Abstract
Ubiquitin-proteasome dependent protein degradation plays a fundamental role in the regulation of the eukaryotic cell cycle. Cell cycle transitions between different phases are tightly regulated to prevent uncontrolled cell proliferation, which is characteristic of cancer cells. To understand cell cycle phase specific regulation of the 26S proteasome and reveal the molecular mechanisms underlying the ubiquitin-proteasome degradation pathway during cell cycle progression, we have carried out comprehensive characterization of cell cycle phase specific proteasome interacting proteins (PIPs) by QTAX analysis of synchronized yeast cells. Our efforts have generated specific proteasome interaction networks for the G1, S, and M phases of the cell cycle and identified a total of 677 PIPs, 266 of which were not previously identified from unsynchronized cells. On the basis of the dynamic changes of their SILAC ratios across the three cell cycle phases, we have employed a profile vector-based clustering approach and identified 20 functionally significant groups of PIPs, 3 of which are enriched with cell cycle related functions. This work presents the first step toward understanding how dynamic proteasome interactions are involved in various cellular pathways during the cell cycle.
Keywords: Proteasome interaction network, QTAX, in vivo cross-linking, quantitative mass spectrometry, 26S proteasome, cell cycle, PIP
Introduction
The ubiquitin-proteasome system (UPS) represents the major pathway for regulated degradation of intracellular proteins in eukaryotes1–3 and helps control and integrate many essential physiological processes in cells including cell cycle progression, apoptosis, DNA repair, and chromosome maintenance. Disruption of the normal UPS has been implicated in the pathogenesis of a number of human diseases including neurodegenerative disorders and cancer.4–8 In recent years, it has been recognized that proteasomes and components of the UPS represent a class of attractive “drugable” targets for cancer treatments.9,10
The 26S proteasome is a protein complex consisting of at least 33 subunits11–13 and is responsible for degradation of polyubiquitinated substrates.1–3 It is composed of two sub-complexes: the 20S core particle (CP) and the 19S regulatory particle (RP).14 The 20S core particle harbors various proteolytic activities and is made up of two copies each of seven different α and seven different β subunits arranged into four stacked rings (α7β7β7α7). The 19S regulatory complex is composed of at least 19 different subunits, which can be further divided into two subcomplexes, the base and the lid.11,15 The base is located proximal to the 20S CP and contains six ATPases (Rpt1–6) plus four non-ATPase subunits (Rpn1, Rpn2, Rpn10, Rpn13). The lid is found distal to the base and contains nine non-ATPase subunits (Rpn3, Rpn5–9, Rpn11–12, Rpn15). The 19S RP is thought to carry out a number of biochemical functions including recognition of polyubiquitinated substrates, cleavage of the polyubiquitin chains to recycle ubiquitin, unfolding of substrates, and assisting in opening the gate of the 20S CP to allow entry of unfolded substrates into the catalytic chamber.2,14,16–18 However, the detailed functions of most 19S subunits and the subunits responsible for recognizing polyubiquitinated substrates still need to be further clarified.
Cell cycle transitions between different phases of the cell cycle are tightly regulated.19 This complex regulatory network is important to prevent uncontrolled cell proliferation, which is characteristic of cancer cells. Similarly, to maintain genome integrity, cells respond to genotoxic stress by inducing strictly controlled cell cycle checkpoint arrests.20 Ubiquitin-proteasome dependent protein degradation plays a fundamental role in the regulation of the eukaryotic cell cycle.19,21–26 The process is highly selective, is precisely timed, and allows an instant switch from one functional program to another. Cell cycle transitions are controlled by two major ubiquitination pathways that are mediated by two types of E3 ubiquitin ligases, SCF (Skp1-Cullin-F-box)27,28 and APC/C (Anaphase-promoting complex/cyclosome) complexes.22,25,29 These pathways trigger most ubiquitination steps during cell cycle checkpoints and the normal progression through the cell division cycle such as protein degradation required for the transition from G1 to S phase,21 sister chromatid separation in anaphase and the exit from mitosis.19,25 APC/C and SCF components have been studied in great detail, and there is a good understanding of how these ubiquitin ligases regulate ubiquitination of diverse cell cycle substrates. However, ubiquitination of cell cycle regulators is only the first step in their degradation, which requires recognition and processing of ubiquitinated cell cycle regulators by the proteasome. Thus, the importance of the 26S proteasome in cell cycle control is evident, but how the proteasome is regulated during the cell cycle and whether the mechanisms underlying substrate recognition and delivery are cell cycle dependent remains unknown.
A broad class of proteasome interacting proteins have been identified, including ubiquitin receptors, ubiquitin ligases, deubiquitinases, proteasome activators and inhibitors, and other types of modulators.11–13,17,30–39 These proteins associate with proteasomes dynamically in response to environmental changes and affect the proteasome function, regulation, assembly, stability, as well as modulation of substrate degradation processes. It is thus reasonable to assume that dynamic changes in proteasome interacting proteins may help govern the cell division cycle. We therefore performed a comprehensive investigation of cell cycle specific proteasome interaction networks.
To capture protein interactions of all natures in a single analysis, we have previously developed an integrated proteomic approach, QTAX, for quantitative analysis of tandem affinity purified in vivo cross-linked (X) protein complexes.38,39 In this approach, in vivo formaldehyde cross-linking was used to freeze protein interactions in the cell prior to lysis and purification, generating a snapshot of the protein interaction network. The cross-linked products were subsequently affinity purified with a “TAP” tag, the His-Bio (HB) tag40 to carry out tandem affinity purification under fully denaturing conditions. The purified protein complexes were then analyzed by SILAC-based mass spectrometry for identification and quantification of specific proteasome interacting proteins (PIPs).38,39 This method is effective for quantitatively characterizing interacting proteins, including weak and transient interactions, and therefore can provide an accurate view of the proteasome complex inside the cell.39 To generate a comprehensive cell cycle specific proteasome interaction network, we have employed the QTAX-based strategy coupled with protein interaction network analysis to analyze synchronized cells. Clustering analysis based on SILAC ratio profiles of PIPs was used to characterize their dynamic changes across G1, S, and M phases. This work presents the first extensive analysis of the proteasome interaction network at different cell cycle phases and the results provide new insights on the link between the proteasome and cell-cycle related signaling pathways.
Experimental Procedures
Chemicals and Reagents
ImmunoPure streptavidin, HRP conjugated antibody, and Super Signal West Pico chemiluminescent substrate were purchased from Pierce Biotechnology (Rockford, IL). Sequencing grade trypsin was from Promega Corp. (Madison, WI). 13C615N4-arginine and 13C615N2-lysine were from Cambridge Isotope Laboratories (Andover, MA). 12C614N4-arginine and 12C614N2-lysine were from Sigma. Anti-Rpt5, anti-Pre10 and PAP-HRP antibodies were purchased from Biomol, Inc. (Plymouth Meeting, PA). All other general chemicals for buffers and culture media were from Fisher Scientific or VWR International.
Yeast Strains and Conditions
Rpn11-HBH and wild type strains used in this study are isogenic to 15 DaubΔ, bar1Δura3Δns, a derivative of BF264–15D, and are auxotrophic for arginine and lysine.39 For validation experiments, a MATa FUS3-TAP strain was used (generous gift from Dr. Haoping Liu at UC, Irvine). Standard yeast growth conditions and media were used41 except in SILAC experiments as described.39
Cell Cycle Arrest and In Vivo Cross-linking
Wild type and RPN11-HBH cells were synchronized in three phases (G1, S, and M) before cross-linking and tandem affinity purification. The yeast strains were grown in 400 mL minimal media containing either 12C14N-Arg/Lys (light) or 13C15N-Arg/Lys (heavy) amino acids at 30 °C to A600 ≈ 0.5. Cells were then cultured in the presence of 200 ng/mL alpha factor (G1 phase arrest), or 15 μg/mL nocodazole (M phase arrest) for 2–3 h(1–1.5 doublings) until complete arrest in G1 phase (for alpha factor treatment) or M phase (for nocodazole treatment) was observed by visual confirmation of cell morphology. Cells were then cross-linked in vivo using formaldehyde and collected as previously described.38 For cell synchronization in S phase, cells were first prearrested in G1 by treatment with 100 ng/mL alpha factor. When uniform cell cycle arrest in G1 was confirmed by microscopic observation of the characteristic cell morphology, cells were washed with 2 culture volumes of medium to remove the alpha-factor and allow cells to re-enter the cell cycle. Cells were then resuspended in 1 culture volume of media and cell growth was monitored by microscopic observation. When the majority of cells had entered S phase, cells were cross-linked and collected as described above.
Analysis of Synchronized Cells by Flow Cytometry
Prior to in vivo cross-linking, 1 mL samples of synchronized cells at each phase were collected for analysis by flow cytometry.42 Briefly, cells were fixed in 70% ethanol and washed with H2O before treatment with RNase A solution (2 mg/mL Ribonuclease A, 50 mM Tris, 15 mM NaCl, pH 8.0) at 37 °C for 2 h or overnight. The RNase A solution was removed, and samples were incubated with 5 mg/mL pepsin/0.45% HCl for 30 min at 37 °C. The cells were washed with H2O and then resuspended in DNA stain (1 μM sytox green in 50 mM Tris pH 7.5) for at least 1 h before analysis. Cells were analyzed for DNA content with a FACScalibur instrument, and data analysis was performed with CellQuest software (BD Biosciences). To optimize the release time, S phase synchronization was monitored by collecting cells at 9 time points with 5 min intervals (5, 10, 15, 20, 25, 30, 35, 40, 45 min) after alpha factor was removed from the culture. Multiple validation experiments were carried out, and similar arrest efficiencies were obtained.
