Abstract
Genes are regulated because their expression involves a fitness cost to the organism. The production of proteins by transcription and translation is a well-known cost factor, but the enzymatic activity of the proteins produced can also reduce fitness, depending on the internal state and the environment of the cell. Here, we map the fitness costs of a key metabolic network, the lactose utilization pathway in Escherichia coli. We measure the growth of several regulatory lac operon mutants in different environments inducing expression of the lac genes. We find a strikingly nonlinear fitness landscape, which depends on the production rate and on the activity rate of the lac proteins. A simple fitness model of the lac pathway, based on elementary biophysical processes, predicts the growth rate of all observed strains. The nonlinearity of fitness is explained by a feedback loop: production and activity of the lac proteins reduce growth, but growth also affects the density of these molecules. This nonlinearity has important consequences for molecular function and evolution. It generates a cliff in the fitness landscape, beyond which populations cannot maintain growth. In viable populations, there is an expression barrier of the lac genes, which cannot be exceeded in any stationary growth process. Furthermore, the nonlinearity determines how the fitness of operon mutants depends on the inducer environment. We argue that fitness nonlinearities, expression barriers, and gene–environment interactions are generic features of fitness landscapes for metabolic pathways, and we discuss their implications for the evolution of regulation.
Author Summary
The levels of protein produced by an organism are likely to change its fitness, potentially driving the evolution of genetic regulation. Importantly, protein expression generates costs as well as benefits. Here, we use a model genetic system, the lac operon of Escherichia coli, to investigate different sources of fitness costs. We find that fitness depends not only on the production rate of proteins but also on their enzymatic activity. A simple quantitative model, which is based on the biophysics of protein production and activity, accurately reproduces the experimental results and provides testable predictions. The model describes a feedback cycle between a molecular pathway and the growth rate of cells: pathway activity impedes growth, but growth itself affects the pathway. This feedback can generate dramatic effects, such as gene expression barriers, fitness cliffs, and population extinctions, which can be triggered by small environmental or genetic changes. Our results disentangle the complex interplay of protein production and activity, and they show how these processes shape the evolution of simple organisms.
Introduction
Gene regulation is a major factor of molecular evolution, and changes in gene expression contribute to phenotypic differences between species [1]. Expression levels are under natural selection, which results from a balance between costs and benefits for the organism. For single-cell organisms, fitness benefits include the ability to digest nutrients in different environments. The cost of gene expression, on the other hand, depends on the biophysics of protein production and of protein activity. The cost of protein production has been studied extensively [2]–[6]. However, enzymatic activities of proteins can also reduce fitness due to energy consumption or toxic effects of the reaction products. What are the relative contributions of these two effects? How do they interact? To address these questions, we have to understand the fitness effects of an entire metabolic pathway, in which protein production is coupled to function and growth. This is the subject of the present paper.
For our analysis, we use the lactose utilization pathway in Escherichia coli, which is one of the best characterized molecular pathways [7]. It is coded in a set of genes referred to as the lac operon. Several studies have addressed fitness effects associated with expression of the lac genes. In particular, production of the lac proteins in the absence of lactose has been shown to involve a fitness cost, that is, to reduce the growth rate of a cell population [2]–[4], [8], [9]. This cost has been ascribed to transcription and translation of the lac genes [10], because toxic effects of the gene products have not been observed. Growth is also reduced by the presence of inducers in the medium, even after the maximum of expression is reached [8]. This fitness cost is likely to arise from inducer transport through the cell membrane [11], [12]. Furthermore, the lac operon has been used to study the interplay of cost and benefit in the evolution of gene expression [9]. Taken together, these observations make the lac operon an ideal system to study the coupled fitness costs of protein production and activity.
Here we determine a fitness landscape of the lac pathway by a combined experimental and theoretical approach. We measure the fitness of different regulatory mutant strains in the presence and absence of the lac inducer IPTG and of the natural sugar lactose. LacY proteins act as transporters (so-called permeases) for IPTG and lactose (i.e., these molecules are substrates of LacY). We develop a quantitative biophysical growth model to disentangle the fitness contributions of protein production (i.e., transcription and translation) and of protein activity (i.e., intra-cellular transport). The model explains the growth rate of all observed mutants in different inducer environments. Its key element is a feedback loop between the lac pathway and fitness: at constant rate of protein production, faster cell growth leads to stronger dilution of proteins and lowers the cost of protein activity. In addition, the rate of lac gene expression itself can depend on growth [13], [14]. Similar growth feedback mechanisms have been argued to play an important role in bacterial drug resistance [15], [16], and to generate diversity in an isogenic population [13], [15], [17].
Our analysis suggests that growth feedback is a pervasive feature of the activity-dependent fitness of metabolic pathways. This feature has important evolutionary consequences. In particular, our model predicts a fitness cliff, beyond which populations cannot maintain viable growth, and an expression barrier, that is, an upper bound for protein production and activity in viable populations. As a consequence, gene regulation in metabolic pathways is likely to be under stronger selection than the mere cost of protein production would suggest.
Results
Fitness effects of lac protein production and activity
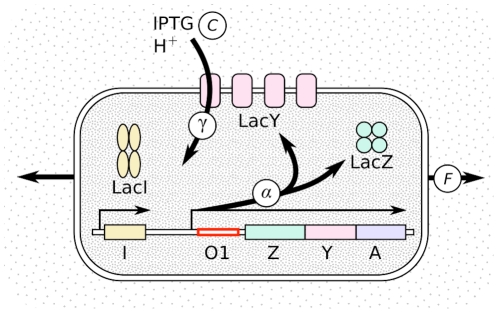
There are two generic sources of fitness cost for a molecular pathway: the cost of protein production and the cost of enzymatic activity [2]–[6]. In the case of the lac pathway, fitness depends strongly on the presence of substrates of the lac proteins, even when these substrates cannot be used as a carbon source [8], [11], [12]. One such substrate is IPTG (isopropyl-1-thio-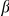 -D-galactoside), which is transported by LacY and induces lac expression (see Figure 1). Hence, there are two potential phenotypes affecting the fitness of the lac operon in an IPTG environment: the rate of lac protein production and the rate of IPTG transport into the cell.
-D-galactoside), which is transported by LacY and induces lac expression (see Figure 1). Hence, there are two potential phenotypes affecting the fitness of the lac operon in an IPTG environment: the rate of lac protein production and the rate of IPTG transport into the cell.
Figure 1. Schematic representation of the lac pathway.
The lac operon is composed of three genes controlled by the same promoter: lac Z, lac Y, and lac A. The lac pathway also involves the constitutively expressed repressor LacI. It represses the transcription from the lac promoter by attaching to the operator sequence lacO1. Inducers, such as IPTG, deactivate the repressor LacI and thus stimulate the synthesis of the gene products LacZ, LacY, and LacA. The rate of production of the three lac proteins is denoted by  and it depends on the sequence of lac O1, on the presence of inducer inside the cell, and on the growth rate. All three lac genes are transcribed with the same rate, hence LacZ can be used as a reporter for the whole operon. LacY transports molecules such as IPTG inside the cell with a rate
and it depends on the sequence of lac O1, on the presence of inducer inside the cell, and on the growth rate. All three lac genes are transcribed with the same rate, hence LacZ can be used as a reporter for the whole operon. LacY transports molecules such as IPTG inside the cell with a rate  , which depends on the concentration of these molecules. One proton
, which depends on the concentration of these molecules. One proton 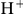 is transported with each substrate molecule [7]. Growth (measured by the Malthusian fitness
is transported with each substrate molecule [7]. Growth (measured by the Malthusian fitness 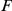 ) dilutes the internal molecules, thus lowering their concentrations. The strains used in this study differ by the lacO1 sequence and are grown in various IPTG concentrations.
) dilutes the internal molecules, thus lowering their concentrations. The strains used in this study differ by the lacO1 sequence and are grown in various IPTG concentrations.
We measure the fitness effects of lac protein production and activity in thirteen regulatory mutants in the lac operon of Escherichia coli. Twelve mutant strains have substitutions in the lacO1 operator region, which affect expression of the lac genes, and one strain has a deletion in the gene of the repressor lacI (see Figure 1 and Text S1 for details). We determine the lac protein concentration and the fitness of these mutants both with and without substrates of the lac permease LacY. Specifically, we compete each mutant strain against a reference strain with deleted lac genes. This assay defines the fitness cost of the lac pathway as the difference in growth rate, or difference in Malthusian fitness, between reference strain and mutant, 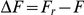 (see Materials and Methods and Text S1 for details).
(see Materials and Methods and Text S1 for details).
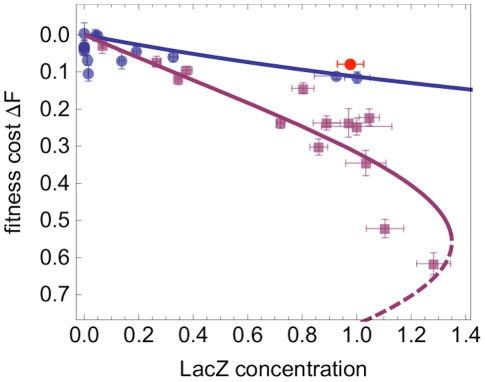
Figure 2 summarizes the results of these experiments. They show that fitness always decreases with increasing concentration of lac proteins inside the cell, but the form of this dependence depends on presence or absence of the substrate. Without substrate, the fitness cost can be fitted to a linear form, which we associate with lac protein production (blue line). When substrate is added, the magnitude of the fitness cost strongly increases and its dependence on concentration becomes nonlinear (purple line). The additional, nonlinear fitness cost in the presence of IPTG can be associated with the transport activity of the LacY proteins. This is shown by a control mutant with deleted lacY gene, for which we only observe the linear cost of protein production (red dot). Deviations of individual data points from the fit curves can be caused by different sources of noise. Competition assays involve experimental errors, in particular for large fitness differences between strains. For example, there can be slight day-to-day differences in medium composition. Furthermore, some of the strains might have acquired mutations with a fitness effect outside the lac operator sequence, although we have controlled for random mutations elsewhere the genome (see Text S1).
Figure 2. Fitness of lac regulatory mutants in different environments.
Measured fitness cost of each mutant strain, plotted against LacZ concentration (normalized to the fully induced wild-type value). Measurements are obtained in minimal medium with 0.1% glycerol in the absence of IPTG (blue dots) and in the same medium with 1 mM IPTG (mauve squares). Fitness is measured by competition against a reference strain which has a deletion of the whole lac locus and of lac I 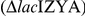 . The fitness cost
. The fitness cost 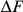 of a given strain is defined as the reduction in growth rate (Malthusian fitness) compared to the reference strain (see Materials and Methods for details). In presence of 1 mM IPTG, a control strain with deleted lac Y gene
of a given strain is defined as the reduction in growth rate (Malthusian fitness) compared to the reference strain (see Materials and Methods for details). In presence of 1 mM IPTG, a control strain with deleted lac Y gene 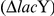 has an expression level comparable to the wild type, but a fitness close to that of constitutive mutants in absence of IPTG (red dot). All points show the average of 12 replicates for fitness and at least 3 replicates for protein concentration, with error bars giving the standard error. Lines show model predictions (the dashed line represents an unstable solution, see main text).
has an expression level comparable to the wild type, but a fitness close to that of constitutive mutants in absence of IPTG (red dot). All points show the average of 12 replicates for fitness and at least 3 replicates for protein concentration, with error bars giving the standard error. Lines show model predictions (the dashed line represents an unstable solution, see main text).
