Abstract
Low-complexity regions (LCRs) within proteins sequences are often considered to evolve neutrally even though recent studies reported evidence for selection acting on some of them. Because of their widespread distribution among eukaryotes genomes and the potential deleterious effect of expansion/contraction of some of them in humans, low-complexity sequences are of major interest and numerous studies have attempted to describe their dynamic between genomes as well as the factors correlated to their variation and to assess their selective value. However, due to the scarcity of individual genomes within a species, most of the analyses so far have been performed at the species level with the implicit assumption that the variation both in composition and size within species is too small relative to the between-species divergence to affect the conclusions of the analysis. Here we used the available genomes of 14 Plasmodium falciparum isolates to assess the relationship between low-complexity sequence variation and factors such as nucleotide polymorphism across strains, sequence composition, and protein expression. We report that more than half of the 7,711 low-complexity sequences found within aligned coding sequences are variable in size among strains. Across strains, we observed an increasing density of polymorphic sites toward the LCR boundaries. This observation strongly suggests the joint effects of lowered selective constraints on low-complexity sequences and a mutagenic effect of these simple sequences.
Keywords: Plasmodium falciparum, low complexity sequences, size variation, polymorphism
Introduction
Low-complexity regions (LCR) are the most commonly shared polypeptide sequences among Eukaryotes. These sequences are characterized by a low information content due to the repetition of few amino acids. Almost 18% of all amino acids in human and more than 50% in the amoeba Dictyostellium discoideum are found within low-complexity sequences (Wootton and Federhen 1993; Haerty and Golding 2010b). These simple sequences are also known to diverge rapidly between species compared with other regions of the same proteins due to both amino acid substitutions and repeats expansion or contraction (Brown et al. 2002; Tompa 2003; Huntley and Clark 2007; Lin et al. 2007; Brown et al. 2010). More generally, several studies reported proteins with LCR to be more diverged between species than proteins deprived of such regions (Brown et al. 2002; Huntley and Golding 2002; Huntley and Clark 2007). As a consequence of their rapid evolution, the absence of stable three-dimensional structure for these peptides, and the lack of functional annotations, simple sequences are considered to evolve nearly neutrally or under relaxed selective constraints (Lovell 2003; Faux et al. 2007; Simon and Hancock 2009).
Even though low-complexity sequences are described to evolve rapidly both between and within species, there is increasing evidence that suggests a functional role for some of these simple sequences and that selection drives their evolution (Huntley and Golding 2006; Haerty and Golding 2010b). Single amino acid repeat size expansion or contraction is directly associated with some genetic disorders in humans (Usdin 2008). Furthermore, the variation in size of two homopolymers within the gene Runx-2 is directly involved in skull morphology variation between dog breeds (Fondon and Garner 2004, 2007), and low-complexity sequence size variation also affects circadian rhythm duration in multiple species and phenotypic variation in mammals, insects, plants, and fungi (Avivi et al. 2001; Galant and Carroll 2002; Lindqvist et al. 2007; Michael et al. 2007; Wang et al. 2009). LCR and single amino acid repeats have been linked to specific molecular functions and biological processes such as development, immunity, reproduction, and cellular localization (Faux et al. 2005; Huntley and Clark 2007; Salichs et al. 2009; Kozlowski et al. 2010).
Many studies that described some of the factors associated with low-complexity sequences variation between species also attempted to assess the nature of the selective forces acting on these simple sequences. However, few studies implemented methods to directly test for selection acting on low-complexity sequences as a consequence of both the repetitive nature of low-complexity sequences and the redundancy of the genetic code (Huntley and Golding 2006; Brown et al. 2010). Therefore, most of the analyses have relied on indirect evidence using the association of LCR with specific functions, their variation in composition and size within and between genomes, as well as the divergence of their flanking sequences between species (Hancock et al. 2001; Alba and Guigo 2004; Faux et al. 2007; Huntley and Clark 2007; Simon and Hancock 2009; Haerty and Golding 2010a; Kozlowski et al. 2010; Mularoni et al. 2010). Although all these analyses revealed a nonrandom distribution, composition, and size variation of low-complexity sequences with respect to protein functions and alternative splicing, they do not provide critical evidence about the low-complexity sequence dynamic at a small time scale as almost all the analyses performed to date involved the comparison of low-complexity sequences within orthologous genes in species diverged up to 75 Ma. In addition, because of the rarity of multiple fully sequenced genomes within a species, a single genome per species was used, thus assuming that the LCR variation within species is negligible in comparison to its variation between species. Although this is likely valid for nucleotide substitutions, this assumption may not be as robust for repeat size variation because of the high mutation rate of repeats due to replication slippage (Li et al. 2002). However, the recent and increasing availability of multiple genomes within the same species in conjunction with genome sequences from closely related species opens new opportunities to gain a better understanding of the evolutionary dynamics of low-complexity sequences and of the genes in which they are found. This can be achieved through the assessment of the factors (nucleotide polymorphism, sequence composition) that affect simple sequences evolution and the comparison across individuals of coding sequence variation classified according to the presence/absence of low-complexity sequences.
A handful of studies have so far analyzed low-complexity sequences using multiple genomes within the same species, two of them focused solely on microsatellites or trinucleotide repeats either within the genomes of four chicken breeds (Brandström and Ellegren 2008 or within two human genomes in addition to the reference genome (Molla et al., 2009). Both studies reported a positive relationship between repeat instability and repeat size as well as an enrichment of coding sequences in trinucleotide repeats in comparison to non-coding DNA. More recently, a study using the sequenced genomes of three strains of Plasmodium falciparum reached similar conclusions. The authors also reported a functional bias associated with tandem repeat number variation across strains and proposed a model to explain tandem repeat expansion/contraction based on the observation of high sequence similarity between the start of the repeat and the 3′ flanking sequence (Tan et al., 2010). However, the authors did not investigate the selective value of low-complexity sequences nor the factors that may relate to tandem repeats variation among P. falciparum strains such as nucleotide polymorphism or sequence composition. Furthermore, perfect or nearly perfect tandem repeats were the main focus of the study, thus excluding aperiodic low-complexity sequences that are known to differ in their pattern of sequence variation among strains in comparison to perfect repeats (Zilversmit et al., 2010).
We used the genomes of 14 strains of P. falciparum to assess the relative importance of factors such as nucleotide and amino acid sequence composition as well as nucleotide polymorphism between strains on low-complexity sequences variation across strains. The main interest of P. falciparum for such a study stems from the richness of its proteome in low-complexity sequences. Depending on the parameters used to define LCR, between 49% to more than 90% of the proteins in P. falciparum have at least one LCR (DePristo et al. 2006). Different functions for the LCRs have been proposed to explain their abundance within the P. falciparum proteome. An adaptive role for low-complexity sequences has been suggested (Hughes 2004) because some LCRs are directly involved in antigen diversification and in some cases the repeated motif is directly responsible for the antibody response (Kemp et al. 1987). LCRs are also proposed to promote genetic diversity by favoring recombination (Ferreira et al. 2003; DePristo et al. 2006; Zilversmit et al. 2010). Other potential functions involve expression regulation or increased messenger RNA (mRNA) stability (Xue and Forsdyke 2003; Frugier et al. 2010). Low-complexity sequences have also been suggested to act as interdomain linkers conferring elasticity to proteins. This last hypothesis is based on the conservation of the physical properties of some low-complexity sequences found between domains despite low amino acid sequence conservation (Xue and Forsdyke 2003; Daughdrill et al. 2007; Rask et al. 2010).
