Abstract
Although there are several inducible expression systems for mammalian cells, the most reliable one is the tetracycline-regulated expression system. This system is well-established and widely used by many researchers. Although Clontech provides several types of cells that stably express reverse tetracycline transactivator (rtTA), the cells that are not provided can be generated with pTet-On-Advanced by first integrating this plasmid into the require type of cell and then introducing the genes of interest. These processes are experimental bottlenecks. To improve this situation, we synthesized an all-in-one vector, termed pMAK17, which enables constitutive expression of puromycin N-acetyltransferase, modified Discosoma red fluorescent protein, and rtTA, as well as PTight-driven enhanced green fluorescent protein (EGFP). The pMAK17-transfected cells could be successfully induced to express EGFP, were selectable by fluorescence-activated cell sorting, and displayed puromycin resistance.
Keywords: DsRED2, Inducible, rtTA, Puromycin, Selection, Single plasmid
Introduction
Although there are several inducible expression systems for mammalian cells, the most reliable one is the tetracycline (Tc)-regulated expression system. This system is based on research conducted by Gossen and Bujard (Gossen and Bujard 1992). Clontech (now acquired by Takara Bio, Japan) has improved the system and released the plasmid pTet-On-Advanced, which enables the constitutive expression of reverse tetracycline transactivator (rtTA) in mammalian cells. A gene of interest driven by the inducible promoter PTight, contained in the plasmid pTRE-Tight (Takara Bio), is thus rarely expressed in the absence of tetracycline in rtTA-expressing cells. However, complex formation between rtTA and tetracycline can strongly activate transcription of the downstream gene of interest.
This system is well-established and widely used by many researchers. The company also provides several types of cells that stably express rtTA. However, in the case of certain cells that are not provided by the company, researchers should first integrate pTet-On-Advanced into such types of cells. After that, they can introduce the gene of interest into the cells. These processes are cumbersome and time-consuming.
In order to improve this situation, it would be desirable to have a single plasmid that constitutively expresses selectable marker(s) and rtTA, and allows the gene of interest to be cloned under the PTight promoter. Therefore, we synthesized an all-in-one vector, termed pMAK17, which enables the constitutive expression of puromycin N-acetyltransferase (PURO), modified Discosoma red fluorescent protein (DsRED2), and rtTA, as well as a PTight-driven enhanced green fluorescent protein (EGFP). Puromycin is an aminonucleoside antibiotic produced by Streptomyces alboniger, which causes premature chain termination during translation in various cell types (De La Luna and Ortin 1992; Vara et al. 1985). PURO inactivates cytotoxic puromycin by acetylating the amino position of its tyrosinyl moiety (Vara et al. 1985). Thus, PURO is a good drug selection marker. DsRED2 expression would facilitate identification of the transfected cells. The results are summarized herein.
Materials and methods
Assembling an all-in-one vector
The plasmid pTet-On-Advanced (Takara Bio), which contains an improved version of rtTA, the cytomegalovirus early enhancer/promoter (PCMV), and the SV40 poly A sequence (PA), was digested with the restriction enzymes XhoI (NEB Japan, Japan) and EcoRI (NEB Japan) to remove PCMV (Fig. 1a). This was used as a template to amplify several components by polymerase chain reaction (PCR) at later steps. All PCR reactions were carried out with Prime STAR polymerase (Takara Bio) according to the manufacturer’s instructions.
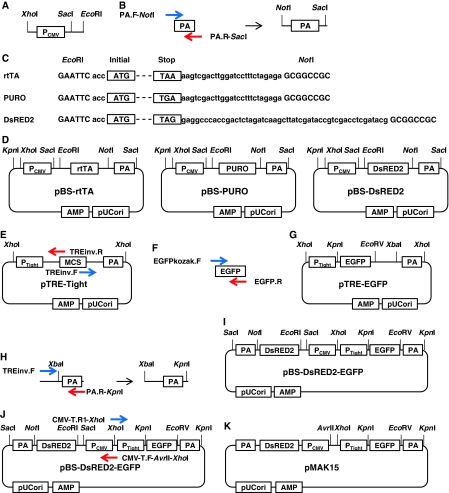
Fig. 1.
