Abstract
Inflammasomes are multi-protein complexes that function as sensors of endogenous or exogenous damage-associated molecular patterns. Here we show that deficiency of NLRP6 in mouse colonic epithelial cells results in reduced IL-18 levels and altered fecal microbiota characterized by expanded representation of the bacterial phyla Bacteroidetes (Prevotellaceae) and TM7. NLRP6 inflammasome-deficient mice were characterized by spontaneous intestinal hyperplasia, inflammatory cell recruitment, and exacerbation of chemical colitis induced by exposure to dextran sodium sulfate (DSS). Cross-fostering and cohousing experiments revealed that the colitogenic activity of this microbiota is transferable to neonatal or adult wild-type mice, leading to exacerbation of DSS colitis via induction of CCL5. Antibiotic treatment and electron microscopy studies further supported the role of Prevotellaceae as a key representative of this microbiota-associated phenotype. Altogether, perturbations in this inflammasome pathway, including NLRP6, ASC, caspase-1 and IL-18 may constitute a predisposing or initiating event in some cases of human IBD.
Keywords: Inflammasome, NLRP6, ASC, IL-18, microbiota, colitis
Introduction
The distal intestine of humans contains tens of trillions of microbes: this community (microbiota) is dominated by members of the domain Bacteria but also includes members of Archaea and Eukarya, and their viruses. The vast repertoire of microbial genes (microbiome) present in the distal gut microbiota performs myriad functions that benefit the host (Qin et al., 2010). The mucosal immune system co-evolves with the microbiota beginning at birth, acquiring the capacity to tolerate components of the microbial community while maintaining the capacity to respond to invading pathogens. The gut epithelium and its overlying mucus provide a physical barrier. Epithelial cell lineages, notably the Paneth cell, sense bacterial products through receptors for microbe-associated molecular patterns (MAMPs), resulting in regulated production of bactericidal molecules (Vaishnava et al., 2008). Mononuclear phagocytes continuously survey luminal contents and participate in maintenance of tissue integrity, and the initiation of immune responses (Macpherson and Uhr, 2004; Niess et al., 2005; Rescigno et al., 2001).
Several families of innate receptors expressed by hematopoietic and non-hematopoietic cells are involved in recognition of MAMPs, such as Toll-like receptors (TLRs), nucleotide-binding oligomerization-domain protein-like receptors (NLRs), and C-type lectin receptors (Geijtenbeek et al., 2004; Janeway and Medzhitov, 2002; Martinon et al., 2002). Inflammasomes are cytoplasmic multi-protein complexes composed of one of several NLR proteins, including NLRP1, NLRP3, and NLRC4, that function as sensors of endogenous or exogenous stress or damage-associated molecular patterns (Schroder and Tschopp, 2010). Upon sensing the relevant signal, they assemble, typically together with the adaptor protein, apoptosis-associated speck-like protein (ASC), into a multi-protein complex that governs caspase-1 activation and subsequent cleavage of effector pro-inflammatory cytokines including pro-IL-1β and pro-IL-18 (Agostini et al., 2004; Martinon et al., 2002).
Several other members of the NLR family, including NLRP6 and NLRP12, possess the structural motifs of molecular sensors, and are recruited to the “specks” formed in the cytosol by ASC oligomerization, leading to pro-caspase-1 activation (Grenier et al., 2002; Wang et al., 2002). However, the triggers and function of NLRP12 are only now being revealed (Arthur et al., 2010), while that of NLRP6 remains unknown. In this study, we describe how the innate immune system regulates the colonic microbiota via a novel mechanism that requires an inflammasome, involving NLRP6, ASC and caspase-1, and leads to the cleavage of pro-IL-18. In mice deficient in NLRP6, ASC, caspase-1, or IL-18, gut microbial ecology is altered, with prominent changes in the representation of members of several bacterial phyla. Strikingly, this altered microbiota is associated with a colitogenic phenotype that is transmissible to cohoused wild-type mice, both early in postnatal life and during adulthood.
Results
ASC-deficient mice develop severe DSS colitis that is transferable to cohoused WT mice
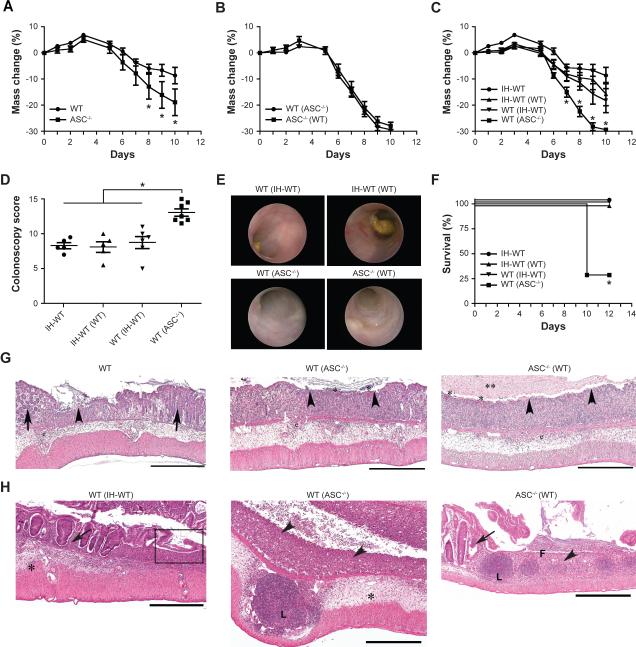
To characterize possible links between inflammasome function and homeostasis achieved between the innate immune system and the gut microbiota we studied mice deficient in ASC. A more severe colitis developed after dextran sodium sulfate (DSS) administration to single-housed ASC−/− mice than in wild-type (WT) mice purchased from a commercial vendor (National Cancer Institute, NCI) (Figure 1A and data not shown). Remarkably, cohousing of adult ASC−/− mice with age-matched WT mice for 4 weeks prior to induction of DSS colitis resulted in development of comparably severe DSS-induced colitis in ASC−/− as well as cohoused WT mice (the latter are designated “WT(ASC−/−)” in Figure 1B).
Figure 1. The increased severity of colitis in ASC-deficient mice is transmissible to cohoused wild-type mice.
To induce colitis, mice were given 2% DSS in their drinking water for 7 days (A) Weight loss of ASC−/− mice and separately housed wild-type (WT) mice. (B) ASC−/− mice and WT mice were cohoused for 4 weeks after which DSS colitis was induced. (C–F) Weight loss (C), colonoscopy severity score at day 7 (D), and survival (F) after induction of DSS colitis of WT mice that were cohoused with (i) in-house WT mice bred for several generations in our vivarium (IH-WT) or (ii) ASC−/− mice (designated WT(IH-WT) and WT(ASC−/−), respectively). (E) Representative images taken during colonoscopy of mice at day 7. (G, H) Representative H&E-stained sections of colons from WT(IH-WT), WT(ASC−/−) and ASC−/−(WT) mice sampled on day 6 (G) and day 12 (H) after the start of DSS exposure. Legend: Epithelial ulceration (arrowheads), severe edema/inflammation (*) with large lymphoid nodules (L), retention/regeneration of crypts (arrows), and evidence of re-epithelialization/repair of the epithelium (box). Scale bars=500 μm. Data are representative for three independent experiments. Error bars represent the SEM of samples within a group. *: p<0.05 by One-way ANOVA.