Using low-complexity sequences within these 14 genomes of P. falciparum isolates, we found significant relationships between protein expression, nucleotide polymorphism, sequence composition, and LCR variation and we assessed the relative importance of each of these factors on low-complexity sequences instability. More generally, we found an increased polymorphism within LCRs and in their vicinity that is characterized by an increased single-nucleotide polymorphism (SNP) density toward the LCR boundaries. These results strongly suggest that low-complexity sequences evolve under relaxed selective constraints in P. falciparum but also that a mutagenic effect is associated with these repeated sequences.
Materials and Methods
Data Collection
Plasmodium falciparum genome sequences were downloaded from the Broad Institute Web site (http://www.broadinstitute.org/science/data#) with the exception of the genome sequence of the strain 3D7, which was retrieved from Ensembl (http://www.ensembl.org, release 5.5).
We retrieved the best reciprocal blast hit (E value 10−10) between transcripts of each strain with genome annotation and the 3D7 strain and performed the nucleotide alignments with tranalign from the EMBOSS package (Rice et al. 2000) after aligning the corresponding proteins with MAFFT (Katoh et al. 2002).
For all the strains without genome assemblies, we used the exon annotations from the P. falciparum sequencing project (strain 3D7) to find the best reciprocal blast hits from the contigs of all the different strains with an E value cutoff of 10−3. Sequences were then aligned using MAFFT (Katoh et al. 2002).
Low-complexity sequences in the five strains with transcript annotations (3D7, HB3 and Dd2, IGH-CR14, RAJ116) were identified using SEG (Wootton and Federhen 1993). SEG allows the identification of all the protein windows with a local amino acid compositional complexity lower than a threshold value. Overlapping windows are merged and extended in both directions until the sequence reach the complexity threshold. Within each extended window, the fragment with the lowest probability of occurrence given its composition is reported. We used the parameters previously defined by Huntley and Golding (2002, window size 15, complexity trigger 1.9) that allow the identification of longer and more repetitive sequences, in comparison to the default parameters (window size 12, complexity trigger 2.2).
For each LCR, using the alignment coordinates corresponding to the longest sequence, the LCR and 100 nucleotides of flanking sequences on both sides of the LCR were collected from the alignments. If an exon boundary was found within the flanking sequences, we discarded the 15 nucleotides closest to the exon boundary to take into account the potential presence of exonic splice enhancers that are known to be under specific evolutionary constraints (Parmley et al. 2006; Warnecke and Hurst 2007). We also rejected flanking sequences that include another LCR. Because many of the genomes are not assembled yet, we used stringent criteria to retain a sequence for analysis. For each strain, we removed any sequence with evidence for a frameshift, in frame stop codon, increased sequence divergence (identity lower than 80%), or with less than 50% aligned nucleotides within the flanking sequence in comparison to the sequence from the 3D7 strain. The longest DNA of the LCR within an alignment was searched for repeated motifs using Tandem Repeat Finder with default values (Benson 1999).
Tan et al. (2010) reported trinucleotide repeats to be enriched within the 5′ end as well as within the midsegment of the proteins in P. falciparum. To examine this, we analyzed the distribution of LCRs within exons. For each LCR, we computed the distance to the closest intron. The observed distribution was compared with the expected distribution based on 1,000 randomizations of the positions of the LCRs within the coding sequences of the reference genome taking into account the LCR size distribution.
We computed the average number of pairwise differences per site (Tajima's π) for each LCR and their flanking sequences separately using the PopGen modules from BioPerl (Stajich and Hahn 2005). We collected a control set to assess the significance of any variation of nucleotide polymorphism within LCR and flanking sequences. The set is composed of sequences gathered within genes with detectable low-complexity sequences. We used exons deprived of simple sequences as well as sequences at least 250 bases distant from the closest LCR. The sequences were randomly selected within the genes, so that their size distribution mirrors the size distribution of the observed sequences.
Because low-complexity sequences are only defined by sequences with an information threshold lower than a set value, and hence includes both single amino acid repeats and repeats of several residues, we also assessed the relationship between low-complexity composition and its variability across strains. We used the Shannon–Weaver index as an estimator of sequence homogeneity.
 |
(1) |
where pi is the frequency of codon i and L the length of the LCR.
The LCR size variation across strains was computed according to Huntley and Clark (2007). We computed the absolute size difference between strains and used the total branch length of a tree built using the FITCH package from PHYLIP (Felsenstein 1989) as a measurement of LCR variation across strains. Because this measurement gives information only on the total variation of the LCR but not on size diversity between strains per se, we used the size of each LCR in each aligned strain to calculate the expected heterozygosity at each loci
 |
(2) |
where pi is the frequency of the ith of m alleles.
Transcript and protein expression data in P. falciparum 3D7 were retrieved from the study of Florens et al. (2002).
Protein Divergence between Plasmodium Species
To assess the effect of low-complexity sequences on protein divergence between species, we aligned the best reciprocal hits orthologs to all the P. falciparum proteins in P. vivax, P. knowlesi, P. chabaudi, P. berghei, and P. yoelii using MAFFT (Katoh et al. 2002). After removal of all the LCR detected using SEG and the previously mentioned parameters, we computed the protein divergence between species as the total branch length of a distance tree based on the JTT matrix using PROTDIST from the PHYLIP package (Felsenstein 1989).
Statistical Analyses
The relationships between LCR size variation and the different factors were tested through partial correlations using the corpcor package from R (written by Schaefer et al. 2010). We assessed the significance of each coefficient of partial correlation through randomization of the values of one parameter keeping the others constant using the boot package from R (Davison and Hinkley 1997).
The comparisons of polymorphism levels or sequence composition between gene categories were performed using a Kruskal–Wallis rank sum test using 10,000 permutations. The analysis of ontology enrichment was performed with AmiGO (http://amigo.geneontology.org/, Carbon et al. 2009). In order to control for a potential bias due to shared ancestry of low-complexity sequences between members of a gene family, we used a single randomly chosen gene from each gene cluster identified using Blastclust (written by Ilya Dondoshansky and Yuri Wolf) with an amino acid identity threshold of 50% and a minimal length of 70%.