As a further experimental step, we test whether these results extend to lactose, which is a natural nutrient of E. coli. The sugar used to support cell growth in the above experiments is glycerol, which is a poor carbon source. Lactose supports faster growth and is known to give an advantage to cells which are able to metabolize it. With 1 mM of lactose, the wild type has a fitness benefit over the reference strain, which amounts to 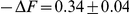 (mean of 4 replicates
(mean of 4 replicates 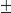 standard error). To assess whether lactose metabolism also involves a cost, we construct a mutant with deleted lacZ and lacI genes. This mutant cannot use lactose and expresses lacY constitutively. In the presence of lactose, it has a fitness cost
standard error). To assess whether lactose metabolism also involves a cost, we construct a mutant with deleted lacZ and lacI genes. This mutant cannot use lactose and expresses lacY constitutively. In the presence of lactose, it has a fitness cost
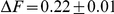 (mean of 12 replicates
(mean of 12 replicates 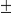 standard error) against the reference strain, which indicates that lactose and IPTG cause a similar decrease in fitness in the presence of the lac permease (see Materials and Methods for details).
standard error) against the reference strain, which indicates that lactose and IPTG cause a similar decrease in fitness in the presence of the lac permease (see Materials and Methods for details).
We conclude that both the rate of protein production and the rate of protein activity (intra-cellular transport by LacY) are phenotypes that affect the fitness cost of the lac pathway. But what is the cause of the fitness nonlinearity in the presence of substrates, and what are its biological consequences? To address these questions, we now describe our experiments in terms of a simple biophysical model.
Fitness model
We use a minimal model of gene expression and inducer transport to disentangle the fitness effects of protein production and activity in a quantitative way. The underlying intra-cellular processes involve transcription and translation, uptake of substrate by active transport, and dilution by cell division. Given the complexity of these processes and their effects on cell growth, our model does not aim at a complete description. However, the model does account for a large part of the fitness variation between strains and between cellular growth conditions. At the same time, it contains only few phenotypes and few parameters, which can be inferred from our fitness measurements.
Within the model, the cost of lac protein production is proportional the production rate  , and we infer this rate from our measurements of fitness and LacZ concentration (see Figure 1 and Materials and Methods). The cost of LacY activity has two different potential contributions: the energy consumption of the transport process (direct transport costs) and growth effects of the molecules transported inside the cell (toxicity costs). Direct transport costs can arise from futile transport cycles: LacY transports one proton with every IPTG molecule, and ATP is consumed to pump the excess protons out of the cell. These costs are proportional to the total LacY pumping rate inside the cell,
, and we infer this rate from our measurements of fitness and LacZ concentration (see Figure 1 and Materials and Methods). The cost of LacY activity has two different potential contributions: the energy consumption of the transport process (direct transport costs) and growth effects of the molecules transported inside the cell (toxicity costs). Direct transport costs can arise from futile transport cycles: LacY transports one proton with every IPTG molecule, and ATP is consumed to pump the excess protons out of the cell. These costs are proportional to the total LacY pumping rate inside the cell, 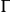 . Toxicity costs are likely to arise from an excess concentration of the transported protons, i.e., a reduction of the intra-cellular pH value [11], [12]. The toxicity of IPTG itself appears to be negligible (see Text S1 and [12]). Toxicity costs are proportional to the steady-state concentration of the toxic molecules, which depends on their uptake rate, the rate of dilution by cell divisions, and the cell volume
. Toxicity costs are likely to arise from an excess concentration of the transported protons, i.e., a reduction of the intra-cellular pH value [11], [12]. The toxicity of IPTG itself appears to be negligible (see Text S1 and [12]). Toxicity costs are proportional to the steady-state concentration of the toxic molecules, which depends on their uptake rate, the rate of dilution by cell divisions, and the cell volume 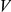 [18]. The excess concentration of protons is, thus, proportional to
[18]. The excess concentration of protons is, thus, proportional to 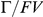 . Furthermore, the steady-state cell volume itself depends on the growth rate,
. Furthermore, the steady-state cell volume itself depends on the growth rate, 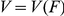 [13].
[13].
The combined fitness cost of protein production and activity in the lac pathway takes the form
| (1) |
in terms of the pathway phenotypes  and
and 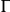 . Here,
. Here, 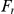 denotes the fitness of the reference strain with deleted lac genes (for which
denotes the fitness of the reference strain with deleted lac genes (for which 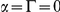 ). Our model contains a feedback loop: fitness depends on the rates
). Our model contains a feedback loop: fitness depends on the rates  and
and 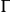 , which in turn depend on fitness. This feedback between pathway phenotypes and fitness is illustrated in Figure 3. It has an important consequence: although the cost contributions in Equation 1 are taken to be additive at any given value of
, which in turn depend on fitness. This feedback between pathway phenotypes and fitness is illustrated in Figure 3. It has an important consequence: although the cost contributions in Equation 1 are taken to be additive at any given value of 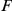 , the resulting dependence of fitness on the pathway phenotypes,
, the resulting dependence of fitness on the pathway phenotypes, 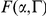 , becomes nonlinear.
, becomes nonlinear.
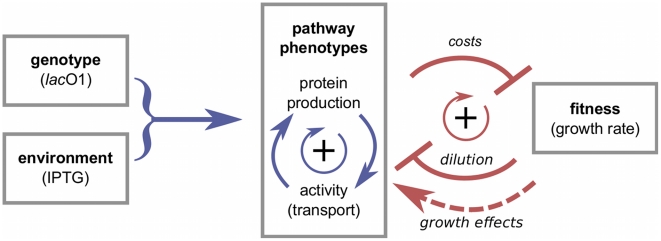
Figure 3. From genotype and environment to pathway phenotypes and fitness.
Environment and genotype determine the function of the lac pathway, which is described by the two phenotypes of protein production and protein (transport) activity. These phenotypes are coupled by a pathway-specific positive feedback loop (blue circle). The pathway itself is coupled to growth (fitness) by a generic positive feedback loop: protein production and protein activity are fitness costs, and cell growth reduces protein concentration and activity by dilution (red circle). In addition, growth can affect the rate of gene expression [13] (dashed arrow). These feedback loops generate strong nonlinearities in the phenotype-fitness map and the genotype-environment-fitness map; see Figure 4 and Figure 5.
By calibrating this model to our experimental data, we can infer the amplitudes  ,
, 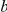 , and
, and  of the different cost factors. Bayesian analysis shows that there are significant fitness contributions of protein production and steady-state concentration (with maximum-likelihood parameter values
of the different cost factors. Bayesian analysis shows that there are significant fitness contributions of protein production and steady-state concentration (with maximum-likelihood parameter values 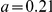 ,
, 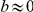 ,
, 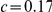 ), but the data are also compatible with a larger direct cost of transport (
), but the data are also compatible with a larger direct cost of transport (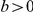 ) (see Materials and Methods and Text S1). As shown in Figure 2, the maximum-likelihood model provides a good fit to the data: the fitness feedback loop quantitatively explains the cost nonlinearity observed in our experiments.
) (see Materials and Methods and Text S1). As shown in Figure 2, the maximum-likelihood model provides a good fit to the data: the fitness feedback loop quantitatively explains the cost nonlinearity observed in our experiments.
We use Equation 1 to derive two representations of a fitness landscape for the lac pathway, which highlight different biological implications of its form. First, we solve this equation to display the dependence of fitness on the pathway phenotypes, 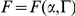 , as shown in Figure 4. Second, we display the dependence of fitness on the external IPTG concentration,
, as shown in Figure 4. Second, we display the dependence of fitness on the external IPTG concentration, 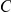 , and on two genotype summary variables, which depend only on lac O1 sequence. As genotype variables, we use the maximal rate of lac protein production at a fixed growth rate of one cell division per hour,
, and on two genotype summary variables, which depend only on lac O1 sequence. As genotype variables, we use the maximal rate of lac protein production at a fixed growth rate of one cell division per hour, 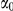 , and the ratio of repressed to unrepressed protein production rates,
, and the ratio of repressed to unrepressed protein production rates, 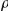 [19]. The resulting function
[19]. The resulting function 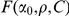 , which is shown in Figure 5, can be called a genotype-environment-fitness map. We note that the change from the phenotype variables
, which is shown in Figure 5, can be called a genotype-environment-fitness map. We note that the change from the phenotype variables 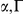 to the genotype-environment variables
to the genotype-environment variables 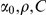 depends itself on fitness. This dependence has two reasons: (i) The LacY pumping rate depends on the production rate, the pumping rate per LacY molecule
depends itself on fitness. This dependence has two reasons: (i) The LacY pumping rate depends on the production rate, the pumping rate per LacY molecule  , and fitness,
, and fitness, 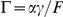 , because LacY molecules are diluted by cell divisions just like the transported molecules. This generic dependence reinforces the basic growth feedback loop by dilution, which also enters Equation 1. (ii) For fixed genotype and environment, the production rate itself can depend on fitness,
, because LacY molecules are diluted by cell divisions just like the transported molecules. This generic dependence reinforces the basic growth feedback loop by dilution, which also enters Equation 1. (ii) For fixed genotype and environment, the production rate itself can depend on fitness, 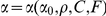 . This growth effect on gene expression generates an additional feedback between the lac pathway and fitness, which is expected under several growth conditions [13], [14]. Including this feedback in our model significantly improves the agreement between data and theory (see Materials and Methods and Text S1 for details). The fitness landscapes of Figure 4 and Figure 5 are obtained from our model using maximum-likelihood parameters, but their shape depends only on the presence of a fitness nonlinearity (
. This growth effect on gene expression generates an additional feedback between the lac pathway and fitness, which is expected under several growth conditions [13], [14]. Including this feedback in our model significantly improves the agreement between data and theory (see Materials and Methods and Text S1 for details). The fitness landscapes of Figure 4 and Figure 5 are obtained from our model using maximum-likelihood parameters, but their shape depends only on the presence of a fitness nonlinearity (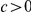 ). We now discuss their form and their biological implications in more detail.
). We now discuss their form and their biological implications in more detail.
Figure 4. Phenotype-fitness map.
The fitness cost 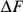 of the lac pathway is shown as a function of the protein production rate
of the lac pathway is shown as a function of the protein production rate  and the transport rate
and the transport rate 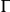 . The fitness landscape obtained from our model (shaded surface) is strongly nonlinear and has two branches. The stable part of the landscape (solid shading) ends at a fitness cliff (solid blue line), beyond which populations cannot maintain growth. The remaining part of the lower fitness branch is unstable (striped shading). Protein expression and activity of viable populations are bounded by a barrier (dotted blue line). Model predictions of pathway phenotypes and fitness for individual strains under varying inducer concentrations are shown as a family of red lines (light red: wild type, dark red: operator mutant strains). Experimental fitness values are shown as dots (the offset from the model surface is marked by gray lines).
