Abstract
Horizontally acquired genes typically function as autonomous units conferring new abilities when introduced into different species. However, we reasoned that proteins preexisting in an organism might constrain the functionality of a horizontally acquired gene product if it operates on an ancestral pathway. Here, we determine how the horizontally acquired pmrD gene product activates the ancestral PmrA/PmrB two-component system in Salmonella enterica but not in the closely related bacterium Escherichia coli. The Salmonella PmrD protein binds to the phosphorylated PmrA protein (PmrA-P), protecting it from dephosphorylation by the PmrB protein. This results in transcription of PmrA-dependent genes, including those conferring polymyxin B resistance. We now report that the E. coli PmrD protein can activate the PmrA/PmrB system in Salmonella even though it cannot do it in E. coli, suggesting that these two species differ in an additional component controlling PmrA-P levels. We establish that the E. coli PmrB displays higher phosphatase activity towards PmrA-P than the Salmonella PmrB, and we identified a PmrB subdomain responsible for this property. Replacement of the E. coli pmrB gene with the Salmonella homolog was sufficient to render E. coli resistant to polymyxin B under PmrD-inducing conditions. Our findings provide a singular example whereby quantitative differences in the biochemical activities of orthologous ancestral proteins dictate the ability of a horizontally acquired gene product to confer species-specific traits. And they suggest that horizontally acquired genes can potentiate selection at ancestral loci.
Author Summary
The traits that distinguish closely related bacterial species are often ascribed to differences in gene content, which arise primarily through horizontal gene transfer. In some instances, the genes mediating a new trait act as independent entities that function in a variety of organisms. However, the ability of a horizontally acquired gene product(s) to operate on an ancestral pathway might be constrained by subtle differences between orthologous ancestral proteins. Here, we examine why the horizontally acquired pmrD gene product post-translationally activates the ancestral PmrA/PmrB two-component system in Salmonella enterica but not in the closely related species Escherichia coli. This allows Salmonella, but not E. coli, to transcribe PmrA-activated genes including those conferring antibiotic resistance when grown in low Mg2+, which is a condition that promotes PmrD expression. We now demonstrate that, paradoxically, the E. coli PmrD protein activates the PmrA/PmrB system in Salmonella even though it fails to do so in E. coli. We establish that quantitative differences in the biochemical activities of the PmrB proteins from Salmonella and E. coli dictate the functionality of PmrD, which protects phosphorylated PmrA from PmrB's phosphatase activity. Our findings demonstrate that ancestral genes can control the ability of horizontally acquired genes to confer species-specific traits upon different organisms.
Introduction
Closely related bacterial species often exhibit significant differences in gene content [1]. Whereas these differences can arise from the duplication and divergence or the loss of ancestral genes [2]–[4], the vast majority of species-specific DNA sequences in bacteria appear to have arisen as a result of horizontal gene transfer events [1], [5]–[7]. Species-specific sequences often endow bacteria with new abilities, which enable them to access unexplored niches such as animal and plant tissues [1], [8]. For example, some genomic islands from pathogenic bacteria function as freestanding virulence “cassettes”, and thus, the pathogenic properties they specify can be “played” effectively when the genomic island is introduced into a different bacterial species, even a non-pathogenic one [9]. Still, other horizontally acquired genes operate on ancestral cellular pathways [10], [11], raising questions as to the changes experienced both by the horizontally acquired DNA and the host bacterial genome that enable an organism to realize the fitness gains mediated by the acquired sequences.
This question can be addressed by first identifying a horizontally acquired gene product that behaves differently in present-day bacterial species, and by subsequently determining the changes in the horizontally acquired locus and/or associated ancestral genes that are responsible for a change in function. Such a strategy can provide insight into the evolutionary events that yielded extant diversity. Here, we provide a singular example of how an ancestral locus controls the ability of a horizontally acquired gene to confer a new ability.
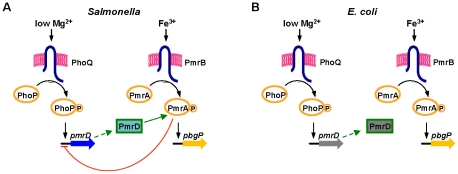
The pmrD gene product enables Salmonella enterica serovar Typhimurium to express loci governing modifications of the lipopolysaccharide (LPS) [12], rendering the organism resistant to killing by the antibiotic polymyxin B (Figure 1A) [13]–[16]. Transcription of these loci is controlled by the PmrA/PmrB two-component regulatory system [13], [17]–[19]. The PhoP-activated pmrD gene is expressed when Salmonella experience low levels of extracytoplasmic Mg2+, which is an inducing signal for the PhoP/PhoQ two-component system [20]. The PmrD protein binds to the phosphorylated form of the DNA binding protein PmrA (PmrA-P), protecting it from dephosphorylation by the sensor PmrB; this enhances PmrA-P levels and promotes transcription of PmrA-dependent genes [21]. Thus, Salmonella can resist killing by polymyxin B not only in response to the Fe3+ signal sensed by PmrB [22] but also in low Mg2+ environments [12].
Figure 1. Model of the interactions between the PhoP/PhoQ and PmrA/PmrB two-component systems in Salmonella and E. coli.
(A) In wild-type Salmonella, transcription of PmrA-activated genes is promoted during growth in low Mg2+ via the PhoP/PhoQ system, the PmrD protein and the PmrA/PmrB system, and in the presence of Fe3+ via the PmrA/PmrB system and independently of PhoP/PhoQ and PmrD. The PmrA protein represses transcription of the pmrD gene. (B) In wild-type E. coli, transcription of PmrA-activated genes is promoted in the presence of Fe3+ via the PmrA/PmrB system. The PmrD protein is produced in low Mg2+ in a PhoP-dependent manner but fails to activate the PmrA/PmrB system.
The closely related species Escherichia coli induces expression of PmrA-regulated genes and resistance to polymyxin B following growth in the presence of Fe3+ (Figure 1B) [14], [16], [23]. However, E. coli does not activate the PmrA/PmrB system in low Mg2+ conditions even though its pmrD gene is expressed in a PhoP/PhoQ-dependent manner [23]. The inability of E. coli to activate the PmrA/PmrB system, and thus, to resist killing by polymyxin B in low Mg2+, has been attributed to its PmrD protein being highly divergent from the Salmonella PmrD [23]. Indeed, replacement of the E. coli pmrD gene by the Salmonella homolog enabled activation of the E. coli PmrA/PmrB system in low Mg2+ [23].
We now show how an ancestral protein can control the ability of a horizontally acquired gene product to confer new traits. We present evidence suggesting that pmrD was acquired by lateral transfer. We demonstrate that the highly divergent E. coli pmrD gene can rescue a Salmonella pmrD mutant to activate the PmrA/PmrB system during growth in low Mg2+. Beyond reinforcing the notion that the E. coli pmrD gene specifies a functional protein [24], our findings indicate that Salmonella and E. coli must differ in an additional component controlling the levels of PmrA-P. We establish that this component is PmrB as the E. coli PmrB exhibits significantly higher phosphatase activity than the Salmonella homolog, and that replacement of the E. coli pmrB gene with the Salmonella homolog is sufficient to restore transcription of PmrA-activated genes and polymyxin B resistance to E. coli grown in low Mg2+. Our findings provide a singular example whereby the ability of a horizontally acquired gene to confer a selective advantage upon a recipient organism is dictated by quantitative differences in the biochemical activities of ancestral proteins.
Results
Evidence That pmrD Is a Horizontally Acquired Gene
Several lines of evidence support the previously suggested hypothesis that pmrD is a horizontally acquired gene [25]. First, a search of the completed genomes in the NCBI microbial genomes database for the presence of sequences homologous to the S. enterica 14028s or E. coli K12 MG1655 PmrD protein (conducted on May 2011) showed that pmrD is limited to enteric bacteria of the Klebsiella, Citrobacter, Salmonella and Escherichia lineages (Figure S1A). Second, a BLAST search identified a pmrD-like gene encoded on plasmid pKP187 from the nitrogen-fixing endophyte K. pneumoniae 342 [26], which specifies a product displaying ∼30% amino acid identity to the Salmonella and E. coli PmrD proteins (Figure S1). The presence of pmrD on a mobile genetic element could provide a means for its transfer among bacteria. Third, phylogenetic analysis of pmrD homologs present in enteric bacteria revealed that the pmrD neighbor-joining tree was similar to the species phylogeny constructed using four housekeeping genes (gapA, groEL, gyrA and pgi). The only exception was the Citrobacter pmrD gene, which clustered with the homolog identified in K. pneumoniae 342 pKP187 (Figure S1A) and this incongruence with the accepted species phylogeny is likely a consequence of horizontal gene transfer. Fourth, the pmrD gene is located in the same chromosomal context in the Citrobacter, Salmonella and Escherichia genomes, which is different from that of the Klebsiella genome (Figure S1B). Collectively, these analyses suggest that the pmrD gene was horizontally acquired by one or several transfer events.
