exported to the image analysis software program Melanie 5 (GeneBio, Geneva, Switzerland). For each gel (three gels per condition), the spots were detected and quantified automatically using the default spot detection parameters. Manual image edition was done when necessary. Automatic spot matching was carried out landmarked by 50 evenly distributed and well-defined spots. Spots were quantified in terms of their relative volume (% vol). Gel reproducibility was evaluated by using the available tools (pair reports and scatter plot analysis). The comparison between two gel groups (e.g. hR3/t16 cell line at initial culture vs. hR3/t16 after 40 days in culture) was based on spot quantification data corresponding to averaged values for the three replicate gels for each condition. To evaluate quantitative changes in protein expression from different comparisons, we selected those spots that were present in all gels analyzed and that also showed quantitative fold changes in their expression ≥3.
Protein identification using 2-DE master gel of host NS0 cell line
Some spots of interest were identified by comparing the obtained gels with a master gel of the host NS0 cell line obtained at our laboratory, in which a set of spots had been identified by MALDI-MS and/or LC–ESI–MS–MS (de la Luz et al. 2008). Spots of interest that had not been yet identified in our master gel were then identified by MALDI-MS analysis of in-gel tryptic digestions.
In-gel digestion
Spots were excised from preparative gels. After excision, the spots of interest were unstained and digested as previously detailed by Shevchenko et al. (1996).
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS)
Following gel digestion, samples were desalted using C-18 ZipTips (Millipore, NY) and 1 μL of the sample was spotted onto a MALDI plate followed immediately by 1 μL of matrix solution composed of 10 mg/mL α-cyano-4-hydroxycinnamic acid (Agilent Technologies, CA) in 0.1% (v/v) trifluoroacetic acid and 60% (v/v) acetonitrile and analyzed on a MALDI-TOF-TOF mass spectrometer (Axima Performance, Shimadzu, Japan), operated in the reflector mode. Spectra were obtained by accumulating 100 consecutive laser shots. Calibration was performed using [M + H] + ions of a mixture of bradykin fragment (m/z 757.39), angiotensin II (m/z 1,046.54) and P14R synthetic peptide (m/z 1,533.85).
Protein identification using database searching algorithms
The resulting peptide masses were subjected to a search against the Swiss-Prot mouse database using the MASCOT search engine. Search parameters included modifications such as propionamide of cysteines and methionine oxidation. Monoisotopic masses of tryptic peptides were used to identify the proteins by peptide mass fingerprinting. Peptide mass tolerance was ±0.8 Da, and no restrictions were imposed on protein molecular mass. One missed cleavage site was allowed in the protein identification. Hits were considered significant if the protein score exceeded the threshold score calculated by the Mascot software assuming p value < 0.05. Proteins identified were also required to meet approximate MW and pI obtained from image analysis.
Results
Culture parameters and recombinant antibody expression
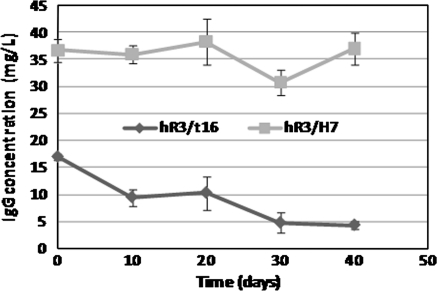
The stability of antibody expression was study in a 40 days culture of the hR3/t16 and hR3/H7 clones. In the case of the hR3/t16 cell line, the recombinant protein production dropped 75% by the end of the study; this drop started from as early as 20 days of culture. In contrast, for the hR3/H7 cell line the levels of secreted recombinant antibody remained unchanged during the study. Therefore, in terms of antibody production, hR3/t16 and hR3/H7 can be classified as unstable and stable cell lines, respectively (Fig. 1).
Fig. 1.
Kinetics of recombinant antibody secretion by hR3/t16 and hR3/H7 clones. The two clones were cultured in T-75 flasks for 40 days. Every 10 days samples were seeded onto 24-well plates, the supernatants collected 10 days later and IgG concentration measured by ELISA. Results represent the mean ± SD of triplicate measurements
The values of maximal cellular concentration (Xv max), specific growth rate (μ), specific production rate (qp) and the maximal IgG concentration at culture start and after 40 days are shown in Table 1. Both cell lines displayed approximately equal growth rates; however, specific production rates were different. In the case of hR3/t16 (unstable clone) the qp value decreased by a half, while it remained constant in the stable clone.