. The fitness landscape obtained from our model (shaded surface) is strongly nonlinear and has two branches. The stable part of the landscape (solid shading) ends at a fitness cliff (solid blue line), beyond which populations cannot maintain growth. The remaining part of the lower fitness branch is unstable (striped shading). Protein expression and activity of viable populations are bounded by a barrier (dotted blue line). Model predictions of pathway phenotypes and fitness for individual strains under varying inducer concentrations are shown as a family of red lines (light red: wild type, dark red: operator mutant strains). Experimental fitness values are shown as dots (the offset from the model surface is marked by gray lines).
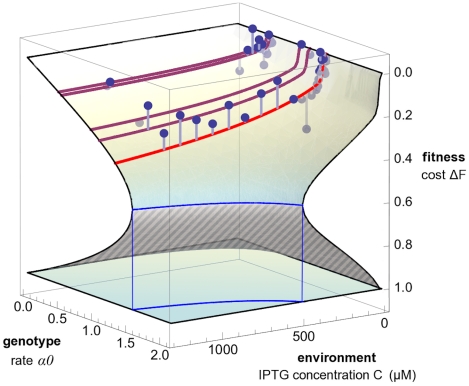
Figure 5. Genotype-environment-fitness map.
The fitness cost 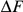 of the lac pathway is shown as a function of the operator genotype summary variable
of the lac pathway is shown as a function of the operator genotype summary variable 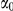 (maximum rate of protein production at a growth of 1 cell division/hr, see text) and the external inducer concentration
(maximum rate of protein production at a growth of 1 cell division/hr, see text) and the external inducer concentration 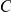 . The model fitness landscape is again strongly nonlinear: it has a stable upper branch (solid shading) and an unstable lower branch (striped shading) separated by a fitness cliff (blue line), similar to the phenotype-fitness map of Figure 4. Model predictions for individual strains under varying inducer concentrations are shown as a family of red lines (light red: wild type; dark red: operator mutant strains with
. The model fitness landscape is again strongly nonlinear: it has a stable upper branch (solid shading) and an unstable lower branch (striped shading) separated by a fitness cliff (blue line), similar to the phenotype-fitness map of Figure 4. Model predictions for individual strains under varying inducer concentrations are shown as a family of red lines (light red: wild type; dark red: operator mutant strains with 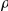 equal to wild type value, see text). Experimental fitness values are shown as dots (the offset from the model surface is marked by gray lines).
equal to wild type value, see text). Experimental fitness values are shown as dots (the offset from the model surface is marked by gray lines).
Phenotype-fitness map
The phenotype-fitness landscape of the lac pathway resulting from our model is shown in Figure 4, together with fitness measurements of different lac O1 operator mutants in different inducer environments. The experimental data are plotted as a function of the pathway phenotypes  and
and 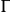 inferred from our model; for each mutant, the dependence of these phenotypes on the IPTG concentration is indicated by a red line. Data and model consistently show that protein production and activity of the lac pathway affect fitness in a highly nonlinear way. Our model explains the nonlinearity in terms of the growth feedback mechanism contained in Equation 1.
inferred from our model; for each mutant, the dependence of these phenotypes on the IPTG concentration is indicated by a red line. Data and model consistently show that protein production and activity of the lac pathway affect fitness in a highly nonlinear way. Our model explains the nonlinearity in terms of the growth feedback mechanism contained in Equation 1.
This form of the phenotype-fitness landscape has two important aspects. First, the nonlinearity of fitness translates into epistatic interactions between the pathway phenotypes: the effect of a change in the production rate  , which is proportional to the slope
, which is proportional to the slope 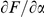 , depends on the transport rate
, depends on the transport rate 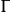 , and vice versa. Second, the fitness landscape is not univalued: for some values of
, and vice versa. Second, the fitness landscape is not univalued: for some values of  and
and 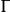 , there are two possible fitness values, for others, there is none. Phenotype values in the no-solution regime cannot be attained by a cell population in steady growth. This regime is bounded by a dotted line in the
, there are two possible fitness values, for others, there is none. Phenotype values in the no-solution regime cannot be attained by a cell population in steady growth. This regime is bounded by a dotted line in the 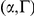 plane, which marks an expression barrier for the lac genes. The barrier occurs at a finite growth rate
plane, which marks an expression barrier for the lac genes. The barrier occurs at a finite growth rate 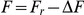 (in contrast to the model of ref. [9]). Double-valued fitness solutions and the existence of an expression barrier for given phenotype values are a direct consequence of the growth feedback loop in Equation 1. The stability analysis described below shows that only the full-shaded part of the landscape describes viable cell populations in stationary growth, whereas the striped part is unstable. Hence, for parameter values between the dotted and the solid lines in the
(in contrast to the model of ref. [9]). Double-valued fitness solutions and the existence of an expression barrier for given phenotype values are a direct consequence of the growth feedback loop in Equation 1. The stability analysis described below shows that only the full-shaded part of the landscape describes viable cell populations in stationary growth, whereas the striped part is unstable. Hence, for parameter values between the dotted and the solid lines in the 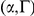 plane, populations can reach two different steady-state growth rates with the same lac pathway phenotypes.
plane, populations can reach two different steady-state growth rates with the same lac pathway phenotypes.
Genotype–environment interactions
We now turn to the dependence of fitness on the lac O1 operator sequence and on the external inducer concentration 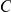 , which are the quantities we manipulate in our experiments. To display the sequence-dependence, we use the genotype summary variables
, which are the quantities we manipulate in our experiments. To display the sequence-dependence, we use the genotype summary variables 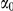 (maximal rate of lac protein production at a fixed growth rate of one cell division per hour) and
(maximal rate of lac protein production at a fixed growth rate of one cell division per hour) and 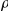 (ratio of repressed to unrepressed protein production rates). These variables reflect the double role of the operator sequence: it acts as a binding site for the repressor LacI, but it also affects other processes that lead to changes in protein production [20]. Figure 5 shows the fitness cost as a function of the maximal production rate and the IPTG concentration,
(ratio of repressed to unrepressed protein production rates). These variables reflect the double role of the operator sequence: it acts as a binding site for the repressor LacI, but it also affects other processes that lead to changes in protein production [20]. Figure 5 shows the fitness cost as a function of the maximal production rate and the IPTG concentration, 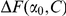 . The ratio
. The ratio 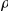 is kept fixed to its wild type value; the figure shows fitness data for the corresponding subset of strains (see Figure S1 and Figure S2 for the full dependence of
is kept fixed to its wild type value; the figure shows fitness data for the corresponding subset of strains (see Figure S1 and Figure S2 for the full dependence of 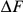 on
on 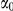 ,
, 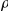 and
and 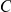 ). Again, cell populations in stationary growth cannot exist for some genotype-environment parameters; this regime is bounded by a blue line in the
). Again, cell populations in stationary growth cannot exist for some genotype-environment parameters; this regime is bounded by a blue line in the 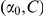 plane.
plane.
The fitness of different mutants as a function of the inducer concentration is again shown as a family of lines. The nonlinearity in the landscape indicates that the inducer environment affects the selective effect of regulatory mutations: higher IPTG concentrations lead to increased fitness differences between mutants. This interaction between genotype and the environment is due to an increase in the pumping rate with increasing IPTG, to the coupling of uptake rate and production rate in the term 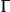 , and to the growth feedback through dilution. Figure S2 further illustrates this interaction. Genotype-environment interactions in the lac operon have been observed previously [21]. Our model shows how such interactions emerge from the basic architecture of metabolic pathways.
, and to the growth feedback through dilution. Figure S2 further illustrates this interaction. Genotype-environment interactions in the lac operon have been observed previously [21]. Our model shows how such interactions emerge from the basic architecture of metabolic pathways.
Nonlinearities generate extinction thresholds
The fitness landscapes of Figure 4 and Figure 5 have a common feature: over a wide range of parameters, there are two possible fitness values 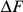 . This double-valued fitness landscape is partitioned into a stable part (full-shaded) and an unstable part (striped); see Text S1, Figure S3, and Figure S4. The stable part of the landscape describes stationary growth of viable populations; i.e., cells with growth rates close to a point on this surface reach a steady state given by a point on the surface. A large part of the lower surface
. This double-valued fitness landscape is partitioned into a stable part (full-shaded) and an unstable part (striped); see Text S1, Figure S3, and Figure S4. The stable part of the landscape describes stationary growth of viable populations; i.e., cells with growth rates close to a point on this surface reach a steady state given by a point on the surface. A large part of the lower surface 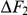 is unstable, i.e., cells with fitness cost
is unstable, i.e., cells with fitness cost 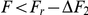 are unable to dilute their proteins and transported molecules fast enough to maintain stable growth. These cells will further decline in fitness, whereas cells with
are unable to dilute their proteins and transported molecules fast enough to maintain stable growth. These cells will further decline in fitness, whereas cells with 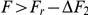 will increase fitness up to the stable value
will increase fitness up to the stable value 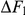 . The stable and the unstable part of the fitness landscape are separated by a fitness cliff, which is shown as a blue line in Figure 4 and 5. The cliff marks an extinction threshold: If a cell population is driven beyond this cliff by mutations or environment changes, it suffers a sudden drop in fitness and cannot maintain a finite growth rate.
. The stable and the unstable part of the fitness landscape are separated by a fitness cliff, which is shown as a blue line in Figure 4 and 5. The cliff marks an extinction threshold: If a cell population is driven beyond this cliff by mutations or environment changes, it suffers a sudden drop in fitness and cannot maintain a finite growth rate.
Existence and position of the fitness cliff depend on the amount of inducer present (see Figure 2, Figure 5, and Figure S2). For IPTG concentrations used in our and other experiments, the cliff is far from the wild type (Figure S2B). We note, however, that lactose is often used in higher concentrations, and lack of growth due to the presence of lactose (lactose killing) has indeed been observed [22].
Discussion
We have shown that in the presence of an inducer, the fitness cost of the lac pathway arises not only from protein production, but also from transport activity of the permease LacY. The cost is governed by a feedback loop, which is the result of two repressive interactions: protein activity results in reduced growth, and growth dilutes proteins as well as transported molecules (see Figure 3). We note that our feedback mechanism does not rely on a limitation of cellular resources to generate a nonlinear relation between lac gene expression and growth (in contrast to the model of ref. [9]). This feedback produces a strongly nonlinear dependence of fitness on pathway phenotypes or on genotype and environment, as shown in the fitness landscapes of Figure 4 and Figure 5. Both landscapes contain a fitness cliff, which is an extinction threshold for cell populations. The nonlinearity of fitness is likely to persist for any substrate of the permease LacY and sets an upper bound for its rates of expression and activity. Thus, changes in lac permease activity or expression can have strong impacts on fitness. This is consistent with the observation that lacY is under particularly strong selection [23], as reflected notably by its low number of synonymous single-nucleotide polymorphisms [24].