The E. coli PmrD Protein Connects the PhoP/PhoQ and PmrA/PmrB Systems in Salmonella But Not in E. coli
To explore the possibility that the E. coli PmrD protein might activate the PmrA/PmrB system in Salmonella experiencing low Mg2+ (even though it does not function in this capacity in E. coli [23]), we engineered a Salmonella strain that expresses a FLAG-tagged E. coli pmrD gene under the control of the Salmonella pmrD promoter in the normal chromosomal location. The resulting strain transcribed the PmrA-activated pbgP gene in low Mg2+, like an isogenic strain expressing Salmonella's own pmrD gene or a FLAG-tagged version of it (Figure 2). As expected, all three strains expressed pbgP to high levels when grown in the presence of Fe3+, which activates the PmrA/PmrB system in a PmrD-independent manner [22], but not under non-inducing (high Mg2+ and no Fe3+) conditions (Figure 2). Moreover, a Salmonella pmrD mutant harboring a plasmid encoding the E. coli pmrD gene or a FLAG-tagged version of it transcribed pbgP in low Mg2+, unlike cells harboring the plasmid vector (Figure S2). These results indicate that the ability of the E. coli PmrD protein to connect the PhoP/PhoQ and PmrA/PmrB systems is context-dependent: it functions in Salmonella, but surprisingly, not in E. coli.
Figure 2. The E. coli PmrD protein enables transcription of PmrA-activated genes in Salmonella experiencing low Mg2+.
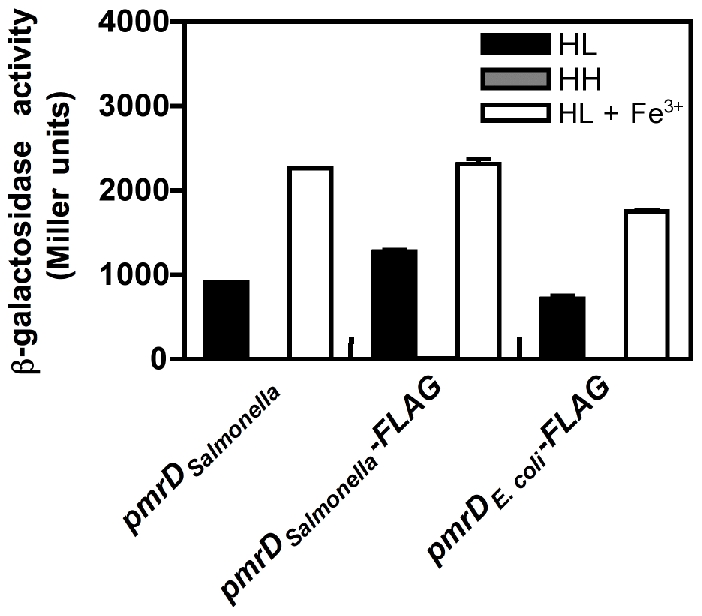
β-galactosidase activity (Miller units) produced by isogenic Salmonella strains harboring a chromosomal pbgP-lac transcriptional fusion and the wild-type Salmonella pmrD gene (EG9241), a 3′ FLAG-tagged Salmonella pmrD gene (EG13942), or a 3′ FLAG-tagged E. coli pmrD gene (EG13941) transcribed from the normal Salmonella pmrD promoter and chromosomal location. Bacteria were grown in N-minimal medium at pH 7.7 with 10 µM Mg2+ (HL), 10 mM Mg2+ (HH) or 10 µM Mg2+ and 100 µM Fe3+ (HL+Fe3+). Data correspond to the mean of three independent experiments performed in duplicate and error bars show standard deviation.
The E. coli PmrB Protein Exhibits Higher Phosphatase Activity towards PmrA-P Than the Salmonella PmrB Protein
Why does expression of the E. coli pmrD gene result in pbgP transcription in Salmonella but not in E. coli during growth in low Mg2+? When bacteria experience inducing conditions for the ancestral PmrA/PmrB system, the sensor PmrB promotes the phosphorylated state of the DNA binding protein PmrA [22], [27], resulting in transcription of PmrA-activated genes [28]. In addition to being an autokinase and phosphotransferase, PmrB is a PmrA-P phosphatase, an activity present primarily under non-inducing conditions [29]. The PmrD protein, which is produced in low Mg2+, promotes the phosphorylated state of PmrA by antagonizing PmrB-mediated dephosphorylation of PmrA-P [21]. Thus, we hypothesized that the orthologous PmrA and/or PmrB proteins of these two species might differ in one or more of their biochemical properties, resulting in lower levels of PmrA-P – the target of PmrD – in E. coli than in Salmonella. To evaluate this possibility, we compared the behaviors of the purified cytoplasmic domain of the PmrB protein (PmrBc) from the two species because it retains all the known enzymatic activities of the full-length PmrB protein, and because PmrBc expression recapitulates the PmrD-dependent activation of PmrA-regulated genes displayed by the full-length PmrB protein in vivo (Figure S3) [21].
First, we examined the autophosphorylation rate of the two proteins and found that the ATP concentration at which it was half maximal was two-fold higher for the E. coli PmrBc protein than for the Salmonella PmrBc protein (Figure S4A; see Table 1 for KM values). In addition, the E. coli PmrBc protein autophosphorylated at a rate that was two-fold higher than the Salmonella PmrBc protein (Figure S4B; see Table 1 for kobserved values). Given that the intracellular ATP concentration is ∼3 mM in E. coli [30], which is >30 times the experimentally determined KM for ATP of the Salmonella and E. coli PmrBc proteins (Table 1), we anticipate that these PmrB orthologs autophosphorylate at similar rates in vivo.
Table 1. Kinetic properties of PmrBc autophosphorylation.
| Parameter | E. coli | Salmonella |
| Apparent KM (µM) | 94±9 | 46±4 |
| kobserved (min-1) | 0.07±0.01 | 0.03±0.003 |
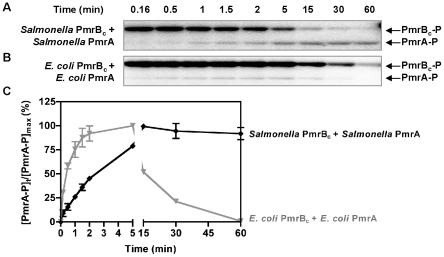
We next analyzed phosphotransfer from the Salmonella PmrBc protein to the Salmonella PmrA protein. The amount of PmrA-P increased steadily, reaching a maximum at 30 min (Figure 3A and 3C). The increase in the levels of PmrA-P was accompanied by a decrease in the levels of PmrBc-P (Figure 3A). By contrast, when the phosphotransfer reaction was performed with the PmrBc-P and PmrA proteins from E. coli, there was a rapid increase in the levels of PmrA-P during the first 5 min, followed by a steady decrease (Figure 3B and 3C). The decrease in PmrA-P levels was not due to a contaminating protease because the total PmrA amounts remained constant throughout the course of the reaction (Figure S4C). The different rates of dephosphorylation of the orthologous PmrA-P proteins after reaching their maximal values reflect the source of the PmrBc proteins (as opposed to resulting from differences in the rate of spontaneous hydrolysis of the phosphoryl group from the PmrA-P proteins) because the stabilities of the Salmonella and E. coli PmrA-P proteins were similar when the PmrBc proteins were omitted from the reactions (Figure S4D). Cumulatively, these experiments suggested that the Salmonella and E. coli PmrBc proteins differ in their phosphatase activity towards PmrA-P.
Figure 3. Phosphotransfer profiles reflect differences between the phosphatase activities of the Salmonella and E. coli PmrBc proteins.
(A–B) Levels of PmrBc-P and PmrA-P following incubation of Salmonella or E. coli PmrBc-P (5 µM) with Salmonella or E. coli PmrA (10 µM) proteins at the times indicated at the top of the figure according to the protocols described in Materials and Methods. (C) Quantitation of the phosphotransfer assays shown in (A–B). The plot depicts the level of PmrA-P relative to the maximum achieved as a function of time. Data correspond to the mean values of three independent experiments and error bars show standard deviation.
We established that the rate with which PmrBc dephosphorylates PmrA-P is 10-fold higher for the reaction with the E. coli PmrBc and PmrA-P proteins than that displayed by the Salmonella PmrBc and PmrA-P proteins (Figure S4E and Table 2). Furthermore, the different rates of PmrBc-promoted dephosphorylation of PmrA-P are largely dependent on the source of PmrBc (rather than on PmrA): the E. coli PmrBc dephosphorylated the PmrA-P proteins from Salmonella and E. coli at rates that were 3–10-fold higher than those with which the Salmonella PmrBc protein dephosphorylated PmrA-P from Salmonella or from E. coli (Figure S4E and Table 2). In sum, the enhanced phosphatase activity of the E. coli PmrBc towards PmrA-P (Figure 3 and Table 2) is predicted to generate lower levels of PmrA-P in E. coli than in Salmonella, which would result in reduced transcription of PmrA-activated genes [31].
Table 2. Rate constants for PmrBc-mediated dephosphorylation of PmrA-P, kobserved (min−1).
| E. coli PmrBc | Salmonella PmrBc | |
| E. coli PmrA | 0.36±0.03 | 0.03±0.001 |
| Salmonella PmrA | 0.14±0.01 | 0.04±0.004 |
Increased Phosphatase Activity in a Chimeric Salmonella PmrB Protein Harboring a Subdomain from the E. coli PmrB Protein
To explore the molecular basis for the differences in phosphatase activity exhibited by the Salmonella and E. coli PmrB proteins, we carried out homology modeling of their cytoplasmic domains using the structure of the sensor kinase HK853 from Thermotoga maritima [32]. This allowed us to predict the secondary structural features of the PmrB homologs and to identify subdomains that differed between the two proteins (Figure S5). Next, we created chimeric PmrBc proteins whereby particular subdomains of the Salmonella PmrBc protein were replaced by amino acid sequences corresponding to those from the E. coli PmrBc protein, aiming to preserve the secondary structure of PmrBc. We then examined the ability of the chimeric proteins to dephosphorylate Salmonella PmrA-P.