Table 1.
Kinetics of growth and productivity parameters of hR3/t16 and hR3/H7 clones
| Cell lines | Xv max (106 cells/mL) | μ (h−1) | qp (pg/cell × h) |
|---|---|---|---|
| hR3/t16 0 | 2.09 ± 0.12 | 0.029 ± 0.005 | 0.154 ± 0.017 |
| hR3/t16 40 | 2.25 ± 0.50 | 0.025 ± 0.006 | 0.062 ± 0.07 |
| hR3/H7 0 | 2.05 ± 0.31 | 0.020 ± 0.004 | 0.241 ± 0.027 |
| hR3/H7 40 | 2.00 ± 0.42 | 0.026 ± 0.003 | 0.239 ± 0.013 |
Results are the mean ± standard deviation of triplicate measurements. Sub indexes 0 and 40 refer to days in culture
Intracellular IgG levels
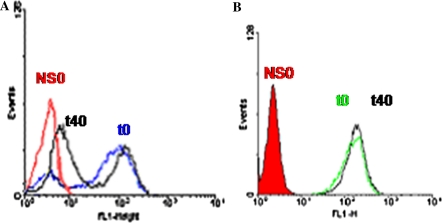
The intracellular IgG levels were determined by FACS analysis (Fig. 2). In spite of being a clone obtained by limiting dilution cloning, hR3/t16 cells showed a bimodal intracellular IgG distribution, a feature that remained over the 40 days of culture. However, by the end of the culture there was an increase in the low-producing population percentage. Of notice, the median fluorescence intensity (MFI) value corresponding to the high-producing population did not change. In other words, instead of a shifting of a single peak to a lower MFI, what we observed was an increase in the frequency of the non-producing cell sub-population. The stable cell line (hR3/H7) was characterized by a homogenous population and the expression levels were similar both at the beginning and at the end of the culture.
Fig. 2.
Intracellular IgG content in hR3/t16 and hR3/H7 clones at the beginning of the culture and after 40 days. Cells were collected at the indicated time points, fixed and analyzed by flow cytometry using a FITC-conjugated anti-human γ chain antibody. a Unstable cell line hR3/t16 at day 0 (blue curve) and 40 (black curve). b Stable cell line hR3/H7 at day 0 (green curve) and 40 (black curve). Red curved: non-transfected host NS0 cell line
Assessment of extra and intracellular heavy and light chain expression
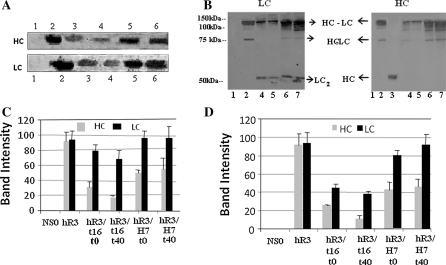
Semi-quantitative assessment of heavy (HC) and light (LC) chain expression was performed by Western blot analysis (Fig. 3). To make comparable the intensity values of both chains, antibodies against HC and LC polypeptides were proportionally diluted according a standard curve (supplementary figure 1). We detected three polypeptide conformations in the supernatant samples, which corresponded to whole antibody (HC2-LC2), heterodimeric “half-antibody” (HC-LC), and the LC homodimer (LC2). Neither HC monomer (HC) nor homodimer (HC2) were detected (Fig. 3b). This result is in agreement with previous reports demonstrating that HC polypeptide is only secreted when linked to a LC polypeptide (Dinnis and James 2005). As shown in Fig. 3a and b and Table 2, the LC polypeptide was more abundant than the HC polypeptide both intracellularly and extracellularly (HC: LC ratio <1).
Fig. 3.