In all, 9 of the 14 strains are not yet fully assembled and have a low sequence coverage, although we have focused on coding sequences in which sequencing and assembly errors are less likely to occur than within noncoding sequences; in order to test the robustness of our results, the analyses were performed again on a restricted data set that includes only the five annotated strains each of which have more than 85% of their bases with a quality score equal to or greater than 40 (http://www.broadinstitute.org/science/data).
Results
Among the five P. falciparum strains with genome annotation, the number of low-complexity sequences detected varies from 4,990 LCRs to 8,986 LCRs in 1,630 and 2,735 proteins, respectively. The large variation in LCR number between strains is most likely due to the difference in the number of annotated proteins (table 1).
Table 1.
Number of Low-Complexity Sequences within Coding Sequences of the Five Annotated Strains of P. falciparum and among Five Sequenced Plasmodium Species
| Speciesa | Strain | Nb | LCR | Proportion of Genes within LCR |
| P. falciparum | 3D7 | 5571 | 8986 (2735) | 0.491 |
| HB3 | 5367 | 8136 (2579) | 0.481 | |
| Dd2 | 4955 | 6733 (2167) | 0.437 | |
| igh-cr14 | 5041 | 8220 (2463) | 0.489 | |
| raj116 | 3180 | 4990 (1630) | 0.513 | |
| P. knowlesi | — | 5102 | 2392 (1413) | 0.277 |
| P. vivax | — | 5050 | 3854 (1817) | 0.360 |
| P. chabaudi | — | 5137 | 2900 (1350) | 0.263 |
| P. berghei | — | 7353 | 3829 (1384) | 0.188 |
| P.yoelii | — | 9821 | 1983 (2064) | 0.210 |
NOTE.—The number of protein-coding genes is given in parentheses.
Because P. reichenowi is currently only partially sequenced, no annotations are available.
Number of protein-coding genes.
The observed low-complexity sequences include both single amino acid repeats and repetitions of few amino acids. A total of 5,035 (56.03%) low-complexity sequences in the 3D7 strain have at least one single amino acid repeat of length 5 or more, of which 3,779 are made of the reiteration of a single codon.
After controlling for potential biases due to shared ancestry between LCRs through the random selection of a single gene per gene family, we found that genes with LCR are enriched in genes involved with host interaction and cell adhesion and with helicase activity while being underrepresented in genes found in the cytosol in comparison to genes without detectable LCRs according to the available gene ontology annotations (table 2). The functions of the genes with low-complexity sequences appears to differ between P. falciparum and previously studied eukaryotes in which genes involved in processes such as development or transcription were found to be enriched in simple sequences (Alba and Guigo 2004; Faux et al. 2005; Huntley and Clark 2007). However, it should be noted that only 2,199 genes are currently annotated in the GeneDB (http://www.genedb.org/).
Table 2.
Gene Ontology Annotation Enrichment among Genes with Low-Complexity Sequences in Comparison to the Remaining P. falciparum Genome
| Ontology number | Description | P value |
| Overrepresentation | ||
| Biological process | ||
| GO:0016337 | Cell–cell adhesion | 2.06 × 10−11 |
| GO:0020013 | Rosetting | 9.47 × 10−11 |
| GO:0051701 | Interaction with host | 3.08 × 10−09 |
| GO:0044406 | Adhesion to host | 1.01 × 10−08 |
| GO:0022610 | Biological adhesion | 1.14 × 10−08 |
| Molecular function | ||
| GO:0050839 | Cell adhesion molecule binding | 2.28 × 10−08 |
| GO:0004386 | Helicase activity | 1.42 × 10−03 |
| GO:0003724 | RNA helicase activity | 2.64 × 10−02 |
| GO:0008026 | ATP-dependent helicase activity | 2.75 × 10−02 |
| GO:0005488 | Binding | 3.86 × 10−02 |
| GO:0005515 | Protein binding | 4.93 × 10−02 |
| Cellular component | ||
| GO:0020030 | Infected host cell surface knob | 4.42 × 10−11 |
| GO:0030430 | Host cell cytoplasm | 3.84 × 10−08 |
| GO:0043656 | Intracellular region of host | 3.84 × 10−08 |
| GO:0033655 | Host cell cytoplasm part | 4.58 × 10−08 |
| Underrepresentation | ||
| GO:0005739 | Mitochondrion | 2.27 × 10−05 |
| GO:0030529 | Ribonucleoprotein complex | 6.52 × 10−05 |
| GO:0032991 | Macromolecular complex | 7.16 × 10−05 |
| GO:0005840 | Ribosome | 9.57 × 10−05 |
| GO:0044422 | Organelle part | 1.23 × 10−04 |
| GO:0005829 | Cytosol | 1.79 × 10−04 |
| GO:0044445 | Cytosolic part | 2.40 × 10−04 |
| GO:0044429 | Mitochondrial part | 8.39 × 10−04 |
| GO:0022626 | Cytosolic ribosome | 3.50 × 10−03 |
| GO:0043228 | Non–membrane-bounded organelle | 9.50 × 10−03 |
| GO:0031975 | Envelope | 2.05 × 10−02 |
| GO:0015935 | Small ribosomal subunit | 2.33 × 10−02 |
Low-complexity sequences are strongly enriched in asparagine and aspartic acid in comparison to the other amino acids (fig. 1A). We observed differences in the composition of low-complexity sequences between our results and those reported by DePristo et al. (2006). The differences originate from the use of different parameter values to identify LCR. DePristo et al. (2006) used the default values for SEG (Wootton and Federhen 1993), whereas we chose more stringent values for our analysis as indicated by the reduced number of LCR detected.
FIG. 1.
(A) Comparison of the average amino acid composition of low-complexity sequences and the remaining proteome in five strains of P. falciparum with genome annotations. (B) Average amino acid composition of variable and nonvariable low-complexity sequences. The error bars represent the standard error.
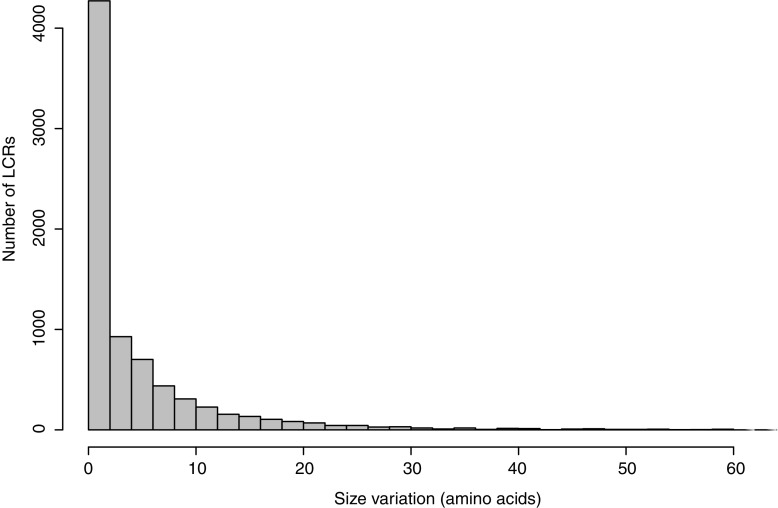
Using the genomes of 14 strains of P. falciparum, we generated alignments for a total of 7,711 low-complexity sequences with a median number of 7 aligned strains per LCR (minimum: 4 strains, maximum: 13 strains) that covers 859,774 nucleotides of the reference genome in 2,404 genes. The size of the collected LCRs varies from 7 amino acids to 320 amino acids (median size: 30 aa). Out of the 7,711 LCR, we found a total of 3,371 LCR sequences (43.67% LCR) in 1,705 genes that do not vary in size among P. falciparum strains. The 4,340 remaining sequences can vary up to a total of 120 amino acids between the strains (fig. 2).