The PmrBc-α2 chimera, which differs from the wild-type Salmonella PmrBc sequence in five residues located in alpha helix 2 (Figure S5), dephosphorylated Salmonella PmrA-P at a higher rate than the Salmonella PmrBc protein, though not as high as the E. coli PmrBc protein (Figure 4). The PmrBc-α2 chimera displayed a phosphotransfer profile that was similar to that of the E. coli PmrB protein: when phosphorylated PmrBc-α2 protein was incubated with Salmonella PmrA protein, the levels of PmrA-P increased between 0–5 min and then decreased (Figure S6). By contrast, five other chimeric PmrBc proteins exhibited similar or lower levels of phosphatase activity towards Salmonella PmrA-P compared to the Salmonella PmrBc protein (Figure S7). As expected, an E. coli PmrBc-α2 chimeric protein harboring the Salmonella PmrB α2 subdomain dephosphorylated E. coli PmrA-P at a lower rate than the E. coli PmrBc (Figure S8). Our results indicate that residues in alpha helix 2 are critical modulators of PmrB's phosphatase activity.
Figure 4. The PmrBc-α2 chimera exhibits higher phosphatase activity than the Salmonella PmrBc protein.
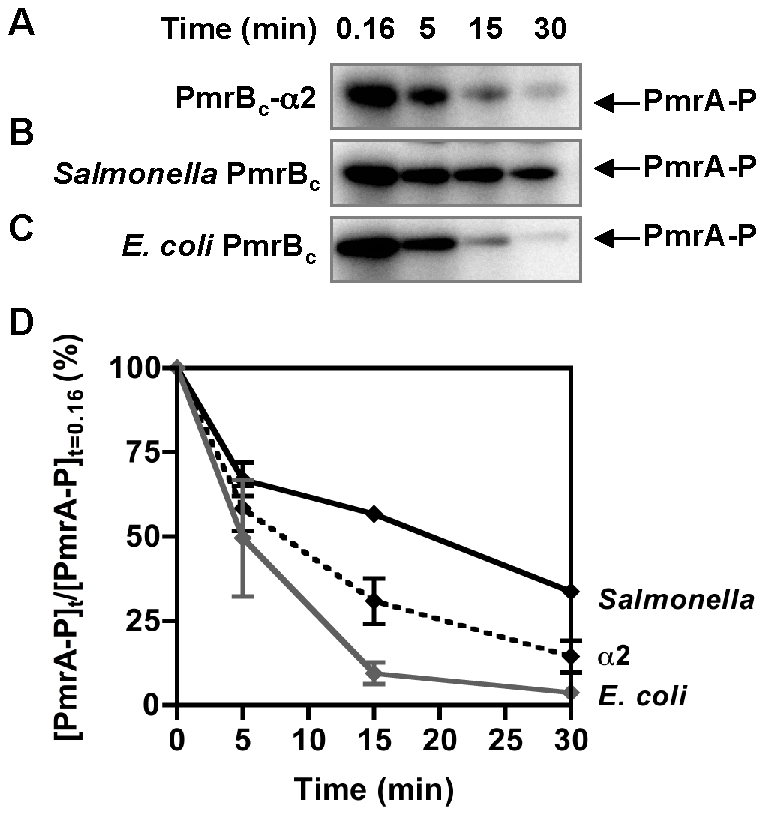
(A–C) Levels of PmrA-P following incubation of Salmonella PmrA-P (2.5 µM) with the PmrBc-α2 chimera (A), the Salmonella PmrBc (B), or the E. coli PmrBc (C) (5 µM) proteins for the indicated times. (D) Quantitation of the phosphatase assays shown in (A–C). The graph depicts the level of PmrA-P at the indicated times relative to levels at the start of the reaction. Data correspond to the mean values of three independent experiments and error bars show standard deviation.
An E. coli Strain Expressing the Salmonella pmrB Gene Transcribes PmrA-Activated Genes and Displays Resistance to Polymyxin B When Grown in Low Mg2+
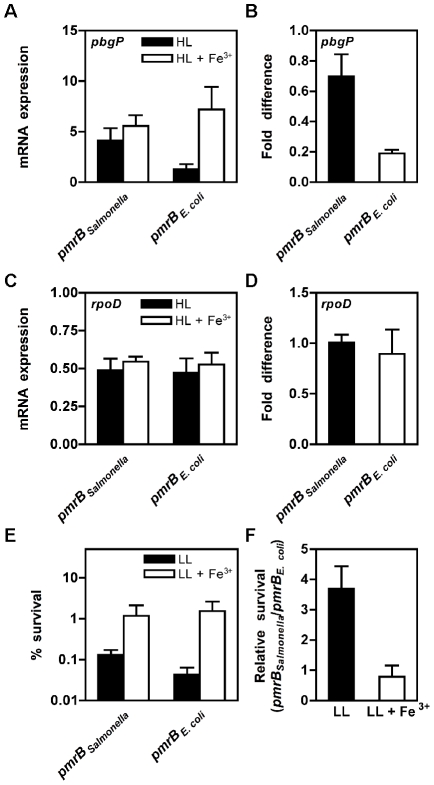
We hypothesized that the heightened phosphatase activity of the E. coli PmrB protein may hinder accumulation of high enough levels of PmrA-P, which constitutes the target of the PmrD protein [21], [23], thereby preventing transcription of PmrA-activated genes and resistance to polymyxin B during growth in low Mg2+. To test this hypothesis, we engineered isogenic E. coli strains expressing either the Salmonella or E. coli pmrB genes from the normal E. coli chromosomal location. We then investigated the PmrD-mediated connectivity between the PhoP/PhoQ and PmrA/PmrB systems by determining the ratio of the mRNA levels corresponding to the PmrA-activated genes pbgP, pmrC and pmrG produced during growth in low Mg2+ and those produced following growth in the presence of Fe3+ [23], which are PmrD-dependent and -independent processes in Salmonella.
PmrD connectivity was higher in the E. coli strain expressing the Salmonella pmrB gene than in the isogenic strain expressing E. coli's own pmrB gene (Figure 5A, 5B; Figure S9A and S9B). An E. coli strain harboring the Salmonella pmrB-α2 gene also exhibited higher PmrD connectivity than the strain expressing the E. coli pmrB gene, though not to the level displayed by the strain with the Salmonella homolog (Figure S9B). As expected, these strains displayed a similar ratio (∼1) for the PmrA-independent rpoD gene (Figure 5C and 5D), which was used as a control. The low PmrD connectivity in the E. coli strain harboring its own pmrB gene is ascribed to its higher phosphatase activity because an E. coli strain expressing the PmrB T156R protein, which dephosphorylates PmrA-P at a lower rate than the E. coli PmrB protein in vitro (Figure S4F), transcribes PmrA-activated genes in low Mg2+ (Figure S9C). The disparate PmrD connectivities were not due to differences in the responses of the orthologous PmrB proteins to Fe3+ because these strains displayed similar levels of iron-promoted pbgP and pmrC transcription (Figure S9D). The heightened expression displayed by the engineered E. coli strain harboring the Salmonella pmrB gene required the PmrD protein because there was no significant transcription of PmrA-activated genes in a pmrD mutant following growth in low Mg2+ (Figure S9E).
Figure 5. E. coli expressing the Salmonella pmrB gene display low Mg2+-promoted pbgP transcription and resistance to polymyxin B.
(A–D) The mRNA levels of the PmrA-activated pbgP gene (A) or the control rpoD gene (C) produced during growth in low Mg2+ and those produced following growth in the presence of Fe3+ was determined in E. coli strains expressing either the Salmonella pmrB gene (DC3) or its own pmrB gene (DC5). The ratio between the pbgP (B) or rpoD (D) mRNA levels produced during growth in low Mg2+ and those produced following growth in the presence of Fe3+ was determined in DC3 or in DC5. RNA samples were prepared from bacteria grown in N-minimal media, pH 7.7 with 10 µM Mg2+ (HL) or 10 µM Mg2+ and 100 µM Fe3+ (HL+Fe3+) to OD600 0.5 and the levels of pbgP or rpoD mRNA were determined by reverse-transcription-qPCR analysis. Data correspond to the mean of at least three independent experiments and error bars represent standard deviation. (E–F) Survival of isogenic E. coli strains expressing either the Salmonella pmrB gene (DC3) or the E. coli pmrB gene (DC5) following incubation in the presence of polymyxin B (2.5 µg/ml). Bacteria were grown in N-minimal medium, pH 5.8, containing 38 mM glycerol with 10 µM Mg2+ (LL) or 10 µM Mg2+ and 100 µM Fe3+ (LL+Fe3+). (E) Percentage survival of bacteria following treatment with polymyxin B relative to those incubated in the presence of PBS. (F) Relative survival of E. coli strain expressing the Salmonella pmrB gene (DC3) to the isogenic E. coli strain that expressing E. coli's own pmrB gene (DC5). Data correspond to the mean of three independent experiments performed in duplicate and error bars show standard deviation.
E. coli displays resistance to polymyxin B when grown in the presence of Fe3+ (but not following growth in low Mg2+) in a process that requires the pmrA and pbgP genes [23]. We reasoned that the E. coli strain expressing the Salmonella pmrB gene should display resistance towards polymyxin B when experiencing low Mg2+ because PmrA-activated genes are transcribed (Figure 5A). Indeed, the E. coli strain expressing the Salmonella pmrB gene was ∼4-fold more resistant to polymyxin B than the strain expressing the E. coli pmrB gene when bacteria were grown in low Mg2+ (Figure 5E and 5F). As expected, both strains displayed similar levels of polymyxin B resistance following growth in the presence of Fe3+ (Figure 5E and 5F). Collectively, these findings demonstrate that the source of the pmrB gene (i.e., Salmonella or E. coli) determines the ability of E. coli to transcribe PmrA-activated genes and to exhibit polymyxin B resistance in response to low Mg2+.