Immunoblotting analysis of intracellular (A) and extracellular (B) HC and LC polypeptides in hR3/t16 and hR3/H7 clones at the beginning of the culture and after 40 days. Light and heavy chains were detected in membranes using a horseradish peroxidase-conjugated goat anti-human κ chain antibody or an alkaline phosphatase-conjugated anti-human γ chain antibody, respectively. In panel A lanes correspond to: (1) non-transfected host NS0 cell line; (2) non-reduced, purified hR3 antibody; (3) reduced, purified hR3/t16 cell line extract at day 0; (4) hR3/t16 cell line extract at day 40; (5) hR3/H7 cell line extract at day 0; (6) hR3/H7 cell line extract at day 40. In panel B lanes correspond to: (1) non-transfected host NS0 cell line; (2) non-reduced, purified hR3 antibody; (3) reduced, purified hR3 antibody; (4) hR3/t16 cell line extract at day 0; (5) hR3/t16 cell line extract at day 40; (6) hR3/H7 cell line extract at day 0; (7) hR3/H7 cell line extract at day 40. Panels (C) and (D) represent the densitometry analysis of bands in A and B, respectively. Band intensity is reported in arbitrary intensity units as the mean ± standard deviation of triplicate measurements
Table 2.
Intra and extracellular HC/LC polypeptides ratio, measured by Western blot
| Cell lines | Intracellular | Extracellular |
|---|---|---|
| NS0 | 0.00 | 0.00 |
| hR3 | 0.98 | 0.98 |
| hR3/t16 t0 | 0.39 | 0.53 |
| hR3/t16 t40 | 0.25 | 0.40 |
| hR3/H7 t0 | 0.52 | 0.53 |
| hR3/H7 t40 | 0.57 | 0.50 |
Values of band intensity in arbitrary intensity units are shown. Results are the mean of triplicate measurements
Comparison of the unstable cell line (hR3/t16) at the beginning and after 40 days in culture revealed a decrease in the intracellular HC and LC expression, which correlated with the lower antibody levels found in culture supernatants at the end of the study. The HC and LC expression levels remained relatively constant in the stable cell line (hR3/H7) at both culture times. In addition, we found that even from the beginning of the culture the intracellular HC/LC ratio was lower in the unstable cell line when compared to the stable one (Table 2). The fact that the HC2-LC2 polypeptide conformation was the most abundant one suggests that it is predominant.
Proteomic characterization of cell lines
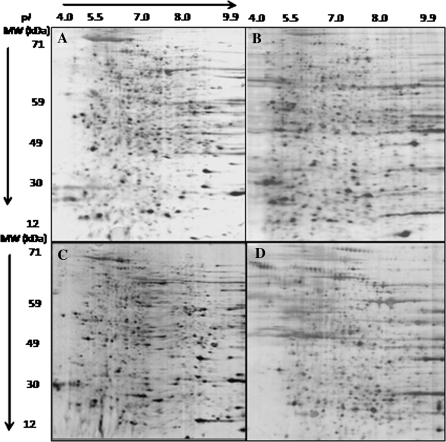
We compared 2-DE gels in a pI range of 3–10, working with whole cell extracts from hR3/t16 and hR3/H7 cell lines taken at the beginning of the culture and after 40 days (Fig. 4). During sample preparation, experimental conditions were controlled to maintain reproducibility. Minor horizontal streaking was observed on the gels for proteins with a pI higher than eight, but this phenomenon is common among proteins focusing in the basic range (Alete et al. 2005). The whole cell extract proteome yielded between 524 and 825 discrete protein spots (mean of 639 spots ± 136) based on twelve gels.
Fig. 4.
Representative 2-DE gels obtained in the pI range of 3–10 and 15% acrylamide with the cell extracts from hR3/t16 and hR3/H7 clones at the beginning of the culture and after 40 days. a hR3/t16 cell line extract at day 0. b hR3/t16 cell line extract at day 40. c hR3/H7 cell line extract at day 0. d hR3/H7 cell line extract at day 40
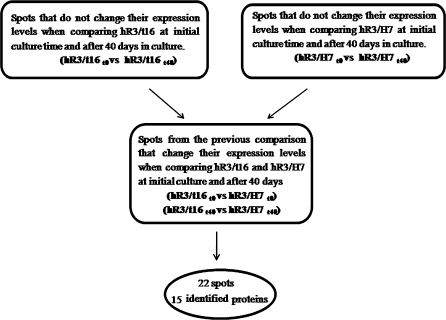
Aiming to identify proteins that could be involved in the transition from the unstable (hR3/t16) to the stable (hR3/H7) phenotypes, we selected a group of spots that matched the following criteria: (1) did not change their expression when comparing either hR3/t16 or hR3/H7 with themselves at the beginning of the culture and after 40 days, and (2) were differentially expressed when comparing hR3/t16 with hR3/H7 at the beginning of the culture and after 40 days (22 spots; Fig. 5).