The nonlinearity of fitness and its consequences are expected to hold in the presence of lactose. If the benefit conferred by lactose (or other sugars) also depends on its internal concentration, we expect an effect of diminishing return: the faster a cell grows, the more it will dilute lactose, which leads to a sublinear increase of fitness with lactose concentration. Hence, combining costs and benefits of the lac proteins will lead to more complex fitness landscapes; their detailed dependence on pathway phenotypes will be addressed in a future study. Importantly, the full landscapes are expected to have a fitness cliff similar to the cost landscapes derived in this paper. This might explain why induced cells grown in a chemostat die after exposure to high concentrations of lactose, a phenomenon known as lactose killing [22]. Moreover, many other metabolic pathways in microorganisms contain a membrane pump or transporter accumulating substrates inside the cell, which often uses the proton motive force as an energy source. Our results are expected to apply to these pathways as well. In particular, we note the similarity of our fitness landscapes and those of the glucose utilization pathway in yeast [25] (see Figure 5 and Figure S2). Other protein activities such as hydrolysis of substrates can produce the same type of feedback, because they also depend on internal concentrations of molecules.
The shape of the fitness landscape described here has various implications for the genomic evolution of the lac pathway. Our fitness model of protein production and activity contains two types of epistasis on the operator lac O1. Within the operator, the fitness reduction caused by two mutations that increase expression is larger than the sum of the fitness costs of either one (see Figure 5). Furthermore, the selection pressure on expression depends on the protein activity rate and, hence, on the sequence of the downstream gene lac Y. The total pumping rate of the cell also depends on the concentration of LacY substrates in the environment, which generates fitness interactions between the operator genotype and the inducer environment. In a broader context, the costs of gene expression due to protein activity and due to protein production affect the evolution of regulatory systems in a different way. Taking into account only protein synthesis, we expect the length of genes to be the main determinant of the fitness cost of gene expression. Including protein activity, however, the selective pressure against expression of a gene can depend primarily on the coding sequence of functional domains and on the environment. For the lac pathway, the cost contributions of protein production and of protein activity are of similar magnitude, and both effects contribute to selection on regulatory sequences.
Generalizing the results of this study, we expect the full landscape of a metabolic network to be filled with cliffs and valleys, whose importance depends on which pathways are more active in a given environment. In addition, a metabolic pathway with growth feedback generates ubiquitous epistasis. For example, any mutation under selection has fitness interactions with mutations in the lac operon: In the presence of IPTG, deleterious (beneficial) mutations outside the lac pathway affect the protein production rate  and the transport rate
and the transport rate 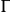 , and hence increase (reduce) the fitness cost of lac activity-enhancing mutations. Thus, higher-dimensional fitness landscapes including more and more metabolic phenotypes are expected to be increasingly rugged.
, and hence increase (reduce) the fitness cost of lac activity-enhancing mutations. Thus, higher-dimensional fitness landscapes including more and more metabolic phenotypes are expected to be increasingly rugged.
Previous experiments have produced fitness landscapes as a function of genotype (see for example [26], [27]). This kind of fitness landscapes omits the intermediate level of phenotypes, which describes how genotype changes affect biophysical functions. Here, we record fitness as a function of well-defined phenotypes of a metabolic pathway. These can be connected to a biophysical model, which describes the dependence of fitness on the operator sequence and on the inducer concentration. Phenotype- and model-based fitness landscapes are predictive: Once the model constants are fixed by one set of measurements, the model predicts the outcome of further experiments with different input parameters. In this study, the most striking model prediction is the extinction of populations beyond a fitness cliff.
Our fitness landscape also differs from previous phenotype-fitness maps, perhaps the most popular of which is Fisher's geometric model [28]. Fitting this model to fitness data is a method to infer distributions of fitness effects of mutations and of epistasic effects between mutations [29], [30]. Fisher's geometric model contains an a priori arbitrary number of unknown molecular phenotypes. In contrast, our model contains a small number of known phenotypes associated to a specific pathway, which are shown to capture salient features of fitness variation between populations (clearly, this does not rule out further phenotypes of this pathway affecting fitness). In the classical geometric model, the fitness landscape is assumed to be smooth, and different phenotypes to contribute additively to fitness. Our fitness landscape contradicts both of these assumptions: there is strong epistasis and ruggedness. These features have been extensively analyzed for genotype-fitness maps (a well-known example is the NK model [31]), but the dependence of fitness on quantitative phenotypes is generally assumed to be smoother. Our study shows that strong epistasis and ruggedness can persist in phenotype-fitness landscapes. It calls for new statistical models of such landscapes, which address their broad consequences for speed and constraints of molecular evolution. An interesting example is a recent extension of the geometric model, which contains epistasis and a fitness cliff [32].
In summary, our measurements and modeling show that the lac pathway of E. coli is governed by a strongly nonlinear fitness landscape depending on phenotypes of protein production and activity. These phenotypes, in turn, depend on the lac operon genotype and on environmental parameters in a coupled way. Fitness nonlinearities and genotype-environment interactions are not specific to the system studied here, but are likely to be general features of metabolic pathways. Thus, the fitness landscape of a metabolic network is much more than a simple superposition of the cost of protein production and the benefit of protein activity. It describes the entire network as a unit of natural selection. Such system-level fitness landscapes emerge already at simplest level of cell growth and metabolism.
Materials and Methods
Strain construction
The background of all strains used in this study is Escherichia coli BW30270  . The lacO1 mutant strains (summarized in Table S1 and Table S2) are constructed as described in [33]. First, the complete lac promoter is deleted and replaced with the chloramphenicol resistance cassette from plasmid pKD3 (see Table S3 for a list of plasmids used in this study). This yields strain S4146 which is
. The lacO1 mutant strains (summarized in Table S1 and Table S2) are constructed as described in [33]. First, the complete lac promoter is deleted and replaced with the chloramphenicol resistance cassette from plasmid pKD3 (see Table S3 for a list of plasmids used in this study). This yields strain S4146 which is  ,
, 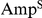 and
and 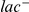 . The full lac promoter and 5′UTR of wild-type Escherichia coli are amplified and cloned (see Table S4 for a list of oligonucleotides used in this study). Specific lac O1 mutations are inserted using PCR mediated mutagenesis [34], and the mutant sequences are cloned in a high-copy-number plasmid (derived from pUC12). The same gene replacement method [33] is then used to replace the chloramphenicol resistance cassette in strain S4146 with the chosen lac promoter and O1 operator. The strains produced in this way are all
. The full lac promoter and 5′UTR of wild-type Escherichia coli are amplified and cloned (see Table S4 for a list of oligonucleotides used in this study). Specific lac O1 mutations are inserted using PCR mediated mutagenesis [34], and the mutant sequences are cloned in a high-copy-number plasmid (derived from pUC12). The same gene replacement method [33] is then used to replace the chloramphenicol resistance cassette in strain S4146 with the chosen lac promoter and O1 operator. The strains produced in this way are all 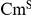 ,
, 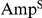 and
and 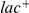 . We noticed that these strains have a general lower fitness than strain BW30270 that cannot be explained by the inserted mutations (see Figure S5) so we use T4GT7 mediated transduction [35] to transfer the lac mutations back to the parent background (BW30270). First, the resistance cassette from strain S4146 is transduced to BW30270, producing strain T218. Then, the mutated lac operon is transduced from each lacO1 mutant to T218. The lac promoter and O1 operator are then sequenced to confirm the correct insertion of the lac operator allele. As a control for the transduction, a wild type construct is obtained in the same way (T273). It has the same fitness as BW30270. The reference strain for the competition
. We noticed that these strains have a general lower fitness than strain BW30270 that cannot be explained by the inserted mutations (see Figure S5) so we use T4GT7 mediated transduction [35] to transfer the lac mutations back to the parent background (BW30270). First, the resistance cassette from strain S4146 is transduced to BW30270, producing strain T218. Then, the mutated lac operon is transduced from each lacO1 mutant to T218. The lac promoter and O1 operator are then sequenced to confirm the correct insertion of the lac operator allele. As a control for the transduction, a wild type construct is obtained in the same way (T273). It has the same fitness as BW30270. The reference strain for the competition 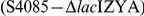 , the lac permease and the lac repressor mutants (
, the lac permease and the lac repressor mutants ( and
and  ) are constructed as described in [33]. Strain
) are constructed as described in [33]. Strain  is constructed by first deleting lac Z following [36] and then deleting lac I following [33].
is constructed by first deleting lac Z following [36] and then deleting lac I following [33].
Media and growth conditions
Unless stated otherwise, all measurements are made in M9 minimal medium with glycerol (0.1% v/v) as carbon source. To distinguish strains in competition, tetrazolium lactose (TL) medium (1% bacto-tryptone, 0.1% yeast extract, 0.5% NaCl, 1% lactose, 0.005% tetrazolium chloride and 1.5% agar) is used. 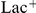 colonies are white and
colonies are white and 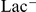 colonies are red in TL plates [37].
colonies are red in TL plates [37]. 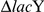 is also white on TL plates. To distinguish this strain from
is also white on TL plates. To distinguish this strain from 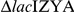 and
and  , LB-XGal-IPTG plates are used (1% bacto-tryptone, 0.5% yeast extract, 0.5% NaCl, 1 mM isopropyl-1-thio-
, LB-XGal-IPTG plates are used (1% bacto-tryptone, 0.5% yeast extract, 0.5% NaCl, 1 mM isopropyl-1-thio-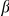 -D-galactoside (IPTG),
-D-galactoside (IPTG), 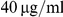 5-Bromo-4-chloro-3-indolyl-
5-Bromo-4-chloro-3-indolyl-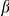 -D-galactoside (X-Gal) and 1.5% agar).
-D-galactoside (X-Gal) and 1.5% agar).
Protein expression
Protein concentration is estimated using a 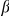 -galactosidase (LacZ) activity assay [38]. Since all our mutants have the same coding sequence for this protein, changes in activity reflect changes in protein concentration inside the cell. The LacZ assays are performed as described in [38]. Overnight cultures are diluted in fresh medium to an optical density at 600 nm (OD600) of 0.05 and harvested after growth in the indicated media at
-galactosidase (LacZ) activity assay [38]. Since all our mutants have the same coding sequence for this protein, changes in activity reflect changes in protein concentration inside the cell. The LacZ assays are performed as described in [38]. Overnight cultures are diluted in fresh medium to an optical density at 600 nm (OD600) of 0.05 and harvested after growth in the indicated media at 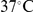 to an OD600 of 0.3. IPTG was added to the overnight culture and to the test cultures in the concentrations mentioned in the text. The enzyme activities are determined from at least three independent cultures. Figure S6 shows the measured LacZ levels for all strains used in this study, in absence and presence of IPTG.
to an OD600 of 0.3. IPTG was added to the overnight culture and to the test cultures in the concentrations mentioned in the text. The enzyme activities are determined from at least three independent cultures. Figure S6 shows the measured LacZ levels for all strains used in this study, in absence and presence of IPTG.