Gel Electrophoresis and Immunoblotting
Cell lysates and elution fractions were separated by SDS-PAGE (7 or 10% gel). Proteins were transferred to a PVDF membrane and analyzed by immunoblotting. The RGS-His6 antibody was used at a 1:2000 dilution (Qiagen). Rpt5 and Pre10 (MCP72) antibodies were used at 1:100 000 and 1:1000 dilutions respectively. TAP-tagged proteins were detected using peroxidase-antiperoxidase (PAP)-HRP conjugate at a 1:5000 dilution.
Tandem Affinity Purification and LC–MS/MS
Purification was carried out similarly as described39 with the following modifications: the lysis buffer contained 8 M urea, 300 mM NaCl, 50 mM NaH2PO4, 0.5% Igepal, 20 mM imidazole, and 1 mM PMSF, pH 8.5. Overnight binding to Ni-Sepharose beads was performed. Beads were then washed twice with 20 bed volumes of lysis buffer, four times with 20 bed volumes of lysis buffer with 10 mM imidazole at pH 6.3. The proteins were eluted from the Ni-Sepharose beads using 3 × 2 bed volumes of lysis buffer at pH 4.3 without imidazole. Streptavidin beads were washed stringently using 40 bed volumes of buffer 2 (8 M urea, 0.2 M NaCl, 2% SDS, 100 mM Tris, pH 7.5). Residual urea and SDS were removed using 40 bed volumes of 25 mM NH4HCO3.
After on-bead trypsin digestion, samples were separated by offline SCX as described,39 but with the following gradient: 0% to 5% B in 2 min, 5% to 30% B in 25 min, 30% to 100% B in 10 min (solvent A: 25% acetonitrile in 5 mM KH2PO4, pH 3; solvent B: solvent A with 350 mM KCl). Approximately 15–20 SCX fractions were collected, desalted using VivaPure C18 microspin columns (Vivascience) and analyzed by LC–MS/MS using LTQ-Orbitrap XL MS (ThermoElectron). The LC analysis was performed using a capillary column (100 μm ID × 150 mm long) packed with C18 resins (GL Sciences) and the peptides were eluted using a linear gradient of 2–35% B in 105 min; (solvent A, 100% H2O/0.1% formic acid; solvent B, 100% acetonitrile/0.1% formic acid). A cycle of one full FT scan mass spectrum (350–1800 m/z, resolution of 60,000 at m/z 400) was followed by ten data-dependent MS/MS acquired in the linear ion trap with normalized collision energy (setting of 35%). Target ions selected for MS/MS were dynamically excluded for 30 s.
Protein Identification and Quantification by Database Searching
Monoisotopic masses of parent ions and corresponding fragment ions, parent ion charge states and ion intensities from LC–MS/MS spectra were extracted using in-house software based on Raw_Extract script from Xcalibur v2.4. The data were searched using the Batch-Tag within the developmental version (v 5.0.1) of Protein Prospector against a decoy database consisting of a normal SGD yeast database concatenated with its reversed version. The mass accuracy for parent ions and fragment ions were set as ± 20 ppm and 0.5 Da, respectively. The proteins were identified by at least two peptides with a false-positive rate ≤0.5%. Determination of the mean SILAC ratio (i.e., Light(L)/Heavy(H)) and standard deviation for a PIP in each QTAX experiment was calculated and manually validated using Search Compare as described.39 The SILAC ratios were normalized to correct variation in mixing prior to purification. The standard deviations of SILAC ratios listed in Supplemental Table 1 (Supporting Information) were less than 35%. For any PIP found in both experimental repeats the average of the normalized SILAC ratio was used to eliminate experimental variations including cell cycle synchronization. Only proteins with validated SILAC ratios above 1.5 for all phases in which they were identified with less than 35% deviation between two biological replicates were considered for final analysis (Supplemental Table 1, Supporting Information).
Protein Interaction Network Analysis
The PIPs identified in each phase were analyzed against the physical yeast protein–protein interaction databases from the following sources: BIOGRID (44 923 interactions among 5347 proteins) (http://www.thebiogrid.org), Collins et al. (top 12 035 most-confident interactions among 1921 proteins using the interaction confidence threshold of 0.38),43 MIPS (7385 interactions among 4223 proteins) (http://mips.gsf.), SGD (44 812 interactions among 5269 proteins) (http://www.yeastgenome.org), and the union of CCSB-YI1, Ito-core and Uetz-screen (2705 interactions among 2018 proteins).44 The list of potential ubiquitinated substrates was extracted from proteomic profiling of global ubiquitination using mass spectrometry.40,45–49 Physical interactions were mapped and visualized using cytoscape version 2.6.3. (http://www.cytoscape.org).
Clustering Analysis of PIPs
In this study, a PIP could be present in a maximum of three phases (i.e., G1, S, and M), and might have up to three phase specific SILAC ratios. Therefore, each PIP could be represented by at most a 3-dimensional “SILAC ratio profile vector”, which was used to describe a PIP’s characteristic changes in SILAC ratios across three phases and as the basis for assigning a PIP to a cluster. If a PIP changed its SILAC ratio in 2 or higher fold between any two phases, this would be considered as a “significant” change. For proteins present in all three phases, 13 possible “profiles” in their SILAC ratio changes can be deduced. The schematic representation of possible profile changes is illustrated in Supplemental Figure 1, Supporting Information. With the profile vector-based approach, a finite number of possible clusters were obtained in this work.
For each cluster, the enrichment of biological functions, protein complexes, and pathways was measured. To evaluate the enrichments, the hit-rate was defined as the percentage of proteins in a cluster out of the total number of annotated proteins in the cluster that share a common function, complex, or pathway. Biological function and protein complex annotations were downloaded from MIPS database (ftp://ftpmips.gsf.de/yeast/), and pathway memberships were downloaded from the KEGG database (http://www.genome.jp/kegg/). The statistical significance of observing a hit-rate was computed as the probability P that the cluster was enriched with the given hit-rate purely by chance. This was done by applying a commonly used model of hypergeometric distribution and by considering the total number of annotated proteins in a data set (N), the number of annotated proteins in a given cluster (n), the number of proteins out of n proteins in the cluster that are annotated with the function/complex/pathway of interest (k), and the number of proteins out of N proteins that are annotated with the function/complex/pathway of interest (K).50,51
All enrichments with p-values higher than 0.1 were discarded from any further analysis.
Validation of the Selected PIP
Reciprocal coimmunoprecipitation and immunoblot analysis using a FUS3-TAP strain was carried out similarly as described38,39 except that cells were treated and synchronized as described above. Co-purification of proteasome subunits were confirmed by specific antibodies of a 19S subunit (Rpt5) and a 20S subunit (Pre10).
Results
1. Cell Synchronization
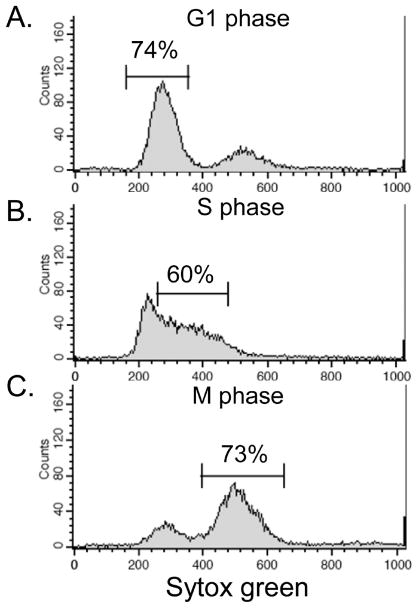
To map the cell cycle specific proteasome interaction network, we first synchronized yeast cells at 3 different phases in the cell cycle: G1, S, and M phases. G1 cells were arrested with the mating pheromone (alpha factor), S phase cells were collected from a synchronized population that was released from pheromone arrest, and nocodazole arrested cells served as the M phase sample. Various concentrations of alpha factor were tested and optimized for obtaining consistent cell synchronization at G1 and S phases, respectively. Release from G1 arrest was monitored over time to warrant collecting cells at peak entrance into S phase, typically around 35 min after release. Cell synchrony was verified by examining cell cycle phase specific morphological changes in the yeast cells and by analyzing DNA content using flow cytometry. Flow cytometry showed the following cell synchrony: 74.4% G1 phase arrest, 60.1% S phase, and 73.2% M phase arrest (Figure 1).
Figure 1.
Cell synchrony verified by flow cytometry. Following cell cycle synchronization, cells were fixed with ethanol and prepared for FACS analysis. Synchronization efficiency: (A) G1 arrest, 74.38%; (B) S phase population, 60.06%; and (C) M arrest, 73.15%.