Fig. 5.
Scheme to illustrate the algorithm used in the proteomic study. Spots with quantitative changes ≥3 fold were selected as differentials
Identification of differentially expressed proteins
In order to guarantee the correspondence of the selected spots with the proteins identified on the master gel, we calibrated internally all gels with the following proteins: heat-shock cognate 71 kDa protein, heterogeneous nuclear ribonucleoprotein L, transketolase, tumor protein p53-inducible nuclear protein 2, and 10 kDa heat shock protein, mitochondrial. These proteins were selected as calibrators because of their distribution in the gel. Each association was verified by manual image edition.
Some spots of interest that had not been previously described from our master gel (de la Luz et al. 2008) were now identified by MALDI-MS (Table 3). From the spots detected above we identified fifteen proteins potentially related to the stable producer phenotype (Table 4). The rest of selected spots were due to protein degradation. Among the molecular functions where the identified proteins participate we found DNA transcription and structure, mRNA processing, protein synthesis and folding, vesicular transport and cytoskeleton structure, and energy production.
Table 3.
Proteins not previously described in the NS0 cell line master gel
| Protein identity | Swiss-Prot accession number | Coverage %/Mascot score | Mw (theor.) Da/pI (theor.) |
|---|---|---|---|
| Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) | Q9CUU3 | 34/66 | 172,122.76/8.19 |
| Poly [ADP-ribose] polymerase | P11103 | 62/70 | 113,100/9.05 |
| Cell division cycle-associated 7-like protein | Q922M5 | 66/59 | 50,217/7.83 |
| Kinesin-like protein KIF2C | Q922S8 | 45/58 | 81,085/8.22 |
| Structural maintenance of chromosomes protein 1A | Q9CU62 | 79/60 | 143,216/7.08 |
| Myosin-Va | Q99104 | 66/76 | 215,595/8.84 |
Protein spots were analyzed by MS (MALDI-TOF) and identified with the Swiss-Prot protein database using MASCOT as search engine
Table 4.
Proteins related to stability of antibody production differentially expressed in the hR3/t16 and hR3/H7 clones
| Protein name | Molecular function | Fold change (hR3/H7 hR3/t16) |
|---|---|---|
| Peptidyl-prolyl cis–trans isomerase A | Protein synthesis and folding | +5.8 |
| T-complex protein 1 | +9.5 | |
| 60 kDa heat shock protein, mitochondrial precursor | +5.76 | |
| Heat shock protein 75 kDa, mitochondrial precursor | +3.4 | |
| Histone H2B F | DNA transcription and structure | +3.8 |
| Histone H4 | +5.6 | |
| Synaptonemal complex protein 2 (SCP-2) | +11.18 | |
| Poly [ADP-ribose] polymerase | + 4.0 | |
| Cell division cycle-associated 7-like protein | −4.21 | |
| Heterogeneous nuclear ribonucleoprotein L | RNAm processing | +5.14 |
| Heterogeneous nuclear ribonucleoprotein A2/B1 | −3.1 | |
| Heterogeneous nuclear ribonucleoprotein H | +3.47 | |
| Tubulin beta 1 chain | Vesicular transport and cytoskeleton structure | +3.0 |
| Tubulin beta 3 chain | +3.05 | |
| Transketolase | Carbohydrate metabolism and energy production | +3.29 |
Protein spots were analyzed by MS (MALDI-TOF) and identified with the Swiss-Prot protein database using MASCOT as search engine. Proteins are grouped according to molecular function
Discussion
The regulatory events in the recombinant protein production process can be viewed to occur within three distinct phases: plasmid integration and chromosomal environment; mRNA stability and processing; and translation and secretory events (Barnes and Dickson 2006). Most reports have studied at the molecular level the phenomenon of production instability in GS-NS0 expression system through the comparison between producer and non-producer cell lines.
