1. INTRODUCTION
Immunoglobulin (Ig) heavy and light chain loci undergo V, (D) and J gene recombination in bone marrow, giving rise to the diverse pre-immune repertoire of V(D)J segments of B cell receptors (BCR). After encountering antigen, naïve B cells divide and differentiate in germinal centers of secondary lymphoid organs. Here, Ig V(D)J gene segments are the targets of point-mutations at a rate of 10−3 change per base per cell division. This rate is a million-fold higher than that of mutations occurring spontaneously in the genome at large; hence, the term somatic hypermutation (SHM). Mutated Ig V(D)J regions provide the structural substrate for positive selection by antigen of high affinity mutants, which are characteristic of a mature antibody response. The Ig locus of germinal center B cells also undergoes class switch DNA recombination (CSR), which replaces the constant μ (Cμ) region of the heavy chain with a downstream Cγ, Cα, Cε region, thereby endowing antibodies with new biological effector functions.
Activation-induced cytidine deaminase (AID) initiates SHM and CSR (Muramatsu et al., 2000; Revy et al., 2000) by directly deaminating cytidine in DNA (dC), thereby yielding premutagenic uracil DNA lesions (dU) (Petersen-Mahrt et al., 2002; Rada et al., 2004; Neuberger et al., 2005). SHM preferential targets the RGYW/WRCY (R = A or G, Y = C or T, W = A or T) mutational hotspot, which contains the preferred AID deamination motif WRC (Petersen-Mahrt and Neuberger, 2003; Pham et al., 2003; Yu et al., 2004). dU is a non-bulky DNA lesion that, if not repaired, can pair with dA without blocking the DNA replication fork, thereby giving rise to dG → dA and dC → dT transitions. Consistent with this prediction, mice with a double deficiency of Ung, a uracil-DNA glycosylase (UDG), and MutantS homolog 2 (Msh2), a mismatch sensor and initiator of the mismatch repair (MMR) cascade, display only dG → dA and dC → dT transitions, likely due to ablation of both the Ung-mediated base excision repair (BER) and the Msh2-mediated MMR pathways (Rada et al., 2004). Accordingly, in contrast to other DNA lesions, such as abasic sites or pyrimidine dimers, dU in the DNA template does not block transcriptional elongation by human RNA polymerase II, leading to incorporation of either G or A in nascent transcripts (Kuraoka et al., 2003). In B cells, this would lead to the introduction of same mutations in antibodies as those that would be generated from mutated DNA templates carrying dG → dA and dC → dT transitions. Thus, dU repair in Ig V(D)J DNA is not under the feedback pressure from faulty transcription; instead, it is dealt with by Ung and Msh2 in an actively mutagenic fashion during DNA synthesis, as effected by error-prone lesion-bypass or translesion DNA synthesis (TLS) polymerases. Here, we will discuss DNA repair factors and their assembly into a putative “mutasome”, which is centered on the proliferating cell nuclear antigen (PCNA), and their role in the SHM process.
2. ABASIC SITES GENERATED BY UNG STALL THE REPLICATION FORK
During T cell-dependent immune responses, B cells are activated by cognate antigen and CD40 ligand (CD154) expressed on T cell surface to blast with an average cell division time of eight hours. In mice immunized with carrier-conjugated hapten or sheep red blood cells (SRBC), germinal centers appear within four days (Jacob et al., 1993; Wang and Carter, 2005) and expression of AID, which is induced by CD154 and/or cytokines such as IL-4 and TGF-β, begins within hours and peaks at day five (Muramatsu et al., 1999). In addition, expression of TLS polymerases critical to SHM, such as DNA polymerase (pol) θ, pol η and pol ζ, is significantly upregulated in B cells by the same stimuli that induce AID expression and SHM (Poltoratsky et al., 2001; Zan et al., 2001; Zeng et al., 2001; Zan et al., 2005), suggesting that SHM is a tightly regulated active process (Figure 1). Mutations in V(D)J DNA are detected thereafter (Jacob et al., 1993), supporting the contention that after generation of initial dU lesions by AID, B cell division and accompanying DNA replication are necessary for mutations to be fixed in the genome. Ung, but not other UDGs, such as SMUG1, contributes most, if not all, of the dU deglycosylation activity in SHM, as suggested by the greatly diminished expression of SMUG1 in germinal center B cells (Kavli et al., 2005; Di Noia et al., 2006). Ung can deglycosylate dU generated by AID in three different contexts: i) as dU in single-stranded DNA (ssDNA); ii) as dU:dG in double-stranded DNA (dsDNA); or iii) as dU:dA in dsDNA after one round of DNA replication that places a dA opposite a dU which escaped repairing. The deglycosylation activity of Ung on dU in ssDNA or dU:dG is 6- and 4-fold higher than that on dU:dA, respectively (Kavli et al., 2002), suggesting that the first two events generate major mutagenic intermediates in SHM. As such, they will be the main focus of our discussion.
Figure 1.
Induction of AID and error-prone TLS polymerases by the same stimuli that are required for SHM induction. BCR crosslinking and T:B cell interaction through CD154:CD40, CD28:CD80 and/or CD28:CD86 coengagement are required for SHM (Zan et al., 1999; Zan et al., 2000). CD154:CD40 engagement induces AID expression, which is further augmented by T cell-secreted cytokines (Muramatsu et al., 1999). Both BCR crosslinking and CD154:CD40 engagement upregulate error-prone pol θ and the Rev3 catalytic subunit of pol ζ in both human and mice. Pol η is upregulated in germinal center B cells in mice, but not in humans (Poltoratsky et al., 2001; Zan et al., 2001; Zeng et al., 2001; Zan et al., 2005).