2. QTAX-Based Purification and Identification of Cell Cycle Specific PIPs
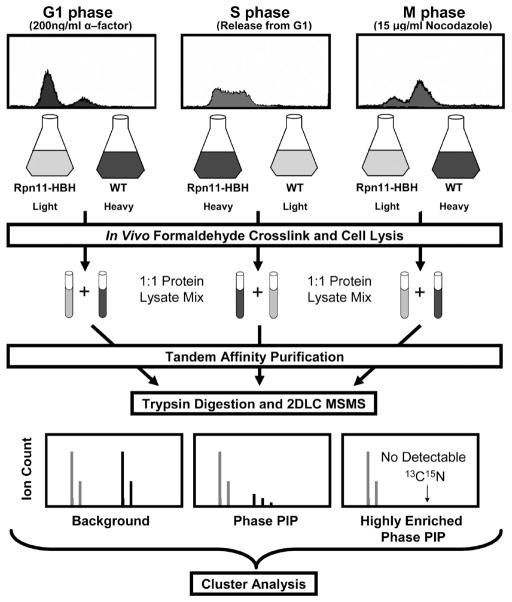
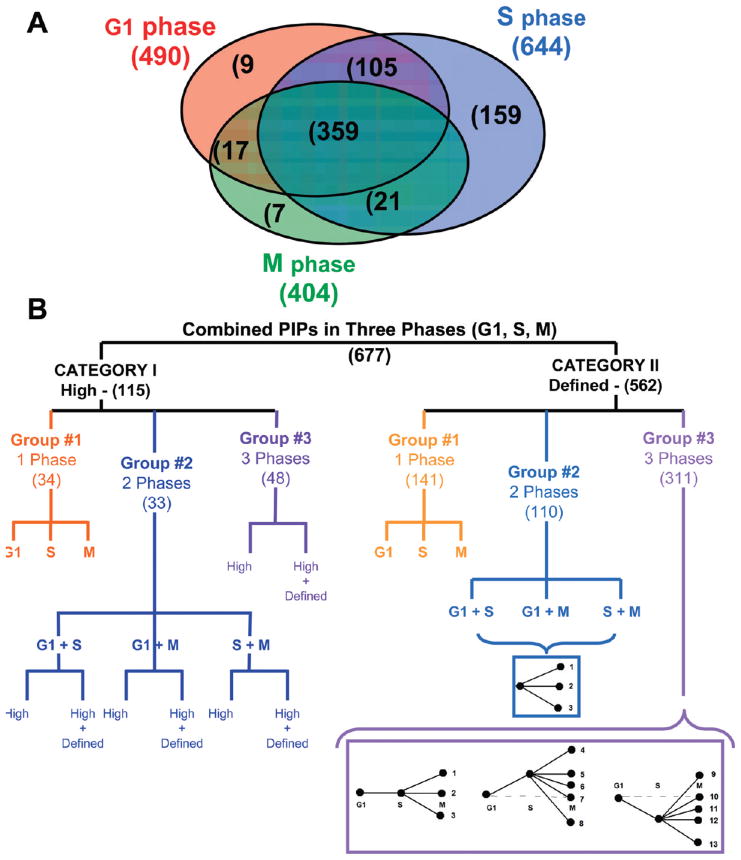
The QTAX method has been employed to identify PIPs that are enriched at a specific cell cycle phase (G1, S or M). In this work, we used a yeast strain expressing HBH-tagged Rpn11 from the endogenous locus to purify PIPs from cells synchronized at different cell cycle phases. We chose Rpn11-HBH as our bait for purification because we previously found that Rpn11-HBH allows the capture of the largest number of PIPs in unsynchronized cells.39 To identify specific PIPs in the different cell cycle phases we first analyzed SILAC-labeled samples from identically synchronized Rpn11-HBH and untagged wild type (wt) cells as outlined in Figure 2. After HB-tag based tandem affinity purification, the isolated in vivo cross-linked protein complexes were subjected to trypsin digestion and analyzed by LC–MS/MS using LTQ-Orbitrap XL MS.52 These analyses resulted in lists of proteasome interacting proteins in G1, S, and M phases of the cell cycle. Comparison of the resultant lists allowed for the identification of cell cycle specific interactors. Combining the results from all of the experiments collected for the three cell cycle phases (G1, S, and M), 677 putative PIPs were identified with manually validated and reproducible SILAC ratios >1.5 as summarized in Supplemental Table 1, Supporting Information. As shown in Figure 3A, 359 PIPs are present in all three phases, 143 in any 2 phases and 175 in only one phase.
Figure 2.
QTAX strategy for quantifying cell-cycle specific proteasome interacting proteins (PIPs) in yeast. Specific PIPs were identified by comparing samples from identically synchronized RPN11-HBH and wt (untagged) yeast cells. A total of three cell cycle synchronization treatments were analyzed: alpha factor induced G1 phase arrest; synchronized release into S phase from alpha factor arrest; nocodazole induced M phase arrest. Light, 12C14N-Arg/Lys; Heavy, 13C15N-Arg/Lys.
Figure 3.
Classification of PIPs. (A) Venn diagram of PIPs identified in G1, S and M phases. (B) Clustering schemes of PIPs in Category I and Category II.
In comparison to our results from unsynchronized cells, 97% of PIPs previously captured using Rpn11-HBH as the bait,38,39 and 93% of the PIPs previously identified by the Tag-team based QTAX strategy using multiple different HBH-tagged proteasome subunits39 were present among the putative PIPs obtained from synchronized cells in this work. This reproducibility reflects the high robustness of the QTAX-based experimental approaches. In addition, 266 PIPs were only identified in this work, most likely due to cell synchronization and better MS instrumentation used for LC–MS/MS analysis (LTQ-Orbitrap XL MS vs QSTAR XL MS).
3. Protein Interaction Network Analysis of the Identified PIPs
Protein interaction network analysis was performed to validate the identified PIPs and understand the biological relevance of their interactions with proteasomes. Only known physical interactions between identified PIPs and proteasome subunits and those among the identified PIPs were extracted from multiple public databases and literature references43,44 for our network analysis. The graphic illustrations of G1, S, and M phase specific proteasome interaction networks based on known protein interactions are displayed in Supplemental Figure 2, Supporting Information. Inclusive of proteasome complex subunits, a total of 2901 interactions among 433 proteins are found in G1 phase, 3918 interactions among 576 proteins in S phase, and 2516 interactions among 354 proteins in M phase. In addition, 80% of the identified PIPs from G1, 82% PIPs from S, and 75% PIPs from M phase have been found to be connected to the proteasome complex either directly or indirectly, supporting the validity of our results. PIPs per each cell cycle phase that did not map onto the proteasome interaction network were considered as putative novel inter-actors including 99 of 490 (G1), 112 of 644 (S), and 98 of 404 (M) PIPs. In comparison to the novel PIPs identified previously in unsynchronized yeast cells,39 83 novel interactors were identified in this work.
4. Functional Categorization and Clustering Analysis of the Identified PIPs
To categorize the identified PIPs and explore their cell cycle dependent dynamic changes, the PIPs were clustered based on their SILAC ratio profiles across the cell cycle phases. We have previously shown that SILAC ratios obtained from QTAX experiments can distinguish between classes of PIPs, with “high” ratio PIPs being either proteasome components or specifically interacting proteins, while “defined” ratio PIPs are less specifically interacting proteins.39 In each phase (i.e., G1, S or M), the identified PIPs were thus classified into two categories according to the characteristics of their SILAC ratios: (I) “high” ratio PIPs, and (II) defined ratio PIPs. In this study, 115 PIPs had high SILAC ratios in at least one phase, while 562 PIPs had defined SILAC ratios in all phases in which they were detected (Figure 3B). Additionally, each identified PIP has at most three cell cycle phase specific SILAC ratios, which can be used for further classification. We have clustered the PIPs based on the similarity in their SILAC ratio profiles across cell cycle phases. By nature, the SILAC ratios for PIPs in Category I and II are not equivalent and therefore we have clustered them differently as shown in Figure 3B and explained below.
4a. Clustering Analysis of the “high” Ratio PIPs in Category I
The PIPs in Category I have “high” SILAC ratios in at least one phase. High SILAC ratios cannot be calculated due to lack of detection for the corresponding heavy peak in the MS spectra. These proteins were only purified from the tagged sample, similar to the proteasome subunits. These PIPs appear to have more direct interactions with the proteasome and are likely key components of the ubiquitin-proteasome degradation pathway such as ubiquitin receptors, regulators or new proteasome subunits.39 To cluster the 115 PIPs in Category I, we first divided them into three groups based on their presence in only one phase, i.e. G1, S, or M (Group #1); in two phases, that is, G1_S, G1_M, or S_M (Group #2); or in all three phases, that is, G1_S_M (Group #3) (Figure 3B). As shown in Supplemental Table 2 (Supporting Information), 34 PIPs were detected only in one phase, 33 PIPs in two phases, and 48 PIPs were in three phases. In each group, the PIPs were further separated into clusters: (1) PIPs with only high ratios in all of the phases in which they were detected; (2) PIPs with mixed SILAC ratios at different phases, meaning they have at least one high and one defined SILAC ratio (see Figure 3B). Therefore, the PIPs in each group were split into “only high” or ‘high and defined’ clusters, totaling 17 clusters (Supplemental Table 2, Supporting Information).
To understand the biological relevance of the identified PIPs, we have further analyzed each cluster to identify any enrichment in biological functions, protein complexes and biological pathways. This information can help isolate cellular processes that proteasomes may be associated with in a cell cycle dependent manner, thus allowing identification of biologically important protein interactions. To this end, the annotated functions, protein complexes and pathways of the identified PIPs were extracted from public protein databases. For each cluster, enrichments were assessed by the hit-rate as well as the statistical significance of observing the hit-rate (p-value).50,51 At least one statistically significantly enriched function, complex, or pathway was found in 8 high ratio clusters as summarized in Table 1.
Table 1.
List of Category I (“High”) PIP Clusters with Functional, Complex, and Pathway Enrichments
| subgroupa | cluster ID | cluster sizeb | enrichment type | enrichment description | hit-rate (%) | p-value (10−2)c |
|---|---|---|---|---|---|---|
| #1 | [S] | 30 | Function | Metabolism | 61.5 | 5.1 |
| #2 | [G1_S]all_high | 13 | Complex | Transcription complexes/Transcriptosome | 75.0 | 3.8 |
| Function | Transcription | 40.0 | 5.4 | |||
| Pathway | Basal Transcription factors | 33.3 | 5.1 | |||
| [G1_S]h_dd | 8 | Function | Metabolism | 75.0 | 8.7 | |
| #3 | [G1_S_M]h_d_dd | 7 | Function | Metabolism | 83.3 | 6.8 |
| [G1_S_M]h_d_hd | 7 | Function | Regulation of metabolism and protein function | 42.9 | 3.1 | |
| [G1_S_M)]h_h_dd | 5 | Function | Cell rescue, defense and virulence | 80.0 | 1.3 | |
| Protein synthesis | 60.0 | 1.6 | ||||
| [G1_S_M]d_d_hd | 13 | Complex | Fatty acid synthetase, cytoplasmic | 25.0 | 7.4 | |
| Phenylalaninine-tRNA-ligase | 25.0 | 7.4 | ||||
| Function | Protein synthesis | 30.8 | 7.0 | |||
| Pathway | Fatty acid biosynthesis | 28.6 | 2.6 | |||
| Phenylalanine, tyrosine and tryptophan biosynthesis | 28.6 | 2.6 | ||||
| Aminoacyl-tRNA biosynthesis | 28.6 | 2.6 | ||||
| [G1_S_M]all_high | 12 | Function | Cell cycle and DNA processing | 58.3 | 0.9 | |
| Pathway | Cell cycle-yeast | 40.0 | 6.6 |
Subgroups categorize PIPs based on their presence in one phase (#1), two phases (#2), or three phases (#3).