To assess the possibility that differences in colitis severity observed between groups of single-housed ASC−/− and WT mice were indeed driven by differences in their intestinal microbiota, WT mice were cohoused for 4 weeks with either ASC−/− mice (WT(ASC−/−)) or WT mice that had been bred in our vivarium for >10 generations (in-house mice (IH-WT), WT(IH-WT)). The severity of DSS-induced colitis was similar among NCI-WT (data not shown), IHWT and WT(IH-WT) as well as IH-WT(WT) as judged by weight loss (Figure 1C); colitis severity score (defined by colonoscopy; Figure 1D, E); and survival (Figure 1F). In contrast, WT(ASC−/−) and ASC−/− mice were characterized by an equally increased severity of disease compared to these other groups at both early and late stages (Figure 1C–H, Supplemental Figure 1A–D).
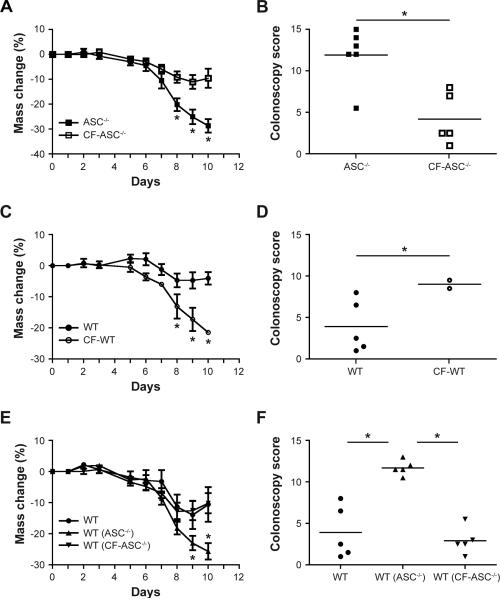
To further establish the role of the intestinal microbiota, we performed cross-fostering experiments. Newborn ASC−/− mice cross-fostered (CF) at birth with in-house WT mothers (CFASC−/−) exhibited milder colitis compared to non-cross-fostered ASC−/− mice (Figure 2A, B). In contrast, newborn WT mice cross-fostered with ASC−/− mothers (CF-WT) developed severe colitis in comparison to non-cross fostered WT mice (Figure 2C, D). Moreover, CF-ASC−/− mice were no longer able to transmit enhanced colitis to cohoused WT mice (Figure 2E, F).
Figure 2. Maternal transmission of an exacerbated DSS colitis phenotype.
Newborn ASC−/− and WT mice were swapped between their respective mothers (cross-fostered), followed by induction of acute DSS colitis at 8 weeks of age. Body weight and colonoscopy severity score were measured in ASC−/− mice and ASC−/− mice cross-fostered with WT mothers (CF-ASC−/−, A,B); WT mice and WT mice cross-fostered with ASC−/− mothers (CF-WT, C,D); WT mice cohoused with ASC−/− or cross-fostered ASC−/− mice for 4 weeks (E,F). Data are representative of 3 independent experiments. Error bars represent the SEM of samples within a group. *: p<0.05 by One-way ANOVA.
Separation of cohoused WT mice from ASC−/− mice and subsequent housing with naïve WT mice resulted in a gradual partial reduction in colitis severity compared to WT(ASC−/−) mice that were not exposed to a WT microbiota (Supplemental Figure 1E–G). Together, these results demonstrate that the ASC−/− microbiota is a dominant colitogenic factor, transmissible early in life to WT mice, and that this colitogenic activity is sustainable in recipient mice for prolonged periods of time. Nonetheless, exposure of an established transferred ASC−/−-derived microbiota in a WT mouse to WT microbiota ameliorates its colitogenic potential, suggesting that the latter community can displace the former and diminish its disease-promoting properties in WT mice.
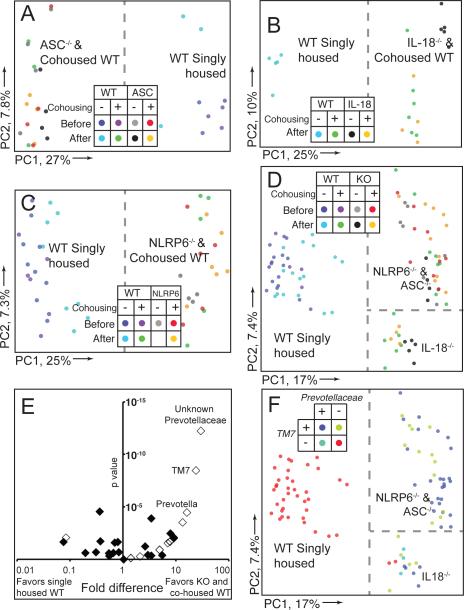
Culture-independent methods were subsequently employed to compare the gut microbial communities. PCR was used to amplify variable region 2 (V2) of bacterial 16S rRNA genes present in fecal samples collected from ASC−/− and WT mice just prior to and 28d following cohousing. The amplicons generated were subjected to multiplex pyrosequencing and the resulting chimera-checked and filtered datasets compared using UniFrac (mean of 3,524±1,023 (SD) 16S rRNA reads/sample; see Methods for details). Figure 3A shows a clear difference in fecal bacterial phylogenetic architecture in WT versus ASC−/− mice. Moreover, after 4 weeks of cohousing, the fecal bacterial communities of WT(ASC−/−) mice clustered together with communities from their ASC−/− cagemates. In addition, the bacterial component of the fecal microbiota of these cohoused ASC−/− mice was similar to ASC−/− mice that never had been cohoused.
Figure 3. Bacterial 16S rRNA-based analysis the fecal microbiota of WT and NLRP6−/− mice.
(A–D) Unweighted UniFrac PCoA of fecal microbiota harvested from WT mice single-housed or cohoused with ASC−/− (A), IL-18−/− (B), NLRP6−/− (C) or all (D) mice. Samples from mice shown in (A) and (C) were taken just prior to cohousing and 28 days later. Dashed line illustrates separation of samples along PC1. (E) Distribution of family-level phylotypes in ASC-, IL-18-, NLRP6-deficient and cohoused WT mice, compared to single-housed WT mice. The horizontal axis shows the fold representation (defined as the ratio of the percentage of samples with genera present in knockout or cohoused mice versus single-housed WT mice); the left side of the axis indicates taxa whose representation is greater in single-housed WT mice; the right denotes taxa whose representation is greater in knockout or cohoused WT mice; the origin represents equivalent recovery of taxa in both groups. The vertical axis shows the calculated p value for each taxa as defined by G-test. Open diamonds represent taxa that were found only in KO/cohoused WT or single-housed WT mice, but where recovery was assumed to be 1 to calculate fold-representation. (F) Unweighted UniFrac PCoA demonstrating presence or absence of TM7 and Prevotellaceae in each sample. Dashed lines show separation of single-housed WT and cohoused WT and knockout mice on PC1. PC2 in panels (D) and (F) shows separation of communities based on host genotype/cohousing.
NLRP6-deficiency produces a microbiota-mediated phenotype that resembles that of ASC-deficiency
To assess whether ASC's function as adaptor protein for inflammasome formation is linked to the changes in gut bacterial community structure and function observed, we cohoused WT mice with caspase-1−/− mice, and these too exhibited more severe DSS-induced colitis compared to single-housed WT mice (Supplemental Figure 2A–E). Similar to WT mice cohoused with ASC−/− mice, WT mice cohoused with caspase-1−/− mice evolved their intestinal bacterial communities to a phylogenetic configuration that was very similar to that of their caspase-1−/− cagemates (Supplemental Figure 2F). These results point to the involvement of an inflammasome in this phenotype.