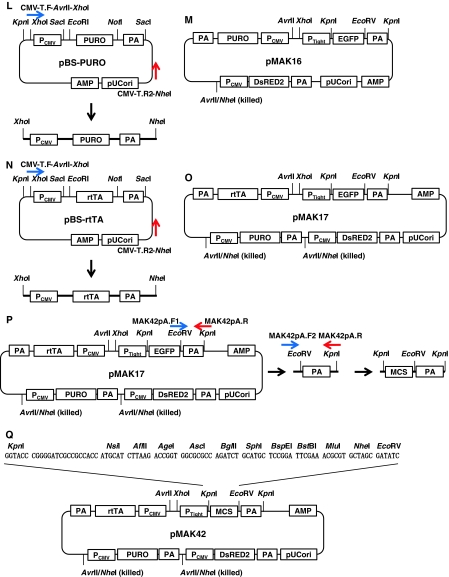
Scheme for pMAK17 construction. a Preparation of pCMV. b Preparation of PA. c, d Preparation of rtTA, PURO, and DsRED2, which were then sub-cloned into pBS. e An inverse PCR to generate pTRE-Tight. f Preparation of EGFP. g Preparation of pTRE-EGFP. h Preparation of PA to change a restriction enzyme site from XhoI to KpnI. i Sub-cloning EGFP with PTight and PA to pBS-DsRED2. j An inverse PCR for pMAK15. k Preparation of pMAK15. l Preparation of PURO with PCMV and PA for pMAK16. m Preparation of pMAK16. n Preparation of PURO with rtTA and PA for pMAK17. o Preparation of pMAK17. p Preparation of MCSs for pMAK42. q Preparation of pMAK42
The PA sequence was obtained by PCR with the following primers, using pTet-On-Advanced as a template: PA.F-NotI, 5′-gagaGCGGCCGCagcttgaacttgtttattgcagc-3′ (the NotI site is capitalized); PA.R-SacI, 5′-gctggGAGCTCacccgatccagacatgataagatac-3′ (the SacI site is capitalized). The PCR amplicon (168 bp) was digested with the restriction enzymes NotI (NEB, Japan) and SacI (NEB, Japan) (Fig. 1b).
The cDNAs encoding for rtTA, PURO, and DsRED2 were digested from the already subcloned plasmids pTet-On-Advanced, pCoPURO (Iwaki and Castellino 2008; Iwaki et al. 2003), and pDsRED2 (Takara Bio), respectively, with the restriction enzymes EcoRI and NotI (Fig. 1c). These fragments were ligated together into pBSIISK (Agilent Technologies, La Jolla, CA) and digested with the restriction enzymes XhoI and SacI; these were then used to generate pBS-rtTA, pBS-PURO, and pBS-DsRED2 (Fig. 1d).
An inverse PCR was performed with the following primers, using pTRE-tight (Takara Bio) as a template (Fig. 1e): TREinvF, 5′-ggccgcatcgataagcttgtcgacg-3′; TREinvR, 5′-gatccccgggtaccgagctcg-3′.
The cDNA coding for enhanced green fluorescent protein was amplified by PCR, using the following primers and pCAG-EGFP (Addgene) as a template (Fig. 1f): EGFPkozakF, 5′-GCCGCCACCatggtgagcaagggcgaggagc-3′ (the Kozak sequence is capitalized); EGFP.R, 5′-ttacttgtacagctcgtccatgc-3′. These PCR amplicons (2578 bp and 729 bp, respectively) were ligated together to generate pTRE-EGFP. The plasmid was digested with the restriction enzymes XhoI and XbaI (NEB, Japan) (Fig. 1g). The PA sequence in pTRE-Tight was amplified by PCR, using following primers (Fig. 1h): TREinvF, 5′-ggccgcatcgataagcttgtcgacg-3′; PA.R-KpnI, 5′-gcatGGTACCgaggcagtgaaaaaaatgctttatttgtg-3′ (the KpnI site is capitalized). The PCR amplicon (238 bp) was digested with the restriction enzymes XbaI and KpnI (Fig. 1h). The XhoI- and XbaI-digested fragment from pTRE-EGFP, and the XbaI- and KpnI-digested PA sequence, were subcloned into pBS-DsRED2 that had been previously digested with XhoI and KpnI (pBS-DsRED2-EGFP; Fig. 1i).
In order to introduce a recognition site for the restriction enzyme AvrII, an inverse PCR was performed using the following primers and pBS-DsRED2-EGFP as a template (Fig. 1j): CMV-T.F-XhoI-AvrII, 5′-taaaCTCGAGgagcCCTAGGttggcccattgcatacgttgtatccatatc-3′ (the XhoI and AvrII sites are capitalized in the said order); CMV-T.R1-XhoI, 5′-gctcCTCGAGtttactccctatcagtgatagagaac-3′ (the XhoI site is capitalized). The inverse PCR amplicon was digested with the restriction enzyme XhoI and then self-ligated, to generate pMAK15 (Fig. 1k).