Cluster Size indicates the number of PIPs present in each cluster; Only clusters with 4 or more PIPs were considered for enrichment analysis.
Forty-seven percent of cluster enrichments have p-values < 0.05; 47% have p-values between 0.05 and 0.075; 5.9% have p-values between 0.075 and 0.1.
h = High and d = Defined for the respective phases, that is, [G1_S_M]h_h_d = G1 (high); S (high); M (defined).
Since the ubiquitin-proteasome degradation pathway is highly active during cell cycle progression,25,53–55 we suspect that some of the identified PIPs may be ubiquitinated substrates. Therefore, we have attempted to identify potential ubiquitinated substrates among the identified PIPs in this work based on the list of candidate ubiquitinated substrates previously determined from global ubiquitination profiling experiments using affinity purification and mass spectrometry.38,46–49,56 67 PIPs from Category I were found to be potential ubiquitinated substrates, among which, 16 are present in only one phase, 17 in two phase, and 34 in three phase clusters. However, this analysis cannot distinguish mono- and poly ubiquitinated substrates, and many regulators of the UPS are themselves ubiquitinated, including proteasome subunits and ubiquitin receptors.38,45–49 Therefore, it is unclear whether the potential ubiquitinated proteins identified as PIPs in our study are substrates trapped at the proteasome or are regulators or modulators of protein degradation. However, it is worth noting that only K48-linked polyubiquitin chains were detected in our analysis, suggesting that protein ubiquitination captured in our preparations are most likely targeted for protein degradation.
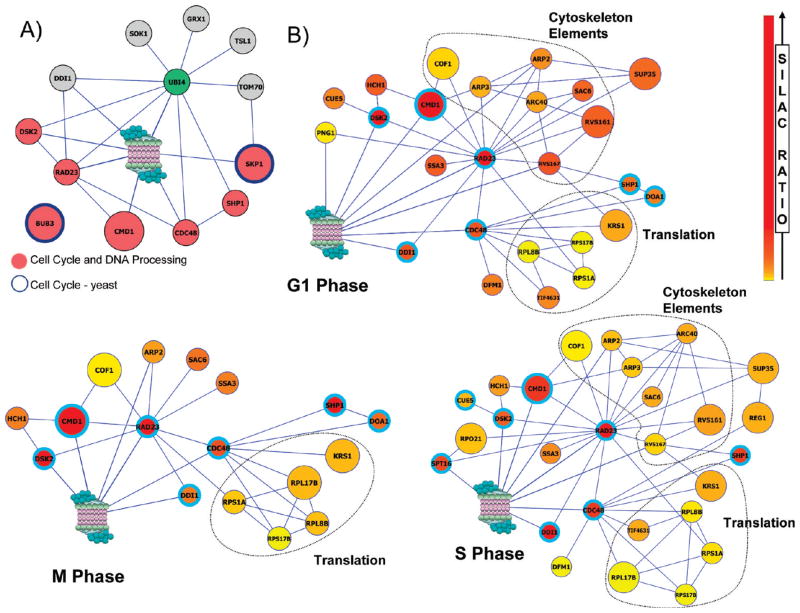
Due to a high number of potential ubiquitinated substrates present in the identified PIPs, ubiquitin was also included for generating proteasome interaction maps with PIPs in each cluster. As an example, Figure 4A displays the proteasome interaction map of the 12 PIPs in the high cluster [G1_S_M]-all_high, in which all of the PIPs have high ratios in all three phases. In this cluster, all PIPs but one have been mapped to the interaction network and 50% of the PIPs have direct interactions with the proteasome. In comparison to global ubiquitination profiling data,38,45–49 9 out of 12 PIPs have been found as potential ubiquitinated substrates. The isolated node is Bub3, which has been shown to be a specific PIP in unsynchronized cells from our previous report.39 In addition, 58% of the PIPs in this cluster have known functions in cell cycle and DNA processing. Interestingly, this cluster contains all of the known ubiquitin receptors, Rad23, Dsk2 and Ddi1, as well as Cdc48, a shuttling factor responsible for delivering misfolded proteins for proteasomal degradation.17 To explore whether these ubiquitin receptors share similar interactions among identified PIPs, we have mapped the direct physical interactions of Rad23, Dsk2, Ddi1 and Cdc48 with proteasome subunits, ubiquitin, and all of the PIPs (Category I and II) identified in G1, S and M, respectively (Figure 4B). As illustrated, Rad23 interacted directly with 15 PIPs in G1, 16 PIPs in S, and 10 PIPs in M phase, while Cdc48 interacted directly with 8 PIPs in G1, 9 PIPs in S and 7 PIPs in M phase. In comparison, Dsk2 and Ddi1 have very few direct physical interactors among the identified PIPs (Figure 4). Several cytoskeleton elements and regulators interact with Rad23, and Rad23 interactions displayed a more or less cell cycle dependent interaction profile, whereas Cdc48 interactors are enriched for proteins involved in translation and show constant interaction with the proteasome throughout the cell cycle (Figure 4B). This implies that Rad23 may play an important role in regulating cell cycle progression. Additionally, we have compared this set of PIPs to the candidate ubiquitinated substrates identified in ubiquitin affinity purification experiments.38,45–49 Interestingly, the percentage of candidate ubiquitinated substrate PIPs that interact with the Ub-receptors changes throughout the cell cycle; 76.2% of the PIPs that directly interact with Ub-receptor proteins in G1 phase are candidate substrates, compared to 68.1% in S phase, and only 36.8% in M phase. Together, this suggests that proteins in this cluster are more likely to be key components of proteasomal degradation pathway and may be involved in controlling cell cycle progression.
Figure 4.
Protein Interaction analysis of the [G1_S_M]all_high cluster. (A) Proteasome interaction map with PIPs in the [G1_S_M]all_high cluster. This cluster is enriched for cell cycle and DNA processing functions (pink nodes), and yeast cell cycle pathways (dark blue borders). (B) Cell cycle phase specific protein interaction map of ubiquitin receptors with their direct interactions and the proteasome. Rad23 interacts directly with a significant portion of cytoskeleton elements during G1 and S phase, but not during M phase, whereas Cdc48 interacts with many translation proteins, which are present in all three phases. Heat map indicates SILAC ratio intensity. Bright blue border indicates “highly enriched” status. Large nodes denote noncandidate ubiquitin substrates. Smaller nodes indicate PIPs as candidate ubiquitin substrates. This is based on global uniquitination profiling reported in literatures.
4b. Clustering Analysis of the PIPs in Category II
We first divided the 562 Category II PIPs with defined SILAC ratios into three groups based on their presence in one, two or three phases for clustering analysis similarly as described above for the Category I PIPs (Figure 3B). 141 PIPs were found in group #1, which were assigned to one of the three one-phase-only clusters: G1 (7 PIPs), S (129 PIPs), or M (5 PIPs) clusters (Supplemental Table 2, Supporting Information). Group #2 and #3 PIPs were further divided based on how their SILAC ratios changed between any of the two phases. To this end, 2-dimensional and 3-dimensional profile vectors were configured to cluster PIPs in groups #2 and #3 respectively (Figure 3B and Supplemental Figure 1, Supporting Information). A SILAC ratio change (increase or decrease) of 2 or higher fold of a PIP between two phases was considered as a significant change. Using this analysis, 8 of 9 possible clusters for 110 PIPs in group #2 and 8 of 13 possible clusters for 311 PIPs in group #3 were generated (Supplemental Table 2, Supporting Information). This analysis led to the formation of 19 defined ratio PIP clusters as summarized in Supplemental Table 2 (Supporting Information). Figure 5 displays the graphical representation of six selected clusters. As shown, each cluster describes a group of PIPs that have similar dynamic changes in their interactions with proteasome complexes during the cell cycle and each cluster has distinct and characteristic changes in the PIPs’ SILAC ratio profiles. To understand the biological relevance of these interactions and recognize novel connections between the proteasome and specific biological processes and protein complexes, we have identified biological functions, protein complexes, and pathways that are statistically significantly enriched as described above. As shown in Table 2, we have found at least one statistically significantly enriched function, complex, or pathway in 12 out of 19 defined ratio clusters.