PmrB Determines the Expression Kinetics of PmrA-Activated Genes
Fe3+ activates the PmrA/PmrB system independently of the PmrD protein both in Salmonella and E. coli [22], [23]. In Salmonella, it has been demonstrated that such activation gives rise to a surge in the mRNA levels of PmrA-activated genes whereby the mRNAs increase, peak and then decrease to reach new steady-state levels [27], and that this reflects changes in the amount of PmrA-P protein [27]. Because the E. coli PmrB protein displays higher phosphatase activity than the Salmonella homolog (Table 2), we wondered whether the levels of PmrA-P, and thus the kinetics of PmrA-dependent gene expression, might differ temporally in E. coli when it expresses the Salmonella (instead of the E. coli) pmrB gene.
When bacteria were shifted to media containing Fe3+, transcription of PmrA-activated genes increased during the first 30 min and then decreased to reach steady-state levels in the E. coli strain harboring its own pmrB gene (Figure 6). By contrast, in the E. coli strain expressing the Salmonella pmrB gene, the mRNA levels of PmrA-activated genes reached a peak at 45 min followed by a decrease to attain steady-state levels (Figure 6). To determine whether the distinct temporal transciption profiles of PmrA-activated genes in these strains were due to differences in the rates of PmrB-mediated PmrA-P dephosphorylation, we examined mRNA levels in an E. coli strain expressing the E. coli PmrB T156R protein, which is compromised in its phosphatase activity (Figure S4F). The mRNA levels of PmrA-activated genes were high even when bacteria were grown high Mg2+ and no Fe3+, consistent with the notion that PmrB functions primarily as a PmrA-P phosphatase under these conditions (Figure 6) [21]. When bacteria were shifted to media containing Fe3+, transcription of PmrA-activated genes increased and peaked at 45 min before decreasing to reach steady-state levels (Figure 6). Altogether, our findings suggest that the higher phosphatase activity displayed by the E. coli PmrB protein results in PmrA-dependent transcription being turned off at an earlier timepoint in the E. coli strain expressing its own pmrB gene than in the isogenic strains harboring the Salmonella homolog or the E. coli pmrB T156R gene.
Figure 6. The source of pmrB determines activation kinetics of PmrA-controlled genes.
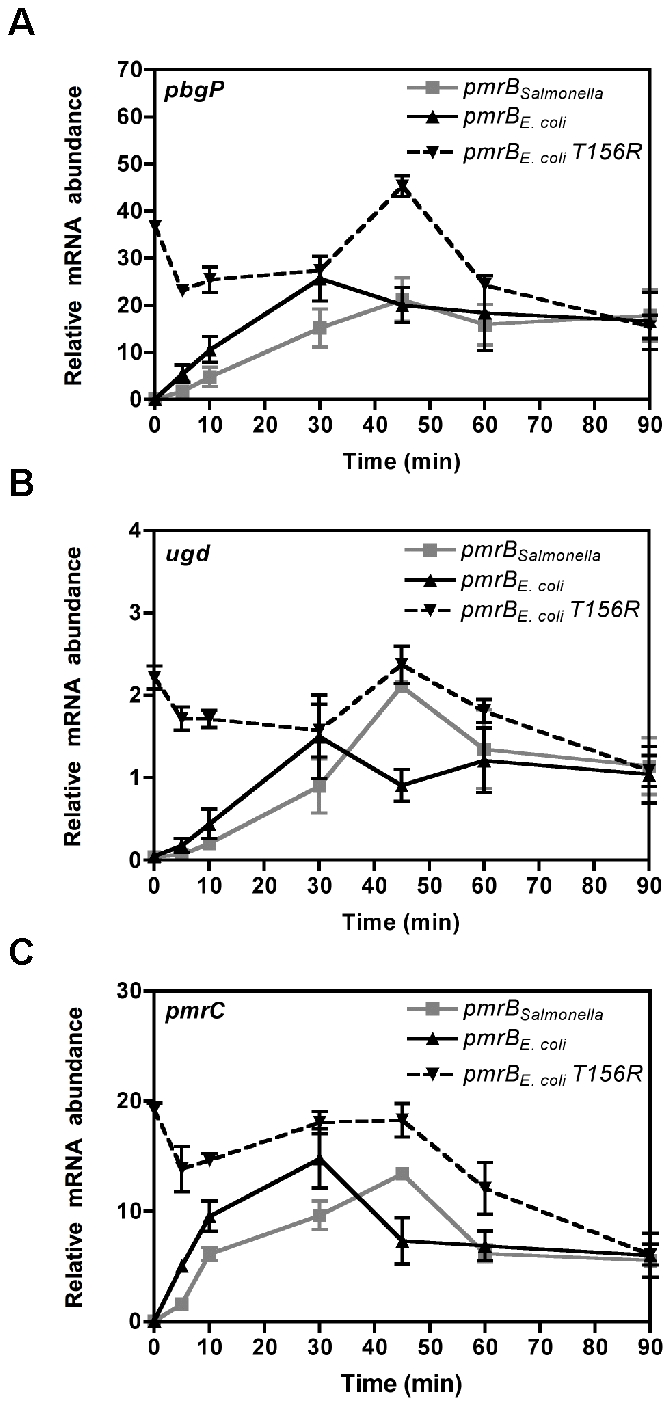
The mRNA levels of the PmrA-activated pbgP (A), pmrC (B) and ugd (C) genes from isogenic E. coli strains deleted in the pmrD gene and expressing either the Salmonella pmrB gene (DC9), the E. coli pmrB gene (DC11) or the E. coli pmrB T156R gene (DC116) were determined by reverse-transcription-qPCR analysis. We used RNA prepared from bacteria grown in medium containing 10 mM Mg2+, shifted to medium containing 10 µM Mg2+ and 100 µM Fe3+ and harvested at the designated times. Expression levels were normalized to those of the 16S ribosomal RNA gene. Data correspond to at least four independent experiments and error bars show standard deviation.
Discussion
Ancestral Genes Can Dictate the Functionality of Horizontally Acquired Genes
Horizontal gene transfer can endow bacteria with new capabilities, such as the ability to resist killing by an antibiotic or to gain access to a eukaryotic cell, and thus, it can potentially expand their ecological niches [1], [5]–[7]. We have now established that variation in the amino acid sequences of the Salmonella and E. coli PmrB sensors results in quantitative differences in their biochemical activities (Figure 3 and Table 2) and governs the ability of these bacteria to survive killing by polymyxin B in low Mg2+ (Figure 5). We proposed that, by generating different amounts of PmrA-P, the different phosphatase activities of the PmrB orthologs determine the ability of PmrD to activate the PmrA/PmrB system (Figure 2 and Figure 5) [23]. Therefore, unlike self-contained horizontally acquired gene clusters that readily confer a new trait upon introgression [1], [8], [9], the functionality of a horizontally acquired gene product operating on ancestral pathways depends largely upon the biochemical properties of existing ancestral proteins, which can act in a variety of ways [33]–[35].
The effect of genetic context on protein function is not limited to horizontally acquired genes in bacteria. A recent genome-wide survey of genes that were essential in one budding yeast strain but not in another strain demonstrated that this essential character was due to distinct genetic interactions, which differed in a strain-specific manner [36]. In higher eukaryotes, the development of complex traits involves multiple interacting genetic loci; thus, the effect of a certain gene may require other genes controlling the same trait [37]. Taken together, the contribution of a given gene to a particular phenotype is largely determined by its interactions with other genes functioning in the same genetic network.
Particular Combinations of Gene Interactions Underlie Qualitatively Similar Traits
Given that the heightened phosphatase activity of the E. coli PmrB protein prevents the E. coli PmrD protein from promoting the accumulation of sufficient levels of PmrA-P (Figure 3 and Figure 5, Table 2), why is it that expression of the Salmonella pmrD gene allows transcription of PmrA-activated genes in E. coli experiencing low Mg2+ [23]? Sequence analysis of the pmrD genes suggested that selection and divergence between the Salmonella and E. coli pmrD loci [23] allowed the Salmonella PmrD, but not the E. coli ortholog, to connect the PhoP/PhoQ and PmrA/PmrB systems in E. coli. These results indicate that particular combinations of pmrD and pmrB alleles dictate whether bacteria experiencing low Mg2+ accumulate sufficient levels of PmrA-P to promote expression of PmrA-dependent genes and to resist killing by polymyxin B (Figure 2 and Figure 5) [23]. Our work, together with studies of gene interactions in model organisms such as budding yeast [36] and analyses of the existing phenotypic variation within natural populations [38], demonstrate that different combinations of particular alleles within a genetic network can lead to qualitatively similar phenotypes in related organisms [37]. Therefore, understanding the phenotypic variation that exists within or between species demands going beyond the study of single-gene effects to investigating how genetic variation at different loci influence one another.
Quantitative Differences in the Phosphatase Activity of Sensor Proteins Determine Expression Kinetics
Organisms respond to a stress condition by altering their gene expression programs and/or behaviors with temporal dynamics specified by their signaling systems [27], [39], [40]. The distinct dynamics that distinguish the responses of closely related organisms are typically ascribed to differences in the genetic architecture of their signaling systems [41]. For example, the enteric species Yersinia pestis, K. pneumoniae and S. enterica utilize different genetic architectures to promote transcription of the pbgP gene when experiencing low Mg2+, resulting in distinct expression profiles [25], [42], [43]. We now provide a singular example where variation in the biochemical activity of orthologous signaling proteins can affect expression dynamics in genetic systems with identical architectures. The identified differences in phosphatase activities exhibited by the Salmonella and E. coli PmrB proteins lead to dissimilar kinetics of PmrA-dependent gene expression (Figure 6, Table 2), which could affect resistance to Fe3+ [44] and contribute to the distinct ability of these two species to survive in soil [45].