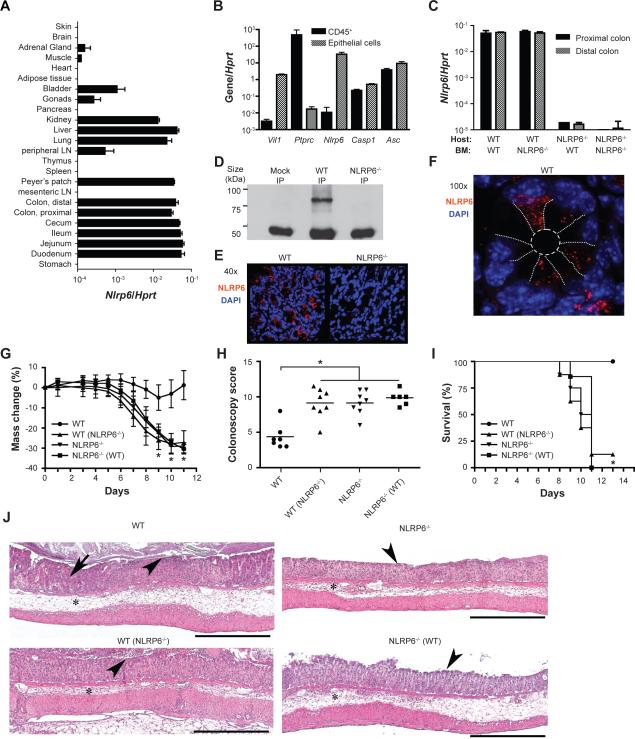
We next sought to identify the NLR(s) upstream of ASC and caspase-1 leading to the phenotype. qRT-PCR analysis of 24 tissues in WT mice revealed that NLRP6, which forms an ASC-dependent inflammasome (Grenier et al., 2002), is most highly expressed in the gastrointestinal tract, and at lower levels in lung, kidney and liver (Figure 4A). Further, we isolated RNA prepared from colonic epithelium and sorted colonic CD45+ hematopoietic cells and found that ASC and caspase-1 are highly expressed in both compartments. NLRP6 expression, in contrast, was essentially limited to the epithelial compartment (Figure 4B). Indeed, in bone marrow transfer experiments NLRP6 was almost undetectable in NLRP6−/− mice (Supplemental Figure 3A, B) receiving WT bone marrow (Figure 4C). Follow up immunoprecipitation (Figure 4D) and immunofluorescence assays (Figure 4E, F) both showed that NLRP6 protein is expressed in primary colonic epithelial cells of WT mice, where it mainly appears within speckled cytoplasmic aggregates, while it was absent in NLRP6−/− mice. WT and NLRP6−/− mice were then single-housed or cohoused for 4 weeks, followed by exposure to DSS. Single-housed NLRP6−/− mice developed more severe colitis compared to single-housed WT mice (Figure 4G–J). The more severe colitis phenotype was transferable to cohoused WT mice (WT(NLRP6−/−), Figure 4G–J, Supplemental Figure 3C–G). 16S rRNA analysis of fecal bacterial communities demonstrated a clear difference in the bacterial community structure between single-housed adult WT mice versus age-matched WT mice cohoused for 4 weeks with NLRP6-deficient mice (Figure 3C). Fecal bacterial communities of WT mice clustered together with communities from their NLRP6−/− cagemates whose microbiota in turn was similar to NLRP6−/− mice that never had been cohoused (Figure 3C).
Figure 4. NLRP6-deficient mice harbor a transmissible colitogenic gut microbiota.
(A) Analysis of NLRP6 expression in various organs, and (B) in colonic epithelial and hematopoietic (CD45+) cells. The purity of the sorted populations in panel B was analyzed using vil1 and ptprc as markers for epithelial and hematopoietic cells, respectively. (C) Bone-marrow chimeras were generated using WT and NLRP6−/− mice as host and bone marrow donor. NLRP6 expression in the colon was analyzed 8 weeks after bone marrow transplantation. (D) Analysis of NLRP6 protein expression was performed by immunoprecipitation using an NLRP6 antibody and lysates of primary colonic epithelial cells isolated from WT and NLRP6−/− mice. (E, F) Representative confocal images of colonic sections analyzed for expression of NLRP6 (red) and counterstained with DAPI. (E) 40×, (F) 100×, white dotted lines were drawn to illustrate the epithelial cell boundaries (G–J) Acute DSS colitis was induced in single-housed WT mice, in WT mice cohoused for 4 weeks with NLRP6−/− mice (WT(NLRP6−/−), the corresponding cohoused NLRP6−/− mice (NLRP6−/−(WT)) and single-housed NLRP6−/− mice (NLRP6−/−). Weight (G), colonoscopy severity score at day 8 (H), and survival (I) of single-housed versus cohoused WT and NLRP6−/− mice. (J) Representative H&E stained sections of colons on day 8 after initiation of DSS exposure. Legend: Edema/inflammation (*), ulceration (arrowheads) and loss of crypts (arrow). Scale bars=500 μm. Data are representative of 3 independent experiments. Error bars represent the SEM of samples within a group. *: p<0.05 by One-way ANOVA.
To ascertain the specificity of this phenotype, we cohoused WT mice with mice that lacked other NLR family members and inflammasome-forming protein AIM2, all shown by qRT-PCR analysis to be expressed in the colon (Supplemental Figure 4A)(Kufer and Sansonetti, 2011; Schroder and Tschopp, 2010). Adult, conventionally-raised, specific pathogen-free knockout mice were either obtained from the same source as NLRP6−/− mice (Millenium, NLRP3−/−, NLRC4−/−, NLRP12−/−), generated in our own laboratory (NLRP10−/−), or obtained from other laboratories (AIM2−/−, K Fitzgerald, U. Massachusetts). NLRP3−/− mice cohoused with WT mice for 4 weeks featured attenuated colitis as compared to their WT cagemates and mild transferability of colitis, suggesting that NLRP3's major effect in this system is negative regulation of the inflammatory process itself (data not shown). Importantly, none of the other above mentioned mouse strains transferred microbiota with increased colitogenic properties to WT mice upon cohousing (Supplemental Figure 4 B–I). Likewise, 16S rRNA analysis of these strains revealed a distinct configuration of their microbiota population as compared to NLRP6 inflammasome deficient mice (Supplemental Table 1). Together, these findings indicate that NLRP6 forms an intestinal epithelial inflammasome that regulates functional properties of the microbiota, and that loss of NLRP6, and the known inflammasome constituents, ASC and caspase-1, leads to the specific development of a transmissible, more colitogenic microbiota.