FIG. 2.
Distribution of LCR size variation across P. falciparum strains indicating a highly skewed distribution with many invariant LCRs and a large tail of extremely variable LCRs.
Factors Associated with LCR Size Variation across P. falciparum Strains
LCR Size and Composition.
Several different factors have previously been proposed to explain low-complexity sequence instability. Therefore, we used partial correlations to assess the relationship between low-complexity variation within strains of P. falciparum and several variables associated with the composition of low-complexity sequences and their flanking regions (fig. 3) and the relative importance of each of those in predicting low-complexity sequence instability. We found a positive association between the length of the low-complexity sequence and its variability approximated by the total length of the tree based on a matrix of pairwise LCR size differences between strains (ρ = 0.21589, P < 0.001; fig. 3). In addition, the low-complexity sequences size variation negatively correlates with the Shannon–Weaver index (ρ = −0.2549, P < 0.001; fig. 3), which indicates that the more homogeneous the low-complexity sequences, the more variable in size they are. This observation holds when using codon composition to compute the Shannon–Weaver index instead of the amino acid composition (ρ = −0.3195, P < 0.001). The same conclusions are reached when using the five annotated strains only (Supplementary figure 1). Because the number of aligned strains may vary between LCRs, we reanalyzed our data by dividing for each LCR the estimated size variation by the number of aligned strains. We also reanalyzed the data using only the LCRs for which all the strains were aligned. For both analyses, our conclusions remained the same.
FIG. 3.
Relationship between low-complexity sequences variation across strains and different factors. Only significant partial correlations are shown. The thickness of the arrows is proportional to the coefficient of partial correlation. Red arrows indicate positive partial correlations whereas blue arrows represent negative relationships between factors.
We found LCRs to be significantly more distant from introns than expected under a random distribution of low-complexity sequences within the coding sequence (P < 0.001).
Because of the large number of low-complexity sequences that do not vary in size between P. falciparum strains (3,371), we compared their composition with the remaining variable LCRs. Nonvariable LCRs are characterized by a smaller size, a more heterogeneous amino acid composition, a higher G + C content (P < 2.2 × 10−16 in all comparisons). The DNA sequences of nonvariable LCRs are mostly aperiodic as evidenced by an underrepresentation of repeated motifs detected with Tandem Repeat Finder in comparison to variable LCR (35.14% vs. 80.97%, χ2 test, P < 0.001). Within nonvariable LCRs, lysine and leucine are overrepresented compared with variable LCR (P < 0.001 in both comparisons; fig. 1B). In contrast, asparagine and aspartic acid are enriched among variable LCRs. The paucity of lysine among variable LCRs is most likely the consequence of the action of selection. Because this amino acid is mostly encoded by the AAA codon in P. falciparum, expansion or contraction within this repeat due to replication slippage is likely to generate frameshifts and hence leads to nonfunctional proteins due to premature stop codons. The same should also apply to the phenylalanine that is predominantly encoded by the TTT codon. This conclusion is also supported by the observation of a smaller variability of polK homopolymers in comparison to polyN and polyD homopolymers (Kruskal--Wallis ranked sum test P < 2.2 × 10−16 in both comparisons). PolyN and polyD vary in a similar fashion (P = 0.225) (supplementary fig. 2, Supplementary Material online). In a previous study, Simon and Hancock (2009) found leucine repeats to be highly conserved between eukaryotic species, which may be because these repeats act as signal peptides.
LCR Size Variation and Protein Expression.
It is now well established that gene expression level imposes strong selective constraints on coding sequences (Drummond et al. 2005, 2006; Larracuente et al. 2008). In a previous study, we showed that variation in exposure to selection associated with alternative splicing significantly affects the size, composition, and variation across species of single amino acid repeats (Haerty and Golding 2010a). Therefore, we summed the size variation of all LCRs across a protein to assess the total size variation of a protein among strains due to LCR instability and assessed how it relates to protein expression level. We found a significant negative correlation between protein expression level and total size variation (ρ = −0.4247, P < 2.2 × 10−16). Because there is also a negative correlation between protein size and protein expression level, we used the proportion of variation with respect to the protein size. We observed once again the negative relationship between LCR instability and protein expression (ρ = −0.1941, P = 2.085 × 10−11). We found the same negative relationship between LCR size variation and protein expression when using the five annotated strains only (supplementary table 1, Supplementary Material online).
Low-Complexity Sequences and Nucleotide Polymorphism
Increased Nucleotide Polymorphism within LCR and Their Flanking Sequences.
Previous studies reported an increased divergence between species in the flanking sequences of LCRs (Faux et al. 2007; Huntley and Clark 2007; Simon and Hancock 2009) and more generally in the vicinity of insertions/deletions (Tian et al. 2008). We compared nucleotide polymorphism within LCRs and their flanking sequences to sequences randomly selected within the same genes and at least 250 bp from any LCR. We found that LCR have a significantly increased nucleotide polymorphism compared with the control sequences (Kruskal–Wallis ranked sum test, P < 2.2 × 10−16). Although the difference in polymorphism between flanking and control sequences is less dramatic than in LCR, it is still significant (P = 0.024). Once again we reached the same conclusions when using the five annotated strains only.
We found a positive relationship between nucleotide polymorphism within LCRs and within the flanking sequences (ρ=0.201, P< 0.001; figure 3). Although there is a strong correlation between the LCR size and nucleotide polymorphism occurring within it (ρ = 0.243, P < 0.001), the correlation between LCR size variation and polymorphism within the LCR is much weaker (ρ = 0.0964, P < 0.001; figure 3).
From the Plasmodium genome database, we collected 48,020 SNPs found within coding sequences and compared the distribution of their distance to the closest LCR to an expected distribution based on a random distribution of the SNPs in the genome. The expected distribution was built from 1,000 random resamplings. The analysis confirms the excess of SNPs within low-complexity sequences in comparison to a random distribution of the SNPs (P < 0.001). We also found an increasing density of SNPs within 150 bp of the LCR boundary (fig. 4). Out of the five annotated strains, only SNPs described between three strains (3D7, DD2, and HB3) are currently available in the Plasmodium database (Plasmodb, http://www.plasmodb.org; Aurrecoechea et al. 2009). Using this restricted data set, we reached the same conclusions as before (supplementary fig. 3, Supplementary Material online). This analysis also allowed us to confirm the observed correlation between LCR size variation and polymorphism within flanking sequences. The comparison of the SNP distribution relative to LCRs classified according to their variability across strains revealed an increasing SNP density close to variable LCRs (fig. 5).