In this work we followed a different approach. In our experience, NS0 clones expressing recombinant antibodies are usually unstable along the time in culture. The progressive decrease in productivity may be due to some epigenetic regulation mechanism that arose during the selection process of the non-immunoglobulin secreting host NS0 cell line (Whitford 2003). However, we have demonstrated that stable producer clones can be obtained from long-term cultures of unstable ones (Castillo et al. 2005). Thus, the phenomenon of the emergency of stable producer clones might be due to a reversal of the putative epigenetic negative regulation mechanism. To identify molecular processes potentially responsible for the stability of recombinant antibody production, we selected as models two different producing clones: a parental unstable clone and a sub-clone from the former with a stable producer phenotype. We carried out a kinetic and proteomic characterization of such clones to try to identify proteins involved in the unstable to stable phenotypic transition.
In our case, the loss of antibody production capacity by the unstable producer clone is due to the emergence of a non-producer cell sub-population, but not to the gradual reduction in the production capacity of every single cell. The increase of non-producer cell sub-populations, resulting in a decrease of qp value, has been previously reported for hybridoma cell lines (Barnes et al. 2003). Two possible explanations exist for such phenomenon: either a single cell that lost antibody production overgrows in the culture due to a higher proliferation rate, or a constant probability to loose antibody production exists for every cell leading to an increase in the non-producer cell sub-population. No differences in μ and Xvmax values were found for the unstable cell line at the beginning of the culture and after 40 days, although different proportions of producer and non-producer cells exist at those times. This suggests that there is not a major difference between the proliferation rates of producer and non-producer cells, although a more conclusive experiment would require the isolation of a non-producer clone to compare its kinetic parameters with those of the producer one. However, previous results with other non-producer recombinant NS0 clones have shown no major changes in proliferation rates (our unpublished results). At present, we support the hypothesis of a gradual loss of antibody production in the cell population due to a constant probability of phenotypic transition from producer to non-producer for every cell.
We determined the expression at protein level of the recombinant immunoglobulin chains and found that the LC polypeptide was more abundant than the HC polypeptide (HC:LC ratio <1). This finding is consistent with previous reports suggesting that HC levels could limit productivity while LC excess is required for efficient antibody folding and assembly (Smales et al. 2004; Schlatter et al. 2005; Dorai et al. 2006). During folding and assembly of IgG in the endoplasmic reticulum of mammalian cells, immunoglobulin chain polypeptides sequentially interact with a range of molecular chaperones, foldases, and oxidoreductases present in macromolecular complexes in the endoplasmic reticulum such as BiP (immunoglobulin binding protein) and PDI (protein disulfide isomerase) (Dinnis and James 2005).
The abundance of heavy and light chains was higher in the stable cell line as compared to the unstable one, similarly to the results obtained by Barnes et al. (2004) at mRNA expression levels. Failures in the secretion mechanism do not seem to account for production instability, because the lower abundance of HC and LC polypeptides in the cell culture supernatant were proportional to their relative abundance inside the cells. In addition, HC/LC polypeptide intracellular content ratio was lower in the unstable than in the stable clone, suggesting a predominant negative regulation on the HC polypeptide expression.
The use of 2-DE and mass spectrometry for proteomics for this kind of studies has been widely reported by other authors (Chevalier et al. 2000; Alete et al. 2005; de la Luz et al. 2008). In our case, we used a Master Gel previously reported by our group to identify differential spots, and this work contributed to the identification of six new proteins. Despite the number of identified proteins in this study is not enough to establish the exact molecular mechanisms to explain the unstable to stable phenotypic transition, they indicate a way to describe the processes involved in this phenomenon.
The fifteen proteins whose differential expression was maintained along the culture time could be related to five cellular processes: protein synthesis and folding, DNA transcription and structure, mRNA processing, vesicular transport and cytoskeleton structure and carbohydrate metabolism and energy production. Only two of such proteins showed a decreased expression in the stable clone.