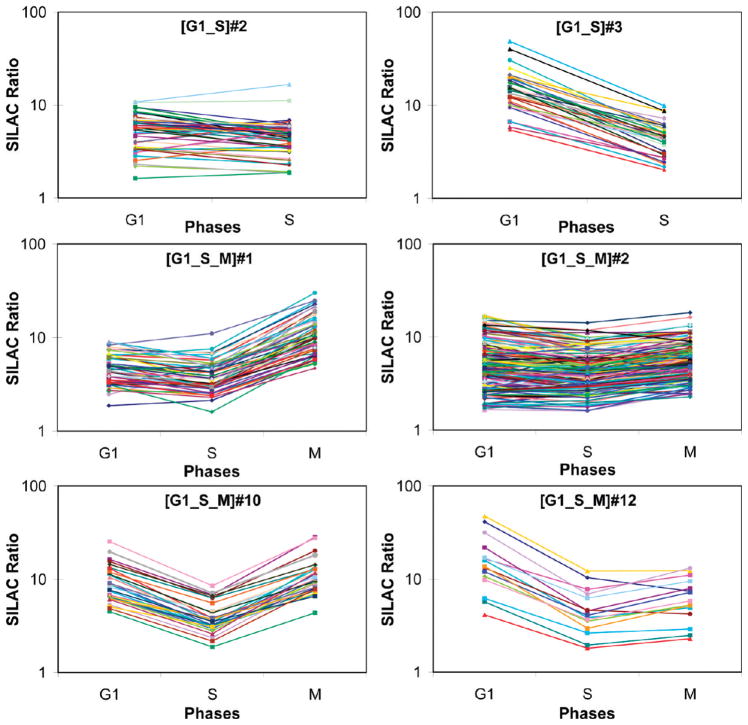
Figure 5.
SILAC ratio profiles of six representative defined ratio clusters.
Table 2.
List of Categroy II (defined) PIP Clusters with Functional, Complex, and Pathway Enrichments
| subgroupa | cluster ID | cluster sizeb | enrichment type | enrichment description | hit-rate (%) | p-value (10−2)c |
|---|---|---|---|---|---|---|
| #1 | [S] | 129 | Complex | Respiration chain complexes | 12.5 | 2.1 |
| RNA processing complexes | 12.5 | 1.0 | ||||
| Function | Protein with binding function or cofactor requirement (structural or catalytic) | 28.8 | 9.9 | |||
| Transcription | 22.9 | 0.0 | ||||
| Biogenesis of cellular components | 16.9 | 4.1 | ||||
| Pathway | Purine metabolism | 23.7 | 1.0 | |||
| Citrate cycle (TCA cycle) | 10.5 | 1.8 | ||||
| Urea cycle and metabolism of amino groups | 10.5 | 4.3 | ||||
| [M] | 5 | Function | Cellular transport, transport facilities and transport routes | 60.0 | 4.6 | |
| #2 | [G1_S]#2 | 46 | Complex | Chaperonine containing T-complex TRiC (TCP RING Complex) | 11.8 | 4.7 |
| Function | Metabolism | 59.1 | 2.5 | |||
| Biogenesis of cellular components | 20.5 | 6.2 | ||||
| Cell type differentiation | 15.9 | 2.7 | ||||
| Pathway | Glutamate metabolism | 20.0 | 0.1 | |||
| Pyrimidine metabolism | 16.0 | 3.5 | ||||
| Histidine metabolism | 12.0 | 2.1 | ||||
| Phenylalanine, tyrosine and tryptophan biosynthesis | 12.0 | 4.5 | ||||
| [G1_S]#3 | 33 | Complex | Intracellular transport complexes | 57.1 | 0.1 | |
| Function | Cellular transport, transport facilities and transport routes | 30.0 | 8.2 | |||
| Cell cycle and DNA processing | 20.0 | 5.8 | ||||
| Interaction with the environment | 16.7 | 3.7 | ||||
| Regulation of metabolism and protein function | 10.0 | 6.2 | ||||
| Cellular communication/signal transduction mechanism | 10.0 | 8.2 | ||||
| [G1_M]#1 | 4 | Function | Cellular transport, transport facilities and transport routes | 75.0 | 2.1 | |
| Biogenesis of cellular components | 50.0 | 7.1 | ||||
| [G1_M]#2 | 8 | Function | Metabolism | 75.0 | 7.7 | |
| Biogenesis of cellular components | 37.5 | 5.8 | ||||
| Cell fate | 25.0 | 8.5 | ||||
| Pathway | Fatty acid metabolism | 40.0 | 0.6 | |||
| [S_M]#2 | 9 | Function | Cell rescue, defense and virulence | 33.3 | 8.2 | |
| #3 | [G1_S_M]#1 | 50 | Function | Cellular transport, transport facilities and transport routes | 28.6 | 4.9 |
| Pathway | Valine, leucine and isoleucine degradation | 9.7 | 3.9 | |||
| Butanoate metabolism | 9.7 | 5.7 | ||||
| Synthesis and degradation of ketone bodies | 6.5 | 1.4 | ||||
| Lysine degradation | 6.5 | 7.1 | ||||
| [G1_S_M]#2 | 207 | Complex | Translation complexes | 67.2 | 0.0 | |
| Function | protein synthesis | 29.4 | 0.2 | |||
| Transposable elements, viral and plasmid proteins | 22.1 | 0.0 | ||||
| Pathway | Ribosome | 34.0 | 0.0 | |||
| Glycolysis/Gluconeogenesis | 15.1 | 2.0 | ||||
| Alanine and aspartate metabolism | 6.6 | 5.7 | ||||
| Galactose metabolism | 5.7 | 5.2 | ||||
| Drug metabolism-other enzymes | 4.7 | 1.1 | ||||
| Pantothenate and CoA biosynthesis | 2.8 | 6.7 | ||||
| [G1_S_M]#3 | 5 | Function | Transcription | 40.0 | 9.5 | |
| Regulation of metabolism and protein function | 40.0 | 0.9 | ||||
| [G1_S_M]#10 | 30 | Complex | Cytoskeleton | 33.3 | 4.3 | |
| Function | Metabolism | 67.9 | 0.8 | |||
| Protein with binding function or cofactor requirement (structural or catalytic) | 39.3 | 4.6 | ||||
| Cell rescue, defense and virulence | 32.1 | 0.3 | ||||
| Pathway | Biosynthesis of steroids | 11.8 | 5.2 | |||
| [G1_S_M]#12 | 15 | Function | protein synthesis | 53.3 | 0.8 | |
| Pathway | Methionine metabolism | 25.0 | 2.1 | |||
| Aminoacyl-tRNA biosynthesis | 25.0 | 9.9 |
Subgroups categorize PIPs based on their presence in one phase (#1), two phases (#2), or three phases (#3).
Cluster Size indicates the number of PIPs present in each cluster; Only clusters with 4 or more PIPs were considered for enrichment analysis.
Sixty-four and eight-tenths percent of cluster enrichments have p-values <0.05; 20.4% have p-values between 0.05 and 0.075; 14.8% have p-values between 0.075 and 0.1.
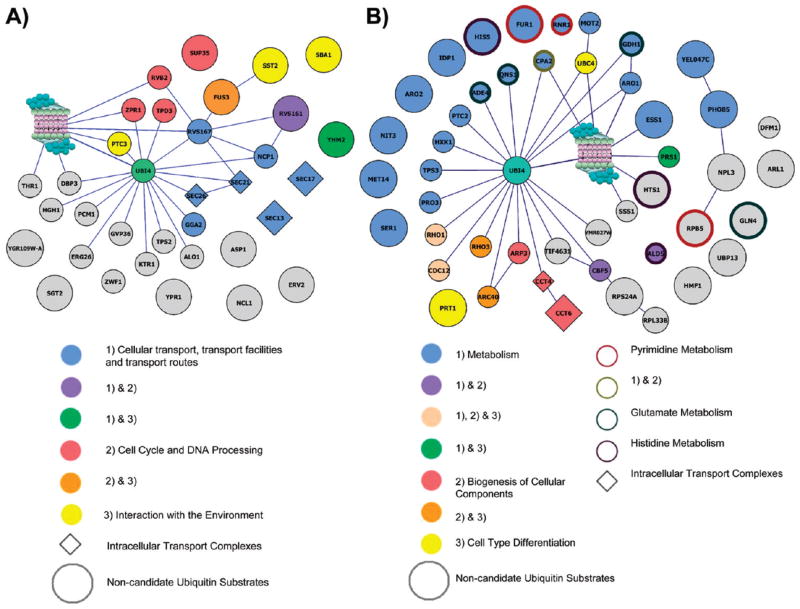
Figure 6A shows the proteasome interaction map with the 33 PIPs from cluster [G1_S]#3, in which interactions were only captured at G1 and S phases. Among them, 21 PIPs have been mapped to the proteasome interaction network derived from the known interaction databases and 6 PIPs were considered as novel interactors that have not been reported before.39 In comparison with global ubiquitination profiling results,38,45–49 nearly 60% of PIPs in cluster [G1_S]#3 had been previously identified as potential ubiquitinated substrates (indicated by small nodes or diamonds in Figure 6A), including all of the PIPs that map to the proteasome exclusively through ubiquitin. Among the candidate ubiquitinated substrates are Gvp36 and Rvs167, which both have confirmed ubiquitination sites.40,47 Importantly, this cluster is enriched with protein functions pertaining to cell cycle regulation (Table 2). One of the PIPs from this cluster, Fus3, is a mitogen activated serine/threonine kinase (i.e., MAP kinase) and directly links receptor activation to the control of cell proliferation.57 Fus3 is the down stream effector of the pheromone signaling pathway and triggers cell cycle arrest in G1 phase. Besides Fus3, two of its substrates Rvs167 (and interacting protein Rvs161) and Sst2, as well as Fus3 interacting protein Tpd358,59 were enriched in this cluster (Figure 6A). Given that these PIPs are involved in pheromone response and that they interact with the proteasome in a G1 and S phase specific manner, these data suggest that the proteasome plays a major role in the MAP kinase signaling pathways. These interesting findings provide a strong basis for future studies to understand whether and how these two pathways crosstalk with proteasomal degradation pathway.
Figure 6.
Protein interaction maps of the defined ratio clusters, (A) [G1_S]#3 and (B) [G1_S]#2. Node color indicates function, border color indicates protein complex, node shape indicates pathway of a particular node (see inset for details). Smaller nodes represent candidate ubiquitin substrates. Gray nodes indicate the PIP has no known annotated function in the protein interaction databases.