Fitness measurements
Fitness is measured in head to head competition as described in [39]. Briefly, frozen cultures (stored at 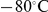 ) are streaked on a Luria broth agar plate and grown over night at
) are streaked on a Luria broth agar plate and grown over night at 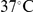 . An isolated colony is randomly selected and grown overnight in 3 ml of the same medium used in the competition, in particular with the same amount of IPTG. Both the reference strain (
. An isolated colony is randomly selected and grown overnight in 3 ml of the same medium used in the competition, in particular with the same amount of IPTG. Both the reference strain (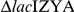 , unless stated otherwise) and assay strains are treated in this way separately. The strains are then mixed and diluted in saline solution (10 mM
, unless stated otherwise) and assay strains are treated in this way separately. The strains are then mixed and diluted in saline solution (10 mM 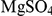 and 0.85% NaCl), such that about 50,000 colony forming units (CFUs) of each strain are used to start the competition. The mixed dilutions are also used to count the starting titer. Cultures are grown for 24 h on 96 deep-well plates in 1 ml of medium, shaken at 150 RPM, reaching saturation (
and 0.85% NaCl), such that about 50,000 colony forming units (CFUs) of each strain are used to start the competition. The mixed dilutions are also used to count the starting titer. Cultures are grown for 24 h on 96 deep-well plates in 1 ml of medium, shaken at 150 RPM, reaching saturation (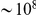 CFUs). They are then diluted and plated on TL or LB-XGal-IPTG medium.
CFUs). They are then diluted and plated on TL or LB-XGal-IPTG medium.
We measure the Malthusian fitness 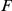 , i.e., the growth rate, of each strain in units of the growth rate of the reference strain (such that
, i.e., the growth rate, of each strain in units of the growth rate of the reference strain (such that 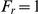 ). The fitness value of a mutant is inferred from a competition experiment with the reference strain,
). The fitness value of a mutant is inferred from a competition experiment with the reference strain,
where 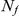 ,
, 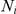 are the final and initial number of mutant CFUs after and before the competition, and
are the final and initial number of mutant CFUs after and before the competition, and 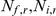 are the corresponding numbers for the reference strain. The growth rate of the reference strain is not affected by IPTG (see Figure S7). Thus, the doubling time of the reference strain is a fixed time unit and fitness measurements across environments are directly comparable. We report the fitness cost of a mutant compared to the reference strain,
are the corresponding numbers for the reference strain. The growth rate of the reference strain is not affected by IPTG (see Figure S7). Thus, the doubling time of the reference strain is a fixed time unit and fitness measurements across environments are directly comparable. We report the fitness cost of a mutant compared to the reference strain, 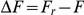 (which is proportional to its selection coefficient measured in units of doubling time,
(which is proportional to its selection coefficient measured in units of doubling time, 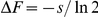 [40]).
[40]).
The  strain has the same phenotype as the reference strain (both are red on TL plates and white on LB-XGal-IPTG plates), so the two cannot be competed directly. Instead, we measure the fitness of this strain by competing it with
strain has the same phenotype as the reference strain (both are red on TL plates and white on LB-XGal-IPTG plates), so the two cannot be competed directly. Instead, we measure the fitness of this strain by competing it with 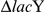 .
. 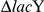 has the same fitness as the reference strain in competition in glycerol minimal medium with 1 mM lactose (
has the same fitness as the reference strain in competition in glycerol minimal medium with 1 mM lactose (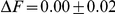 ).
).
Dependence of pathway phenotypes on genotype and environment
As explained in the Results section, the protein production rate  and the transport rate
and the transport rate 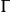 can be expressed in terms of genotypic and environmental parameters, and fitness. This map relates the fitness landscapes of Figure 4 and Figure 5 and can be obtained as follows.
can be expressed in terms of genotypic and environmental parameters, and fitness. This map relates the fitness landscapes of Figure 4 and Figure 5 and can be obtained as follows.
The first phenotype of the lac pathway, the protein production rate  , has two main components: one is independent of the lac repressor (LacI) and the other depends on the probability of the repressor to bind the operator. The independent component is given by the direct effect of the operator sequence (quantified by the first genetic component
, has two main components: one is independent of the lac repressor (LacI) and the other depends on the probability of the repressor to bind the operator. The independent component is given by the direct effect of the operator sequence (quantified by the first genetic component 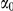 ) and by the growth rate
) and by the growth rate 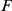 (through a function
(through a function 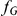 specified below). The LacI-dependent component of
specified below). The LacI-dependent component of  depends on the affinity of the operator sequence (measured by the second genetic component
depends on the affinity of the operator sequence (measured by the second genetic component 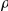 ) and on the concentration of inducer
) and on the concentration of inducer 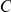 in the environment. The dependence on
in the environment. The dependence on 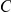 has the form of a Hill function
has the form of a Hill function 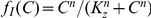 with parameters
with parameters 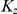 (the half saturation constant, taken to be
(the half saturation constant, taken to be 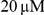 ) and
) and  (the Hill coefficient, taken to be
(the Hill coefficient, taken to be 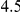 ) [18]. The protein production rate is then
) [18]. The protein production rate is then 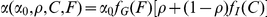 . We now derive the form of
. We now derive the form of 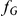 , and estimate
, and estimate 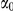 and
and 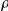 .
.
As mentioned before, 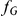 is the dependence of the production rate on the growth rate
is the dependence of the production rate on the growth rate 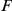 . Following [13], [14],
. Following [13], [14], 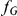 is expressed relative to the fitness of a strain growing at the rate of 1 doubling/hour,
is expressed relative to the fitness of a strain growing at the rate of 1 doubling/hour, 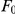 , such that
, such that 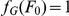 . Note that the reference strain has a growth rate of
. Note that the reference strain has a growth rate of 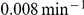 , so
, so 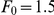 . The parameter
. The parameter 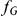 reflects the following observation: When the growth rate changes due to nutrient quality, there is a linear inverse correlation between protein concentration (
reflects the following observation: When the growth rate changes due to nutrient quality, there is a linear inverse correlation between protein concentration (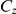 ) and growth rate [14],
) and growth rate [14], 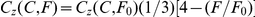 (see Figure S8A). This relationship can be extended to the protein production rate
(see Figure S8A). This relationship can be extended to the protein production rate  , because
, because 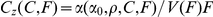 at steady state. We choose a linear dependence of the cell volume
at steady state. We choose a linear dependence of the cell volume 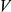 on the fitness,
on the fitness, 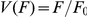 ; see Text S1 and Figure S8B. Using the dependences inferred above and assuming
; see Text S1 and Figure S8B. Using the dependences inferred above and assuming 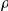 to be independent of
to be independent of 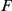 , the dependence of protein production on growth rate can be estimated:
, the dependence of protein production on growth rate can be estimated: 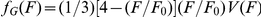 . We have verified that including
. We have verified that including 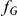 significantly improves the agreement between model and data (see Text S1), although it is not obvious a priori that a correlation between
significantly improves the agreement between model and data (see Text S1), although it is not obvious a priori that a correlation between 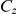 and growth rate is relevant in the context of our experiments.
and growth rate is relevant in the context of our experiments.
The two genetic components, 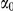 (the maximal protein production rate at fixed growth rate) and
(the maximal protein production rate at fixed growth rate) and 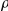 (the ratio of repressed to unrepressed protein production rates), depend only on the genotype and were calculated for each strain separately.
(the ratio of repressed to unrepressed protein production rates), depend only on the genotype and were calculated for each strain separately. 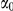 can be derived from the protein concentration and fitness measured at a concentration
can be derived from the protein concentration and fitness measured at a concentration 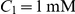 IPTG, where the LacI proteins cannot bind DNA (
IPTG, where the LacI proteins cannot bind DNA (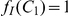 ). As explained above, the cell volume
). As explained above, the cell volume 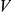 , the growth rate
, the growth rate 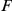 , and the effects of growth on expression
, and the effects of growth on expression 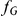 affect
affect 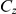 , such that
, such that 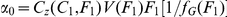 , where
, where 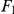 is the measured growth rate at 1 mM of IPTG. Similarly,
is the measured growth rate at 1 mM of IPTG. Similarly, 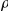 can be estimated using
can be estimated using 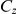 and
and 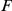 measured at 0 mM of IPTG:
measured at 0 mM of IPTG: 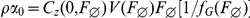 , where
, where 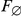 is fitness in the absence of IPTG. Both
is fitness in the absence of IPTG. Both 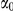 and
and 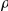 are independent of the model in Equation 1 and of the growth-dependence of the volume. Inferred values of
are independent of the model in Equation 1 and of the growth-dependence of the volume. Inferred values of 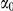 and
and 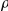 are shown in Figure S9 and Table S1. The parameter
are shown in Figure S9 and Table S1. The parameter 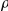 is related to the “repression level”
is related to the “repression level” 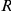 defined by Müller-Hill and co-workers as the ratio of LacZ activity between strains differing only by the presence/absence of the lac repressor,
defined by Müller-Hill and co-workers as the ratio of LacZ activity between strains differing only by the presence/absence of the lac repressor, 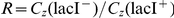 [19]. Neglecting the growth difference between both strains, these quantities are inversely related,
[19]. Neglecting the growth difference between both strains, these quantities are inversely related, 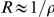 .
.
The second phenotype of the lac pathway, the total transport rate 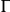 , is the product of the number of LacY molecules in the cell and the transport rate per LacY molecule,
, is the product of the number of LacY molecules in the cell and the transport rate per LacY molecule, 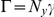 . The number
. The number 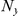 is equal to
is equal to 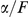 , with
, with  the protein production rate and
the protein production rate and 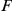 the growth rate, because LacY molecules are diluted by cell divisions. Note that
the growth rate, because LacY molecules are diluted by cell divisions. Note that  is measured for LacZ, but all proteins of the operon are produced proportionally. The ratio of LacY molecules per LacZ molecule, which is close to 3 [10], and other numerical constants are absorbed in the coefficients
is measured for LacZ, but all proteins of the operon are produced proportionally. The ratio of LacY molecules per LacZ molecule, which is close to 3 [10], and other numerical constants are absorbed in the coefficients  ,
, 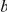 and
and  . The transport rate
. The transport rate  depends on the external IPTG concentration,
depends on the external IPTG concentration, 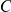 , and on the half-saturation constant for inducer uptake,
, and on the half-saturation constant for inducer uptake, 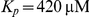 [41]. An expression for
[41]. An expression for  can be derived from the known functioning of the permease [42], with efflux neglected (see Text S1). We obtain
can be derived from the known functioning of the permease [42], with efflux neglected (see Text S1). We obtain 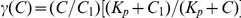 , normalizing
, normalizing  to
to 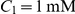 of IPTG.
of IPTG.
The uncertainties on  and
and  are obtained by standard error propagation, assuming independent experimental errors on
are obtained by standard error propagation, assuming independent experimental errors on  and
and 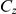 (see Text S1). A possible error in the IPTG concentration
(see Text S1). A possible error in the IPTG concentration 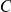 is not considered, because it is expected to be small.
is not considered, because it is expected to be small.