AID effectively deaminates dC in ssDNA (Pham et al., 2003), which transiently exists in the transcription bubble or immediately upstream of the replication fork. A coupling of deamination and deglycosylation of dU in such a context would explain the ready induction of mutations in BL2 Burkitt’s lymphoma B cells induced by antibodies to CD19, CD21 and BCR (Faili et al., 2002). Interestingly, in such B cells, about 50% of the mutations were dC/dG transitions, likely resulting from either “replicating over” of dU or the insertion of dA across the abasic sites generated by Ung (see below), two events that require at least one round of DNA replication.* DNA strands unwound by a helicase during replication may serve as the substrate for both AID and Ung that localizes at replication foci (Otterlei et al., 1999). The coupling of AID-mediated deamination with Ung-mediated deglycosylation of dU is also implied by the failure to remove dU from ssDNA in B cells from a patient with hyper-IgM syndrome expressing a mutated and non-functional Ung (Kavli et al., 2005). One possible mechanism for such a coupling to occur is through replication protein A (RPA), which has been shown to bind both phosphorylated AID (Chaudhuri et al., 2004) and Ung (Otterlei et al., 1999). In an in vitro experiment, half of AGCA in a single-stranded oligonucleotide was deaminated by AID and deglycosylated by Ung within 5 minutes (Yu et al., 2004), suggesting that Ung, if at a high local concentration, as effected by either a high protein level or a high kcat/Km ratio, would be able to process most of dU in ssDNA.
Recognition and deglycosylation of dU:dG by Ung has been hypothesized to occur mainly in the nucleoplasm (Krokan et al., 2002). Here, the resulting abasic sites would be primarily repaired in an error-free fashion through a short patch BER process, which is independent of DNA replication and underpinned by nicking of DNA by apurinic/apyrimidinic endonucleases (APE) and insertion of a single nucleotide by the high fidelity DNA pol β (Krokan et al., 2002). Such abasic sites could also be processed through the long-patch BER process, effected by structure-specific flap endonuclease 1 (Fen1) and gap-filling of 2-8 nucleotides by the high fidelity pol β or pol δ (Klungland and Lindahl, 1997). Alternatively, dU:dG deglycosylation by Ung localized in DNA replication foci would generate abasic sites (Otterlei et al., 1999), which, if not processed by APE, would effectively block nucleotide incorporation by the high fidelity replicative pol δ and pol ε and halt the DNA replication fork. Similarly, deglycosylation of dU in a ssDNA template during DNA synthesis, as discussed above, would also block the replication fork (Figure 2). The processive nature of AID, as shown by its deamination of a series of dC in the same ssDNA (Pham et al., 2003), suggests that AID and Ung can generate a high number of abasic sites in the template strands. These abasic sites need to be bypassed or repaired efficiently in dividing germinal center B cells. Interestingly, in the absence of APEs, Ung remains tightly bound to abasic sites with an affinity even higher than to the dU in dsDNA (Parikh et al., 1998), thereby possibly playing an additional role in recruiting DNA repair factors to bypass abasic sites.
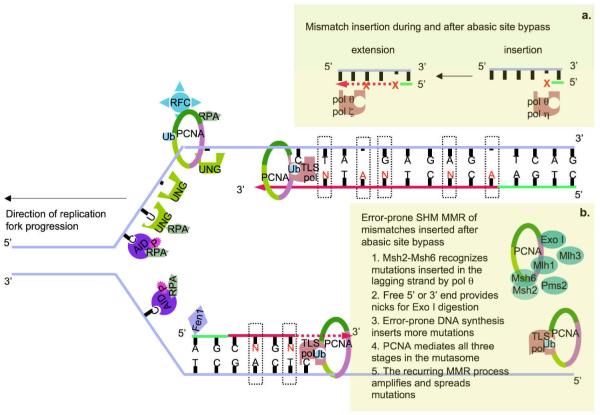
Figure 2.
An integrated model of SHM during DNA replication. The depicted model is based on the assumption that AID mediates SHM through deamination on both DNA strands. Coupling of dC deamination and dU deglycosylation generates abasic sites that stall replication fork progression in DNA replication foci. During DNA replication, template strands are transient unwound by the DNA helicase, yielding ssDNA stabilized by RPA, which would recruit phosphorylated AID to deaminate dC. dU:dG generated by AID during V gene transcription would be unwound and result in dU in ssDNA as well (not depicted here). Coupling of dC deamination and dU deglycosylation generates abasic sites, depicted as shorter bars without a base, that stall replication fork progression in DNA replication foci. Loading of the ubiquitinated (Ub) PCNA trimeric ring, possibly facilitated by Ung and dependent on RFC and RPA, initiates the assembly of a mutasome that introduces mutations during repair of abasic sites. a. Abasic site bypass is effected through polymerase switch in both the leading and lagging strands. Monoubiquitinated PCNA plays a central role in the switch from the high fidelity pol δ and pol ε to TLS polymerases. These TLS polymerases are able to incorporate nucleotides across abasic sites and continue synthesizing DNA. Two different TLS enzymatic activities are necessary to bypass abasic sites, first, insertion, as performed by pol θ, pol η, Rev1 and, perhaps, pol ι, and, then, extension, as performed by pol θ and pol ζ. Further, because of the unique dual function of pol θ, i.e., inserting mismatches and extending past inserted mismatches, pol θ would generate additional mutations, depicted as “N”, when copying undamaged template after abasic sites bypass. b. Error-prone MMR of mismatches generated during the extension stage after abasic site bypass. Mismatches are recognized by Msh2-Msh6, possibly aided by PCNA, and MutL dimers are recruited to initiate the DNA excision stage. Strand nicks necessary for the entry and activity of the exonuclease I (Exo I) are provided by 3′-end of the leading strand and both 5′-end and 3′-end of the Okazaki fragments in the lagging strand. Subsequent patch DNA re-synthesis by error-prone TLS polymerases will generate additional mutations which would initiate another round of MMR. Recurring SHM MMR would spreads the mutations along the V(D)J DNA. A similar SHM MMR process initiated by dU:dG in the template DNA strand would also, possibly aided by the Mre11-Rad50-Nbs1 (MRN) complex, amplify mutations. dU:dG that is not repaired will be replicated over to generate dC → dT and dG → dA transition mutations.