PCR was carried out using either pBS-PURO or pBS-rtTA as a template and the following primers, and the amplicons were digested with the restriction enzymes XhoI and NheI (NEB Japan) (Fig. 1l and 1n, respectively):
CMV-T.F-XhoI-AvrII, as above; CMV-T.R2-NheI, 5′-gatcGCTAGCcatgttctttcctgcgttatcccctgattctgtg-3′ (the NheI site is capitalized).
The fragment for the PURO cassette was subcloned into pMAK15, which was digested with the restriction enzymes XhoI and AvrII (NEB, Japan), to generate pMAK16 (Fig. 1m). The fragment for the rtTA cassette was then subcloned into pMAK16 and digested with the restriction enzymes XhoI and AvrII, to finally generate pMAK17 (Fig. 1o). To replace EGFP with multiple cloning sites (MCSs) in pMAK17, a PCR was performed using pMAK17 as a template with the following primers (Fig. 1p): MAK42pA.F1, 5′-cgccagatctgcatgctccggattcgaaacgcgtgctagcGATATCtctagaggatcataatcagcca-3′ (the EcoRV site is capitalized); MAK42pA.R, 5′-gggcgaattgGGTACCgaggcagtgaaaaaaatgctttatttgtg-3′ (the KpnI site is capitalized). The PCR amplicon (260 bp) was then used as a template for a further PCR, using the following primers; the amplicon (315 bp) was then digested with the restriction enzyme KpnI (Fig. 1p): MAK42pA.F2, 5’-attcgagctcGGTACCcggggatcgccgccaccatgcatcttaagaccggtggcgcgccagatctgcatg-3’ (the KpnI site is capitalized); MAK42pA.R, as above. This KpnI-digested fragment was inserted into KpnI-digested pMAK17, to generate pMAK42 (Fig. 1q).
Cell transfection and fluorescence detection
Non-transfected HEK293 cells were generated by transforming human embryonic kidney cell cultures with sheared adenovirus 5 DNA (Riken Bio resource center, Japan), and OK-P cells were derived from the kidney of an adult female North American opossum (a kind gift from Dr. H. Yamamoto). Cell cultures were maintained in Dulbecco’s modified Eagle’s medium (D-MEM) (Sigma–Aldrich Japan, Japan) containing 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA), 1 × antibiotic/antimycotic solution (Sigma–Aldrich, Japan), and 2 mM l-glutamine solution (Sigma–Aldrich, Japan) (to give complete medium) at 37 °C in 5% CO2. HEK293 and OK-P cells (2 × 105) were seeded into each well of a 6-well plate containing 2 mL of complete medium. Stable transformants were produced using a Calcium Phosphate Transfection kit (Invitrogen) according to the manufacturer’s instructions. Briefly, 300 μL of solution containing 20 μg pMAK17 and 240 mM CaCl2 was slowly mixed with 300 μL 2 × HBS. The calcium phosphate-DNA precipitate was incubated with the cells for 16 h, after which the cells were washed twice with 2 mL of Hanks’ Balanced Salt Solution (HBSS) (Sigma–Aldrich Japan). Cells were detached from the wells with 0.5 mL of Accutase (Innovative Cell Technologies, San Diego, CA), and 0.1 mL of cell suspension was transferred to each well of a 4-well chamber slide (Nalge Nunc International, Naperville, IL), with 0.5 mL complete medium containing doxycycline (Dox) (Takara Bio) at a final concentration of 1 μg/mL. Following a 24 h induction, the cells were fixed in 4% paraformaldehyde in HBSS for 30 min, after which the slides were mounted with ProLong Gold Antifade Reagent with DAPI (Invitrogen).
Purification of positive cells by fluorescence-activated cell sorting
Transfected cells were gradually expanded, using the complete medium. Once the cell density had reached more than 80% confluency in a 175 cm2 tissue culture flask, the cells were detached with 5 mL of Accutase. The cells were gently pelleted for 2 min at 2000 rpm, and then resuspended in 3 mL of complete medium for fluorescence-activated cell sorting (FACS) analysis. FACS analysis was carried out using the FACSAria cytometer (BD, Becton Drive, NJ). The sorted cells were cultured in an appropriate tissue culture flask and expanded as described above. This step was repeated again.
Puromycin killing curve assay
In order to determine the suitable concentration of puromycin, non-transfected HEK293 and OK-P cells (2 × 105) were seeded in each well of a 6-well plate containing 2 mL of complete medium for 16 h. After the initial incubation, various concentrations of puromycin (ranging between 0.2 and 5 μg/mL) were applied to the cells. The cells were then incubated for 72 h.