Evidence that NLRP6 affects the gut microbiota via IL-18
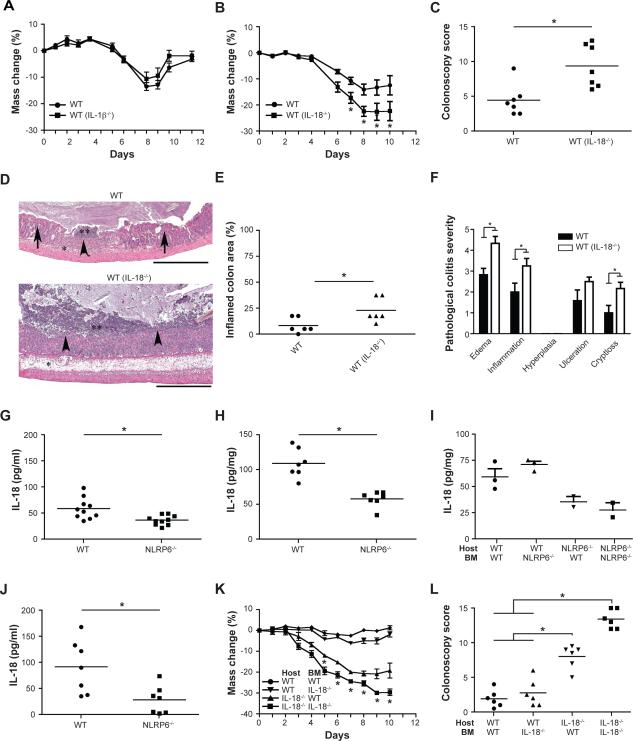
Activation of inflammasomes results in multiple downstream effects, including proteolytic cleavage of pro-IL-1β and pro-IL-18 to their active forms (Schroder and Tschopp, 2010). To test whether the effect of NLRP6-deficiency is mediated via IL-1β or IL-18 deficiency, we cohoused adult WT mice with either IL-1β−/− (Figure 5A) or IL-1R−/− mice (Supplemental Figure 4J, K). Cohousing WT mice with these strains did not result in any significant changes in the severity of DSS colitis compared to single-housed WT mice, excluding a major contribution of the IL-1 axis. In contrast, IL-18−/− mice, and more importantly WT mice cohoused with them, exhibited a significant exacerbation of colitis severity, compared to single-housed WT mice (Figure 5B–F).
Figure 5. Processing of IL-18 by NLRP6 inflammasome suppresses colitogenic microbiota.
WT mice were cohoused with IL-1β−/− mice or IL-18−/− mice for 4 weeks and colitis was subsequently induced with DSS. Comparison of weight loss (A) in single-housed WT mice and in WT mice previously cohoused with IL-1β−/− mice (WT(IL-1β−/−)). Weight loss (B) and colonoscopy severity score at day 7 (C) for single-housed WT mice and WT mice previously cohoused with IL-18−/− mice (WT(IL-18−/−)). (D–F) Representative H&E-stained sections (D) and pathologic quantitation of disease severity (E, F) of colons from single-housed WT mice and WT mice cohoused with IL18−/− mice sampled 6 days after the start of DSS administration. Scale bars=500 μm. (G, H) IL-18 levels measured in sera (G) and colon explants (H) obtained from WT and NLRP6-deficient mice without treatment. (I) Bone-marrow chimeras were generated using both WT and NLRP6−/− mice as host and bone marrow donor. IL-18 production by colon explants was analyzed 8 weeks after bone marrow transplantation. (J) IL-18 concentrations in the serum 5 days after induction of DSS colitis. (K, L) Bone-marrow chimeras were generated using WT and IL-18−/− mice as host and bone marrow donor: weight (K) and colonoscopy severity scores at day 7 (L) of mice with acute DSS colitis are shown. Data in panels A–E are representative of at least 3 experiments, data in panels I–L are representative of two experiments (n=6 mice/samples analyzed per group). *: p<0.05 by One-way ANOVA.
In the steady state, single-housed NLRP6−/− mice had significantly reduced serum levels of IL-18 compared to their WT counterparts, and reduced production of this cytokine in their colonic explants (Figure 5G, H). To study the relative contribution of hematopoietic and nonhematopoietic NLRP6 deficiency to this reduction in active IL-18, we measured IL-18 protein levels in colonic explants prepared from chimeric mice that had received bone marrow transplants from NLRP6−/− or WT donors. Significantly lower IL-18 protein levels were noted only in explants prepared from mice with NLRP6 deficiency in the non-hematopoietic compartment (Figure 5I). This result indicates that NLRP6 expressed in a non-hematopoietic component of the colon, likely the epithelium, is a major contributor to production of active IL-18. Furthermore, in contrast to WT mice, NLRP6−/− mice failed to significantly upregulate IL-18 in the serum and in tissue explants following induction of DSS colitis (Figure 5J and data not shown).
To study whether IL-18 production by non-hematopoietic cells is the major contributor to the microbiota-associated enhanced colitogenic phenotype, we performed a bone marrow transfer experiment using IL-18−/− and WT mice as both recipients and donors. Indeed, mice deficient in IL-18 in the non-hematopoietic compartment exhibited more severe disease compared to mice that were sufficient for IL-18 in the non-hematopoietic compartment (Figure 5K, L). Bacterial 16S rRNA studies demonstrated that the fecal microbiota of WT mice exposed to IL-18−/− mice changed its phylogenetic configuration to resemble that of IL-18−/− cagemates (Figure 3B). Interestingly, as seen in the PC2 axis in the PCoA plot of unweighted UniFrac distances, the fecal microbiota of ASC−/− and NLRP6−/− mice were distinct from IL-18−/− mice, possibly reflecting the existence of additional NLRP6 inflammasome-mediated IL-18-independent mechanisms of microflora regulation (Figure 3D). Together, these results led us to conclude that the decrease in colonic epithelial IL-18 production in mice deficient in components of the NLRP6 inflammasome is critically involved in the enhanced colitogenic properties of the microbiota.
The gut microbiota from NLRP6-inflammasome deficient mice induces CCL5 production and immune cell recruitment leading to spontaneous inflammation
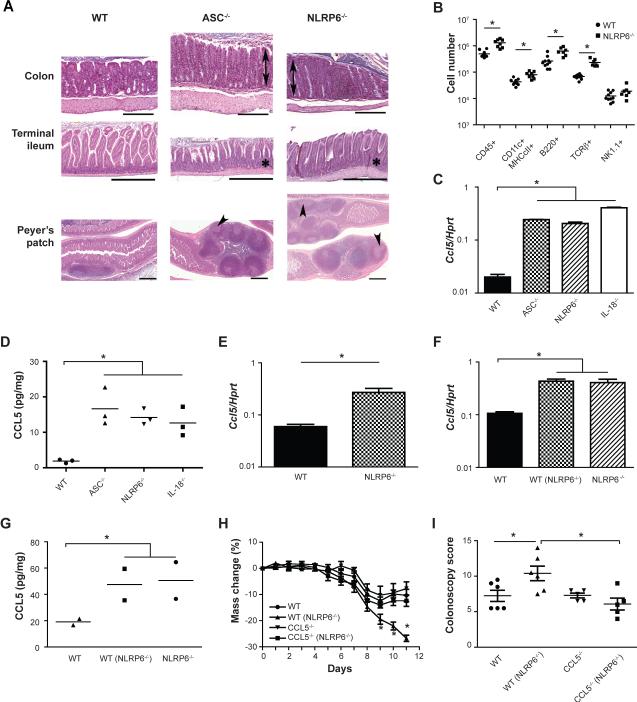
We next examined the intestines of untreated ASC−/− and NLRP6−/− mice for signs of spontaneous pathological changes. The colons, terminal ileums and Peyer's patches of ASC−/− and NLRP6−/− mice exhibited colonic crypt hyperplasia, changes in crypt-to-villus ratios in the terminal ileum, and enlargement of Peyer's patches with formation of germinal centers (Figure 6A and Supplemental Figure 5A, B). NLRP6 inflammasome-deficient mice also had significantly elevated serum IgG2c and IgA levels, as did cohoused WT mice (Supplemental Figure 5C–F). In addition, we recovered significantly more CD45+ cells from colons of NLRP6−/− mice compared to WT controls (Figure 6B). These results prompted us to investigate downstream effector mechanisms by which the altered microbiota could induce this immune cell infiltration. Multiplex analysis of cytokine and chemokine production by tissue explants (Supplemental Figure 5G), followed by validation at the RNA (Figure 6C) and protein levels (Figure 6D) indicated that CCL5 levels were significantly elevated in single-caged untreated ASC−/−, NLRP6−/− and IL-18−/− compared to WT mice. Furthermore, CCL5 mRNA upregulation was found to originate from epithelial cells (Figure 6E). Moreover, CCL5 levels were induced in WT mice upon cohousing (Figure 6F, G) showing that this property was specified by the microbiota and not the mutated inflammasome per se. Notably, in the steady state, CCL5−/− mice and WT mice featured a comparable representation of immune subsets with the exception of slight reduction in γδ TCR+ lymphocytes, indicating that CCL5 is not generally required for immune cell recruitment to the colon (Supplemental Figure 5H).