FIG. 4.
Distribution of the distances between SNPs and the closest low-complexity sequences (gray). The black bars represent the expected median proportion of SNPs within a distance bin. The expected distribution (black bars, dashed lines) was build using 1000 randomizations of the SNPs positions in the coding sequences. The errors bars represent the 5th and 95th percentiles. The inset shows the SNP distribution 1 kb upstream and 1 kb downstream of low-complexity sequences.
FIG. 5.
Distribution of the observed distances between SNPs and the closest low-complexity sequences classified according to their variability between isolates. Distances to nonvariable LCRs is plotted in green and distances to LCRs with increasing size variation across strains are represented by the blue, red, and orange, respectively.
In addition, the protein size variation due to LCRs negatively correlates to the average number of pairwise differences per site at the full coding sequence level (ρ = −0.2039, P < 2.2 × 10−16). Although not as large, the negative relationship is still significant when using the normalized LCR variation (ρ = −0.0971, P = 2.51 × 10−6). We also observed this negative correlation protein size variation due to LCR and nucleotide polymorphism when we performed the analyses on the restricted data set (ρ = −0.1376, P = 5.79 × 10−8, and ρ = −0.04995, P = 0.04979, respectively, supplementary table 1, Supplementary Material online).
LCR, Antigenic Variation, and Host Interaction
The negative correlation between protein expression levels and LCR size variation across strains strongly suggests that the LCR instability is affected by the selective pressures associated with the protein expression level. Therefore, we tested how selection, more specifically positive selection, affects low-complexity sequence variation across strains.
Escape from the host immune system by generating antigen diversity is one of the suggested functions of low-complexity sequences in P. falciparum. This potential association is based on the observation of an antibody response to the repeated motif within the circumsporozoite protein (Kemp et al. 1987). Adaptive selection to escape the immune response as well as balancing selection are assumed to be the main forces shaping their evolution for many of the genes involved with host interaction (Nygaard et al. 2010; Weedall and Conway 2010). We used Plasmodb to compile a list of 510 genes annotated as being involved in antigenic response, drug resistance, or expressed on the surface of the parasite (i.e., merozoite surface proteins, erythrocyte membrane proteins).
Although the gene ontology analysis showed an enrichment of LCRs within genes involved with host interaction, such genes are underrepresented within our data set of aligned low-complexity sequences. This is likely a consequence of the stringent criteria we used to identify our sequences and parse the low-complexity sequences.
Both the flanking and LCR sequences within the coding sequences of these genes involved with host interaction are characterized by a higher nucleotide polymorphism in comparison to flanking and LCR sequences in the remaining coding sequences (Kruskal–Wallis ranked sum test, P = 0.049 and P < 2.2 × 10−16, respectively). We also observed a lower size variation, heterozygosity, and A + T content within LCR in genes potentially involved with the host interactions than in the control set (P = 0.0293, P = 0.0144, and P < 2.2 × 10−16 respectively). Although the significant difference in A + T content still holds when using the five annotated strains only, we observed no difference in LCR size nor LCR size variation between classes (P = 0.1445 and P = 0.6913, respectively). As a result of the difference in A + T content, the LCR composition strongly differ between genes involved with host interaction and the remaining genes. LCR within genes involved with host interactions are enriched in alanine, glutamine, glutamic acid, proline, and glycine, whereas asparagine is underrepresented in comparison to LCR within other genes (supplementary fig. 4, Supplementary Material online).
Discussion
Low-complexity sequences are found within all eukaryotic proteomes studied thus far, and although their relative abundance varies greatly between organisms, there is increasing evidence for a functional role for many of them (Alba and Guigo 2004; Faux et al. 2005; Huntley and Clark 2007; Salichs et al. 2009; Haerty and Golding 2010b).
This study is the first to our knowledge that attempts to assess the factors affecting LCRs evolution at the genome-wide scale using multiple genomes of the same species. Because of the repetitive nature of low-complexity sequences, sequence variation may be overestimated due to both assembly and sequencing errors. Such a bias is expected to be low in our study as we focused solely on coding sequences, and the genomes of P. falciparum have been sequenced through whole-genome shotgun sequencing leading to longer sequences and therefore an increased accuracy of the assembly process. Despite the fact that we used genomes at different stages of completeness, our conclusions are robust to the absence of annotation and assembly as well as potential sequencing errors. We used stringent criteria to retain a sequence for analysis and the analyses restricted to the five annotated strains, each of which have at least 85% of their bases with a quality score greater or equal to 40, to confirm the robustness of our conclusions. Our results show that even at the within-species level, low-complexity sequences can be extremely variable both in size and nucleotide composition. In comparison to the study of Tan et al. (2010), we found a greater number of genes with variable low-complexity sequences (489 vs. 1,784 genes), which is the consequence of using all sequences defined by a low information content instead of perfect or nearly perfect repeats and in addition to a greater number of P. falciparum genomes.
We found that more homogeneous low-complexity sequences are more variable in size within species. The observation of a similar relationship when using codon composition instead of amino acid composition as well as a higher variability of low-complexity sequences including microsatellites compared with those composed of minisatellites supports the importance of replication slippage in the observed LCR size variation among isolates. A similar relationship between homopolymer homogeneity and homopolymer instability has previously been reported using 96 and 52 polyglutamine tracts in human and mouse (Alba et al. 1999). Recombination has also been proposed to explain the size variation of microsatellites (Richard and Paques 2000; Ellegren 2004). A recent study analyzing low-complexity variation within 16 genes in 16 populations of P. falciparum suggested that unequal crossing-overs are likely involved in size variation of low-complexity sequences characterized by both G + C rich content and the presence of minisatellites resulting in the observation of large insertions/deletions (Zilversmit et al. 2010). Among all the LCR we collected, 35 are found among LCR with a G + C content below the 5th percentile and a size variation above the 95th percentile, which according to the criteria proposed by Zilversmit et al. (2010) are suggestive of the action of recombination.
There is a nonrandom distribution of nucleotide variation with respect to low-complexity sequences. Although previous studies based on the analysis of a small number of loci in P. falciparum reported contrasting results pertaining to the density of SNPs within simple sequences (Volkman et al. 2001, 2007), we found that low-complexity sequences as well as their flanking sequences are more variable than regions of the same gene that are not found in the vicinity of LCRs. We also report an increasing density of single-nucleotide polymorphisms toward the LCR boundaries.