The current knowledge of DNA replication and repair has stemmed from studies in E. coli and yeast cells as well as in vitro systems reconstituted with defined DNA templates and purified protein factors. In both E. coli. and yeast cells, stalling of the replication fork by unrepaired abasic site can be overcome through either homologous recombination (HR) or TLS (Otterlei et al., 2000), both of which are functional in mammalian cells. Existing genetic and biochemical evidence strongly suggests that error-prone TLS polymerases bypass abasic sites and insert mismatches in SHM. This is reflected in the upregulation of TLS polymerases, which are needed to cope with the pace of generation of abasic sites by processive AID and Ung in germinal center B cells.
3. ABASIC SITE BYPASS BY TLS AND POLYMERASE SWITCH
At the replication fork stalled by lesions in the template strand, the high fidelity pol δ and pol ε are substituted by TLS polymerases that are able to incorporate nucleotides across and/or past damaged bases, including abasic sites (“polymerase switch”) (Friedberg et al., 2005). The high number of AID-generated abasic sites that need to be bypassed suggests that TLS polymerases bind to template strands during SHM for a much longer time than that during general DNA repair processes, before the reversion to pol δ and pol ε occurs. Two enzymatic activities are essential to carry out the abasic site bypass, one that incorporates nucleotides across from abasic sites (insertion), and another that elongates past the nascent primer end (extension) (Prakash et al., 2005) (Figure 2a). DNA pol η, Rev1, pol ι or even the high fidelity pol δ possess various degrees of the inserting ability, but cannot extend past the inserted nucleotide. Pol ζ cannot insert, rather it efficiently extends nucleotides incorporated across abasic sites. Pol θ is the only TLS polymerase that possesses both enzymatic activities (Seki et al., 2004) (Table 1). The significant upregulation of TLS polymerases by stimuli that also induce SHM suggests that SHM co-opts the otherwise high-fidelity abasic site bypass machinery to actively introduce mismatches. As pol θ could function as both an “inserter” and an “extender”, thereby bypassing abasic sites with a much higher efficiency than other TLS polymerases (Seki et al., 2004), it plays a major role in this process and overall SHM. This has been shown by the more than 70% reduction of the overall mutation frequency in a knockout mouse strain in which pol θ expression is ablated (Zan et al., 2005). In another knockout mouse strain, in which 99 of the 532 amino acids of the polymerase domain of pol θ were deleted and the polymerase activity was inactivated, the mutation frequency at dC/dG decreased by 40% and most of the decrease was focused on the AID deamination motif WRC, strongly suggesting that at least in SHM, bypass of abasic sites depends on pol θ (Masuda et al., 2005). An abasic site is non-instructional and all four nucleotides can be incorporated across it. Nevertheless, dA is preferentially incorporated across abasic sites by pol θ (“A rule”) (Seki et al., 2004), thereby yielding dC → dT and dG → dA transitions. This would partially explain why among dC/dG mutations transitions are more prevalent than transversions, being 50% in vivo and in hypermutating Ramos B cells (Komori et al., 2006) instead of 33% if all four nucleotides are equally incorporated (dG insertion does not generate a mutation). In contrast, Rev1 efficiently inserts dC regardless of the template sequence (Lawrence, 2002), possibly through a mechanism entailed by the high affinity interaction between incoming dCTP and an arginine residue in the active site of Rev1 (Nair et al., 2005). The virtual absence of dC → dG transversions in the non-transcribed strand of the Vλ1 segment DNA in Rev1 deficient mice is consistent with the role of Rev1 in inserting dC across abasic sites during SHM (Jansen et al., 2006).
Table 1. Properties of eukaryotic DNA polymerases and their role in SHM.
| DNA polymerase |
Class | Proposed main function1 |
Fidelity1 | Proce- ssivity2 |
Role in SHM3 |
|---|---|---|---|---|---|
| pol α | B | primase in DNA replication |
10−4-10−5 | moderate | no |
| pol β | X | BER | 10−4-10−5 | low | no |
| pol γ | A | mitochondrial DNA replication |
10−5-10−6 | high | ND4 |
| pol δ | B | DNA replication |
10−5-10−6 | high | no |
| pol ε | B | DNA replication |
10−5-10−6 | high | ND4 |
| pol ζ | B | translesion DNA synthesis |
10−4-10−5 | moderate | mispair extension |
| pol η | Y | translesion DNA synthesis |
10−2-10−3 | low | insertion of mutations at dA/dT |
| pol ι | Y | translesion DNA synthesis |
10−1-10−5 | low | no or marginal |
| pol κ | Y | translesion DNA synthesis |
10−3-10−4 | low | no |
| pol λ | X | NHEJ | 10−4-10−5 | low | no |
| pol μ | X | NHEJ | 10−4-10−5 | low | no |
| Rev1 | Y | translesion DNA synthesis |
ND1 | low | dC insertion across abasic sites |
| pol θ | A | translesion DNA synthesis |
10−2-10−3 | ND4 | insertion across abasic sites; mispair insertion and extension |
ND, not determined.