Figure 6. Microbiota induction of CCL5.
(A) Representative H&E-stained sections of the colon, terminal ileum and Peyer's patches from WT, ASC−/− and NLRP6−/− mice not exposed to DSS. Legend: Mucosal hyperplasia in the colon (double arrows), increased crypt to villus ratio in the terminal ileum (*), and enlargement of Peyer's patches with formation of germinal centers (arrowheads). Scale bars = 500 μm. (B) Enumeration of subsets of hematopoietic cells harvested from the lamina propria of WT and NLRP6−/− mice. (C, D) Analysis of CCL5 colonic mRNA expression (C), and protein expression in colonic explants (D) in WT, ASC−/−, NLRP6−/−, and IL-18−/− mice. (E) CCL5 expression in epithelial cells from the colons of WT and NLRP6−/− mice. (F,G) Analysis of CCL5 colonic mRNA expression (F) and protein expression in colonic explants (G) in single-housed WT mice and WT mice cohoused with NLRP6−/− mice. (H, I) WT and CCL5−/− mice were either single-housed or cohoused for 4 weeks with NLRP6−/− mice followed by exposure to DSS. Weight loss (H) and colonoscopy severity score at day 7 (I) of mice after induction of acute DSS colitis. Data shown in panels A-G are representative of at least two experiments. Data presented in panels H and I are from three experiments (n=5–6 mice). Error bars represent the SEM of samples within a group. *: p<0.05 by One-way ANOVA.
To test the role of CCL5 in mediating the enhanced colitogenic properties of the NLRP6−/− mouse microbiota, we cohoused WT or CCL5−/− mice with NLRP6−/− mice for 4 weeks. We subsequently induced DSS colitis and found comparable colitis severity between single-housed WT and CCL5−/− mice (Figure 6H, I). However, upon cohousing, WT(NLRP6−/−) mice had significantly worse DSS-induced colitis compared to CCL5−/−(NLRP6−/−) mice, despite comparable acquisition of the NLRP6−/− colitogenic flora (Supplemental Figure 5I). These findings support the notion that CCL5 upregulation in response to the altered microbiota is responsible for the exacerbation of colitis that occurs in WT mice cohoused with NLRP6 inflammasome-deficient mice.
Identification of bacterial phylotypes that are markedly expanded in both NLRP6 inflammasome-deficient mice and in cohoused WT mice
To identify whether increased colitis severity is driven by bacterial components, we first treated ASC−/− mice with a combination of antibiotics known to reduce the proportional representation of a broad range of bacterial phylotypes in the gut (Fagarasan et al., 2002; Rakoff-Nahoum et al., 2004). Antibiotic therapy reduced the severity of DSS colitis in ASC−/− mice to WT levels (Supplemental Figure 6A, B). To exclude a possible role for herpesviruses, fungi and parasites, single-housed WT and ASC−/− mice were treated for 3 weeks with either oral gancyclovir, amphotericin, or albendazole and praziquantil, respectively. None of these treatments altered the severity of colitis in ASC-deficient mice (Supplemental Figure 6 C–E). Furthermore, fecal tests for rotavirus, lymphocytic choriomeningitis virus, K87, murine cytomegalovirus, mouse hepatitis virus, mouse parvovirus, reovirus, Theiler's murine encephalomyelitis virus were all negative, and there was no histological evidence of inclusion bodies, characteristic of virally infected colonic epithelial cells (data not shown). Together, these results pointed to bacterial components as being responsible for the transferrable colitis phenotype in NLRP6 inflammasome-deficient mice.
Supplemental Table 1 lists bacterial phylotypes whose presence or absence was significantly different in (i) single-housed WT mice compared to (ii) ASC−/−, and NLRP6−/−, and caspase-1−/−, and IL-18−/−, and all types of cohoused WT mice (all untreated with DSS). Nine genera belonging to four phyla (Firmicutes, Bacteroidetes, Proteobacteria and TM7) satisfied our requirement of having significant differences in their representation in the fecal microbiota in group (i) versus group (ii). The genus-level phylotype most significantly associated with the fecal microbiota of ASC−/−, NLRP6−/−, caspase-1−/−, IL-18−/− and cohoused WT mice was a member of the family Prevotellaceae in the phylum Bacteroidetes. Beyond this unnamed genus in the Prevotellaceae, the next two most discriminatory genus-level taxa belonged to the phylum TM7 and the named genus Prevotella within the Prevotellaceae (Figure 3E, Supplemental Figure 2G). Likewise, Prevotellaceae was absent from single-housed CCL5−/− mice, and highly acquired following cohousing with NLRP6−/− mice (Supplemental Figure 5J, K). Also included in this list was a member of the family Helicobacteraceae (order Campylobacterales); tests for the pathogen Helicobacter hepaticus were consistently negative in these mice (n=6 samples per strain screened with PCR).
Histopathologic analyses of colonic sections stained with hematoxylin and eosin as well as Warthin-Starry stain disclosed microbes with a long branching, striated morphotype closely associated with the crypt epithelium of single-housed ASC−/− and NLRP6−/− mice; these organisms were rare in WT mice (Supplemental Figure 6F and data not shown). This morphotype is consistent with members of TM7 (Hugenholtz et al., 2001). Quadruple antibiotic treatment for 3 weeks eliminated microbes with this morphology from ASC−/− mice as judged by histopathologic analysis (n=5 mice; data not shown).