The Salmonella and E. coli PmrB proteins are the first example, to our knowledge, where amino acid changes between orthologous bifunctional sensor kinases bring about dramatic differences in phosphatase activity (Table 2) without much effect on autokinase activity (Table 1). These two enzymatic activities are carried out by the cytoplasmic portion of these proteins, which consists of the dimerization and histidine phosphotransfer (DHp) domain joined by a flexible linker to the catalytic and ATP-binding (CA) domain [32], [46], [47]. Amino acid residues identified in the DHp [48]–[51] and CA domains [52] of other sensor kinases as being important for phosphatase activity are conserved between the Salmonella and E. coli PmrB proteins. This was expected because mutation of these residues usually abolishes phosphatase activity and both the Salmonella and E. coli PmrB proteins exhibit phosphatase activity towards PmrA-P, albeit to different levels (Table 2). One or all the five amino acids residues differing between the orthologous PmrB proteins in the alpha-2 helix (Figure S5) modulate phosphatase activity (Figure 4 and Figure S8). Some of these amino acids lie within a weakly conserved region (termed X-region) of the DHp domain and linker region that is proposed to stimulate sensor protein phosphatase activity by properly orienting the DHp and CA domains towards each other (Figure S5) [49], [51]. Thus, one or more of these five amino acids might also modulate contacts between the DHp and CA domains of the PmrB proteins.
Evolution of an Ancestral Regulatory System Targeted by a Horizontally Acquired Gene Product
We have established that quantitative differences between the biochemical properties of the orthologous PmrB proteins control the functionality of the pmrD gene product in present-day Salmonella and E. coli. Yet, when did these genetic events (i.e. horizontal acquisition of pmrD and mutational changes in pmrB) occur in the evolutionary history of enteric species? And how might the integration of horizontally acquired genes into ancestral regulatory circuits drive their evolution and facilitate bacterial adaptation to particular environments?
The pmrD gene appears to have been acquired by the common ancestor to the enteric species K. pneumoniae, Citrobacter spp., S. enterica and E. coli and incorporated into the regulatory circuit of the ancestral PhoP/PhoQ system because it is present in these four species but absent from all other examined enteric bacteria (Figure S1) [25]. This ancestral organism likely transcribed PmrA-activated genes in low Mg2+ via the PmrD-mediated pathway as this ability is retained in Klebsiella and Salmonella [12], [25]. After the Salmonella and E. coli lineages diverged, the ability to make low Mg2+-dependent, PmrA-controlled modifications of the LPS was retained by Salmonella but lost in E. coli, perhaps contributing towards their survival and proliferation in distinct host and non-host environments [45]. This difference can be attributed, in part, to selection at the pmrD locus and divergence of the Salmonella and E. coli PmrD orthologs because expression of the Salmonella pmrD gene allows transcription of PmrA-activated genes under low Mg2+-inducing conditions in E. coli [23]. We further propose that selection for a Salmonella PmrB that has low phosphatase activity may have served to maintain the PmrD-mediated connection between the PhoP/PhoQ and PmrA/PmrB systems (Table 2), thus enabling Salmonella to display the LPS modifications, including those resulting in polymyxin B resistance in environments denoted by low Mg2+ [12], [21]. By contrast, the accumulation of PmrA-P in low Mg2+ may be detrimental to E. coli's lifestyle because a mutant that constitutively activates the PmrA/PmrB system exhibits increased susceptibility to the bile detergent deoxycholic acid [53], which E. coli may experience in the mammalian intestine [54]. This may have driven selection for a PmrB protein with a higher phosphatase activity in E. coli (Table 2). Indeed, molecular analysis of the pmrB gene from natural isolates corresponding to the Salmonella reference collection C (SARC) [55] and natural isolates from the E. coli standard reference collection (ECOR) [56] revealed that the pmrB gene is evolving in a non-neutral fashion (Table S1).
In Salmonella, further integration of the pmrD gene into the regulatory circuits of the ancestral PhoP/PhoQ and PmrA/PmrB systems has entailed the evolution of a negative feedback loop where PmrA-P represses PhoP-promoted pmrD transcription, thus preventing excessive expression of PmrA-activated genes (Figure 1A) [29]. This feedback loop is absent in E. coli because the PmrD protein does not activate the PmrA/PmrB system in this species following growth in low Mg2+ (Figure 1B) [23]. Our work, and that of others [57], suggests that changes in ancestral regulatory and metabolic networks arising from horizontal gene transfer contribute to the survival of bacterial species in their particular niches.
Concluding Remarks
In sum, genetic changes are often contingent upon those previously fixed within an evolutionary lineage [58]. For example, evolution of the ability to metabolize citrate in an experimental population of E. coli was shown to require the occurrence of earlier mutations, which potentiated the appearance of the latter mutation giving rise to the novel trait [59]. Our findings now suggest that horizontal acquisition of the pmrD gene might have potentiated selection at the Salmonella and E. coli pmrB loci, further cementing the ability of Salmonella, but not E. coli, to express PmrA-activated genes and to resist killing by polymyxin B in the presence of low Mg2+.
Finally, the behavior of an organism, or its survival in particular ecological niches, is often deduced from the presence or absence of genes and/or the predicted functions of the encoded gene products, which are based on experiments typically carried out with model organisms. However, more complex mechanisms could link an organism's genotype to its phenotype [60]. We have now established that quantitative differences in the biochemical activities of an ancestral protein can determine whether a horizontally acquired gene product connects different regulatory systems, thereby affecting the ability of a bacterial species to survive killing by cationic antimicrobial peptides. Consequently, subtle quantitative differences in the biochemical activities of conserved orthologous proteins, which are not easily predicted by sequence comparison and identification of conserved catalytic residues, can give rise to phenotypic variation between and within closely related species [38], [61]. Our work demonstrates that protein function can only be appreciated in the context of other proteins operating within the same genetic network, and underscores the need to experimentally verify predictions derived from comparative genomics.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
S. enterica serovar Typhimurium strains were derived from the wild-type strain 14028s and E. coli strains from the wild-type strain MG1655. Bacteria were grown at 37°C with aeration in Luria-Bertani (LB) broth or in N-minimal media (pH 7.7), supplemented with 0.1% casamino acids, 38 mM glycerol, 10 µM or 10 mM MgCl2 and 100 µM FeSO4 [62]. When necessary, antibiotics were added at the following final concentrations: ampicillin, 50 µg/ml; chloramphenicol, 20 µg/ml; kanamycin, 50 µg/ml, and tetracycline, 10 µg/ml. P1 transduction of E. coli strains was performed as described [63]. P22 transduction of Salmonella strains was performed as described [64]. E. coli DH5α was used as a host for the preparation of plasmid DNA. We describe the construction of bacterial strains and plasmids as well as other molecular biology procedures in the Text S1. Bacterial strains and plasmids used in this study are listed in Table S2 and S3, respectively.
β-Galactosidase Assay
β-galactosidase assays were carried out in triplicate, and the activity was determined as described [63]. Data correspond to mean values of three independent experiments performed in duplicate.
Autokinase, Phosphotransferase, and Phosphatase Assays
Biochemical assays were carried out as in [21] and a detailed description is provided in Text S1.
RNA Isolation and Real-Time PCR to Determine Transcript Levels
E. coli cells from overnight cultures grown in N-minimal medium at pH 7.7 with 10 mM MgCl2 were washed twice with N-minimal medium containing no Mg2+ and added into the appropriate fresh media with 1∶50 dilution. The bacterial cultures were grown to OD600 0.5 in a shaking water bath at 37°C. 0.5 ml aliquots of cells were removed, mixed with RNAprotect Bacteria Reagent (Qiagen) for stabilization of RNA, and total RNA was isolated using RNeasy Kit (Qiagen) with on-column DNase treatment. cDNA was synthesized using TaqMan (Applied Biosystems) and random hexamers. Quantification of transcripts was carried out by real-time PCR using SYBR Green PCR Master Mix (Applied Biosystems) in an ABI 7500 Sequence Detection System (Applied Biosystems). The relative amount of cDNA was determined using a standard curve obtained from PCR with serially diluted genomic DNA, and results were normalized to the levels of 16S ribosomal RNA. Data shown is an average from at least three independent experiments.
Polymyxin B Susceptibility Assay
Polymyxin B susceptibility assays were carried out as described in [23]. Data correspond to mean values of three independent experiments performed in duplicate.
Homology Searches and Phylogenetic Analyses of the pmrD Gene
To identify homologs of the pmrD gene, the deduced PmrD protein sequences from E. coli K12 MG1655 and S. enterica serovar Typhimurium 14028s were used as query to search the NCBI microbial genomes database as well as the NCBI non-redundant nucleotide database using TBLASTN. In parallel, the UniProt database was searched for homologs using BLASTP [65]. Similar results were obtained in these searches and the protein sequences of the homologs were used for phylogenetic analyses. The sequences of the pmrD gene were aligned using ClustalX2 [66] and phylogenetic trees were constructed using the neighbor-joining algorithm with default parameters and 1000 bootstrap replicates using MEGA 4 [67]. For comparison, the sequences of 4 housekeeping genes (gapA, groEL, gyrA and pgi) [68] were concatenated, aligned using ClustalX2 [66] and used for construction of a species tree.