Pol ζ and pol θ are the only TLS polymerases known to efficiently extend the nascent DNA strand past an abasic site by copying undamaged portion of DNA template, consistent with their major role in SHM (Diaz et al., 2001; Zan et al., 2001; Masuda et al., 2005; Zan et al., 2005). Pol ζ is relatively error-free when copying undamaged DNA template. In contrast, pol θ can insert mismatches opposite an undamaged DNA template and extend past inserted mismatches, consistent with its error-prone function in the extension stage of the abasic site bypass (Figure 2a). Mismatches inserted by pol θ can also be extended by pol ζ. Such mismatches could further trigger an error-prone MMR entailing pol θ, pol ζ, pol η and, perhaps, pol ι, instead of the canonical high fidelity pol δ and pol ε, to spread the mutations (see below). The dominant role of pol θ in SHM likely reflects the involvement of this TLS polymerase in the following crucial stages of the process: mismatch insertion across from abasic sites, mismatch introduction during extension past abasic sites and gap-filling during error-prone MMR.
Evidence in both human and mice indicates that pol η plays a role in SHM. Pol η preferentially introduces mutations at dA/dT (Matsuda et al., 2000). Indeed, in Xeroderma pigmentosum variant (XP-V) patients, who are congenitally deficient in pol η, the mutation spectrum shows a strong bias towards dC/dG (Zeng et al., 2001), and in pol η-deficient mice, the overall frequency of mutations is normal but mutations at dA/dT are decreased by two thirds (Delbos et al., 2005; Martomo et al., 2005). Similar decreased mutations at dA/dT in mice deficient in the MMR proteins Msh2, Msh6 or Exo I suggest that pol η is involved in the error-prone MMR of SHM. As in abasic site bypass, the function of pol η would be restricted to inserting a mismatch that is then extended by either pol θ or pol ζ. Finally, pol ι seems to play only a marginal role in SHM, as mice with pol ι deficiency display a decreased mutation frequency only when crossed with pol η-deficient mice (Delbos et al., 2005).
4. THE MUTASOME: PCNA, MMR AND TLS
The modalities and mechanisms of the polymerase switch to bypass abasic sites in SHM remain to be determined. In E. coli, the β-sliding clamp simultaneously binds both the high fidelity pol III and the low fidelity pol IV in a tool-belt fashion. Pol IV is activated only when pol III stalls (Indiani et al., 2005). Interestingly, while the moving pol III suppresses pol IV, reflecting the copying of an intact DNA template, the active and moving pol IV, as occurring after a bypassed DNA lesion, is quickly taken over by pol III (Indiani et al., 2005). The mammalian β-sliding clamp, PCNA, likely plays an analogous role in polymerase switch. PCNA is the pivotal factor for almost all DNA transactions, including DNA replication and repair (Maga and Hübscher, 2003). PCNA has a unique six-fold head-to-tail trimeric ring structure with an inner side rich in α-helices that allows binding to and sliding along DNA (Figure 3) (Krishna et al., 1994; Gulbis et al., 1996; O’Donnell and Kuriyan, 2006). Each PCNA monomer has two functional domains connected by a coiled loop from Leu121 to Asp132. Such a coiled loop mediates the interaction with pol δ, Ung, Fen1 and DNA ligase I through the QxxI/LxxFF motif in those ligands. The C-terminus of PCNA is important for its binding to pol ε and PCNA loading protein replication factor C (RFC) through the similar QxxI/LxxFF motif (Warbrick, 2000; Maga and Hübscher, 2003).
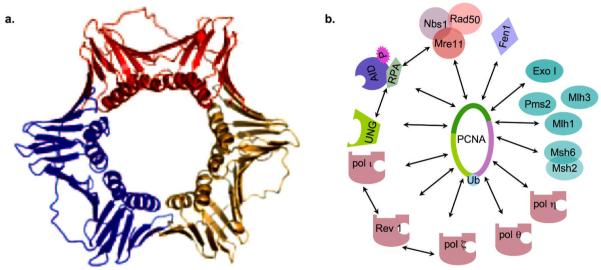
Figure 3.
Structure of PCNA and its interacting proteins in the mutasome. a. Ribbon representations of human PCNA.* b. The mutasome contains PCNA and different DNA replication/repair factors that interact with PCNA and are involved in SHM.
The polymerase switch process is possibly co-opted and subverted by SHM. The PCNA-binding motif QxxI/LxxFF of Ung would recruit PCNA to the replication fork stalled by the Ung-abasic site complex, which in turn recruits TLS pol η, pol ζ and pol θ. The role of Ung in recruiting PCNA in SHM polymerase switch could be analyzed by generating knockin mice that carry a mutated QxxI/LxxFF motif in Ung. The interaction of TLS polymerases with PCNA likely depends on the posttranslational modification of PCNA, as PCNA is ubiquitinated upon UV irradiation, and interaction between pol η and monoubiquitinated PCNA is critical to confer UV resistance in mammalian cells (Kannouche et al., 2004). Monoubiquitinated PCNA Lys164 constitutes another binding motif that is recognized by the ubiquitin-binding domain (UBD) in pol η, pol ι (Bienko et al., 2005) and possibly other TLS polymerases. Further, PCNA, together with RFC and RPA, strongly enhances the insertion activity of human pol η (Haracska et al., 2001a; Haracska et al., 2001b) and PCNA binds to pol ζ and strongly stimulates its TLS activity to copy UV-damaged DNA template in vitro (Garg et al., 2005), suggesting that PCNA functions not only as an “adaptor” but also as an “inducer” of TLS polymerases during SHM. PCNA might also interact with pol θ and augment its abasic site bypassing, mismatch insertion and extension functions. The role of monoubiquitinated PCNA in the SHM polymerase switch can be addressed in knockin mice with a mutation in Lys164 in PCNA. This mutated PCNA cannot be ubiquitinated (Kannouche et al., 2004) and thereby would be unable to activate TLS polymerases (Bienko et al., 2005). PCNA also undergoes polyubiquitination and sumoylation in yeast (Papouli et al., 2005; Pfander et al., 2005), suggesting the existence of more posttranslational modifications of the mammalian PCNA that can play a role in the SHM polymerase switch. Negative regulators of the polymerase switch would include the deubiquitinase USP1, whose degradation correlates with the accumulation monoubiquitinated PCNA in mammalian cells (Huang et al., 2006). The recent report that ubiquitination of PCNA neither decreases the affinity of this adaptor/inducer for the high fidelity pol δ and pol ε nor does it increase the affinity of PCNA for TLS polymerases, suggests the possible existence of an inhibitory intermediate factor (Haracska et al., 2006).