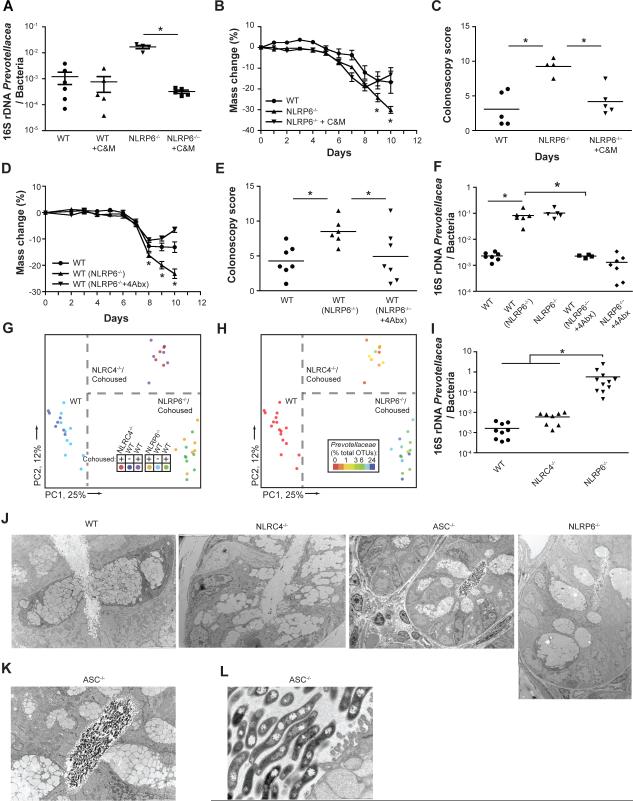
A significant reduction in Prevotellaceae was noted in stools of NLRP6−/− mice treated with the same combination of four antibiotics. The most complete eradication was achieved using a combination of metronidazole and ciprofloxacin, a commonly used regimen for treatment of human IBD (Figure 7A). The severity of DSS colitis was also significantly reduced in antibiotic treated compared to untreated NLRP6−/− mice (Figure 7B, C).
Figure 7. Decreased abundance of Prevotella in antibiotic-treated NLRP6−/− correlates with ameliorated colitogenic microbiota.
(A–C) WT and NLRP6−/− mice were treated with a combination of metronidazole and ciprofloxacin for 3 weeks. Prevotellaceae loads compared to total bacteria (A) were measured in fecal samples at the end of the antibiotic treatment period using qPCR analysis. DSS exposure was begun 3 days later. Weight loss (B) and colonoscopy score at day 7 (C). (D–F) NLRP6−/− mice were treated with a combination of ampicillin, neomycin, vancomycin, and metronidazole for 3 weeks and then cohoused with WT mice for 4 weeks. In parallel, WT mice were cohoused with untreated NLRP6−/− mice. Subsequently, DSS colitis was induced and weight (D) and colonoscopic assessment of mucosal damage at day 7 (E) were recorded. (F) qPCR assay for the abundance of Prevotella in fecal samples obtained after 4 weeks of cohousing. (G, H). WT mice were cohoused for four weeks with either NLRP6−/− or NLRC4−/− mice. (G) Unweighted UniFrac PCoA of fecal microbiota harvested after cohousing. (H) Unweighted UniFrac PCoA colored by relative abundance of Prevotellaceae as percent of total OTUs. (I) Quantification of Prevotellaceae in the crypt compartment, following extensive removal of stool content. (J–L) Representative transmission electron microscopy images taken from colonic sections of WT (J, ×4200), NLRC4−/− (J, ×4200), NLRP6−/−(J, ×2500), and ASC−/− mice (J ×1700; K ×4200; L ×26,000).
Next, we tested whether antibiotic treatment affected the ability of NLRP6−/− mice to transfer the colitogenic microbiota to WT mice. Strikingly, WT mice cohoused with antibiotic-treated NLRP6−/− mice developed significantly less severe DSS colitis compared to WT mice cohoused with untreated NLRP6−/− mice (Figure 7D, E). This reduction in severity correlated with decreased abundance of Prevotellaceae and TM7, but not of Bacteroidetes in WT mice cohoused with antibiotic-treated NLRP6−/− mice (Figure 7F, Supplemental Figure 6G, H). Low level representation of Prevotellaceae was noted in non-phenotypic NLR deficient mice bred for generations in our vivarium (Supplemental Table 1). As representative NLR's, we decided to directly compare the quantititative differences in Prevotellaceae abundance and its impact on transmissibility to WT mice between NLRP6−/− and NLRC4−/− mice, since the latter lacks a closely related colonic- epithelium expressed protein that is also able to form an inflammasome and process IL-18. Indeed, NLRC4−/− and their cohoused WT cagemates featured a clustering pattern in the PCoA plot (Figure 7G) distinct from both single-housed WT mice as well as from NLRP6−/− mice and cohoused WT mice. Specifically, Prevotellaceae was highly abundant in NLRP6−/− mice while low to absent in NLRC4−/− mice, their cohoused WT cagemates, and single-housed WT mice (Figure 7H).
To determine whether NLRP6 deficiency was associated with an alteration in the physical distribution (biogeography) of the microbiota within the gut, we analyzed colon tissue that had been thoroughly washed of fecal matter (see Methods for details). This enabled enhanced detection of bacteria residing in crypts. TM7 and Prevotellaceae were significantly more prevalent in the washed colons of NLRP6−/− mice compared to WT and NLRC4−/− mice (Figure 7I and data not shown). Further, transmission electron microscopy studies revealed multiple monomorphic bacteria in crypt bases of ASC−/− and NLRP6−/− but not WT and NLRPC4−/− mice, featuring an abundance of electron dense intracellular material that is consistent with the pigmentation that is characteristic of many Prevotella species (Figure 7J–L and data not shown). Overall, these findings indicate that the dysbiosis in NLRP6-inflammasome deficient mice may involve aberrant host-microbial cross-talk within the colonic crypt.
Discussion
We describe a novel regulatory sensing system in the colon dependent on the NLRP6 inflammasome. We show that genetic deletion of components of this sensing system has drastic consequences on the composition of the microbial communities leading to a shift towards a pro-inflammatory configuration that drives spontaneous and induced colitis.
On a molecular level, it appears unlikely that the evolutionarily conserved, innate mucosal immune arm possesses the ability to distinctly identify the myriad bacterial, archaeal and eukaryotic microbial phylotypes and virotypes that comprise the gut microbiota and differentiate autochthonous (entrenched) or allochthonous (transient/nomadic) components of this community that act as commensals or mutualists from those that act as pathogens. Rather, this function may be achieved by sensing signals related to tissue integrity, or factors released by tissue damage, that serve as “danger signals” promoting activation of an innate response (Matzinger, 2007). Inflammasomes are capable of fulfilling this task as they can be activated by many microbial ligands, but also by host-derived factors released upon cell or tissue damage, such as uric acid, ATP, and hyaluronan (Schroder and Tschopp, 2010). NLRP6 assembly in the colonic epithelial compartment may be driven by a low level of these substances or by yet unidentified molecules signaling tissue integrity, resulting in local production of IL-18. Interestingly, in the rat, NLRP6, caspase-1, ASC and pro-IL-18 are absent at embryonic day 16 (E16), and first appear at E20, with the processed form of IL-18 emerging in the gut during the early postnatal period (Kempster et al., 2011), coinciding with the time of colonization of the gut ecosystem.
Dysbiosis may contribute to IBD by expansion of colitogenic strains such as enteroinvasive E.coli (Darfeuille-Michaud et al., 2004), by reduction of tolerogenic strains such as Faecalibacterium prausnitzi (Sokol et al., 2008), or through a combination of both mechanisms. In our study, a colitogenic microbiota with altered representation of distinct bacterial members formed in the intestines of NLRP6-deficient mice: this microbiota was transferred across generations within a kinship, and could displace the gut microbiota of cohoused immunocompetent mice. Once this community was horizontally transmitted to suckling or adult WT mice, it could persist. Compared to WT mice, NLRP6-inflammasome deficient mice exhibit both quantitative and qualitative changes in numerous taxa including increased representation of members of Prevotellaceae and TM7, and reductions in members of genus Lactobacillus in the Firmicutes phylum.