Accession Numbers
GenBank accession numbers are available in Table S4.
Supporting Information
Phylogeny of the pmrD gene and its genetic context in enteric bacteria. (A) Neighbor-joining tree of the pmrD genes from Klebsiella, Citrobacter, Salmonella and Escherichia. For comparison, the sequences of four housekeeping loci were concatenated and similarly used for construction of a neighbor-joining tree. Bootstrap support is indicated above each node, with only values >50% being shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. (B) Genomic context of the pmrD gene in representative Klebsiella, Citrobacter, Salmonella and Escherichia strains.
(TIF)
The E. coli PmrD protein enables transcription of the PmrA-activated pbgP gene in Salmonella experiencing low Mg2+. (A) Transcript levels of the PmrA-activated pbgP gene were determined in a Salmonella pmrD strain (EG11491) harboring plasmids pUH-pmrDE. coli-FLAG, pUH-pmrDE. coli or the vector pUHE21-2lacIq. (B) The ratio between the pbgP mRNA levels produced during growth in low Mg2+ and those produced following growth in the presence of Fe3+ was determined in a Salmonella pmrD strain (EG11491) harboring plasmids pUH-pmrDE. coli-FLAG, pUH-pmrDE. coli or the vector pUHE21-2lacIq. Bacteria were grown with 0.5 mM IPTG in N-minimal medium at pH 7.7 containing 10 µM Mg2+ (HL) or 10 µM Mg2+ and 100 µM Fe3+ (HL+Fe3+). Data correspond to the mean of three independent experiments and error bars show standard deviation.
(TIF)
The E. coli PmrB cytoplasmic domain (PmrBc) is sufficient for PmrD-dependent transcription of PmrA-activated genes. β-galactosidase activity (Miller units) from a chromosomal pbgP-lac transcriptional fusion were determined in ΔpmrB (EG10065) and ΔpmrB ΔpmrD (EG12060) Salmonella strains harboring pUH-pmrB, pUH-pmrBc or pUHE21-2lacIq. Bacteria were grown in N-minimal medium at pH 7.7, 0.05 mM IPTG and 50 µg/ml ampicillin with 10 µM Mg2+ (HL) or 10 mM Mg2+ (HH). Data correspond to the mean of three independent experiments performed in duplicate and error bars show standard deviation.
(TIF)
The total levels of PmrBc and PmrA protein remain constant throughout the course of the phosphotransfer reaction and the PmrA-P proteins display similar stabilities in the absence of PmrBc. (A) Levels of Salmonella or E. coli PmrBc-P following incubation of PmrBc (2.5 µM) with ATP at the concentrations indicated at the top of the figure according to the protocols described in Text S1. (B) Levels of Salmonella or E. coli PmrBc-P following incubation of PmrBc (2.5 µM) with 0.75 mM ATP at the times indicated at the top of the figure according to the protocols described in Text S1. (C) Levels of the PmrBc and PmrA proteins from Salmonella and E. coli were determined by SDS-PAGE and Coomassie staining following phosphotransfer from PmrBc-P (5 µM) to PmrA (10 µM) as described in Text S1 using aliquots taken at the indicated times. (D) Levels of PmrA-P proteins from Salmonella and E. coli were determined as described in Text S1 with aliquots taken at the indicated time points. (E) Levels of PmrA-P following incubation of Salmonella or E. coli PmrBc (5 µM) with the Salmonella or E. coli PmrA-P (2.5 µM) proteins for the indicated times according to the protocols described in Text S1. (F) Levels of PmrA-P following incubation of E. coli PmrBc T156R (5 µM) with the E. coli PmrA-P (2.5 µM) protein for the indicated times according to the protocols described in Text S1.
(TIF)
Primary sequence alignment of the Salmonella and E. coli PmrB sensor proteins. Residues that differ between the two proteins are highlighted in green (conserved changes) or red (nonconserved changes). A blue vertical dotted line is present before the first amino acid of the PmrBc cytoplasmic domain. The structures of the cytoplasmic domains of the PmrB proteins were predicted using a homology-modeling program (Phyre) [69] and were based on HK853 sensor histidine kinase cytoplasmic domain from Thermotoga maritima [32]. The dimerization and histidine phosphotransfer (DHp) domain consists of amino acids 128–200, while the catalytic and ATP-binding (CA) domain consists of amino acids 215–356 and 215–363 from the Salmonella and E. coli PmrB proteins, respectively. β sheets and α helices in the Salmonella and E. coli PmrBc proteins that contain non-identical amino acid residues are highlighted by colored boxes, and those that are identical between these proteins are highlighted by grey boxes. The X-region (amino acids 179–202), which is underlined with a black dashed line, was predicted based on sequence alignment to the sensor kinase EnvZ [49], [51].
(TIF)
Phosphotransfer from the PmrBc-α2 chimera to the Salmonella PmrA protein is similar to that displayed by the E. coli PmrBc protein. (A–C) Levels of PmrBc-P and PmrA-P following incubation of PmrBc-P (5 µM) and PmrA (10 µM) as described in Text S1 with aliquots taken at the times indicated at the top of the figure. Three different reactions were set up with the PmrBc-α2 and Salmonella PmrA proteins (A), Salmonella PmrBc and Salmonella PmrA proteins (B), and E. coli PmrBc and Salmonella PmrA proteins (C). (D) Quantitation of the phosphotransfer assays shown in (A–C). The plot depicts the level of PmrA-P relative to the maximum achieved as a function of time.
(TIF)
The PmrBc chimeras exhibit similar or lower levels of phosphatase activity than the Salmonella PmrBc protein. (A–E) The graphs depict the levels of PmrA-P at the indicated times relative to levels at the start of the reaction (t = 0.16 min) following incubation of Salmonella PmrA-P (2.5 µM) with the PmrBc-α1 protein (A) or PmrBc-α3 protein (B) or the PmrBc-β2 protein (C) or the PmrBc-β4 protein (D) or the PmrBc-7aa protein (E). The abilities of the PmrBc chimeric proteins (5 µM) to dephosphorylate PmrA-P were compared to those of the Salmonella or E. coli PmrBc proteins. Data correspond to the mean values of at least three independent experiments and error bars show standard deviation.
(TIF)
The E. coli PmrBc–α2 chimera exhibits a lower level of phosphatase activity than the E. coli PmrBc protein. (A–C) Levels of PmrA-P following incubation of E. coli PmrA-P (2.5 µM) with the E. coli PmrBc-α2 chimera (A), the Salmonella PmrBc (B) or the E. coli PmrBc (C) (5 µM) proteins for the indicated times. (D) Quantitation of the phosphatase assays shown in (A–C). The graph depicts the levels of PmrA-P at the indicated times relative to levels at the start of the reaction (t = 0.16 min). Data correspond to the mean values of three independent experiments and error bars show standard deviation.
(TIF)
The Salmonella pmrB gene restores low Mg2+-promoted transcription of PmrA-activated genes in E. coli. (A–C) The ratio between the mRNA levels of the PmrA-activated genes pmrG, pmrC and pbgP produced during growth in low Mg2+ and those produced following growth in the presence of Fe3+ was determined in E. coli expressing the Salmonella pmrB gene (DC3) or its own pmrB gene (DC5) or the Salmonella pmrB-α2 gene (DC68) or the E. coli pmrB T156R gene (DC116) from the normal chromosomal location. RNA samples were prepared from bacteria grown in N-minimal media, pH 7.7 with 10 µM Mg2+ (HL) or 10 µM Mg2+ and 100 µM Fe3+ (HL+Fe3+) to OD600 of 0.5, and the levels of pmrG, pmrC or pbgP mRNA were determined by reverse-transcription-qPCR analysis. (D) Transcript levels of the PmrA-activated genes pbgP and pmrC were determined in E. coli strains expressing either the Salmonella pmrB gene (DC3) or the E. coli pmrB gene (DC5). Bacteria were grown in N-minimal media, pH 7.7 with 1 mM Mg2+ (HH) or 1 mM Mg2+ and 100 µM Fe3+ (HH+Fe3+) to an OD600 0.5 before RNA samples were prepared for reverse-transcription-qPCR analysis. (E) Transcript levels of the PmrA-activated genes pbgP and pmrC were determined in E. coli strains deleted for the pmrD gene and expressing either the Salmonella pmrB gene (DC9) or the E. coli pmrB gene (DC11). Bacteria were grown in N-minimal media, pH 7.7 with 10 µM Mg2+ (HL) or 10 µM Mg2+ and 100 µM Fe3+ (HL+Fe3+) to an OD600 0.5 before RNA samples were prepared for reverse-transcription-qPCR analysis. Data correspond to the mean of at least three independent experiments and error bars represent standard deviation.
(TIF)
McDonald-Kreitman test for pmrB evolution.
(DOC)
Bacterial strains and plasmids used in this study.
(DOC)
DNA sequences of primers used in this study.
(DOC)
GenBank accession numbers for the pmrB genes from Salmonella and E. coli natural isolates.
(DOC)
Supplementary materials and methods.