Cell division cycle-associated 7-like protein plays a role in transcriptional regulation as a repressor that inhibits monoamine oxidase A activity and gene expression by binding to the promoter. Also, it has an important oncogenic role and it is involved in apoptotic signaling pathways (Ou et al. 2006). However, at present, the relationship between this protein and recombinant cell lines had not been studied. The hnRPN A2/B1 protein has been described to act in a variety of systems as splicing repressors (Hutchison et al. 2002; Nasim et al. 2002), and in human cancer the overexpression of this ribonucleoprotein has been associated to genetic instability (Martínez and Mulshine 2002). In a recent proteomic study of eleven antibody-producing GS-NS0 cell lines, hnRPN A2/B1 protein was downregulated in higher producing clones (Seth et al. 2007). On the contrary, poly [ADP-ribose] polymerase protein, which was up-regulated in the stable producer clone, is involved in the base excision repair pathway (Piskunova et al. 2008), contributing therefore to genome stability.
Among the proteins involved in mRNA processing, heterogeneous nuclear ribonucleoprotein (hnRPN) L, A2/B1 and H are found. Two chaperones were up-regulated in our study: 60 and 75 kDa heat shock proteins, suggesting their involvement in immunoglobulin synthesis. Heat shock proteins represent a group of highly conserved protein species that are located in different cellular compartments and assist newly synthesized proteins in folding or translocation through membranes, stabilizing certain protein conformations and helping to eliminate denatured proteins (Agshe and Hartl 2000). Moreover, peptidyl-prolyl cis–trans isomerase A and T-complex protein 1 were also up-regulated in the stable producer clone. T-complex protein 1 is known to play a role in the folding of actin and tubulin proteins, thus it is noteworthy that tubulin was overexpressed too. Other proteins related to vesicular transport and cytoskeleton structure were also increased. It is well known that components of the cytoskeleton-like microtubules (formed by dimers of tubulin) are involved in many cellular processes including mitosis, cytokinesis, and vesicular transport (Nogales et al. 1998).
Transketolase, an enzyme of the pentose phosphate pathway, was found up-regulated in the stable producer cell line. In mammals, transketolase connects the pentose phosphate pathway to glycolysis (Voet and Voet 1995). It is one of the most important metabolic pathways providing a source of precursors and energy for the cell. It has been previously reported that lymphocyte activation and proliferation are largely dependent on aerobic glycolysis (MacIver et al. 2008). Aerobic glycolysis is a key determinant of anabolic metabolism, a hallmark of proliferating cells (Vander Heiden et al. 2009), and possibly of cells with a high protein secretion activity.
Recombinant antibody production stability involves several metabolic pathways. Our results suggest that epigenetic regulation mechanisms could explain how a stable producer clone can emerge from an unstable one. Specific silencing of key proteins in the cellular processes possibly associated with the stable phenotype would confirm their relative contribution. It is highly unlikely to achieve a stable producer cell phenotype by a single genetic manipulation. However, microRNA molecules can participate in the regulation of protein expression levels involving several metabolic pathways, from which by targeting a specific microRNA molecule, several processes could be affected at the same time. We are currently studying the microRNA expression profile of both the stable and unstable recombinant antibody-producer clones.
The fifteen proteins identified in this work with differential expression between the stable and unstable clones participate in general cellular processes potentially related to the expression and secretion of recombinant proteins by any mammalian cell. Therefore, although the study of additional recombinant NS0 cell lines, now ongoing, is needed to propose those proteins as general predictor factors of productive behavior, the present results are probably representative of what can be expected even for different recombinant proteins-producing cell lines, such as CHO (Chinese Hamster Ovary) cells. In any case, the proteomic approach we used proved to be useful to identify molecular markers that can help to explain the arising of a stable producer phenotype from an unstable clone.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary figure 1. Immunoblot analysis of hR3 antibody chains. HC and LC polypeptides from purified hR3 antibody were detected using an alkaline phosphatase-conjugated anti-human γ-chain antibody or a horseradish peroxidase-conjugated goat anti-human ĸ chain antibody, respectively. (TIFF 166 kb)
Acknowledgments
We thank Dr. Yassel Ramos Gómez from the Center of Genetic Engineering and Biotechnology (CIGB, Havana, Cuba) for his valuable help in the proteomics assays and Dr. Alejandro López-Requena for a careful reading of the manuscript.