Results and discussion
Construction of pMAK17 and transfection to HEK293 and OK-P cells
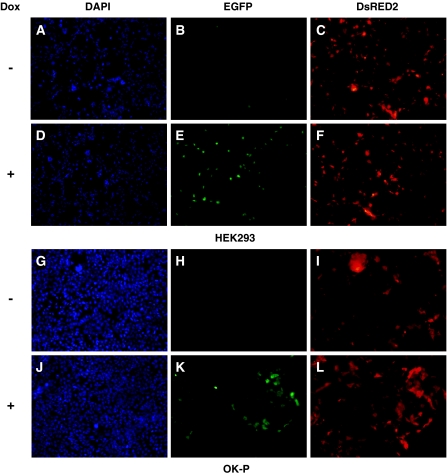
We were able to successfully assemble an all-in-one vector, pMAK17 (Fig. 1o). All of the vital parts, such as the coding sequences for DsRED2, PURO, rtTA, and EGFP, were verified by DNA sequencing (data not shown). After transfection, approximately 2% of HEK293 cells positively expressed DsRED2 (Fig. 2c); these cells rarely expressed EGFP without Dox treatment (Fig. 2b). OK-P cells were relatively resistant to the transfection, with only approximately 0.5% of the cells expressing DsRED2 (Fig. 2i); no EGFP expression was observed in the absence of Dox treatment (Fig. 2h). After Dox treatment, most of the DsRED2-positive HEK293 and OK-P cells positively expressed EGFP (Fig. 2e, k, respectively), without alteration of cell populations.
Fig. 2.
Fluorescent signals from pMAK17-transfected HEK293 (a–f) and OK-P (g–l) cells. (a, d, g, and j) DAPI stain; (b, e, h, and k) EGFP; and (c, f, i, and l) DsRED2. a, b, c, g, h, and i no Dox treatment; d, e, f, j, k, and l with Dox treatment. Original magnification, 100×
FACS analysis and cell purification
Both transfected HEK293 and OK-P cells were sorted using the FACSAria cytometer to select DsRED2-expressing cells. At the first stage, the rate of DsRED2-positive cells for both HEK293 and OK-P cells was about 1–2%. Therefore, the transfection efficiency by the calcium phosphate method for these cells was about 1–2%. After 2 cycles of FACS sorting, the rate of DsRED2-positive cells was around 60%, and most of them were positive for EGFP after Dox treatment (data not shown).
Puromycin resistance
Half of the non-transfected HEK293 cells were eliminated within 3 days when treated with a puromycin concentration of 1 μg/mL (data not shown). A puromycin concentration of 2 μg/mL was required to obtain the same result for OK-P cells (data not shown). However, sorted HEK293 and OK-P cells were highly resistant to a puromycin concentration of 1 μg/mL and 2 μg/mL, respectively. These cells remained in a proliferative state at even higher concentrations of puromycin (up to 5 μg/mL; data not shown).
Conclusion
We have successfully generated pMAK17, a plasmid that allows a gene of interest to be expressed within a Dox-inducible gene expression system, with the constitutive expression of Pac, DsRED2, and rtTA. Typically, it takes up to 2–3 months to establish stable rtTA-expressing cell lines followed by the integration of the gene of interest, using the material provided by the company. Using our system, this time could be reduced remarkably, even to less than a month. This single plasmid transfection system can be easily handled and gives reliable and reproductive results for recombinant protein synthesis. The vectors pMAK17 and pMAK42 are now available from the Riken Bioresource Center (http://www.brc.riken.jp/lab/dna/).
Acknowledgments
We would like to thank Dr. Hironori Yamamoto for providing OK-P cells, and Mr. Kiyoshi Shibata for his technical assistance with the FACS analysis. This work was supported in part by the Japan Society for the Promotion of Science (JSPS) KAKENHI 20890093 and 22790247, and the Uehara Memorial Foundation (to T.I.).
References
- La Luna S, Ortin J. pac gene as efficient dominant marker and reporter gene in mammalian cells. Methods Enzymol. 1992;216:376–385. doi: 10.1016/0076-6879(92)16035-I. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki T, Castellino FJ. A single plasmid transfection that offers a significant advantage associated with puromycin selection in Drosophila Schneider S2 cells expressing heterologous proteins. Cytotechnology. 2008;57:45–49. doi: 10.1007/s10616-008-9129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki T, Figuera M, Ploplis Va, Castellino FJ. Rapid selection of Drosophila S2 cells with the puromycin resistance gene. Biotechniques. 2003;35:482–486. doi: 10.2144/03353bm08. [DOI] [PubMed] [Google Scholar]
- Vara J, Perez-Gonzalez JA, Jimenez A. Biosynthesis of puromycin by Streptomyces alboniger: characterization of puromycin N-acetyltransferase. Biochemistry. 1985;24:8074–8081. doi: 10.1021/bi00348a036. [DOI] [PubMed] [Google Scholar]