(DOC)
Acknowledgments
We thank W.S. Yeo and T. Latifi for technical assistance and discussions; and J. Fay, H. Huang, A. Mitrophanov, H. Ochman, J.C. Perez, and members of the Groisman lab for comments on an earlier version of the manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by grant 42336 from the National Institutes of Health to EAG and by the Howard Hughes Medical Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 2.Kunin V, Ouzounis CA. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 2003;13:1589–1594. doi: 10.1101/gr.1092603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snel B, Bork P, Huynen MA. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 2002;12:17–25. doi: 10.1101/gr.176501. [DOI] [PubMed] [Google Scholar]
- 4.Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 6.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 7.Jain R, Rivera MC, Moore JE, Lake JA. Horizontal gene transfer in microbial genome evolution. Theor Popul Biol. 2002;61:489–495. doi: 10.1006/tpbi.2002.1596. [DOI] [PubMed] [Google Scholar]
- 8.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Micro. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 9.McDaniel TK, Kaper JB. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol Microbiol. 1997;23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 10.Lercher MJ, Pal C. Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Mol Biol Evol. 2008;25:559–567. doi: 10.1093/molbev/msm283. [DOI] [PubMed] [Google Scholar]
- 11.Pal C, Papp B, Lercher MJ. Horizontal gene transfer depends on gene content of the host. Bioinformatics. 2005;21(Suppl 2):ii222–223. doi: 10.1093/bioinformatics/bti1136. [DOI] [PubMed] [Google Scholar]
- 12.Kox LF, Wosten MM, Groisman EA. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 2000;19:1861–1872. doi: 10.1093/emboj/19.8.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunn JS, Lim KB, Krueger J, Kim K, Guo L, et al. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 14.Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CR. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J Biol Chem. 2001;276:43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- 15.Roland KL, Martin LE, Esther CR, Spitznagel JK. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J Bacteriol. 1993;175:4154–4164. doi: 10.1128/jb.175.13.4154-4164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breazeale SD, Ribeiro AA, Raetz CR. Oxidative decarboxylation of UDP-glucuronic acid in extracts of polymyxin-resistant Escherichia coli. Origin of lipid A species modified with 4-amino-4-deoxy-L-arabinose. J Biol Chem. 2002;277:2886–2896. doi: 10.1074/jbc.M109377200. [DOI] [PubMed] [Google Scholar]
- 17.Groisman EA, Kayser J, Soncini FC. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J Bacteriol. 1997;179:7040–7045. doi: 10.1128/jb.179.22.7040-7045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrera CM, Hankins JV, Trent MS. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol Microbiol. 2010;76:1444–1460. doi: 10.1111/j.1365-2958.2010.07150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, Hsu FF, Turk J, Groisman EA. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J Bacteriol. 2004;186:4124–4133. doi: 10.1128/JB.186.13.4124-4133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soncini FC, Garcia Vescovi E, Solomon F, Groisman EA. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato A, Groisman EA. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 2004;18:2302–2313. doi: 10.1101/gad.1230804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wosten MM, Kox LF, Chamnongpol S, Soncini FC, Groisman EA. A signal transduction system that responds to extracellular iron. Cell. 2000;103:113–125. doi: 10.1016/s0092-8674(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 23.Winfield MD, Groisman EA. Phenotypic differences between Salmonella and Escherichia coli resulting from the disparate regulation of homologous genes. Proc Natl Acad Sci U S A. 2004;101:17162–17167. doi: 10.1073/pnas.0406038101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagiwara D, Yamashino T, Mizuno T. A Genome-wide view of the Escherichia coli BasS-BasR two-component system implicated in iron-responses. Biosci Biotechnol Biochem. 2004;68:1758–1767. doi: 10.1271/bbb.68.1758. [DOI] [PubMed] [Google Scholar]
- 25.Mitrophanov AY, Jewett MW, Hadley TJ, Groisman EA. Evolution and dynamics of regulatory architectures controlling polymyxin B resistance in enteric bacteria. PLoS Genet. 2008;4:e1000233. doi: 10.1371/journal.pgen.1000233. doi: 10.1371/journal.pgen.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fouts DE, Tyler HL, DeBoy RT, Daugherty S, Ren Q, et al. Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet. 2008;4:e1000141. doi: 10.1371/journal.pgen.1000141. doi: 10.1371/journal.pgen.1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin D, Lee EJ, Huang H, Groisman EA. A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science. 2006;314:1607–1609. doi: 10.1126/science.1134930. [DOI] [PubMed] [Google Scholar]
- 28.Wosten MM, Groisman EA. Molecular characterization of the PmrA regulon. J Biol Chem. 1999;274:27185–27190. doi: 10.1074/jbc.274.38.27185. [DOI] [PubMed] [Google Scholar]
- 29.Kato A, Latifi T, Groisman EA. Closing the loop: the PmrA/PmrB two-component system negatively controls expression of its posttranscriptional activator PmrD. Proc Natl Acad Sci U S A. 2003;100:4706–4711. doi: 10.1073/pnas.0836837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neuhard J, Nygaard P. Purines and pyrimidines. In: Neidhardt FC, editor. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Washington, D.C.: ASM Press; 1987. pp. 445–473. [Google Scholar]
- 31.Shin D, Groisman EA. Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in vivo. J Biol Chem. 2005;280:4089–4094. doi: 10.1074/jbc.M412741200. [DOI] [PubMed] [Google Scholar]
- 32.Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Champion K, Higgins NP. Growth rate toxicity phenotypes and homeostatic supercoil control differentiate Escherichia coli from Salmonella enterica serovar Typhimurium. J Bacteriol. 2007;189:5839–5849. doi: 10.1128/JB.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchetti M, Capela D, Glew M, Cruveiller S, Chane-Woon-Ming B, et al. Experimental evolution of a plant pathogen into a legume symbiont. PLoS Biol. 2010;8:e1000280. doi: 10.1371/journal.pbio.1000280. doi: 10.1371/journal.pbio.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakata N, Tobe T, Fukuda I, Suzuki T, Komatsu K, et al. The absence of a surface protease, OmpT, determines the intercellular spreading ability of Shigella: the relationship between the ompT and kcpA loci. Mol Microbiol. 1993;9:459–468. doi: 10.1111/j.1365-2958.1993.tb01707.x. [DOI] [PubMed] [Google Scholar]
- 36.Dowell RD, Ryan O, Jansen A, Cheung D, Agarwala S, et al. Genotype to phenotype: a complex problem. Science. 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartman JLt, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- 38.Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science. 2006;313:101–104. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]
- 39.Shih VF, Kearns JD, Basak S, Savinova OV, Ghosh G, et al. Kinetic control of negative feedback regulators of NF-κB/RelA determines their pathogen- and cytokine-receptor signaling specificity. Proc Natl Acad Sci U S A. 2009;106:9619–9624. doi: 10.1073/pnas.0812367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 42.Kato A, Mitrophanov AY, Groisman EA. A connector of two-component regulatory systems promotes signal amplification and persistence of expression. Proc Natl Acad Sci U S A. 2007;104:12063–12068. doi: 10.1073/pnas.0704462104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winfield MD, Latifi T, Groisman EA. Transcriptional regulation of the 4-amino-4-deoxy-L-arabinose biosynthetic genes in Yersinia pestis. J Biol Chem. 2005;280:14765–14772. doi: 10.1074/jbc.M413900200. [DOI] [PubMed] [Google Scholar]
- 44.Chamnongpol S, Dodson W, Cromie MJ, Harris ZL, Groisman EA. Fe(III)-mediated cellular toxicity. Mol Microbiol. 2002;45:711–719. doi: 10.1046/j.1365-2958.2002.03041.x. [DOI] [PubMed] [Google Scholar]
- 45.Winfield MD, Groisman EA. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl Environ Microbiol. 2003;69:3687–3694. doi: 10.1128/AEM.69.7.3687-3694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 47.Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 48.Dutta R, Yoshida T, Inouye M. The critical role of the conserved Thr247 residue in the functioning of the osmosensor EnvZ, a histidine Kinase/Phosphatase, in Escherichia coli. J Biol Chem. 2000;275:38645–38653. doi: 10.1074/jbc.M005872200. [DOI] [PubMed] [Google Scholar]
- 49.Hsing W, Russo FD, Bernd KK, Silhavy TJ. Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J Bacteriol. 1998;180:4538–4546. doi: 10.1128/jb.180.17.4538-4546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsing W, Silhavy TJ. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol. 1997;179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu Y, Qin L, Yoshida T, Inouye M. Phosphatase activity of histidine kinase EnvZ without kinase catalytic domain. Proc Natl Acad Sci U S A. 2000;97:7808–7813. doi: 10.1073/pnas.97.14.7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Y, Inouye M. The role of the G2 box, a conserved motif in the histidine kinase superfamily, in modulating the function of EnvZ. Mol Microbiol. 2002;45:653–663. doi: 10.1046/j.1365-2958.2002.03061.x. [DOI] [PubMed] [Google Scholar]
- 53.Froelich JM, Tran K, Wall D. A pmrA constitutive mutant sensitizes Escherichia coli to deoxycholic acid. J Bacteriol. 2006;188:1180–1183. doi: 10.1128/JB.188.3.1180-1183.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gunn JS. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2000;2:907–913. doi: 10.1016/s1286-4579(00)00392-0. [DOI] [PubMed] [Google Scholar]
- 55.Boyd EF, Wang FS, Whittam TS, Selander RK. Molecular genetic relationships of the salmonellae. Appl Environ Microbiol. 1996;62:804–808. doi: 10.1128/aem.62.3.804-808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ochman H, Selander RK. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pal C, Papp B, Lercher MJ. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet. 2005;37:1372–1375. doi: 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
- 58.Gould SJ. Wonderful Life. 1989. Norton, New York.