The coefficients  ,
, 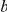 and
and  in Equation 1 are obtained by likelihood analysis of our model and the experimental data. This analysis is based on the dependence
in Equation 1 are obtained by likelihood analysis of our model and the experimental data. This analysis is based on the dependence 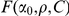 , where
, where 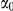 and
and 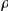 are inferred for each mutant as described above. The fitting procedure and score-based model comparisons are detailed in Text S1 (see also Figure S10 and Figure S11).
are inferred for each mutant as described above. The fitting procedure and score-based model comparisons are detailed in Text S1 (see also Figure S10 and Figure S11).
Supporting Information
Fitness cost 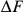 as a function of the IPTG concentration for five strains: (A) the wild type, (B) T274, (C) T320, (D)
as a function of the IPTG concentration for five strains: (A) the wild type, (B) T274, (C) T320, (D)  , (E) T275. The full lines are model predictions; dots show the experimental data (the error bars represent the standard error of the mean on 4 replicates, on 12 replicates for data at 0 and 1 mM IPTG). See Table S1 for a list of strains and the corresponding values of
, (E) T275. The full lines are model predictions; dots show the experimental data (the error bars represent the standard error of the mean on 4 replicates, on 12 replicates for data at 0 and 1 mM IPTG). See Table S1 for a list of strains and the corresponding values of 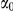 and
and 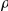 .
.
(TIF)
Fitness cost 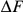 as a function of
as a function of 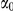 and
and 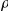 , at fixed external IPTG concentration (A)
, at fixed external IPTG concentration (A) 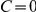 , (B)
, (B) 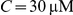 and (C)
and (C) 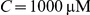 ; as a function of
; as a function of 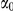 and
and 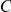 , at fixed (D)
, at fixed (D) 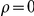 , (E)
, (E) 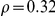 and (F)
and (F) 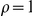 . The dots are the experimental data, the grey vertical bars show the distance between data and model prediction. Strains shown: (A) and (C) all operator mutants, wild type,
. The dots are the experimental data, the grey vertical bars show the distance between data and model prediction. Strains shown: (A) and (C) all operator mutants, wild type,  (D) wild type, T319, T320, T378, T379; (B) wild type, T274, T275, T320,
(D) wild type, T319, T320, T378, T379; (B) wild type, T274, T275, T320,  ; (E) T323; (F) T275,
; (E) T323; (F) T275,  . The light-green surfaces show the stable solutions, the dark gray the unstable one. The blue line marks their boundary: when
. The light-green surfaces show the stable solutions, the dark gray the unstable one. The blue line marks their boundary: when 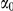 or the external IPTG concentration is increased beyond this “cliff”, the population falls on the no-growth solution, and thus goes to extinction. Panel D is identical to Figure 5 of the main text.
or the external IPTG concentration is increased beyond this “cliff”, the population falls on the no-growth solution, and thus goes to extinction. Panel D is identical to Figure 5 of the main text.
(TIF)
Dynamical analysis. (A) 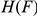 for the wild type at different IPTG concentrations: 0.1 mM (red), 1 mM (blue) and 100 mM (green). The steady-state solutions
for the wild type at different IPTG concentrations: 0.1 mM (red), 1 mM (blue) and 100 mM (green). The steady-state solutions 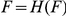 lie at the intersection of
lie at the intersection of 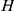 with the first bisecting line (black line); if it crosses it from above, the solution is stable (full dot), otherwise it is unstable (empty dot). (B) Time evolution of the growth rate of the wild type in 1 mM IPTG obtained by a discrete process (Equation 8 of Text S1, with a time step
with the first bisecting line (black line); if it crosses it from above, the solution is stable (full dot), otherwise it is unstable (empty dot). (B) Time evolution of the growth rate of the wild type in 1 mM IPTG obtained by a discrete process (Equation 8 of Text S1, with a time step 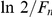 between step
between step 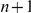 and
and  ; dots) and a continuous-time description (Equation 10 of Text S1; lines), for various initial growth rates. The generation time in minutes is
; dots) and a continuous-time description (Equation 10 of Text S1; lines), for various initial growth rates. The generation time in minutes is 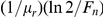 , with
, with 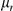 the growth rate of the reference strain measured to be
the growth rate of the reference strain measured to be 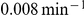 . The full black line shows the stable steady state, the dashed line the unstable one. See Text S1 for definitions.
. The full black line shows the stable steady state, the dashed line the unstable one. See Text S1 for definitions.
(TIF)
Fitness cost 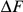 as a function (A) of the protein synthesis rate
as a function (A) of the protein synthesis rate  , (B) of the protein concentration
, (B) of the protein concentration 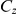 . The dots show the measured fitness cost for different strains, in absence of IPTG (blue circles) and in 1 mM IPTG (mauve squares). The red dot shows the fitness cost measured for
. The dots show the measured fitness cost for different strains, in absence of IPTG (blue circles) and in 1 mM IPTG (mauve squares). The red dot shows the fitness cost measured for  in 1 mM IPTG. The data shown in panel B are the same as those shown in Figure 2 of the main text. Error bars represent the standard error of the mean. The lines are the theoretical prediction, in absence of IPTG (blue) and in 1 mM IPTG (mauve). The dashed lines show the unstable solutions. The gray lines show the correlation of
in 1 mM IPTG. The data shown in panel B are the same as those shown in Figure 2 of the main text. Error bars represent the standard error of the mean. The lines are the theoretical prediction, in absence of IPTG (blue) and in 1 mM IPTG (mauve). The dashed lines show the unstable solutions. The gray lines show the correlation of  and
and 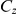 with
with 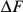 due to growth effects (see Text S1), for different values of
due to growth effects (see Text S1), for different values of 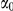 . Starting from an initial selection coefficient (e.g., upon a change of medium), a given strain moves along a gray line toward the stable steady-state solution, and away from the unstable one.
. Starting from an initial selection coefficient (e.g., upon a change of medium), a given strain moves along a gray line toward the stable steady-state solution, and away from the unstable one.
(TIF)
Comparison of protein expression (left) and fitness cost 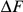 (right) on control strains. BW30270 is the wild type strain, T45 is a direct Datsenko-Wanner wild type construction and T273 is a transduction wild-type construction which went through the same procedures as all the lac operon mutants. Measurements were made in glycerol minimal medium without IPTG (white) and with 1 mM of IPTG (blue). Fitness was measured in competition against
(right) on control strains. BW30270 is the wild type strain, T45 is a direct Datsenko-Wanner wild type construction and T273 is a transduction wild-type construction which went through the same procedures as all the lac operon mutants. Measurements were made in glycerol minimal medium without IPTG (white) and with 1 mM of IPTG (blue). Fitness was measured in competition against 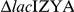 . See Materials and Methods of the main text for a description of the strain constructions and competition experiments. The error bars represent the standard error of the mean.
. See Materials and Methods of the main text for a description of the strain constructions and competition experiments. The error bars represent the standard error of the mean.
(TIF)
Expression levels of the different lac operator mutants. Protein expression was measured as described in Materials and Methods of the main text without IPTG (white) and with 1 mM of IPTG (blue). The error bars represent the standard error of the mean, with at least three replicates in each condition.
(TIF)
Growth rate, in 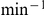 , measured in the same conditions as described for the competition experiments, except each strain was grown separately. Every hour, for 10 hours,
, measured in the same conditions as described for the competition experiments, except each strain was grown separately. Every hour, for 10 hours, 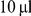 of the culture was taken and diluted appropriately, then plated on LB plates. Their mean lag phase was about 2 hours, therefore points 0, 1 hour and 2 hours were not used to estimate the growth rate. The growth rate
of the culture was taken and diluted appropriately, then plated on LB plates. Their mean lag phase was about 2 hours, therefore points 0, 1 hour and 2 hours were not used to estimate the growth rate. The growth rate 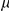 was estimated as the slope of the regression of
was estimated as the slope of the regression of 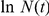 on time
on time  , where
, where 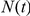 is the population size, such that:
is the population size, such that: 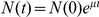 . The error bars represent the standard error of the mean of 3 independent replicates. The Malthusian fitness
. The error bars represent the standard error of the mean of 3 independent replicates. The Malthusian fitness 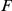 defined in Materials and Methods of the main text is equal to
defined in Materials and Methods of the main text is equal to  , with the growth rate of the reference strain
, with the growth rate of the reference strain 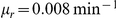 .
.
(TIF)
Growth effects on gene expression and cell volume. (A) The protein concentration 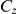 of a constitutively expressed gene has been proposed to correlate linearly with the growth rate
of a constitutively expressed gene has been proposed to correlate linearly with the growth rate 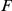 (red line), instead of the hyperbolic dependence dilution alone would induce (black line) [14]. (B) The cell volume
(red line), instead of the hyperbolic dependence dilution alone would induce (black line) [14]. (B) The cell volume 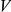 also correlates with
also correlates with 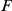 ; dots show experimental data taken from [13]; we choose to represent this correlation via a simple proportional dependence (red line). (C) Both correlations lead to a dependence
; dots show experimental data taken from [13]; we choose to represent this correlation via a simple proportional dependence (red line). (C) Both correlations lead to a dependence 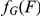 of the rate of protein synthesis on the growth rate
of the rate of protein synthesis on the growth rate 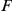 (red line; see Materials and Methods of the main text). Following [13], [14], the dependences are shown relative to the values at a growth rate
(red line; see Materials and Methods of the main text). Following [13], [14], the dependences are shown relative to the values at a growth rate 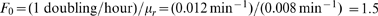 . The highlighted area
. The highlighted area 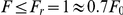 shows the range of growth rates relevant in this study.
shows the range of growth rates relevant in this study.
(TIF)
Estimated maximal rate of expression at 1 doubling/hour 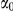 and ratio of repressed to unrepressed rates
and ratio of repressed to unrepressed rates 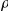 (see Materials and Methods of the main text), for all mutants used in this study. The wild type (red) and
(see Materials and Methods of the main text), for all mutants used in this study. The wild type (red) and  (orange) are barely distinguishable, as expected. In purple, the mutants which have a
(orange) are barely distinguishable, as expected. In purple, the mutants which have a 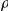 value very close to that of the wild type and are shown on Figure 5 of the main text (these are strains T319, T320, T378 and T379). In yellow, the whole operator mutants (T274, T275, T318). In green, the strain
value very close to that of the wild type and are shown on Figure 5 of the main text (these are strains T319, T320, T378 and T379). In yellow, the whole operator mutants (T274, T275, T318). In green, the strain  . The values of
. The values of 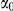 and
and 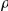 are reported in Table S1. Errors were computed as described in Text S1.
are reported in Table S1. Errors were computed as described in Text S1.
(TIF)
Statistical score of the model for a range of coefficients 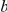 and
and  , with
, with  fixed at its fitted value. The higher the score, the lighter the shading color. The contours are drawn at scores −420, −450, −500, −550, −600, −650, −700, and −750. The highest score −417 is obtained for
fixed at its fitted value. The higher the score, the lighter the shading color. The contours are drawn at scores −420, −450, −500, −550, −600, −650, −700, and −750. The highest score −417 is obtained for 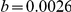 and
and 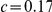 (red dot), significantly better than the best model with
(red dot), significantly better than the best model with 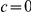 (which has score −426).