The observed increased nucleotide polymorphism associated with the presence of low-complexity sequences could be explained by two nonexclusive processes that involve the relaxation of selective constraints on LCRs and an increased mutation rate associated to the presence of a repeated sequence.
We report several observations that support relaxation of selection on low-complexity sequences. The rapid divergence between Plasmodium species of proteins with LCRs relative to proteins without LCRs and the increased polymorphism within LCRs and their flanking sequences relative to others regions of the same genes are expected under such an evolutionary process. In addition, the greater distance of LCRs to exons is also expected to result from relaxation of selection as previous studies reported sequences near introns to be under strong selective constraints due to the presence of splicing regulatory elements and to the deleterious effects of the disruption of these elements (Pagani et al. 2005; Parmley et al. 2006; Warnecke and Hurst 2007). Previous analyses comparing the divergence of simple sequences flanking regions between human and mouse also associated the observed increased divergence to relaxation of selection (Hancock et al. 2001; Faux et al. 2007). In addition to the increased polymorphism in the vicinity of LCRs, we also found a negative correlation between protein expression levels and size variation. Highly expressed proteins are well known to be under stronger selective pressures than proteins expressed at lower levels and therefore to be significantly less variable across species (Drummond et al. 2005; Larracuente et al. 2008). Several studies showed that low-complexity sequences are preferentially found on the surface of proteins in direct contact with the solvent or within loops between protein domains (Romero et al. 2006; Hegyi et al. 2011). Rapid divergence has been reported for amino acids exposed to the solvent in comparison to buried residues (Lin et al. 2007). Therefore, the observed variation of nucleotide polymorphism between regions within the same protein may also reflect such a phenomenon. Although there is no information relative to alternative splicing in P. falciparum, we previously reported an enrichment of simple sequences within alternatively spliced exons compared with constitutively spliced exons, which is likely the consequence of reduced selective pressures on alternatively spliced exons due to lower inclusion levels in the mature mRNAs. Across several eukaryotic genomes, we observed more homogeneous amino acid composition and more unstable repeats within regions with the lowest selective constraints (Haerty and Golding 2010a). The same observation was made for intrinsically unstructured sequences (Romero et al. 2006; Ridout et al. 2010a).
Even though the observation of a higher nucleotide polymorphism in the vicinity of LCRs is suggestive of relaxation of selection on simple sequences and their neighborhood, it can also be the result of a mutagenic effect associated with repeated sequences. Furthermore, the positive correlation between LCR size variation and nucleotide polymorphism within LCR and the increasing SNP density toward the LCR boundaries support the hypothesis of a mutagenic effect of low-complexity sequences as suggested for the increased SNP density in the vicinity of indels in eukaryotes and the mutational pattern within the flanking sequences of microsatellites (Tian et al. 2008; Amos 2010b; Varela and Amos 2010). Within species, size heterozygosity at the LCR locus is expected to increase not only the LCR instability but also the mutation rate around the simple sequence (Tian et al. 2008; Amos 2010a).
We also report a negative correlation between overall protein size variation due to low-complexity sequences and nucleotide polymorphism. This observation seems to conflict with the observation of increased polymorphism in the vicinity of LCRs. But such phenomenon can be explained by a higher mutation and recombination rates within those regions that lead to decreased sequence repetitiveness and lower opportunity for replication slippage to occur (Kruglyak et al. 1998; Ellegren 2004). A negative association between nucleotide polymorphism and microsatellites instability has previously been reported by Brandström and Ellegren (2008) in chicken. These authors found a strong negative correlation between SNPs density and microsatellite size variation. The same forces are likely acting in the Plasmodium genomes.
Although an adaptive role in escaping host immunity has been proposed for low-complexity sequences, we do not observe an increased variability of these simple sequences within genes potentially involved with host interactions. In fact when analyzing the 14 strains, the opposite result is found. This observation is likely the consequence of the increased polymorphism observed within genes involved with host interaction reducing the opportunity for replication slippage to occur. Therefore, our observations concur with Zilversmit et al. (2010) conclusions stating that although it is likely that some low-complexity sequences have an important role in escaping the host immunity, this does not extend to most of the LCR in P. falciparum.
The observation of a high abundance of low-complexity sequences within proteins has raised numerous questions pertaining to the causes and consequences of their presence. Because of the lack of functional annotation, LCRs have been considered to act as spacers between functional domains (Huntley and Golding 2000). Although some of these LCRs are essential for the function of the protein domains they separate (Clarke et al. 2003) or can provide an increased flexibility to the protein (Faux et al. 2005), the size variation of others does not affect the expression nor the function of the gene hosting them (Muralidharan et al. 2011). Our observations of a higher mutation rate in the vicinity of simple sequences in association with the different factors shown to directly relate to the sequence instability raise several questions pertaining to the evolution of low-complexity sequences. Although the hypothesis is still controversial, there have been multiple reports suggesting that simple sequences can generate evolutionary novelty not only by having a direct effect on the expression of the genes (Fondon and Garner 2004; Kashi and King 2006; Vinces et al. 2009; Haerty and Golding 2010b) but also by directly affecting their surrounding sequences (Hancock et al. 2001; Faux et al. 2007; Huntley and Clark 2007; Varela and Amos 2010). Huntley and Clark (2007) even reported an increased number of sites with evidence for positive selection in the vicinity of single amino acid repeats in Drosophila. However, only the use of an increasing number of fully sequenced individual genomes in combination with genomes of closely related species will allow a more complete test of this hypothesis. Furthermore, in order to be able to identify and tease apart the causes and consequences of the abundance of low-complexity sequences within proteins, gene-specific studies will have to be performed such as those done by Clarke et al. (2003) or Muralidharan et al. (2011).
Supplementary Material
Supplementary figures 1–4 and tables 1 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
We are grateful to Dr Richard Morton for his comments on the early versions of the manuscript. This work was supported by a Natural Sciences and Engineering Research Council of Canada and Canada Research Chair grant to G.B.G.