There are several intriguing links between the abundance of Prevotellaceae and TM7 and human diseases. Prevotellaceae has been implicated in periodontal disease (Kumar et al., 2003), and several reports have documented prominent representation of this group in samples from IBD patients (Kleessen et al., 2002; Lucke et al., 2006). Provetellaceae might disrupt the mucosal barrier function through production of sulfatases that actively degrade mucus oligosaccharides (Wright et al., 2000); these enzymes are elevated in intestinal biopsies from IBD patients (Tsai et al., 1995). While they have not been cultured, members of the TM7 phylum have been identified in 16S rRNA surveys of terrestrial and aquatic microbial communities as well in human periodontal disease (Brinig et al., 2003; Marcy et al., 2007; Ouverney et al., 2003), and in IBD patients (Kuehbacher et al., 2008). Defining the nature of the interactions of Prevotellaceae and TM7 with the NLRP6 inflammasome may provide insights about probiotic interventions that may mitigate microbiota-mediated enhanced inflammatory responses.
Four previous reports indicated that caspase-1, ASC or NLRP3 deficiencies were associated with an increased severity of acute DSS colitis in mice, and suggested that exacerbated disease was mediated in part by a defect in repair of the intestinal mucosa (Allen et al., 2010; Dupaul-Chicoine et al., 2010; Hirota et al., 2010; Zaki et al., 2010). Opposing results were found in two other studies using the same colitis model. The first study to investigate the role of caspase-1 in intestinal auto-inflammation, even prior to the discovery of the inflammasome, found ameliorated acute and chronic colitis in caspase-1−/− mice (Siegmund et al., 2001). More recently, a second study demonstrated reduced severity of disease in NLRP3−/− mice that correlated with decreased levels of pro-inflammatory IL-1β (Bauer et al., 2010). It has been hypothesized that these differences might be the result of distinct roles of inflammasomes in non-hematopoietic versus hematopoietic cells (Siegmund, 2010). The proposed function in epithelial cells is to regulate secretion of IL-18 that stimulates epithelial cell barrier function and regeneration, while in hematopoietic cells inflammasome activation would have a pro-inflammatory effect. Varying degrees of tissue injury and subsequent inflammation may result in shifting the balance between protective and detrimental effects, depending on the experimental condition and the inflammatory context. However, we believe that inflammasome-driven effects on the colonic microbiota, as revealed in our study, add yet another layer of regulation that affects and effects initiation of auto-inflammation. As such, exacerbation in colitis severity in single-housed inflammasome-deficient mice may in fact involve defects in tissue regeneration, but this histopathological process may be dramatically influenced by the effects imposed by altered elements in the microbiota including, for example, the enhanced representation of Prevotellaceae in the crypt. Thus, we propose that the fundamental role of the microbiota in shaping processes related to tissue damage, regeneration and stress response might offer an explanation for the opposing results between these studies. 16S rRNA enumeration studies combined with various permutated cohousing experiments of the type described in this report, coupled with mechanistic molecular studies, would allow this notion to be tested directly. Furthermore, our results suggest that prolonged cohousing (or littermate controls) should be used when NLRs and other innate receptors are studied: this would allow for equilibration of differences in gut microbial ecology that may exist between groups of mice, and allow investigators to determine which features of their phenotypes can be ascribed to the microbiota. Indeed, using co-housing conditions we were able to demonstrate that the NLRC4 inflammasome is a direct negative regulator of colonic epithelial cell tumorigenesis which is not driven by the microbiota (Hu et al., 2010).
Our results show that the resultant aberrant microbiota promotes local epithelial induction of CCL5 transcription as a downstream mechanism, ultimately leading to an exaggerated auto-inflammatory response. CCL5 is potently induced by bacterial and viral infections, and in turn induces massive recruitment of a variety of innate and adaptive immune cells carrying CCR1, CCR3, CCR4 and CCR5 (Mantovani et al., 2004). Interestingly, both NOD2 and TLRs have been shown to induce CCL5 transcription (Berube et al., 2009; Werts et al., 2007). It will be of interest to investigate the crosstalk between these immune recognition systems in future experiments.
Recent studies have highlighted the importance of the gut microbiota in the pathogenesis of various autoimmune disorders that manifest outside of the gastrointestinal tract. In some autoimmune models germ-free conditions or inoculation with a microbiota from healthy mice ameliorates disease (Lee et al., 2010; Mazmanian et al., 2008; Sinkorova et al., 2008; Wu et al., 2010). In contrast, rats with collagen-induced arthritis feature exacerbated disease when reared under germ-free conditions (Breban et al., 1993) while germ-free NOD MyD88−/− mice fail to develop diabetes unlike their colonized counterparts (Wen et al., 2008). In humans, epidemiological evidence points to possible links between dysbiosis and rheumatoid arthritis, asthma, and atopic dermatitis (Bjorksten, 1999; Penders et al., 2007; Vaahtovuo et al., 2008). Our study indicates that deficiencies in the NLRP6 pathway should be added to the list of host genetic factors that may drive disease-specific alterations in the microbiota, which in turn may promote disease in these hosts or in individuals who have been exposed to these microbial communities and who have also experienced disruption in their gut epithelial barrier function due to a variety of insults.
Experimental Procedures
Mice
NLRP6−/− mice were generated by replacing exons 1 and 2 with a neomycin selection cassette (IRESnlslacZ/MC1neo). For cohousing experiments, age- and gender-matched WT and knockout mice were co-housed at 1:1 ratios for 4 weeks.
DSS colitis
Mice were treated with 2% (w/v) DSS (M.W. =36,000–50,000 Da; MP Biomedicals) in their drinking water for 7 days followed by regular access to water.
16S rRNA analyses
Aliquots of frozen fecal samples (n=212) were processed for DNA isolation using a previously validated protocol (Turnbaugh et al., 2009). ~365bp amplicons, spanning variable region 2 (V2) of the 16S rRNA gene were generated by using primer containing barcodes and sequenced on 454 sequencer. Data was processed using the QIIME (Quantitative Insights Into Microbial Ecology) analysis pipeline (Caporaso et al., 2010) and analyzed using UniFrac that defines the similarities and differences between microbial communities based on the degree to which community members share branch length on a bacterial tree of life (Lozupone et al., 2006).
Statistical Analysis
Data are expressed as mean ± SEM. Differences were analyzed by Student's t-test and ANOVA, and post-hoc analysis for multiple group comparison. P values ≤0.05 were considered significant.
Supplementary Material
Acknowledgements
We would like to thank A Hafemann, S. Campton, E. Eynon, J. Alderman, W. Philbrick, C. Zorca, M. Musaheb, J. Stein, A. Ferrandino, F. Manzo and the members of the Flavell lab for technical help and helpful discussions; M. Graham and C. Rahner, CCMI EM Core Facility, Yale School of Medicine; J. Manchester and S. Deng for their assistance with DNA sequencing; V. Nagy for discussion and help with figure preparation; and H. Elinav for important discussions, suggestions and critique. EE is supported by Cancer Research Institute (2010–2012), the Israel-US educational foundation (2009), and is the recipient of the Claire and Emmanuel G. Rosenblatt award from the American Physicians for Medicine in Israel Foundation. ALK is the recipient of a post-doctoral fellowship from the W.M Keck Foundation. JHM is supported by a LLS Postdoctoral Fellowship. This work was supported in part by Howard Hughes Medical Institute (RAF) and the Crohn's and Colitis Foundation of America (J.I.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data deposition 16S rRNA datasets have been deposited in MG-RAST under accession number XXXXXXXX.