Another cluster with an enriched function in cell type differentiation is the defined ratio cluster [G1_S]#2, which contains 46 PIPs with 27 mapping to the proteasome interaction network and 19 isolated nodes (Figure 6B). This cluster represents a group of PIPs enriched for 1 protein complex, 3 functions and 4 biological pathways, which are indicated in Figure 6B. Interestingly, the ubiquitin-conjugating enzyme (E2) Ubc4 is grouped to this cluster. Notably, 56% of PIPs in cluster [G1_S]#2 were previously identified as potential ubiquitinated candidates with Gdh1 having a confirmed ubiquitination site.40,47 Although defined ratio clusters [G1_S]#2 and [G1_S]#3 have PIPs only present in G1 and S phases and also have similar enriched functions, the dynamic changes of PIP interactions with the proteasome appear to be different. As shown in Figure 5, the PIPs in cluster [G1_S]#2 have no significant change of their SILAC ratios between G1 and S phases, whereas the PIPs in cluster [G1_S]#3 are 2-fold or more abundant in G1 phase than that in S phase.
5. Validation of Interacting Protein Fus3 by Coimmuno-precipitation and Immunoblotting
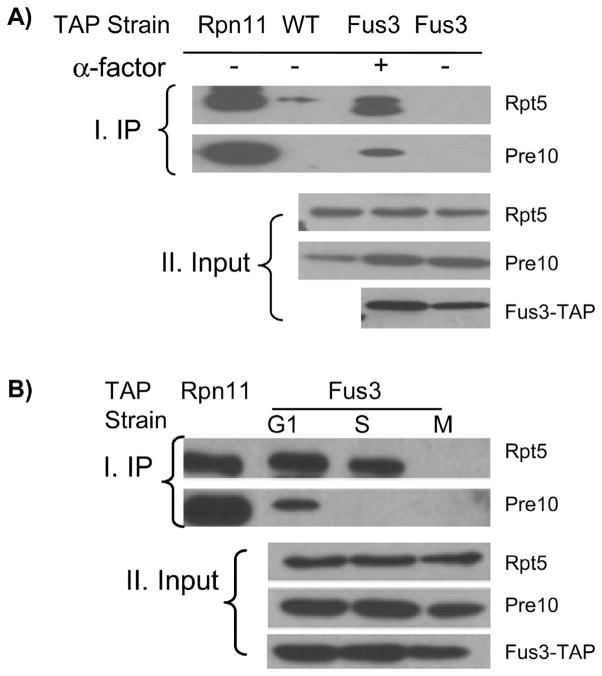
Based on our clustering analysis, the defined cluster [G1_S]#3 is highly interesting due to the enriched functions in cell cycle progression. Therefore we selected a PIP from this cluster, Fus3, for further validation. Our MS analysis has shown that Fus3 is a specific interactor of the proteasome when compared with the untagged control, and that Fus3 interacts with the proteasomes only in G1 and S phases, but not in M phase. To confirm the cell cycle specific interaction of Fus3 with the proteasome, we carried out reciprocal coimmunoprecipitation and immunoblotting using a yeast strain expressing TAP-tagged Fus3 (Figure 7). We first confirmed that Fus3 specifically interacts with the proteasome and this interaction is dependent on alpha factor treatment (Figure 7A). It is also noted that Fus3 interaction with the proteasome was detectable even at 30 min after alpha factor treatment (data not shown). To further verify whether its interaction with the proteasome is specific to cell cycle phases, we synchronized the FUS3-TAP strain at G1, S, and M phase prior to immunoprecipitation. Although Fus3 is expressed at all phases, it is evident that Fus3 interaction with the proteasome only occurs in G1 and S phases, but not in M phase (Figure 7B). In addition, consistent with the MS results, the interaction is strongest in the G1 phase. Fus3 copurified with both 19S and 20S subunits in G1 phase however in S-phase we were only able to detect interaction with the 19S subunit. Since Fus3 displays decreased interaction with the proteasome during S phase, loss of copurification with Pre10 could be due to the limited sensitivity of Pre10 antibody (Figure 7B). Previous studies exploring genetic interactions have shown that FUS3 interacts with two 19S proteasome subunits, RPN6 and SEM1.43,60 Our results provided physical support for these genetic data by demonstrating that Fus3, a key regulator in pheromone signaling, physically interacts with the proteasome in a cell-cycle dependent manner.
Figure 7.
Validation of Fus3 interaction with the proteasome using coimmunoprecipitation. Affinity purification of Fus3 interacting proteins using Fus3-TAP (A) in unsynchronized cells (no treatment) and in cells treated with 200 ng/mL alpha factor for 3 h; (B) from synchronized cells at G1, S, and M phases. Rpn11-TAP yeast cells served as a positive control and wt cells as a negative control. Proteasome subunits Rpt5 (19S) and Pre10 (20S) were probed with their specific antibodies.
Discussion
In this study we have investigated the cell cycle specific interaction networks of the yeast 26S proteasome by applying the QTAX strategy38,39 to the analyses of samples from synchronized cell populations. A total of 677 PIPs have been captured, identified and quantified from three cell cycle phases (G1, S, and M). In comparison to the results from unsynchronized cells,38,39 266 additional proteins were identified. To further understand the identified proteasome interactions, we have clustered the PIPs by the characteristics of their cell cycle phase dependent SILAC ratios using a profile vector-based clustering approach.61–63 This method separates PIPs based on a set of constraints that we have defined rather than by using the known heuristic clustering algorithms such as hierarchical and k-means methods.64,65 Although both the hierarchical and k-means clustering methods produced PIP clusters with assorted enrichments, the profile vector-based method produced more mid to high density clusters and had significantly more enrichments, suggesting that more biological relevant information was extracted by this approach. In total, we have identified 36 function, 9 complex, and 26 pathway enrichments from 20 total clusters, including notable functional enrichments in signal transduction and cell fate as well as complexes involved in intracellular transport and transcription. Among the 20 clusters, 3 different PIP clusters were found to be enriched with biological functions and pathways pertaining to cell cycle (Tables 1 and 2).
In this study, it is noted that the synchronization efficiency for S phase cells was limited. This is due to the fact that we had to release the cells from G1 phase arrest to obtain the S phase samples since the common chemical to induce S phase arrest, that is, hydroxyurea, can not be used due to its interference with formaldehyde cross-linking in QTAX experiments. In contrast to S phase arrest using cell cycle checkpoint induction by hydroxyurea, the arrest-release strategy allows for obtaining an unperturbed S phase population. However, the arrest-release strategy has limitations as it is necessary to compromise between complete release from G1 and progression of cells too far into S-phase or G2. Despite the limited cell synchronization in S phase, our results remain valid since we have clustered proteins based on changes in their interactions with proteasome among three different phases. Since all samples were subjected to the same synchronization, proteins with similar interaction changes should still be grouped/clustered together based on their SILAC ratio profiles. Although better synchronization might increase the ratio changes of a protein between two phases, proteins interacting similarly with proteasomes at three different phases would still have similar SILAC ratio profiles and thus allow them being clustered together.
In this work, our analysis identified clusters enriched with a total of 71 functions/complexes/pathways (i.e., 36 function, 9 complex, and 26 pathway enrichments) that have p-values below 0.1 (Tables 1 and 2). Among these enrichments, 43 (60.5%) have p-values lower than 0.05, 19 (26.8%) have p-values in the 0.05–0.075 range, and only 9 (12.7%) have p-values in the 0.075–0.1 range. We use p-value <0.1 as the threshold for cluster enrichment in functions/complexes/pathways due to the fact that the biological experiments and the PPI network data used in this study are not perfect and do contain some noise. In addition, many genes and their protein products do not have functional/complex/pathway annotations in current databases. Therefore, the lower the p-value threshold used, the more likely one is to omit results that are of biological importance but are not statistically significant. Moreover, this threshold allows increased flexibility and balance between removing those enrichments that are very likely to occur at random and keeping potentially biologically interesting but statistically nonsignificant results. This is important, since it has been shown that statistically significant results may not be scientifically or biologically significant and that nonsignificant results may turn out to be very important.66
One of the interesting clusters identified is the defined ratio cluster [G1_S]#3, which has enriched functions in cell cycle and DNA processing (Figure 6A). Five PIPs in this cluster, that is, Fus3, Rvs161, Rvs167, Sst2, and Tpd3, are known to be involved in the yeast mating signaling pathway that induces cell cycle arrest.58,59 Upon pheromone binding to the G-protein-coupled-receptor (GPCR) on the cell-surface a series of events occur, which ultimately lead to up- and down-regulation of transcription of many genes, arrest in G1 phase, and bud tip formation and elongation.58 In the presence of a suitable mating partner, the fusion of the two plasma membranes and nuclei also occurs.58,67 There are many proteins involved in this signal cascade including GPCRs and several effector kinases such as Fus3.58,59 Fus3 is involved in several processes including control of pheromone induced gene expression by phosphorylation of transcription factors, induction of cell cycle arrest in the G1 phase by phosphorylating the cyclin-dependent kinase inhibitor Far1, and pathways related to membrane fusion.58,68–70 Reciprocal co-IP and Western blot analyses have confirmed the cell cycle specific interaction of Fus3 with the proteasome during G1 and S phase (Figure 7). Rvs161, an amphiphysin homologue, is localized at the shmoo tip during mating and is involved in actin cytoskeleton organization, regulation of cell polarity, bud formation, and cell fusion.71–73 Rvs161 interacts and functions with another amphiphysin like protein, Rvs167, another PIP in this cluster.72,74,75 Additionally, the stability of Rvs161 is largely dependent upon the presence of Rvs167.72 Interestingly, Rvs167 is also a Fus3 substrate subsequent to pheromone signaling.76 Another Fus3 substrate, Sst2, is also a member of this cluster. Sst2 localizes to the plasma membrane and acts as a GTPase activating protein, thus desensitizing the cell to pheromone signaling.77 Tpd3 is a component of the yeast PP2A Ser/Thr phosphatase and has been shown to be involved in mediating G1 arrest in response to ceramide.78 Its presence in this cluster suggests that Tpd3 may be associated with α-factor induced G1 phase arrest. Taken together, these results have provided the first physical evidence directly linking the proteasome to pheromone signaling pathway in yeast and indicated that the proteasome may regulate the cell cycle through this signaling pathway. However, whether the proteasome is the target of pheromone signaling pathway or conversely a potential regulator of this pathway needs to be further investigated.