References
- Alba MM, Guigo R. Comparative analysis of amino acid repeats in rodents and humans. Genome Res. 2004;14(4):549–554. doi: 10.1101/gr.1925704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba MM, Santibanez-Koref MF, Hancock JM. Conservation of polyglutamine tract size between mice and humans depends on codon interruption. Mol Biol Evol. 1999;16(11):1641–1644. doi: 10.1093/oxfordjournals.molbev.a026077. [DOI] [PubMed] [Google Scholar]
- Amos W. Heterozygosity and mutation rate: evidence for an interaction and its implications: the potential for meiotic gene conversions to influence both mutation rate and distribution. Bioessays. 2010a;32(1):82–90. doi: 10.1002/bies.200900108. [DOI] [PubMed] [Google Scholar]
- Amos W. Mutation biases and mutation rate variation around very short human microsatellites revealed by human-chimpanzee-orangutan genomic sequence alignments. J Mol Evol. 2010b;71(3):192–201. doi: 10.1007/s00239-010-9377-4. [DOI] [PubMed] [Google Scholar]
- Aurrecoechea C, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37(Database issue):D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avivi A, et al. Biological clock in total darkness: the Clock/MOP3 circadian system of the blind subterranean mole rat. Proc Natl Acad Sci U S A. 2001;98(24):13751–13756. doi: 10.1073/pnas.181484498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandström M, Ellegren H. Genome-wide analysis of microsatellite polymorphism in chicken circumventing the ascertainment bias. Genome Res. 2008;18(6):881–887. doi: 10.1101/gr.075242.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Johnson AK, Daughdrill GW. Comparing models of evolution for ordered and disordered proteins. Mol Biol Evol. 2010;27(3):609–621. doi: 10.1093/molbev/msp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Takayama S, Campen AM, Vise P, Marshall TW, Oldfield CJ, Williams CJ, Dunker AK. Evolutionary rate heterogeneity in proteins with long disordered regions. J Mol Evol. 2002;55(1):104–110. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
- Carbon S, et al. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25(2):288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JL, Sodeinde O, Mason PJ. A unique insertion in Plasmodium berghei glucose-6-phosphate dehydrogenase-6-phosphogluconolactonase: evolutionary and functional studies. Mol Biochem Parasitol. 2003;127(1):1–8. doi: 10.1016/s0166-6851(02)00298-0. [DOI] [PubMed] [Google Scholar]
- Daughdrill GW, Narayanaswami P, Gilmore SH, Belczyk A, Brown CJ. Dynamic behavior of an intrinsically unstructured linker domain is conserved in the face of negligible amino acid sequence conservation. J Mol Evol. 2007;65(3):277–88. doi: 10.1007/s00239-007-9011-2. [DOI] [PubMed] [Google Scholar]
- Davison A, Hinkley D, editors. 1997. Bootsrap methods and their applications. Cambridge (UK): Cambridge University Press. [Google Scholar]
- DePristo MA, Zilversmit MM, Hartl DL. On the abundance, amino acid composition, and evolutionary dynamics of low-complexity regions in proteins. Gene. 2006;378:19–30. doi: 10.1016/j.gene.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 2005;102(40):14338–14443. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Raval A, Wilke CO. A single determinant dominates the rate of yeast protein evolution. Mol Biol Evol. 2006;23(2):327–337. doi: 10.1093/molbev/msj038. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5(6):435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- Faux NG, et al. Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res. 2005;15(4):537–551. doi: 10.1101/gr.3096505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faux NG, et al. RCPdb: an evolutionary classification and codon usage database for repeat-containing proteins. Genome Res. 2007;17(7):1118–1127. doi: 10.1101/gr.6255407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Ferreira MU, Ribeiro WL, Tonon AP, Kawamoto F, Rich SM. Sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-1 (MSP-1) of Plasmodium falciparum. Gene. 2003;304:65–75. doi: 10.1016/s0378-1119(02)01180-0. [DOI] [PubMed] [Google Scholar]
- Florens L, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419(6906):520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- Fondon JW, III, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci U S A. 2004;101(52):18058–63. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondon JW, III, Garner HR. Detection of length-dependent effects of tandem repeat alleles by 3-D geometric decomposition of craniofacial variation. Dev Genes Evol. 2007;217(1):79–85. doi: 10.1007/s00427-006-0113-4. [DOI] [PubMed] [Google Scholar]
- Frugier M, Bour T, Ayach M, Santos MA, Rudinger-Thirion J, Theobald-Dietrich A, Pizzi E. Low complexity regions behave as tRNA sponges to help co-translational folding of plasmodial proteins. FEBS Lett. 2010;584(2):448–454. doi: 10.1016/j.febslet.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Galant R, Carroll SB. Evolution of a transcriptional repression domain in an insect Hox protein. Nature. 2002;415(6874):910–913. doi: 10.1038/nature717. [DOI] [PubMed] [Google Scholar]
- Haerty W, Golding GB. Genome-wide evidence for selection acting on single amino acid repeats. Genome Res. 2010a;20(6):755–760. doi: 10.1101/gr.101246.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty W, Golding GB. Low-complexity sequences and single amino acid repeats: not just ”junk” peptide sequences. Genome. 2010b;53(10):753–762. doi: 10.1139/g10-063. [DOI] [PubMed] [Google Scholar]
- Hancock JM, Worthey EA, Santibanez-Koref MF. A role for selection in regulating the evolutionary emergence of disease-causing and other coding CAG repeats in humans and mice. Mol Biol Evol. 2001;18(6):1014–23. doi: 10.1093/oxfordjournals.molbev.a003873. [DOI] [PubMed] [Google Scholar]
- Hegyi H, Kalmar L, Horvath T, Tompa P. Verification of alternative splicing variants based on domain integrity, truncation length and intrinsic protein disorder. Nucleic Acids Res. 2011;39(4):1208–1219. doi: 10.1093/nar/gkq843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. The evolution of amino acid repeat arrays in Plasmodium and other organisms. J Mol Evol. 2004;59(4):528–535. doi: 10.1007/s00239-004-2645-4. [DOI] [PubMed] [Google Scholar]
- Huntley M, Golding GB. Evolution of simple sequence in proteins. J Mol Evol. 2000;51(2):131–140. doi: 10.1007/s002390010073. [DOI] [PubMed] [Google Scholar]
- Huntley MA, Clark AG. Evolutionary analysis of amino acid repeats across the genomes of 12 Drosophila species. Mol Biol Evol. 2007;24(12):2598–2609. doi: 10.1093/molbev/msm129. [DOI] [PubMed] [Google Scholar]
- Huntley MA, Golding GB. Simple sequences are rare in the Protein Data Bank. Proteins. 2002;48(1):134–140. doi: 10.1002/prot.10150. [DOI] [PubMed] [Google Scholar]
- Huntley MA, Golding GB. Selection and slippage creating serine homopolymers. Mol Biol Evol. 2006;23(11):2017–2025. doi: 10.1093/molbev/msl073. [DOI] [PubMed] [Google Scholar]
- Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22(5):253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp DJ, Coppel RL, Anders RF. Repetitive proteins and genes of malaria. Annu Rev Microbiol. 1987;41:181–208. doi: 10.1146/annurev.mi.41.100187.001145. [DOI] [PubMed] [Google Scholar]