The homotrimeric nature and different interaction motif/domains of PCNA suggest that multiple proteins could bind to PCNA simultaneously. However, the plethora of PCNA-binding proteins would be tightly regulated for their interaction with PCNA and there may exist a dynamic change in the protein factors bound to PCNA. Indeed, PCNA could function as a “docking bay” for DNA replication/repair factors, such as RPA, Ung, TLS polymerases, Msh2-Msh6 heterodimer, Exo I and the MRN complex. Therefore, a term mutasome has been proposed to describe a protein complex localized in the DNA replication foci, in which PCNA plays a central role in coordinating the functions of different protein factors in the polymerase switch during abasic site bypass and subsequent error-prone MMR (see below), thereby effecting SHM (Casali et al., 2006).
The docking bay function of PCNA depends on its mobility between the nucleoplasm and replication/repair foci. Whereas during G1/G2 phase, most PCNA is diffused and mobile, PCNA in S phase is transiently enriched and immobilized to the replication foci, effectively raising its local concentration that is critical for DNA transaction (Essers et al., 2005). DNA damage would further increase the residence time of PCNA, allowing it to be engaged in more time-consuming DNA repair processes, such as abasic sites bypass or MMR in SHM. UV-induced DNA damage results in PCNA accumulation within the replication/repair foci, suggesting a DNA replication-dependent lesion repair mediated by PCNA (Essers et al., 2005). Such a PCNA-based DNA replication/repair process would likely occur in mutasome during SHM. Depending on different types of DNA damage, PCNA carrying the Lys164 mutation and deficient in ubiquitination, displays different kinetic distribution patterns (Solomon et al., 2004; Essers et al., 2005), suggesting that monoubiquitinated PCNA plays specific roles in the repair of different DNA lesions, including abasic sites and mismatches around the stalled replication fork in Ig V(D)J DNA. Based on analysis of the cell cycle-dependence of SHM-specific dsDNA breaks (DSB), it is suggested that SHM primarily occurs in the S/G2 phase when DNA replication is about to end or has ended (Papavasiliou and Schatz, 2000; Zan et al., 2003). While the exact role of DSB in SHM awaits definition, monoubiquitinated PCNA specifically recruited to the V(D)J DNA may be a good indicator for the crucial DNA lesions and cell cycle-dependency of SHM.
5. ERROR-PRONE MMR AMPLIFIES MUTATIONS IN THE MUTASOME
In the mutasome, the mismatches inserted by pol θ during the extension stage following abasic site bypass would be subjected to the surveillance of the Msh2-Msh6 heterodimer, the sensor for single-nucleotide mismatches in MMR. In general, MMR corrects misincorporated nucleotides in the newly synthesized strand during DNA replication (Kunkel and Erie, 2005). However, in the mutasome, an environment with a high local concentration of error-prone TLS polymerases and monoubiquitinated PCNA, the MMR process would be co-opted to become highly mutagenic. The late phylogenic emergence of SHM, appearing first in sharks (Flajnik and Du Pasquier, 2004), suggests that “SHM MMR” (Figure 2b) likely evolved from the error-free MMR process that appeared earlier in E. coli. and yeast cells.
PCNA in the mutasome would play a role in all three stages of SHM MMR; i) mismatch recognition; ii) mismatch excision; and iii) re-synthesis of DNA patch. In mammals, MMR is mediated by proteins homologous to prokaryotic MMR factors, such as Msh2, Msh3 and Msh6 and MutL homologues (Mlh) 1, Mlh3, postmeiotic segregation increased (Pms)1 and Pms2. Consistent with the role of Msh2-Msh6 as the sensor for single-nucleotide mismatches (Kunkel and Erie, 2005; Zhang et al., 2005), mice deficient in Msh2 and/or Msh6 exhibit an overall decreased frequency of mutations. In these mice, residual mutations show a significantly altered spectrum, with decreased mutations at dA/dT and concurrent increased mutations at dC/dG (Table 2) (Rada et al., 1998; Martin and Scharff, 2002; Li et al., 2004; Martomo et al., 2004; Rada et al., 2004). SHM MMR, as occurring in the mutasome, could also access AID-generated dU:dG mispairs that have escaped Ung-mediated deglycosylation in DNA template strands in proximity of a stalled replication fork. Msh2-Msh6 binds to dU:dG with an affinity comparable to that to dT:dG in vitro (Wilson et al., 2005). PCNA plays a role in the mismatch recognition stage by binding to Msh6 and increasing its affinity to the mismatches (Kunkel and Erie, 2005). PCNA also interacts with Mlh1, which heterodimerizes with one of its three partners, Pms2, Pms1 or Mlh3. Mlh1, Pms2 and Mlh3 are involved in SHM, albeit in different ways, as mice deficient in Mlh1, Pms2 or Mlh3 display different spectra of mutations (Li et al., 2006; Stavnezer and Schrader, 2006; Wu et al., 2006). The Mlh dimer, as recruited by the Msh2-Msh6, would coordinate the mismatch recognition stage and the subsequent strand excision stage, although a MutL-independent pathway possibly exists in mlh1−/− mice, as suggested by the moderate increase in dC/dG mutations as compared to msh2−/− and msh6−/− mice (Stavnezer and Schrader, 2006).
Table 2.