(which has score −426).
(TIF)
(A) Fitness cost 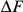 as a function of
as a function of 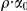 , in absence of IPTG. (B) Fitness cost
, in absence of IPTG. (B) Fitness cost 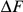 as a function of
as a function of 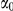 , in 1 mM IPTG. Dots show the selection coefficient measured for different strains (error bars represent the standard error of the mean), lines are model predictions. In presence of IPTG, the stable solution shown as a full line in panel B was used to compute the score
, in 1 mM IPTG. Dots show the selection coefficient measured for different strains (error bars represent the standard error of the mean), lines are model predictions. In presence of IPTG, the stable solution shown as a full line in panel B was used to compute the score 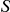 and fit the data.
and fit the data. 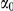 and
and 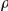 are estimated for each strain as explained in Materials and Methods of the main text. Errors were computed as described in Text S1.
are estimated for each strain as explained in Materials and Methods of the main text. Errors were computed as described in Text S1.
(TIF)
List of the strain studied, their lac O1 alleles and sequences (starting at the +1 site; underlined: mutations with respect to the wild type). The estimated values for the maximum rate of protein synthesis at 1 doubling/hour 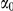 and ratio of repressed to unrepressed rates
and ratio of repressed to unrepressed rates 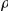 are also shown (see Materials and Methods of the main text). Errors were computed as described in Text S1.
are also shown (see Materials and Methods of the main text). Errors were computed as described in Text S1.
(PDF)
List of the strains used in this study, their genotype and the way they were constructed.
(PDF)
List of the plasmids used in this study, their relevant traits and the way they were obtained.
(PDF)
List of the oligonucleotides used in this study, their sequence and the strain for the construction of which they were used.
(PDF)
Supplementary Material.
(PDF)
Acknowledgments
We thank I. Bouchara and T. Stratmann for strains S4085 and S4197; O. Tenaillon, T. Hwa and an anonymous reviewer for helpful comments.
Footnotes
The authors have declared that no competing interests exist.
This work was funded by the Deutsche Forschungsgemeinschaft grant SFB 680 and Fundação para a Ciência e Tecnologia grant SFRH\BPD\43224\2008 (LP). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 2.Zamenhof S, Eichhorn HH. Study of microbial evolution through loss of biosynthetic functions: establishment of ‘defective’ mutants. Nature. 1967;216:456–458. doi: 10.1038/216456a0. [DOI] [PubMed] [Google Scholar]
- 3.Andrews KJ, Hegeman GD. Selective disadvantage of non-functional protein synthesis in Escherichia coli. J Mol Evol. 1976;8:317–328. doi: 10.1007/BF01739257. [DOI] [PubMed] [Google Scholar]
- 4.Koch AL. The protein burden of lac operon products. J Mol Evol. 1983;19:455–462. doi: 10.1007/BF02102321. [DOI] [PubMed] [Google Scholar]
- 5.Vind J, Sørensen MA, Rasmussen MD, Pedersen S. Synthesis of proteins in Escherichia coli is limited by the concentration of free ribosomes. Expression from reporter genes does not always reect functional mRNA levels. J Mol Biol. 1993;231:678–688. doi: 10.1006/jmbi.1993.1319. [DOI] [PubMed] [Google Scholar]
- 6.Dong H, Nilsson L, Kurland CG. Gratuitous overexpression of genes in Escherichia coli leads to growth inhibition and ribosome destruction. J Bacteriol. 1995;177:1497–1504. doi: 10.1128/jb.177.6.1497-1504.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müller-Hill B. The lac operon: a short history of a genetic paradigm. Walter de Gruyter; 1996. [Google Scholar]
- 8.Novick A, Weiner M. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci U S A. 1957;43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- 10.Stoebel DM, Dean AM, Dykhuizen DE. The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics. 2008;178:1653–1660. doi: 10.1534/genetics.107.085399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.v Hofsten B. The inhibitory effect of galactosides on the growth of Escherichia coli. Biochimica et Biophysica Acta. 1961;48:164–171. doi: 10.1016/0006-3002(61)90528-5. [DOI] [PubMed] [Google Scholar]
- 12.Wilson DM, Putzrath RM, Wilson TH. Inhibition of growth of Escherichia coli by lactose and other galactosides. Biochim Biophys Acta (BBA) - Biomembranes. 1981;649:377–384. doi: 10.1016/0005-2736(81)90427-2. [DOI] [PubMed] [Google Scholar]
- 13.Klumpp S, Zhang Z, Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell. 2009;139:1366–1375. doi: 10.1016/j.cell.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. Interdependence of Cell Growth and Gene Expression: Origins and Consequences. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- 15.Elf J, Nilsson K, Tenson T, Ehrenberg M. Bistable bacterial growth rate in response to antibiotics with low membrane permeability. Phys Rev Lett. 2006;97:258104. doi: 10.1103/PhysRevLett.97.258104. [DOI] [PubMed] [Google Scholar]
- 16.Fange D, Nilsson K, Tenson T, Ehrenberg M. Drug efflux pump deficiency and drug target resistance masking in growing bacteria. Proc Natl Acad Sci U S A. 2009;106:8215–8220. doi: 10.1073/pnas.0811514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan C, Marguet P, You L. Emergent bistability by a growth-modulating positive feedback circuit. Nat Chem Biol. 2009;5:842–848. doi: 10.1038/nchembio.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhlman T, Zhang Z, Saier MH, Hwa T. Combinatorial transcriptional control of the lactose operon of Escherichia coli. Proc Natl Acad Sci U S A. 2007;104:6043–6048. doi: 10.1073/pnas.0606717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehming N, Sartorius J, Niemöller M, Genenger G, von Wilcken-Bergmann B, et al. The interaction of the recognition helix of lac repressor with lac operator. EMBO J. 1987;6:3145–3153. doi: 10.1002/j.1460-2075.1987.tb02625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yarchuk O, Jacques N, Guillerez J, Dreyfus M. Interdependence of translation, transcription and mRNA degradation in the lacZ genes. J Mol Biol. 1992;226:581–596. doi: 10.1016/0022-2836(92)90617-s. [DOI] [PubMed] [Google Scholar]
- 21.Dean AM. A molecular investigation of genotype by environment interactions. Genetics. 1995;139:19–33. doi: 10.1093/genetics/139.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dykhuizen D, Hartl D. Transport by the lactose permease of Escherichia coli as the basis of lactose killing. J Bacteriol. 1978;135:876–882. doi: 10.1128/jb.135.3.876-882.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dykhuizen DE, Dean AM, Hartl DL. Metabolic ux and fitness. Genetics. 1987;115:25–31. doi: 10.1093/genetics/115.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner RR, Riley MA. Low synonymous site variation at the lacY locus in Escherichia coli suggests the action of positive selection. J Mol Evol. 1996;42:79–84. doi: 10.1007/BF02198831. [DOI] [PubMed] [Google Scholar]
- 25.Youk H, van Oudenaarden A. Growth landscape formed by perception and import of glucose in yeast. Nature. 2009;462:875–879. doi: 10.1038/nature08653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 27.de Visser JAGM, Park SC, Krug J. Exploring the effect of sex on empirical fitness landscapes. Am Nat. 2009;174(Suppl 1):S15–S30. doi: 10.1086/599081. [DOI] [PubMed] [Google Scholar]
- 28.Fisher RA. The Genetical Theory of Natural Selection. Oxford University Press; 1930. [Google Scholar]
- 29.Orr HA. The distribution of fitness effects among beneficial mutations in Fisher's geometric model of adaptation. J Theor Biol. 2006;238:279–285. doi: 10.1016/j.jtbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Martin G, Elena SF, Lenormand T. Distributions of epistasis in microbes fit predictions from a fitness landscape model. Nat Genet. 2007;39:555–560. doi: 10.1038/ng1998. [DOI] [PubMed] [Google Scholar]
- 31.Kauffman S, Levin S. Towards a general theory of adaptive walks on rugged landscapes. J Theor Biol. 1987;128:11–45. doi: 10.1016/s0022-5193(87)80029-2. [DOI] [PubMed] [Google Scholar]
- 32.Gros PA, Nagard HL, Tenaillon O. The evolution of epistasis and its links with genetic robustness, complexity and drift in a phenotypic model of adaptation. Genetics. 2009;182:277–293. doi: 10.1534/genetics.108.099127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Russell D. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 35.Wilson GG, Young KY, Edlin GJ, Konigsberg W. High-frequency generalised transduction by bacteriophage T4. Nature. 1979;280:80–82. doi: 10.1038/280080a0. [DOI] [PubMed] [Google Scholar]
- 36.Hamilton CM, Aldea M, Washburn BK, Babitzke P, Kushner SR. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen TN, Phan QG, Duong LP, Bertrand KP, Lenski RE. Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol Biol Evol. 1989;6:213–225. doi: 10.1093/oxfordjournals.molbev.a040545. [DOI] [PubMed] [Google Scholar]
- 38.Miller JH. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 39.Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- 40.Chevin LM. On measuring selection in experimental evolution. Biol Lett. 2011;7:210–213. doi: 10.1098/rsbl.2010.0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng B, Fournier RL, Relue PA, Schisler J. An experimental and theoretical study of the inhibition of Escherichia coli lac operon gene expression by antigene oligonucleotides. Biotechnology and bioengineering. 2001;74:220–229. doi: 10.1002/bit.1111. [DOI] [PubMed] [Google Scholar]
- 42.Noel JT, Pilyugin SS, Narang A. The diffusive inux and carrier efflux have a strong effect on the bistability of the lac operon in Escherichia coli. J Theor Biol. 2009;256:14–28. doi: 10.1016/j.jtbi.2008.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fitness cost 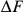 as a function of the IPTG concentration for five strains: (A) the wild type, (B) T274, (C) T320, (D)
as a function of the IPTG concentration for five strains: (A) the wild type, (B) T274, (C) T320, (D)  , (E) T275. The full lines are model predictions; dots show the experimental data (the error bars represent the standard error of the mean on 4 replicates, on 12 replicates for data at 0 and 1 mM IPTG). See Table S1 for a list of strains and the corresponding values of
, (E) T275. The full lines are model predictions; dots show the experimental data (the error bars represent the standard error of the mean on 4 replicates, on 12 replicates for data at 0 and 1 mM IPTG). See Table S1 for a list of strains and the corresponding values of 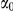 and
and 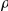 .
.