Another interesting cluster is the [G1_S_M]all_high cluster, in which the PIPs have high ratios in all three phases, the same characteristic SILAC ratios of proteasome subunits,39 suggesting their highly specific binding to the proteasome particles. In addition to the known ubiquitin receptors (Rad23, Dsk2 and Ddi1), a shuttling protein Cdc48 and its adaptor protein, Shp1, are grouped into this cluster. Shp1 has both a UBA (ubiquitin -associated) and a UBX (ubiquitin regulatory X) domain and has been shown to bind to ubiquitinated substrates in vivo and to be involved in proteasome dependent degradation.79 The [G1_S_M]all_high cluster also contained Skp1, a core component of SCF ubiquitin ligases.28,80 However, other components of SCF ligases were not among the members of this cluster, suggesting that the Skp1 interacted independently of its SCF ligase binding partners. Interestingly, Skp1 has also been identified as a component of the yeast kinetochore independently of its function in the SCF ubiquitin ligase complex,81 and another protein linked to the kinetochore, Bub3, was placed in the [G1_S_M]all_high cluster. Bub3 is involved in the mitotic spindle checkpoint, and was identified as a “high” ratio PIP from unsynchronized cells previously.39,82,83 The other four PIPs in this cluster are Cmd1, Sok1, Grx1, Tsl1, and Tom70, which do not have confirmed physical interactions with any of the proteasome subunits except Sok1. Sok1 was identified as a high ratio PIP in our previous study using unsynchronized yeast cells.39 Although the biological functions of these PIPs’ (i.e., Cdm1, Bub3, Sok1, Grx1, Tsl1, and Tom3) interactions with the proteasome has yet to be determined, the fact that these PIPs have grouped to this cluster that harbors all known ubiquitin receptors and several key players in the proteasomal degradation pathway strongly suggests that these high ratio PIPs most likely play critical roles in assisting proteasomal degradation.
Although proteasome subunits including Rpn11 have stable expression throughout the cell cycle, it has been suggested that about 15% of budding yeast genes are subjected to transcriptional regulation during the cell cycle based on several genome-wide transcript measurements.84–86 Therefore, some of the identified PIPs whose cell cycle-dependent dynamic interactions with the proteasome may be the results of their cell cycle regulated expression changes. However, direct comparison of our data and the reported transcriptome data84–86 is not feasible due to differences in experimental conditions and the fact that protein expression level does not correlate well with its mRNA level.87 Future studies targeting at complete proteomic profiling of cell cycle regulated protein abundance changes will make it possible for us to further characterize these PIPs.
In summary, the QTAX approach has proven to be a robust and effective strategy for studying proteasome interacting proteins throughout the cell cycle. These results describe the first extensive proteomic analysis of the cell cycle specific proteasome interaction networks. Protein network analysis combined with cluster analysis has led to the direct physical connection between the proteasome and the pheromone induced signaling pathway. This study represents the first step toward understanding of how dynamics of the proteasome complex itself is involved in regulating cell cycle progression.
Supplementary Material
Acknowledgments
We thank Prof. A. L. Burlingame, Peter Baker, and Aenoch Lynn for using Protein Prospector and members of the Huang laboratory for their help during this study. We thank Dr. Cortnie Guerrero for her initial work on the cell cycle study. This work was supported by National Institutes of Health grants (GM-74830 to L.H. and GM-66164 to P.K.) and by an NSF CAREER grant (IIS-0644424 to N.P.).
Abbreviations
- QTAX
quantitative analysis of tandem affinity purified in vivo cross-linked (X) protein complexes
- UPS
ubiquitin-proteasome system
- CP
core particle
- RP
regulatory particle
- PIP
proteasome interacting protein
- HBH
histidine-biotin-histidine tag
- SILAC
stable isotope labeling of amino acids in cell culture
- wt
wild type
- L
light
- H
heavy
- SILAC ratios (i.e., L/H)
relative abundance ratio of light labeled to heavy labeled peptides
- SCX
strong cation exchange
Footnotes
Supporting Information Available: Supplemental Table 1. List of 677 putative PIPs identified in three cell cycle phases. Supplemental Table 2. Classification of the putative PIPs using profile vector-based clustering analysis. Supplemental Figure 1. Clustering schemes of the PIPs using a SILAC ratio profile vector-based approach. (A) Three possible SILAC ratio profile vectors for PIPs present in any two phases, that is, G1_S, S_M, or G1_M. (B) Thirteen possible SILAC ratio profile vectors for PIPs present in three phases (G1_S_M). Each cluster is represented by 2–3 nodes, and each of the nodes describes a phase (x-axis) and relative SILAC ratio (y-axis). Note: It is possible that between the three phase specific SILAC ratios of a PIP, the difference between two of the phase ratios could be within 2-fold, but the third phase ratio could be 2-fold higher or lower than either the first or second (irrespective of the order of phases). To eliminate any ambiguity, we assigned three generic numbers X, Y, and Z to the three ratios such that X ≥ Y ≥ Z (regardless of which one corresponds to which phase); then, if Y ≥ (X + Z)/2, we set Y = X; otherwise, if Y < (X + Z)/2, then Y = Z. We identified the corresponding phases and their trends among three phases. Supplemental Figure 2. Cell cycle specific protein interaction networks of the yeast 26S protea-some, i.e. proteasome interacting networks at the G1, S and M phases. Individual PIPs were represented as nodes, and interactions between PIPs represented by blue lines. Heat map indicates SILAC ratio intensity with light blue (lower ratio) and dark blue (higher ratio). “High ratio” PIPs are indicated by red node borders. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Goldberg AL. Nobel committee tags ubiquitin for distinction. Neuron. 2005;45(3):339–44. doi: 10.1016/j.neuron.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5(3):177–87. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 3.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 4.Nalepa G, Harper JW. Therapeutic anti-cancer targets upstream of the proteasome. Cancer Treat Rev. 2003;29(suppl 1):49–57. doi: 10.1016/s0305-7372(03)00083-5. [DOI] [PubMed] [Google Scholar]
- 5.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458(7237):438–44. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 6.Ciechanover A. Intracellular protein degradation from a vague idea through the lysosome and the ubiquitin-proteasome system and on to human diseases and drug targeting: Nobel Lecture, December 8, 2004. Ann NY Acad Sci. 2007;1116:1–28. doi: 10.1196/annals.1402.078. [DOI] [PubMed] [Google Scholar]
- 7.Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans. 2007;35(Pt 1):12–7. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- 8.Dahlmann B. Role of proteasomes in disease. BMC Biochem. 2007;8(Suppl 1):S3. doi: 10.1186/1471-2091-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah JJ, Orlowski RZ. Proteasome inhibitors in the treatment of multiple myeloma. Leukemia. 2009;23(11):1964–79. doi: 10.1038/leu.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5(5):417–21. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt M, Hanna J, Elsasser S, Finley D. Proteasome-associated proteins: regulation of a proteolytic machine. Biol Chem. 2005;386(8):725–37. doi: 10.1515/BC.2005.085. [DOI] [PubMed] [Google Scholar]
- 12.Gomes AV, Zong C, Edmondson RD, Li X, Stefani E, Zhang J, Jones RC, Thyparambil S, Wang GW, Qiao X, Bardag-Gorce F, Ping P. Mapping the murine cardiac 26S proteasome complexes. Circ Res. 2006;99(4):362–71. doi: 10.1161/01.RES.0000237386.98506.f7. [DOI] [PubMed] [Google Scholar]
- 13.Hanna J, Meides A, Zhang DP, Finley D. A ubiquitin stress response induces altered proteasome composition. Cell. 2007;129(4):747–59. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 14.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–68. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 15.Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18(6):3149–62. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kloetzel PM. Antigen processing by the proteasome. Nat Rev Mol Cell Biol. 2001;2(3):179–87. doi: 10.1038/35056572. [DOI] [PubMed] [Google Scholar]
- 17.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rockel B, Baumeister W. A tale of two giant proteases. Ernst Schering Found Symp Proc. 2008;(1):17–40. doi: 10.1007/2789_2008_099. [DOI] [PubMed] [Google Scholar]
- 19.Peters JM, King RW, Hoog C, Kirschner MW. Identification of BIME as a subunit of the anaphase-promoting complex. Science. 1996;274(5290):1199–201. doi: 10.1126/science.274.5290.1199. [DOI] [PubMed] [Google Scholar]
- 20.Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246(4930):629–34. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 21.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8(6):438–49. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hershko A. Roles of ubiquitin-mediated proteolysis in cell cycle control. Curr Opin Cell Biol. 1997;9(6):788–99. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 23.Reed SI. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat Rev Mol Cell Biol. 2003;4(11):855–64. doi: 10.1038/nrm1246. [DOI] [PubMed] [Google Scholar]
- 24.Tyers M, Jorgensen P. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr Opin Genet Dev. 2000;10:54–64. doi: 10.1016/s0959-437x(99)00049-0. [DOI] [PubMed] [Google Scholar]
- 25.Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13(16):2039–58. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 26.Deshaies RJ. Phosphorylation and proteolysis: partners in the regulation of cell division in the budding yeast. Curr Opin Genet Dev. 1997;7:7–16. doi: 10.1016/s0959-437x(97)80103-7. [DOI] [PubMed] [Google Scholar]