MMR proteins and the impact of their deficiencies on SHM1
| Frequency of mutations2,3 |
Spectrum of mutations |
||
|---|---|---|---|
| dC/dG:dA/dT3 | ts:tv at dC/dG3 | ||
| MutS homologs | |||
| Msh2 | ↓ | ↑ | ↑ |
| Msh3 | = | = | = |
| Msh6 | ↓ | ↑ | ↑ |
| MutL homologs | |||
| Mlh1 | ↓ 4 | ↑ = |
= |
| Mlh3 | = | ↑ | = |
| Pms2 | ↓4 = |
↑ = |
= |
| Exonuclease | |||
| Exo I | ↓ | ND5 | ND5 |
Data are from the references quoted in the Table 1 of Wu et al. (2006).
Mutations in the JH4 intronic DNA were analyzed unless noted otherwise.
↑ increase; ↓ decrease; = no change; ts, transition; tv, transversion.
Mutations in Ig V(D)J DNA were analyzed.
Not determined.
A hallmark of MMR is the exclusive targeting of the nascent DNA strand during the DNA excision stage. In E. coli, only the nascent and transiently unmethylated strand is nicked by the MutH endonuclease (Kunkel and Erie, 2005). The resulting nick provides the entry point for Exo I to excise the mismatch-containing DNA segment (Schofield and Hsieh, 2003; Kunkel and Erie, 2005). Consistent with the role of Exo I in mammalian MMR, this exonuclease contributes to SHM, as suggested by the altered mutation spectrum in Exo I-deficient mice (Bardwell et al., 2004). The 3′-end of the leading strand and both 3′-end and Fen1-processed 5′-end at each Okazaki fragment on the lagging strand can be used as entry points for Exo I in eukaryotes, in which an endonuclease that nicks nascent DNA strands has yet to be identified. However, in the template DNA strand, the intervention of an endonuclease is necessary to create an entry point for Exo I to excise dU, which escaped deglycosylation. Interestingly, Mre11-Rad50, which are part of the MRN complex, possesses an evolutionarily conserved lyase activity (Larson et al., 2005), which could nick a nearby abasic site generated by Ung in the template strand, thereby providing the entry point for Exo I. Consistent with its role in nicking abasic sites, Mre11, but not the ubiquitous APE, is enriched on V(D)J DNA of hypermutating B cells and the MRN complex promotes SHM (Larson et al., 2005; Yabuki et al., 2005), suggesting that at least some SHM MMR is initiated by this complex. Both Exo I and MRN bind to PCNA (Maser et al., 2001; Nielsen et al., 2004), indicating that PCNA is also involved in DNA strand excision.
DNA digestion by Exo I would yield ssDNA which would be “coated” and protected by RPA before re-synthesis of the DNA patch. The TLS polymerases would be recruited in the mutasome by the monoubiquitinated PCNA to effect DNA synthesis in an error-prone fashion. Indeed, the interaction between pol η and Msh2-Msh6 in vivo is possibly facilitated by PCNA (Wilson et al., 2005). Pol θ, pol η, Rev1 and, perhaps, pol ι could insert even more mismatches, which are extended by either pol θ or pol ζ, thereby spreading the mutations along the whole V(D)J DNA. The newly introduced mutations at dA/dT would be subject to a new round of Msh2-Msh6-initiated MMR, thereby giving rise to an amplification loop that would lead to further introduction of mutations. Such an amplification loop could also explain the ready emergence of mutations that are not transitions at dC/dG in BL2 cells induced by BCR/CD19/CD21 crosslinking (Faili et al., 2002). Finally, consistent with the mutation spectrum in Msh2-deficient mice, in which mutations are more focused on dC/dG in the mutation hotspot RGYW/WRCY and the putative AID deamination motif WRC (Rada et al., 1998), multiple rounds of error-prone synthesis by TLS polymerases would be instrumental in spreading mutations farther away from the original dU lesion site.
7. CONCLUSIONS
It is clear that the AID-generated dU lesion is the initiator of a cascade of DNA repair events which are mediated by either Ung or Msh2 and eventually introduce mutations in the Ig locus (Neuberger et al., 2005). However, the relationship between these events and DNA replication is not clearly defined. The involvement of DNA repair factors, such as PCNA, RPA and RFC, which are also crucial DNA replication factors, and Ung or Msh2 in the correction of nucleotides misincorporated during DNA synthesis strongly suggests a tight correlation between SHM and DNA replication. Based on existing genetic and biochemical data, we propose a mechanistic model explaining how mutations would be introduced with a high frequency in the Ig locus (Figure 2). Central to this model are the multiple functions of PCNA and the intervention of TLS polymerases, whose levels are greatly upregulated in germinal center B cells. In the mutasome, abasic site bypass and error-prone MMR are the main processes responsible for the introduction of mutations. Further, the highly regulated, both spatially and temporally, functions of Ung and Msh2 would also explain why SHM is mainly confined to the fast dividing germinal center B cell, a differentiation stage entailing a high DNA synthesis rate. Finally, the amplification of mutations by the error-prone SHM MMR would further upregulate the rate of mutations in Ig V(D)J DNA to the high levels that are the hallmark of SHM.
8. ACKNOWLEDGEMENTS
This work was supported by NIH grants AR 40908, AI 45011 and AI 60573.
Footnotes
PCR amplification of dU generated from dC deamination would also give rise to apparent dC→dT and dG → dA transitions. However, this seems unlikely in this study because under PCR conditions, incorporation of dA across dU by high fidelity DNA polymerases, such as Pfu® (Invitrogen Corp.) used in this study, is inefficient (our unpublished data).
Reprinted from Current Opinion in Structural Biology, Volume 16, Mike O’Donnell and John Kuriyan, Clamp loaders and replication initiation, Pages 35-41, Copyright 2006, with permission from Elsevier.