(TIF)
Fitness cost 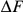 as a function of
as a function of 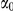 and
and 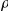 , at fixed external IPTG concentration (A)
, at fixed external IPTG concentration (A) 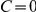 , (B)
, (B) 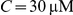 and (C)
and (C) 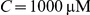 ; as a function of
; as a function of 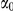 and
and 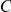 , at fixed (D)
, at fixed (D) 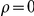 , (E)
, (E) 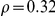 and (F)
and (F) 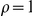 . The dots are the experimental data, the grey vertical bars show the distance between data and model prediction. Strains shown: (A) and (C) all operator mutants, wild type,
. The dots are the experimental data, the grey vertical bars show the distance between data and model prediction. Strains shown: (A) and (C) all operator mutants, wild type,  (D) wild type, T319, T320, T378, T379; (B) wild type, T274, T275, T320,
(D) wild type, T319, T320, T378, T379; (B) wild type, T274, T275, T320,  ; (E) T323; (F) T275,
; (E) T323; (F) T275,  . The light-green surfaces show the stable solutions, the dark gray the unstable one. The blue line marks their boundary: when
. The light-green surfaces show the stable solutions, the dark gray the unstable one. The blue line marks their boundary: when 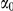 or the external IPTG concentration is increased beyond this “cliff”, the population falls on the no-growth solution, and thus goes to extinction. Panel D is identical to Figure 5 of the main text.
or the external IPTG concentration is increased beyond this “cliff”, the population falls on the no-growth solution, and thus goes to extinction. Panel D is identical to Figure 5 of the main text.
(TIF)
Dynamical analysis. (A) 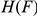 for the wild type at different IPTG concentrations: 0.1 mM (red), 1 mM (blue) and 100 mM (green). The steady-state solutions
for the wild type at different IPTG concentrations: 0.1 mM (red), 1 mM (blue) and 100 mM (green). The steady-state solutions 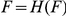 lie at the intersection of
lie at the intersection of 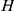 with the first bisecting line (black line); if it crosses it from above, the solution is stable (full dot), otherwise it is unstable (empty dot). (B) Time evolution of the growth rate of the wild type in 1 mM IPTG obtained by a discrete process (Equation 8 of Text S1, with a time step
with the first bisecting line (black line); if it crosses it from above, the solution is stable (full dot), otherwise it is unstable (empty dot). (B) Time evolution of the growth rate of the wild type in 1 mM IPTG obtained by a discrete process (Equation 8 of Text S1, with a time step 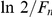 between step
between step 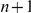 and
and  ; dots) and a continuous-time description (Equation 10 of Text S1; lines), for various initial growth rates. The generation time in minutes is
; dots) and a continuous-time description (Equation 10 of Text S1; lines), for various initial growth rates. The generation time in minutes is 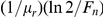 , with
, with 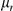 the growth rate of the reference strain measured to be
the growth rate of the reference strain measured to be 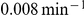 . The full black line shows the stable steady state, the dashed line the unstable one. See Text S1 for definitions.
. The full black line shows the stable steady state, the dashed line the unstable one. See Text S1 for definitions.
(TIF)
Fitness cost 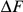 as a function (A) of the protein synthesis rate
as a function (A) of the protein synthesis rate  , (B) of the protein concentration
, (B) of the protein concentration 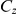 . The dots show the measured fitness cost for different strains, in absence of IPTG (blue circles) and in 1 mM IPTG (mauve squares). The red dot shows the fitness cost measured for
. The dots show the measured fitness cost for different strains, in absence of IPTG (blue circles) and in 1 mM IPTG (mauve squares). The red dot shows the fitness cost measured for  in 1 mM IPTG. The data shown in panel B are the same as those shown in Figure 2 of the main text. Error bars represent the standard error of the mean. The lines are the theoretical prediction, in absence of IPTG (blue) and in 1 mM IPTG (mauve). The dashed lines show the unstable solutions. The gray lines show the correlation of
in 1 mM IPTG. The data shown in panel B are the same as those shown in Figure 2 of the main text. Error bars represent the standard error of the mean. The lines are the theoretical prediction, in absence of IPTG (blue) and in 1 mM IPTG (mauve). The dashed lines show the unstable solutions. The gray lines show the correlation of  and
and 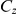 with
with 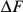 due to growth effects (see Text S1), for different values of
due to growth effects (see Text S1), for different values of 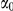 . Starting from an initial selection coefficient (e.g., upon a change of medium), a given strain moves along a gray line toward the stable steady-state solution, and away from the unstable one.
. Starting from an initial selection coefficient (e.g., upon a change of medium), a given strain moves along a gray line toward the stable steady-state solution, and away from the unstable one.
(TIF)
Comparison of protein expression (left) and fitness cost 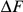 (right) on control strains. BW30270 is the wild type strain, T45 is a direct Datsenko-Wanner wild type construction and T273 is a transduction wild-type construction which went through the same procedures as all the lac operon mutants. Measurements were made in glycerol minimal medium without IPTG (white) and with 1 mM of IPTG (blue). Fitness was measured in competition against
(right) on control strains. BW30270 is the wild type strain, T45 is a direct Datsenko-Wanner wild type construction and T273 is a transduction wild-type construction which went through the same procedures as all the lac operon mutants. Measurements were made in glycerol minimal medium without IPTG (white) and with 1 mM of IPTG (blue). Fitness was measured in competition against 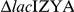 . See Materials and Methods of the main text for a description of the strain constructions and competition experiments. The error bars represent the standard error of the mean.
. See Materials and Methods of the main text for a description of the strain constructions and competition experiments. The error bars represent the standard error of the mean.
(TIF)
Expression levels of the different lac operator mutants. Protein expression was measured as described in Materials and Methods of the main text without IPTG (white) and with 1 mM of IPTG (blue). The error bars represent the standard error of the mean, with at least three replicates in each condition.
(TIF)
Growth rate, in 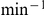 , measured in the same conditions as described for the competition experiments, except each strain was grown separately. Every hour, for 10 hours,
, measured in the same conditions as described for the competition experiments, except each strain was grown separately. Every hour, for 10 hours, 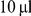 of the culture was taken and diluted appropriately, then plated on LB plates. Their mean lag phase was about 2 hours, therefore points 0, 1 hour and 2 hours were not used to estimate the growth rate. The growth rate
of the culture was taken and diluted appropriately, then plated on LB plates. Their mean lag phase was about 2 hours, therefore points 0, 1 hour and 2 hours were not used to estimate the growth rate. The growth rate 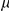 was estimated as the slope of the regression of
was estimated as the slope of the regression of 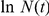 on time
on time  , where
, where 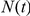 is the population size, such that:
is the population size, such that: 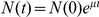 . The error bars represent the standard error of the mean of 3 independent replicates. The Malthusian fitness
. The error bars represent the standard error of the mean of 3 independent replicates. The Malthusian fitness  defined in Materials and Methods of the main text is equal to
defined in Materials and Methods of the main text is equal to 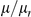 , with the growth rate of the reference strain
, with the growth rate of the reference strain 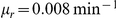 .
.
(TIF)
Growth effects on gene expression and cell volume. (A) The protein concentration 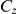 of a constitutively expressed gene has been proposed to correlate linearly with the growth rate
of a constitutively expressed gene has been proposed to correlate linearly with the growth rate 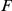 (red line), instead of the hyperbolic dependence dilution alone would induce (black line) [14]. (B) The cell volume
(red line), instead of the hyperbolic dependence dilution alone would induce (black line) [14]. (B) The cell volume 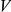 also correlates with
also correlates with 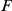 ; dots show experimental data taken from [13]; we choose to represent this correlation via a simple proportional dependence (red line). (C) Both correlations lead to a dependence
; dots show experimental data taken from [13]; we choose to represent this correlation via a simple proportional dependence (red line). (C) Both correlations lead to a dependence 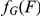 of the rate of protein synthesis on the growth rate
of the rate of protein synthesis on the growth rate 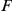 (red line; see Materials and Methods of the main text). Following [13], [14], the dependences are shown relative to the values at a growth rate
(red line; see Materials and Methods of the main text). Following [13], [14], the dependences are shown relative to the values at a growth rate 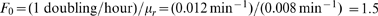 . The highlighted area
. The highlighted area 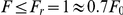 shows the range of growth rates relevant in this study.
shows the range of growth rates relevant in this study.
(TIF)
Estimated maximal rate of expression at 1 doubling/hour  and ratio of repressed to unrepressed rates
and ratio of repressed to unrepressed rates  (see Materials and Methods of the main text), for all mutants used in this study. The wild type (red) and
(see Materials and Methods of the main text), for all mutants used in this study. The wild type (red) and  (orange) are barely distinguishable, as expected. In purple, the mutants which have a
(orange) are barely distinguishable, as expected. In purple, the mutants which have a 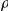 value very close to that of the wild type and are shown on Figure 5 of the main text (these are strains T319, T320, T378 and T379). In yellow, the whole operator mutants (T274, T275, T318). In green, the strain
value very close to that of the wild type and are shown on Figure 5 of the main text (these are strains T319, T320, T378 and T379). In yellow, the whole operator mutants (T274, T275, T318). In green, the strain  . The values of
. The values of 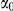 and
and 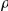 are reported in Table S1. Errors were computed as described in Text S1.
are reported in Table S1. Errors were computed as described in Text S1.
(TIF)
Statistical score of the model for a range of coefficients 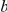 and
and  , with
, with  fixed at its fitted value. The higher the score, the lighter the shading color. The contours are drawn at scores −420, −450, −500, −550, −600, −650, −700, and −750. The highest score −417 is obtained for
fixed at its fitted value. The higher the score, the lighter the shading color. The contours are drawn at scores −420, −450, −500, −550, −600, −650, −700, and −750. The highest score −417 is obtained for 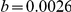 and
and 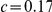 (red dot), significantly better than the best model with
(red dot), significantly better than the best model with 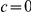 (which has score −426).
(which has score −426).
(TIF)
(A) Fitness cost 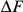 as a function of
as a function of 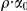 , in absence of IPTG. (B) Fitness cost
, in absence of IPTG. (B) Fitness cost 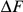 as a function of
as a function of 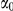 , in 1 mM IPTG. Dots show the selection coefficient measured for different strains (error bars represent the standard error of the mean), lines are model predictions. In presence of IPTG, the stable solution shown as a full line in panel B was used to compute the score
, in 1 mM IPTG. Dots show the selection coefficient measured for different strains (error bars represent the standard error of the mean), lines are model predictions. In presence of IPTG, the stable solution shown as a full line in panel B was used to compute the score 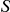 and fit the data.
and fit the data. 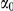 and
and 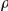 are estimated for each strain as explained in Materials and Methods of the main text. Errors were computed as described in Text S1.
are estimated for each strain as explained in Materials and Methods of the main text. Errors were computed as described in Text S1.
(TIF)
List of the strain studied, their lac O1 alleles and sequences (starting at the +1 site; underlined: mutations with respect to the wild type). The estimated values for the maximum rate of protein synthesis at 1 doubling/hour 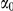 and ratio of repressed to unrepressed rates
and ratio of repressed to unrepressed rates 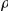 are also shown (see Materials and Methods of the main text). Errors were computed as described in Text S1.
are also shown (see Materials and Methods of the main text). Errors were computed as described in Text S1.
(PDF)
List of the strains used in this study, their genotype and the way they were constructed.
(PDF)
List of the plasmids used in this study, their relevant traits and the way they were obtained.
(PDF)
List of the oligonucleotides used in this study, their sequence and the strain for the construction of which they were used.
(PDF)
Supplementary Material.
(PDF)