A detailed description of materials and methods used in this paper can be found in the supplemental information.
References
- Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB, Herfarth HH, Jobin C, Ting JP. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Lich JD, Ye Z, Allen IC, Gris D, Wilson JE, Schneider M, Roney KE, O'Connor BP, Moore CB, et al. Cutting edge: NLRP12 controls dendritic and myeloid cell migration to affect contact hypersensitivity. J Immunol. 2010;185:4515–4519. doi: 10.4049/jimmunol.1002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C, Duewell P, Mayer C, Lehr HA, Fitzgerald KA, Dauer M, Tschopp J, Endres S, Latz E, Schnurr M. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut. 2010;59:1192–1199. doi: 10.1136/gut.2009.197822. [DOI] [PubMed] [Google Scholar]
- Berube J, Bourdon C, Yao Y, Rousseau S. Distinct intracellular signaling pathways control the synthesis of IL-8 and RANTES in TLR1/TLR2, TLR3 or NOD1 activated human airway epithelial cells. Cell Signal. 2009;21:448–456. doi: 10.1016/j.cellsig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Bjorksten B. The environmental influence on childhood asthma. Allergy. 1999;54(Suppl 49):17–23. doi: 10.1111/j.1398-9995.1999.tb04383.x. [DOI] [PubMed] [Google Scholar]
- Breban MA, Moreau MC, Fournier C, Ducluzeau R, Kahn MF. Influence of the bacterial flora on collagen-induced arthritis in susceptible and resistant strains of rats. Clin Exp Rheumatol. 1993;11:61–64. [PubMed] [Google Scholar]
- Brinig MM, Lepp PW, Ouverney CC, Armitage GC, Relman DA. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Appl Environ Microbiol. 2003;69:1687–1694. doi: 10.1128/AEM.69.3.1687-1694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, Colombel JF. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, McIntire CR, LeBlanc PM, Meunier C, Turbide C, Gros P, Beauchemin N, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–378. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek TB, van Vliet SJ, Engering A, t Hart BA, van Kooyk Y. Self- and nonself-recognition by C-type lectins on dendritic cells. Annu Rev Immunol. 2004;22:33–54. doi: 10.1146/annurev.immunol.22.012703.104558. [DOI] [PubMed] [Google Scholar]
- Grenier JM, Wang L, Manji GA, Huang WJ, Al-Garawi A, Kelly R, Carlson A, Merriam S, Lora JM, Briskin M, et al. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-kappaB and caspase-1. FEBS Lett. 2002;530:73–78. doi: 10.1016/s0014-5793(02)03416-6. [DOI] [PubMed] [Google Scholar]
- Hirota SA, Ng J, Lueng A, Khajah M, Parhar K, Li Y, Lam V, Potentier MS, Ng K, Bawa M, et al. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm Bowel Dis. 2010 doi: 10.1002/ibd.21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Elinav E, Huber S, Booth CJ, Strowig T, Jin C, Eisenbarth SC, Flavell RA. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1016814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Tyson GW, Webb RI, Wagner AM, Blackall LL. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl Environ Microbiol. 2001;67:411–419. doi: 10.1128/AEM.67.1.411-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA, Jr., Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Kempster SL, Belteki G, Forhead AJ, Fowden AL, Catalano RD, Lam BY, McFarlane I, Charnock-Jones DS, Smith GC. Developmental control of the Nlrp6 inflammasome and a substrate, IL-18, in mammalian intestine. Am J Physiol Gastrointest Liver Physiol. 2011;300:G253–263. doi: 10.1152/ajpgi.00397.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleessen B, Kroesen AJ, Buhr HJ, Blaut M. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand J Gastroenterol. 2002;37:1034–1041. doi: 10.1080/003655202320378220. [DOI] [PubMed] [Google Scholar]
- Kuehbacher T, Rehman A, Lepage P, Hellmig S, Folsch UR, Schreiber S, Ott SJ. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol. 2008;57:1569–1576. doi: 10.1099/jmm.0.47719-0. [DOI] [PubMed] [Google Scholar]
- Kufer TA, Sansonetti PJ. NLR functions beyond pathogen recognition. Nat Immunol. 2011;12:121–128. doi: 10.1038/ni.1985. [DOI] [PubMed] [Google Scholar]
- Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–344. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Microbes and Health Sackler Colloquium: Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucke K, Miehlke S, Jacobs E, Schuppler M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J Med Microbiol. 2006;55:617–624. doi: 10.1099/jmm.0.46198-0. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci U S A. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- Matzinger P. Friendly and dangerous signals: is the tissue in control? Nat Immunol. 2007;8:11–13. doi: 10.1038/ni0107-11. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- Ouverney CC, Armitage GC, Relman DA. Single-cell enumeration of an uncultivated TM7 subgroup in the human subgingival crevice. Appl Environ Microbiol. 2003;69:6294–6298. doi: 10.1128/AEM.69.10.6294-6298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J, Stobberingh EE, van den Brandt PA, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy. 2007;62:1223–1236. doi: 10.1111/j.1398-9995.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Siegmund B. Interleukin-18 in intestinal inflammation: friend and foe? Immunity. 2010;32:300–302. doi: 10.1016/j.immuni.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Siegmund B, Lehr HA, Fantuzzi G, Dinarello CA. IL-1 beta -converting enzyme (caspase-1) in intestinal inflammation. Proc Natl Acad Sci USA. 2001;98:13249–13254. doi: 10.1073/pnas.231473998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkorova Z, Capkova J, Niederlova J, Stepankova R, Sinkora J. Commensal intestinal bacterial strains trigger ankylosing enthesopathy of the ankle in inbred B10.BR (H-2(k)) male mice. Hum Immunol. 2008;69:845–850. doi: 10.1016/j.humimm.2008.08.296. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HH, Dwarakanath AD, Hart CA, Milton JD, Rhodes JM. Increased faecal mucin sulphatase activity in ulcerative colitis: a potential target for treatment. Gut. 1995;36:570–576. doi: 10.1136/gut.36.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaahtovuo J, Munukka E, Korkeamaki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol. 2008;35:1500–1505. [PubMed] [Google Scholar]
- Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Manji GA, Grenier JM, Al-Garawi A, Merriam S, Lora JM, Geddes BJ, Briskin M, DiStefano PS, Bertin J. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. J Biol Chem. 2002;277:29874–29880. doi: 10.1074/jbc.M203915200. [DOI] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werts C, le Bourhis L, Liu J, Magalhaes JG, Carneiro LA, Fritz JH, Stockinger S, Balloy V, Chignard M, Decker T, et al. Nod1 and Nod2 induce CCL5/RANTES through the NF-kappaB pathway. Eur J Immunol. 2007;37:2499–2508. doi: 10.1002/eji.200737069. [DOI] [PubMed] [Google Scholar]
- Wright DP, Rosendale DI, Robertson AM. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol Lett. 2000;190:73–79. doi: 10.1111/j.1574-6968.2000.tb09265.x. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.