- Kozlowski P, de Mezer M, Krzyzosiak WJ. Trinucleotide repeats in human genome and exome. Nucleic Acids Res. 2010;38(12):4027–4039. doi: 10.1093/nar/gkq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak S, Durrett RT, Schug MD, Aquadro CF. Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proc Natl Acad Sci U S A. 1998;95(18):10774–10778. doi: 10.1073/pnas.95.18.10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente AM, et al. Evolution of protein-coding genes in Drosophila. Trends Genet. 2008;24(3):114–123. doi: 10.1016/j.tig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Li YC, Korol AB, Fahima T, Beiles A, Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol. 2002;11(12):2453–2465. doi: 10.1046/j.1365-294x.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- Lin YS, Hsu WL, Hwang JK, Li WH. Proportion of solvent-exposed amino acids in a protein and rate of protein evolution. Mol Biol Evol. 2007;24(4):1005–1011. doi: 10.1093/molbev/msm019. [DOI] [PubMed] [Google Scholar]
- Lindqvist C, Laakkonen L, Albert VA. Polyglutamine variation in a flowering time protein correlates with island age in a Hawaiian plant radiation. BMC Evol Biol. 2007;7:105. doi: 10.1186/1471-2148-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell SC. Are non-functional, unfolded proteins (“junk proteins”) common in the genome? FEBS Lett. 2003;554(3):237–239. doi: 10.1016/s0014-5793(03)01223-7. [DOI] [PubMed] [Google Scholar]
- Michael TP, et al. Simple sequence repeats provide a substrate for phenotypic variation in the Neurospora crassa circadian clock. PLoS One. 2007;2(8):e795. doi: 10.1371/journal.pone.0000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla M, Delcher A, Sunyaev S, Cantor C, Kasif S. Triplet repeat length bias and variation in the human transcriptome. Proc Natl Acad Sci U S A. 2009;106(40):17095–17100. doi: 10.1073/pnas.0907112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mularoni L, Ledda A, Toll-Riera M, Alba MM. Natural selection drives the accumulation of amino acid tandem repeats in human proteins. Genome Res. 2010;20(6):745–754. doi: 10.1101/gr.101261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan V, Oksman A, Iwamoto M, Wandless TJ, Goldberg DE. Asparagine repeat function in a Plasmodium falciparum protein assessed via a regulatable fluorescent affinity tag. Proc Natl Acad Sci U S A. 2011;108(11):4411–4416. doi: 10.1073/pnas.1018449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard S, et al. Long- and short-term selective forces on malaria parasite genomes. PLoS Genet. 2010 doi: 10.1371/journal.pgen.1001099. 6(9):e1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani F, Raponi M, Baralle FE. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc Natl Acad Sci U S A. 2005;102(18):6368–6372. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmley JL, Chamary JV, Hurst LD. Evidence for purifying selection against synonymous mutations in mammalian exonic splicing enhancers. Mol Biol Evol. 2006;23(2):301–309. doi: 10.1093/molbev/msj035. [DOI] [PubMed] [Google Scholar]
- Rask TS, Hansen DA, Theander TG, Gorm Pedersen A, Lavstsen T. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes—divide and conquer. PLoS Comput Biol. 2010 doi: 10.1371/journal.pcbi.1000933. 6(9): e1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Richard GF, Paques F. Mini- and microsatellite expansions: the recombination connection. EMBO Rep. 2000;1(2):122–126. doi: 10.1093/embo-reports/kvd031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridout KE, Dixon CJ, Filatov DA. Positive selection differs between protein secondary structure elements in Drosophila. Genome Biol Evol. 2010;2:166–179. doi: 10.1093/gbe/evq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero PR, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci U S A. 2006;103(22):8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salichs E, Ledda A, Mularoni L, Alba MM, de la Luna S. Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment. PLoS Genet. 2009;5(3):e1000397. doi: 10.1371/journal.pgen.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer J, Opgen-Rhein R, Strimmer K. 2010. corpcor: Efficient Estimation of Covariance and (Partial) Correlation. http://strimmerlab.org/software/corpcor/ [Google Scholar]
- Simon M, Hancock JM. Tandem and cryptic amino acid repeats accumulate in disordered regions of proteins. Genome Biol. 2009;10(6):R59. doi: 10.1186/gb-2009-10-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Hahn MW. Disentangling the effects of demography and selection in human history. Mol Biol Evol. 2005;22(1):63–73. doi: 10.1093/molbev/msh252. [DOI] [PubMed] [Google Scholar]
- Tan JC, Tan A, Checkley L, Honsa CM, Ferdig MT. Variable numbers of tandem repeats in Plasmodium falciparum genes. J Mol Evol. 2010;71(4):268–278. doi: 10.1007/s00239-010-9381-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, et al. Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature. 2008;455(7209):105–108. doi: 10.1038/nature07175. [DOI] [PubMed] [Google Scholar]
- Tompa P. Intrinsically unstructured proteins evolve by repeat expansion. Bioessays. 2003;25(9):847–855. doi: 10.1002/bies.10324. [DOI] [PubMed] [Google Scholar]
- Usdin K. The biological effects of simple tandem repeats: lessons from the repeat expansion diseases. Genome Res. 2008;18(7):1011–1019. doi: 10.1101/gr.070409.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela MA, Amos W. Heterogeneous distribution of SNPs in the human genome: microsatellites as predictors of nucleotide diversity and divergence. Genomics. 2010;95(3):151–159. doi: 10.1016/j.ygeno.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324(5931):1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman SK, et al. Recent origin of Plasmodium falciparum from a single progenitor. Science. 2001;293(5529):482–484. doi: 10.1126/science.1059878. [DOI] [PubMed] [Google Scholar]
- Volkman SK, et al. Genomic heterogeneity in the density of noncoding single-nucleotide and microsatellite polymorphisms in Plasmodium falciparum. Gene. 2007;387(1-2):1–6. doi: 10.1016/j.gene.2006.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. Adaptive evolution of 5’HoxD genes in the origin and diversification of the cetacean flipper. Mol Biol Evol. 2009;26(3):613–622. doi: 10.1093/molbev/msn282. [DOI] [PubMed] [Google Scholar]
- Warnecke T, Hurst LD. Evidence for a trade-off between translational efficiency and splicing regulation in determining synonymous codon usage in Drosophila melanogaster. Mol Biol Evol. 2007;24(12):2755–62. doi: 10.1093/molbev/msm210. [DOI] [PubMed] [Google Scholar]
- Weedall GD, Conway DJ. Detecting signatures of balancing selection to identify targets of anti-parasite immunity. Trends Parasitol. 2010;26(7):363–369. doi: 10.1016/j.pt.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Statistics of local complexity in amino acid sequences and sequence databases. Comput Chem. 1993;17(2):149–163. [Google Scholar]
- Xue HY, Forsdyke DR. Low-complexity segments in Plasmodium falciparum proteins are primarily nucleic acid level adaptations. Mol Biochem Parasitol. 2003;128(1):21–32. doi: 10.1016/s0166-6851(03)00039-2. [DOI] [PubMed] [Google Scholar]
- Zilversmit MM, et al. Low-complexity regions in Plasmodium falciparum: missing links in the evolution of an extreme genome. Mol Biol Evol. 2010;27(9):2198–209. doi: 10.1093/molbev/msq108. [DOI] [PMC free article] [PubMed] [Google Scholar]