- 59.Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323:746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Storz JF, Runck AM, Sabatino SJ, Kelly JK, Ferrand N, et al. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc Natl Acad Sci U S A. 2009;106:14450–14455. doi: 10.1073/pnas.0905224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller JH. 1972. Experiments in molecular genetics: Cold Spring Harbour Laboratory Press, Cold Spring Harbour, NY.
- 64.Davis RW, Bolstein D, Roth JR. 1980. Advanced bacterial genetics: Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 65.The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 67.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 68.Wertz JE, Goldstone C, Gordon DM, Riley MA. A molecular phylogeny of enteric bacteria and implications for a bacterial species concept. J Evol Biol. 2003;16:1236–1248. doi: 10.1046/j.1420-9101.2003.00612.x. [DOI] [PubMed] [Google Scholar]
- 69.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogeny of the pmrD gene and its genetic context in enteric bacteria. (A) Neighbor-joining tree of the pmrD genes from Klebsiella, Citrobacter, Salmonella and Escherichia. For comparison, the sequences of four housekeeping loci were concatenated and similarly used for construction of a neighbor-joining tree. Bootstrap support is indicated above each node, with only values >50% being shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. (B) Genomic context of the pmrD gene in representative Klebsiella, Citrobacter, Salmonella and Escherichia strains.
(TIF)
The E. coli PmrD protein enables transcription of the PmrA-activated pbgP gene in Salmonella experiencing low Mg2+. (A) Transcript levels of the PmrA-activated pbgP gene were determined in a Salmonella pmrD strain (EG11491) harboring plasmids pUH-pmrDE. coli-FLAG, pUH-pmrDE. coli or the vector pUHE21-2lacIq. (B) The ratio between the pbgP mRNA levels produced during growth in low Mg2+ and those produced following growth in the presence of Fe3+ was determined in a Salmonella pmrD strain (EG11491) harboring plasmids pUH-pmrDE. coli-FLAG, pUH-pmrDE. coli or the vector pUHE21-2lacIq. Bacteria were grown with 0.5 mM IPTG in N-minimal medium at pH 7.7 containing 10 µM Mg2+ (HL) or 10 µM Mg2+ and 100 µM Fe3+ (HL+Fe3+). Data correspond to the mean of three independent experiments and error bars show standard deviation.
(TIF)
The E. coli PmrB cytoplasmic domain (PmrBc) is sufficient for PmrD-dependent transcription of PmrA-activated genes. β-galactosidase activity (Miller units) from a chromosomal pbgP-lac transcriptional fusion were determined in ΔpmrB (EG10065) and ΔpmrB ΔpmrD (EG12060) Salmonella strains harboring pUH-pmrB, pUH-pmrBc or pUHE21-2lacIq. Bacteria were grown in N-minimal medium at pH 7.7, 0.05 mM IPTG and 50 µg/ml ampicillin with 10 µM Mg2+ (HL) or 10 mM Mg2+ (HH). Data correspond to the mean of three independent experiments performed in duplicate and error bars show standard deviation.
(TIF)
The total levels of PmrBc and PmrA protein remain constant throughout the course of the phosphotransfer reaction and the PmrA-P proteins display similar stabilities in the absence of PmrBc. (A) Levels of Salmonella or E. coli PmrBc-P following incubation of PmrBc (2.5 µM) with ATP at the concentrations indicated at the top of the figure according to the protocols described in Text S1. (B) Levels of Salmonella or E. coli PmrBc-P following incubation of PmrBc (2.5 µM) with 0.75 mM ATP at the times indicated at the top of the figure according to the protocols described in Text S1. (C) Levels of the PmrBc and PmrA proteins from Salmonella and E. coli were determined by SDS-PAGE and Coomassie staining following phosphotransfer from PmrBc-P (5 µM) to PmrA (10 µM) as described in Text S1 using aliquots taken at the indicated times. (D) Levels of PmrA-P proteins from Salmonella and E. coli were determined as described in Text S1 with aliquots taken at the indicated time points. (E) Levels of PmrA-P following incubation of Salmonella or E. coli PmrBc (5 µM) with the Salmonella or E. coli PmrA-P (2.5 µM) proteins for the indicated times according to the protocols described in Text S1. (F) Levels of PmrA-P following incubation of E. coli PmrBc T156R (5 µM) with the E. coli PmrA-P (2.5 µM) protein for the indicated times according to the protocols described in Text S1.
(TIF)
Primary sequence alignment of the Salmonella and E. coli PmrB sensor proteins. Residues that differ between the two proteins are highlighted in green (conserved changes) or red (nonconserved changes). A blue vertical dotted line is present before the first amino acid of the PmrBc cytoplasmic domain. The structures of the cytoplasmic domains of the PmrB proteins were predicted using a homology-modeling program (Phyre) [69] and were based on HK853 sensor histidine kinase cytoplasmic domain from Thermotoga maritima [32]. The dimerization and histidine phosphotransfer (DHp) domain consists of amino acids 128–200, while the catalytic and ATP-binding (CA) domain consists of amino acids 215–356 and 215–363 from the Salmonella and E. coli PmrB proteins, respectively. β sheets and α helices in the Salmonella and E. coli PmrBc proteins that contain non-identical amino acid residues are highlighted by colored boxes, and those that are identical between these proteins are highlighted by grey boxes. The X-region (amino acids 179–202), which is underlined with a black dashed line, was predicted based on sequence alignment to the sensor kinase EnvZ [49], [51].
(TIF)
Phosphotransfer from the PmrBc-α2 chimera to the Salmonella PmrA protein is similar to that displayed by the E. coli PmrBc protein. (A–C) Levels of PmrBc-P and PmrA-P following incubation of PmrBc-P (5 µM) and PmrA (10 µM) as described in Text S1 with aliquots taken at the times indicated at the top of the figure. Three different reactions were set up with the PmrBc-α2 and Salmonella PmrA proteins (A), Salmonella PmrBc and Salmonella PmrA proteins (B), and E. coli PmrBc and Salmonella PmrA proteins (C). (D) Quantitation of the phosphotransfer assays shown in (A–C). The plot depicts the level of PmrA-P relative to the maximum achieved as a function of time.
(TIF)
The PmrBc chimeras exhibit similar or lower levels of phosphatase activity than the Salmonella PmrBc protein. (A–E) The graphs depict the levels of PmrA-P at the indicated times relative to levels at the start of the reaction (t = 0.16 min) following incubation of Salmonella PmrA-P (2.5 µM) with the PmrBc-α1 protein (A) or PmrBc-α3 protein (B) or the PmrBc-β2 protein (C) or the PmrBc-β4 protein (D) or the PmrBc-7aa protein (E). The abilities of the PmrBc chimeric proteins (5 µM) to dephosphorylate PmrA-P were compared to those of the Salmonella or E. coli PmrBc proteins. Data correspond to the mean values of at least three independent experiments and error bars show standard deviation.
(TIF)
The E. coli PmrBc–α2 chimera exhibits a lower level of phosphatase activity than the E. coli PmrBc protein. (A–C) Levels of PmrA-P following incubation of E. coli PmrA-P (2.5 µM) with the E. coli PmrBc-α2 chimera (A), the Salmonella PmrBc (B) or the E. coli PmrBc (C) (5 µM) proteins for the indicated times. (D) Quantitation of the phosphatase assays shown in (A–C). The graph depicts the levels of PmrA-P at the indicated times relative to levels at the start of the reaction (t = 0.16 min). Data correspond to the mean values of three independent experiments and error bars show standard deviation.
(TIF)
The Salmonella pmrB gene restores low Mg2+-promoted transcription of PmrA-activated genes in E. coli. (A–C) The ratio between the mRNA levels of the PmrA-activated genes pmrG, pmrC and pbgP produced during growth in low Mg2+ and those produced following growth in the presence of Fe3+ was determined in E. coli expressing the Salmonella pmrB gene (DC3) or its own pmrB gene (DC5) or the Salmonella pmrB-α2 gene (DC68) or the E. coli pmrB T156R gene (DC116) from the normal chromosomal location. RNA samples were prepared from bacteria grown in N-minimal media, pH 7.7 with 10 µM Mg2+ (HL) or 10 µM Mg2+ and 100 µM Fe3+ (HL+Fe3+) to OD600 of 0.5, and the levels of pmrG, pmrC or pbgP mRNA were determined by reverse-transcription-qPCR analysis. (D) Transcript levels of the PmrA-activated genes pbgP and pmrC were determined in E. coli strains expressing either the Salmonella pmrB gene (DC3) or the E. coli pmrB gene (DC5). Bacteria were grown in N-minimal media, pH 7.7 with 1 mM Mg2+ (HH) or 1 mM Mg2+ and 100 µM Fe3+ (HH+Fe3+) to an OD600 0.5 before RNA samples were prepared for reverse-transcription-qPCR analysis. (E) Transcript levels of the PmrA-activated genes pbgP and pmrC were determined in E. coli strains deleted for the pmrD gene and expressing either the Salmonella pmrB gene (DC9) or the E. coli pmrB gene (DC11). Bacteria were grown in N-minimal media, pH 7.7 with 10 µM Mg2+ (HL) or 10 µM Mg2+ and 100 µM Fe3+ (HL+Fe3+) to an OD600 0.5 before RNA samples were prepared for reverse-transcription-qPCR analysis. Data correspond to the mean of at least three independent experiments and error bars represent standard deviation.
(TIF)
McDonald-Kreitman test for pmrB evolution.
(DOC)
Bacterial strains and plasmids used in this study.
(DOC)
DNA sequences of primers used in this study.
(DOC)
GenBank accession numbers for the pmrB genes from Salmonella and E. coli natural isolates.
(DOC)
Supplementary materials and methods.
(DOC)