9. REFERENCE
- Bardwell PD, Woo CJ, Wei K, Li Z, Martin A, S.Z S, Parris T, Edelmann W, Scharff MD. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat. Immunol. 2004;5:224–229. doi: 10.1038/ni1031. [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I. Ubiquitin-binding domains in y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Casali P, Pal Z, Xu Z, Zan H. DNA repair in antibody somatic hypermutation. Trends Immunol. 2006;27 doi: 10.1016/j.it.2006.05.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- Delbos F, De Smet A, Faili A, Aoufouchi S, Weill JC, Reynaud CA. Contribution of DNA polymerase η to immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 2005;201:1191–1196. doi: 10.1084/jem.20050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Rada C, Neuberger MS. SMUG1 is able to excise uracil from immunoglobulin genes: insight into mutation versus repair. EMBO J. 2006;25:585–595. doi: 10.1038/sj.emboj.7600939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M, Casali P. Somatic immunoglobulin hypermutation. Curr Opin Immunol. 2002;14:235–240. doi: 10.1016/s0952-7915(02)00327-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M, Verkoczy LK, Flajnik MF, Klinman NR. Decreased frequency of somatic hypermutation and impaired affinity maturation but intact germinal center formation in mice expressing antisense RNA to DNA polymerase ζ. J. Immunol. 2001;167:327–335. doi: 10.4049/jimmunol.167.1.327. [DOI] [PubMed] [Google Scholar]
- Essers J, Theil AF, Baldeyron C, van Cappellen WA, Houtsmuller AB, Kanaar R, Vermeulen W. Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell. Biol. 2005;25:9350–9359. doi: 10.1128/MCB.25.21.9350-9359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faili A, Aoufouchi S, Gueranger Q, Zober C, Leon A, Bertocci B, Weill JC, Reynaud CA. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat. Immunol. 2002;3:815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]
- Flajnik MF, Du Pasquier L. Evolution of innate and adaptive immunity: can we draw a line. Trends Immunol. 2004;25:640–644. doi: 10.1016/j.it.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis. Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Garg P, Stith CM, Majka J, Burgers PM. Proliferating cell nuclear antigen promotes translesion synthesis by DNA polymerase ζ. J. Biol. Chem. 2005;280:23446–23450. doi: 10.1074/jbc.C500173200. [DOI] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S. Physical and functional interactions of human DNA polymerase η with PCNA. Mol. Cell. Biol. 2001a;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol. Cell. 2001b;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc. Natl. Acad. Sci. USA. 2006 doi: 10.1073/pnas.0510924103. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D’Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 2006;8:341–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- Indiani C, McInerney P, Georgescu R, Goodman MF, O’Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol. Cell. 2005;19:805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Jacob J, Przylepa J, Miller C, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J. Exp. Med. 1993;178:1293–1307. doi: 10.1084/jem.178.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J. Exp. Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- Kavli B, Andersen S, Otterlei M, Liabakk NB, Imai K, Fischer A, Durandy A, Krokan HE, Slupphaug G. B cells from hyper-IgM patients carrying UNG mutations lack ability to remove uracil from ssDNA and have elevated genomic uracil. J. Exp. Med. 2005;201:2011–2021. doi: 10.1084/jem.20050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, Slupphaug G. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 2002;277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori A, Xu Z, Wu X, Zan H, Casali P. Biased dA/dT somatic hypermutation as regulated by the heavy chain intronic iEmu enhancer and 3′Ealpha enhancers in human lymphoblastoid B cells. Mol. Immunol. 2006;43:1817–1826. doi: 10.1016/j.molimm.2005.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Krokan HE, Drablos F, Slupphaug G. Uracil in DNA--occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Kuraoka I, Endou M, Yamaguchi Y, Wada T, Handa H, Tanaka K. Effects of endogenous DNA base lesions on transcription elongation by mammalian RNA polymerase II. Implications for transcription-coupled DNA repair and transcriptional mutagenesis. J. Biol. Chem. 2003;278:7294–7299. doi: 10.1074/jbc.M208102200. [DOI] [PubMed] [Google Scholar]
- Larson ED, Cummings WJ, Bednarski DW, Maizels N. MRE11/RAD50 cleaves DNA in the AID/UNG-dependent pathway of immunoglobulin gene diversification. Mol. Cell. 2005;20:367–375. doi: 10.1016/j.molcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Lawrence CW. Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair. 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- Li Z, Peled JU, Zhao C, Svetlanov A, Ronai D, Cohen PE, Scharff MD. A role for Mlh3 in somatic hypermutation. DNA Repair (Amst) 2006 doi: 10.1016/j.dnarep.2006.02.003. in press. [DOI] [PubMed] [Google Scholar]
- Li Z, Scherer SJ, Ronai D, Iglesias-Ussel MD, Peled JU, Bardwell PD, Zhuang M, Lee K, Martin A, Edelman M, Scharff MD. Examination of Msh6- and Msh3-deficient mice in class switching reveals overlapping and distinct roles of MutS homologues in antibody diversification. J. Exp. Med. 2004;200:47–59. doi: 10.1084/jem.20040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G, Hübscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell. Sci. 2003;116:3150–3160. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- Martin A, Scharff MD. AID and mismatch repair in antibody diversification. Nat. Rev. Immunol. 2002;2:605–614. doi: 10.1038/nri858. [DOI] [PubMed] [Google Scholar]