References
- Agshe VR, Hartl F-U. Roles of molecular chaperones in cytoplasmic protein folding. Cell Dev Biol. 2000;11:15–25. doi: 10.1006/scdb.1999.0347. [DOI] [PubMed] [Google Scholar]
- Alete D, Racher AJ, Birch JR, Stansfield SH, James D, Smales CM. Proteomic analysis of enriched microsomal fractions from GS-NS0 murine myeloma cells with varying secreted recombinant monoclonal antibody productivities. Proteomics. 2005;5:4689–4704. doi: 10.1002/pmic.200500019. [DOI] [PubMed] [Google Scholar]
- Barnes L, Dickson A. Mammalian cell factories for efficient and stable protein expression. Curr Opin Biotechnol. 2006;17:381–386. doi: 10.1016/j.copbio.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Barnes L, Bentley C, Dickson A. Advances in animal cell recombinant protein production: GS-NS0 expression system. Cytotechnology. 2000;32:109–123. doi: 10.1023/A:1008170710003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes L, Bentley CM, Dickson A. Stability of protein production from recombinant mammalian cells. Biotechnol Bioeng. 2003;81:631–639. doi: 10.1002/bit.10517. [DOI] [PubMed] [Google Scholar]
- Barnes L, Bentley C, Dickson A. Molecular definition of predictive indicators of stable protein expression in recombinant NS0 myeloma cells. Biotechnol Bioeng. 2004;85:115–121. doi: 10.1002/bit.10893. [DOI] [PubMed] [Google Scholar]
- Barnes L, Bentley CM, Moy N, Dickson AJ. Molecular analysis of successful cell line selection in transfected GS-NS0 myeloma cells. Biotechnol Bioeng. 2007;96:337–348. doi: 10.1002/bit.21119. [DOI] [PubMed] [Google Scholar]
- Castillo A, Victores S, Faife E, Rabasa Y, de la Luz KR (2005) Development of an integrated strategy for recombinant cell line selection. In: Gódia F, Fussenegger M (ed) Animal cell technology meets genomics. Springer, Berlin, pp 505–507
- Chevalier S, MacDonald N, Tonge R, Rayner S, Rowlinson R, Shaw J, Young J, Davison M, Roberts R. Proteomic analysis of differential protein expression in primary hepatocytes induced by EGF, tumour necrosis factor alpha or the peroxisome proliferators nafenopin. Eur J Biochem. 2000;267:4624–4634. doi: 10.1046/j.1432-1327.2000.01487.x. [DOI] [PubMed] [Google Scholar]
- de la Luz KR, Rojas L, Victores S, Lage A, Eyers C, Hart S, Castellanos L, Castillo A, Gaskell S (2008) Proteomic analysis of the adaptation of the host NS0 myeloma cell line to a protein-free medium. Biotecnología Aplicada 24:215–223
- Dinnis D, James D. Engineering mammalian cell factories for improved recombinant monoclonal antibody production: lessons from nature? Biotechnol Bioeng. 2005;91:180–189. doi: 10.1002/bit.20499. [DOI] [PubMed] [Google Scholar]
- Dorai H, Csirke B, Scallon B, Ganguly S. Correlation of heavy and light chain mRNA copy numbers to antibody productivity in mouse myeloma production cell lines. Hybridoma. 2006;25:1–9. doi: 10.1089/hyb.2006.25.1. [DOI] [PubMed] [Google Scholar]
- Faife E, Rodríguez T, Rabaza Y, Badía T, Victores S, Castillo A. Effects of selection and adaptation of NS0 cells to protein-free medium on the properties, affinity and biological activity of a monoclonal antibody. Biotecnología Aplicada. 2008;25:247–253. [Google Scholar]
- Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gels: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Hutchison S, LeBel C, Blanchette M, Chabot B. Distinct sets of adjacent heterogeneous nuclear ribonucleoprotein (hnRNP) A1/A2 binding sites control 5′ splice site selection in the hnRNP A1 mRNA precursor. J Biol Chem. 2002;277:29745–29752. doi: 10.1074/jbc.M203633200. [DOI] [PubMed] [Google Scholar]