- 27.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 28.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 29.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 30.Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11(10):3425–39. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10(3):495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 32.Meng F, Forbes AJ, Miller LM, Kelleher NL. Detection and localization of protein modifications by high resolution tandem mass spectrometry. Mass Spectrom Rev. 2005;24(2):126–34. doi: 10.1002/mas.20009. [DOI] [PubMed] [Google Scholar]
- 33.Hartmann-Petersen R, Gordon C. Proteins interacting with the 26S proteasome. Cell Mol Life Sci. 2004;61(13):1589–95. doi: 10.1007/s00018-004-4132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127(1):99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 35.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127(7):1401–13. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Chen CF, Baker PR, Chen PL, Kaiser P, Huang L. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46(11):3553–65. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Huang L. Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol Cell Proteomics. 2008;7(1):46–57. doi: 10.1074/mcp.M700261-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Guerrero C, Tagwerker C, Kaiser P, Huang L. An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol Cell Proteomics. 2006;5(2):366–78. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Guerrero C, Milenkovic T, Przulj N, Kaiser P, Huang L. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc Natl Acad Sci USA. 2008;105(36):13333–8. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol Cell Proteomics. 2006;5(4):737–48. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. Academic Press, Inc; San Diego, CA: 1991. [Google Scholar]
- 42.Haase SB, Reed SI. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle. 2002;1(2):132–6. [PubMed] [Google Scholar]
- 43.Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6(3):439–50. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet AS, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabasi AL, Tavernier J, Hill DE, Vidal M. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322(5898):104–10. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayor T, Lipford JR, Graumann J, Smith GT, Deshaies RJ. Analysis of polyubiquitin conjugates reveals that the Rpn10 substrate receptor contributes to the turnover of multiple proteasome targets. Mol Cell Proteomics. 2005;4(6):741–51. doi: 10.1074/mcp.M400220-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Mayor T, Graumann J, Bryan J, MacCoss MJ, Deshaies RJ. Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol Cell Proteomics. 2007;6(11):1885–95. doi: 10.1074/mcp.M700264-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21(8):921–6. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 48.Seyfried NT, Xu P, Duong DM, Cheng D, Hanfelt J, Peng J. Systematic approach for validating the ubiquitinated proteome. Anal Chem. 2008;80(11):4161–9. doi: 10.1021/ac702516a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137(1):133–45. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol Syst Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milenkovic T, Przulj N. Uncovering Biological Network Function via Graphlet Degree Signatures. Cancer Informatics. 2008;4:257–73. [PMC free article] [PubMed] [Google Scholar]
- 52.Fang L, Wang X, Yamoah K, Chen PL, Pan ZQ, Huang L. Characterization of the human COP9 signalosome complex using affinity purification and mass spectrometry. J Proteome Res. 2008;7(11):4914–25. doi: 10.1021/pr800574c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Craig KL, Tyers M. The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog Biophys Mol Biol. 1999;72(3):299–328. doi: 10.1016/s0079-6107(99)00010-3. [DOI] [PubMed] [Google Scholar]
- 54.Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: it’s not just for mitosis any more. Genes Dev. 2002;16(17):2179–206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- 55.Reed SI. The ubiquitin-proteasome pathway in cell cycle control. Results Probl Cell Differ. 2006;42:147–81. doi: 10.1007/b136681. [DOI] [PubMed] [Google Scholar]
- 56.Mayor T, Lipford JR, Graumann J, Smith GT, Deshaies RJ. Analysis of poly-ubiquitin conjugates reveals that the Rpn10 substrate receptor contributes to the turnover of multiple proteasome targets. Mol Cell Proteomics. 2005;4(6):741–51. doi: 10.1074/mcp.M400220-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Boulton TG, Yancopoulos GD, Gregory JS, Slaughter C, Moomaw C, Hsu J, Cobb MH. An insulin-stimulated protein kinase similar to yeast kinases involved in cell cycle control. Science. 1990;249(4964):64–7. doi: 10.1126/science.2164259. [DOI] [PubMed] [Google Scholar]
- 58.Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2005;26(2):339–50. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dohlman HG. G proteins and pheromone signaling. Annu Rev Physiol. 2002;64:129–52. doi: 10.1146/annurev.physiol.64.081701.133448. [DOI] [PubMed] [Google Scholar]
- 60.Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, Silva AC, Shales M, Collins SR, van Wageningen S, Kemmeren P, Holstege FC, Weissman JS, Keogh MC, Koller D, Shokat KM, Krogan NJ. Functional organization of the, S. cerevisiae phosphorylation network. Cell. 2009;136(5):952–63. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Milenkovic T, Lai J, Przulj N. GraphCrunch: a tool for large network analyses. BMC Bioinformatics. 2008;9:70. doi: 10.1186/1471-2105-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Przulj N, Corneil DG, Jurisica I. Modeling interactome: scale-free or geometric. Bioinformatics. 2004;20(18):3508–15. doi: 10.1093/bioinformatics/bth436. [DOI] [PubMed] [Google Scholar]
- 63.Przulj N. Biological network comparison using graphlet degree distribution. Bioinformatics. 2007;23(2):e177–83. doi: 10.1093/bioinformatics/btl301. [DOI] [PubMed] [Google Scholar]
- 64.Xu R, Wunsch D., 2nd Survey of clustering algorithms. IEEE Trans Neural Netw. 2005;16(3):645–78. doi: 10.1109/TNN.2005.845141. [DOI] [PubMed] [Google Scholar]
- 65.Sardiu ME, Florens L, Washburn MP. Evaluation of clustering algorithms for protein complex and protein interaction network assembly. J Proteome Res. 2009;8(6):2944–52. doi: 10.1021/pr900073d. [DOI] [PubMed] [Google Scholar]
- 66.Motulsky H. Intuitive Biostatistics. Oxford University Press; New York: 1995. [Google Scholar]
- 67.Melloy P, Shen S, White E, McIntosh JR, Rose MD. Nuclear fusion during yeast mating occurs by a three-step pathway. J Cell Biol. 2007;179(4):659–70. doi: 10.1083/jcb.200706151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63(5):999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- 69.Gartner A, Nasmyth K, Ammerer G. Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes Dev. 1992;6(7):1280–92. doi: 10.1101/gad.6.7.1280. [DOI] [PubMed] [Google Scholar]
- 70.Gartner A, Jovanovic A, Jeoung DI, Bourlat S, Cross FR, Ammerer G. Pheromone-dependent G1 cell cycle arrest requires Far1 phosphorylation, but may not involve inhibition of Cdc28-Cln2 kinase, in vivo. Mol Cell Biol. 1998;18(7):3681–91. doi: 10.1128/mcb.18.7.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sivadon P, Bauer F, Aigle M, Crouzet M. Actin cytoskeleton and budding pattern are altered in the yeast rvs161 mutant: the Rvs161 protein shares common domains with the brain protein amphiphysin. Mol Gen Genet. 1995;246(4):485–95. doi: 10.1007/BF00290452. [DOI] [PubMed] [Google Scholar]
- 72.Lombardi R, Riezman H. Rvs161p and Rvs167p, the two yeast amphiphysin homologs, function together in vivo. J Biol Chem. 2001;276(8):6016–22. doi: 10.1074/jbc.M008735200. [DOI] [PubMed] [Google Scholar]
- 73.Brizzio V, Gammie AE, Rose MD. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J Cell Biol. 1998;141(3):567–84. doi: 10.1083/jcb.141.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Navarro P, Durrens P, Aigle M. Protein-protein interaction between the RVS161 and RVS167 gene products of Saccharomyces cerevisiae. Biochim Biophys Acta. 1997;1343(2):187–92. doi: 10.1016/s0167-4838(97)00108-8. [DOI] [PubMed] [Google Scholar]
- 75.Balguerie A, Sivadon P, Bonneu M, Aigle M. Rvs167p, the budding yeast homolog of amphiphysin, colocalizes with actin patches. J Cell Sci. 1999;112(Pt 15):2529–37. doi: 10.1242/jcs.112.15.2529. [DOI] [PubMed] [Google Scholar]
- 76.Friesen H, Murphy K, Breitkreutz A, Tyers M, Andrews B. Regulation of the yeast amphiphysin homologue Rvs167p by phosphorylation. Mol Biol Cell. 2003;14(7):3027–40. doi: 10.1091/mbc.E02-09-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dohlman HG, Song J, Ma D, Courchesne WE, Thorner J. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein alpha subunit) Mol Cell Biol. 1996;16(9):5194–209. doi: 10.1128/mcb.16.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nickels JT, Broach JR. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 1996;10(4):382–94. doi: 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]
- 79.Schuberth C, Richly H, Rumpf S, Buchberger A. Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep. 2004;5(8):818–24. doi: 10.1038/sj.embor.7400203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86(2):263–74. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 81.Kaplan KB, Hyman AA, Sorger PK. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell. 1997;91(4):491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 82.Brady DM, Hardwick KG. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr Biol. 2000;10(11):675–8. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- 83.Hoyt MA, Totis L, Roberts BTS. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66(3):507–17. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 84.Pramila T, Wu W, Miles S, Noble WS, Breeden LL. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20(16):2266–78. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9(12):3273–97. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, Davis RW. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2(1):65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 87.Griffin TJ, Gygi SP, Ideker T, Rist B, Eng J, Hood L, Aebersold R. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics. 2002;1(4):323–33. doi: 10.1074/mcp.m200001-mcp200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.