- Martomo SA, Yang WW, Gearhart PJ. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J. Exp. Med. 2004;200:61–68. doi: 10.1084/jem.20040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martomo SA, Yang WW, Wersto RP, Ohkumo T, Kondo Y, Yokoi M, Masutani C, Hanaoka F, Gearhart PJ. Different mutation signatures in DNA polymerase η- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc. Natl. Acad. Sci. USA. 2005;102:8656–8661. doi: 10.1073/pnas.0501852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Mirzoeva OK, Wells J, Olivares H, Williams BR, Zinkel RA, Farnham PJ, Petrini JH. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 2001;21:6006–6016. doi: 10.1128/MCB.21.17.6006-6016.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, Kawamura K, Tagawa M, Tokuhisa T, Azuma T, J OW. DNA polymerase θ contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc. Natl. Acad. Sci. USA. 2005;102:13986–13891. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase η. Nature. 2000;404:1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- Neuberger MS, Di Noia JM, Beale RC, Williams GT, Yang Z, Rada C. Somatic hypermutation at A.T pairs: polymerase error versus dUTP incorporation. Nat. Rev. Immunol. 2005;5:171–178. doi: 10.1038/nri1553. [DOI] [PubMed] [Google Scholar]
- Nielsen FC, Jager AC, Lutzen A, Bundgaard JR, Rasmussen LJ. Characterization of human exonuclease 1 in complex with mismatch repair proteins, subcellular localization and association with PCNA. Oncogene. 2004;23:1457–1468. doi: 10.1038/sj.onc.1207265. [DOI] [PubMed] [Google Scholar]
- O’Donnell M, Kuriyan J. Clamp loaders and replication initiation. Curr. Opin. Struct. Biol. 2006;16:35–41. doi: 10.1016/j.sbi.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Otterlei M, Kavli B, Standal R, Skjelbred C, Bharati S, Krokan HE. Repair of chromosomal abasic sites in vivo involves at least three different repair pathways. EMBO J. 2000;19:5542–5551. doi: 10.1093/emboj/19.20.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakke O, Krokan HE. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Neuberger MS. In vitro deamination of cytosine to uracil in single-stranded DNA by apolipoprotein B editing complex catalytic subunit 1 (APOBEC1) J. Biol. Chem. 2003;278:19583–19586. doi: 10.1074/jbc.C300114200. [DOI] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Poltoratsky V, Woo CJ, Tippin B, Martin A, Goodman MF, Scharff MD. Expression of error-prone polymerases in BL2 cells activated for Ig somatic hypermutation. Proc. Natl. Acad. Sci. USA. 2001;98:7976–7981. doi: 10.1073/pnas.141222198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L. Eukaryotic Translesion Synthesis DNA Polymerases: Specificity of Structure and Function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Rada C, Ehrenstein MR, Neuberger MS, Milstein C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- Rattray AJ, Strathern JN. Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu. Rev. Genet. 2003;37:31–66. doi: 10.1146/annurev.genet.37.042203.132748. [DOI] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- Seki M, Gearhart PJ, Wood RD. DNA polymerases and somatic hypermutation of immunoglobulin genes. EMBO Rep. 2005;6:1143–1148. doi: 10.1038/sj.embor.7400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Cardoso MC, Knudsen ES. Dynamic targeting of the replication machinery to sites of DNA damage. J. Cell. Biol. 2004;166:455–463. doi: 10.1083/jcb.200312048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Schrader CE. Mismatch repair converts AID-instigated nicks to double-strand breaks for antibody class-switch recombination. Trends Genet. 2006;22:23–28. doi: 10.1016/j.tig.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Wang Y, Carter RH. CD19 regulates B cell maturation, proliferation, and positive selection in the FDC zone of murine splenic germinal centers. Immunity. 2005;22:749–761. doi: 10.1016/j.immuni.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Warbrick E. The puzzle of PCNA’s many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Wilson TM, Vaisman A, Martomo SA, Sullivan P, Lan L, Hanaoka F, Yasui A, Woodgate R, Gearhart PJ. MSH2-MSH6 stimulates DNA polymerase η, suggesting a role for A:T mutations in antibody genes. J. Exp. Med. 2005;201:637–645. doi: 10.1084/jem.20042066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Tsai CY, Patam MB, Zan H, Chen JP, Lipkin SM, Casali P. A Role for the MutL Mismatch Repair Mlh3 Protein in Immunoglobulin Class Switch DNA Recombination and Somatic Hypermutation. J. Immunol. 2006;176:5426–5437. doi: 10.4049/jimmunol.176.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Fulop Z, Zhong Y, Evinger AJ, Zan H, Casali P. DNA lesions and repair in immunoglobulin class switch recombination and somatic hypermutation. Ann. N.Y. Acad. Sci. 2005;1050:146–162. doi: 10.1196/annals.1313.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki M, Fujii MM, Maizels N. The MRE11-RAD50-NBS1 complex accelerates somatic hypermutation and gene conversion of immunoglobulin variable regions. Nat. Immunol. 2005;6:730–736. doi: 10.1038/ni1215. [DOI] [PubMed] [Google Scholar]
- Yu K, Huang FT, Lieber MR. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 2004;279:6496–6500. doi: 10.1074/jbc.M311616200. [DOI] [PubMed] [Google Scholar]
- Zan H, Cerutti A, Dramitinos P, Schaffer A, Li Z, Casali P. Induction of Ig somatic hypermutation and class switching in a human monoclonal IgM+ IgD+ B cell line in vitro: definition of the requirements and modalities of hypermutation. J. Immunol. 1999;162:3437–3447. [PMC free article] [PubMed] [Google Scholar]
- Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik MF, Diaz M, Casali P. The translesion DNA polymerase ζ plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 2001;14:643–653. doi: 10.1016/s1074-7613(01)00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Li Z, Yamaji K, Dramitinos P, Cerutti A, Casali P. B cell receptor engagement and T cell contact induce Bcl-6 somatic hypermutation in human B cells: identity with Ig hypermutation. J. Immunol. 2000;165:830–839. doi: 10.4049/jimmunol.165.2.830. [DOI] [PubMed] [Google Scholar]
- Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger AJI, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase θ plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Wu X, Komori A, Holloman WK, Casali P. AID-dependent generation of resected double-strand DNA breaks and recruitment of Rad52/Rad51 in somatic hypermutation. Immunity. 2003;18:727–738. doi: 10.1016/s1074-7613(03)00151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ. DNA polymerase η is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]