- Köhler G, Milstein C. Derivation of specific antibody-producing tissue culture and tumour lines by cell fusion. Eur J Inmunol. 1976;6:511–519. doi: 10.1002/eji.1830060713. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- MacIver NJ, Jacobs SR, Wieman HL, Wofford JA, Coloff JL, Rathmell JC. Glucose metabolism in lymphocytes is a regulated process with significant effects on immune cell function and survival. J Leukoc Biol. 2008;84:949–957. doi: 10.1189/jlb.0108024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez A, Mulshine JL. Heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP A2/B1) as a marker of preinvasive lung cancer. J Clin Ligand Assay. 2002;25:100–103. [Google Scholar]
- Mateo C, Moreno E, Amour K, Lombardero J, Harris W, Pérez R. Humanization of mouse monoclonal antibody that block the EGF-R: recovery of antagonistic activity. Immunotechnology. 1997;3:71–81. doi: 10.1016/S1380-2933(97)00065-1. [DOI] [PubMed] [Google Scholar]
- Nasim FU, Hutchison S, Cordeau M, Chabot B. High-affinity hnRNP A1-binding sites and duplex–forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression model. RNA. 2002;8:1078–1089. doi: 10.1017/S1355838202024056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Ou XM, Chen K, Shih JC. Monoamine oxidase A and repressor R1 are involved in apoptotic signaling pathway. Proc Natl Acad Sci USA. 2006;103:10923–10928. doi: 10.1073/pnas.0601515103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskunova TS, Yurova MN, Ovsyannikov AI, Semenchenko AV, Zabezhinski MA, Popovich IG, Wang ZQ, Anisimov VN (2008) Deficiency in poly (ADP-ribose) polymerase-1 (PARP-1) accelerates aging and spontaneous carcinogenesis in mice. Curr Gerontol Geriatr Res 754190. Epub Apr 14. PMID: 19415146 [DOI] [PMC free article] [PubMed]
- Rojas LE, de la Luz KR, Victores S, Castellanos L, Gaskell S, Castillo A, Pérez R (2005) Comparative proteomic study of mechanisms involved in the expression of recombinant monoclonal antibody in non amplified NS0 myeloma cell line. In: Smith R (ed) Cell technology for cell products. Springer Publisher, Berlin, pp 715–721
- Schlatter S, Stansfield S, Dinnis DM, Racher A, Birch J, James D. On the optimal ratio of heavy to light chain genes for efficient recombinant antibody production by CHO cells. Biotechnol Prog. 2005;21:122–133. doi: 10.1021/bp049780w. [DOI] [PubMed] [Google Scholar]
- Seth G, Philp RJ, Lau A, Jiun KY, Yap M, Hu WS. Molecular portrait of high productivity in recombinant NS0 cells. Biotechnol Bioeng. 2007;97(4):933–951. doi: 10.1002/bit.21234. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Smales CM, Dinnis DM, Stansfield SH, Alete D, Sage EA, Birch JR, Racher AJ, Marshall CT, James DC. Comparative proteomic analysis of GS-NS0 murine myeloma cell lines with varying recombinant monoclonal antibody production rate. Biotechnol Bioeng. 2004;88:474–488. doi: 10.1002/bit.20272. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voet D, Voet JG (1995) Translation. In: Rose N (ed) Biochemistry, second edition. Wiley, New York, pp 997–1003
- Wallrath LL. Unfolding the mysteries of heterochromatin. Curr Opin Genet Dev. 1998;8:147–153. doi: 10.1016/S0959-437X(98)80135-4. [DOI] [PubMed] [Google Scholar]
- Whitford W (2003) NS0 serum-free culture and applications. BioProcess Int 12:36–47
- Wilson C, Bellen HJ, Gehring WJ. Position effects on eukaryotic gene expression. Annu Rev Cell Biol. 1990;6:679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
- Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cell. Nat Biotechnol. 2004;22:1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Supplementary figure 1. Immunoblot analysis of hR3 antibody chains. HC and LC polypeptides from purified hR3 antibody were detected using an alkaline phosphatase-conjugated anti-human γ-chain antibody or a horseradish peroxidase-conjugated goat anti-human ĸ chain antibody, respectively. (TIFF 166 kb)