Abstract
Lantibiotics are small peptide antibiotics that contain the characteristic thioether amino acids lanthionine and methyllanthionine. As ribosomally synthesized peptides, lantibiotics possess biosynthetic gene clusters which contain the structural gene (lanA) as well as the other genes which are involved in lantibiotic modification (lanM, lanB, lanC, lanP), regulation (lanR, lanK), export (lanT(P)) and immunity (lanEFG). The lantibiotic mersacidin is produced by Bacillus sp. HIL Y-85,54728, which is not naturally competent.
Methodology/Principal Findings
The aim of these studies was to test if the production of mersacidin could be transferred to a naturally competent Bacillus strain employing genomic DNA of the producer strain. Bacillus amyloliquefaciens FZB42 was chosen for these experiments because it already harbors the mersacidin immunity genes. After transfer of the biosynthetic part of the gene cluster by competence transformation, production of active mersacidin was obtained from a plasmid in trans. Furthermore, comparison of several DNA sequences and biochemical testing of B. amyloliquefaciens FZB42 and B. sp. HIL Y-85,54728 showed that the producer strain of mersacidin is a member of the species B. amyloliquefaciens.
Conclusions/Significance
The lantibiotic mersacidin can be produced in B. amyloliquefaciens FZB42, which is closely related to the wild type producer strain of mersacidin. The new mersacidin producer strain enables us to use the full potential of the biosynthetic gene cluster for genetic manipulation and downstream modification approaches.
Introduction
The lantibiotic (i.e. lanthionine-containing antibiotic) mersacidin is an antimicrobial peptide that consists of 20 amino acids. The producer strain of mersacidin, Bacillus sp. HIL Y-85,54728 [1], has not yet been closely characterized. The structural gene (mrsA) and the genes for modification enzymes, transporters and producer self-protection are encoded on a 12.3 kb biosynthetic gene cluster on the chromosome of the producer strain [2]. Moreover, three regulatory genes are present in the gene cluster. The two-component regulatory system MrsR2/K2 is mainly involved in immunity and induction of mersacidin biosynthesis in the presence of mersacidin by a quorum sensing mechanism [3]. A further single regulatory protein, MrsR1, is encoded downstream of mrsA and is essential for mersacidin production [4]. Mersacidin inhibits the growth of gram-positive bacteria by binding to the cell wall precursor lipid II and thereby inhibiting cell wall biosynthesis [5].
Production of mersacidin and genetically engineered mersacidin peptides has so far been performed in variants of the original producer strain, Bacillus sp. HIL Y-85,54728. Production of engineered peptides was obtained either in trans after inactivation of mrsA by introduction of a stop codon [6] or in cis after double homologous recombination [7]. However, transformation of the producer strain with exogenous plasmids could only be achieved by protoplast transformation or electroporation and both methods yielded only low transformation frequencies. Therefore, the aim of these studies was to build an expression system for mersacidin in a naturally competent Bacillus strain in synthetic medium and exploit competence transformation as an efficient method for the transfer of the biosynthetic gene cluster and plasmids harboring mrsA.
Successful heterologous production has previously been shown for several lantibiotics, e. g. subtilin and nisin by B. subtilis 168 [8], [9], lacticin 3147 by Enterococcus faecalis [10], or epicidin 280 by Staphylococcus carnosus [11]. Very recently, production of active lichenicidin has even been achieved in Escherichia coli [12]. However, heterologous production of a lantibiotic cannot be taken for granted and remains difficult. For example, epicidin 280 and Pep5 are two closely related lantibiotics, but Pep5 shows a higher antibacterial activity than epicidin 280. In contrast to epicidin 280 [11], Pep5 cannot be expressed in S. carnosus, which is susceptible to this agent in the nanomolar range, indicating that the toxicity of the product may be a problem here (G. Bierbaum, unpublished data). Therefore, for successful and high-level expression of mersacidin in a heterologous host, two conditions had to be met, functional producer self-protection and transfer of the biosynthetic gene cluster to the new host. The genome of Bacillus amyloliquefaciens FZB42, a competent plant-growth promoting rhizobacterium, has recently been sequenced. The part of the mersacidin biosynthetic gene cluster, that is devoted to producer self-protection, is already present in this organism [13] making it an ideal candidate for the transfer of the biosynthetic genes. Here we show that production of mersacidin is possible in B. amyloliquefaciens FZB42 and that the mersacidin producer strain itself is a member of this species.
Results and Discussion
B. amyloliquefaciens FZB42 is immune to mersacidin
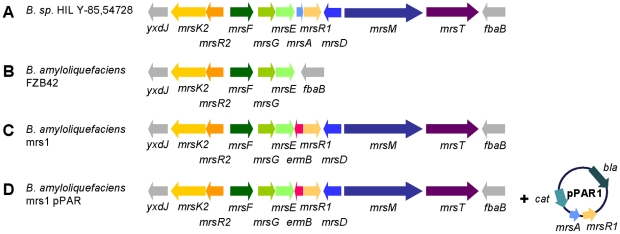
Analysis of the complete genome sequence of B. amyloliquefaciens FZB42 [13] had shown, that mrsFGE, which encode an ABC transporter that inhibits binding of mersacidin to the cells [4], and mrsK2R2, the genes of a two component system that induces expression of mrsFGE in the presence of mersacidin [3], are present in this organism (Fig. 1). These genes are located at the same site as in the original producer strain of mersacidin, i. e. between ycdJ and fbaB [2]. A detailed comparison showed that the encoded proteins share at least 98 % amino acid identity (two exchanges in MrsE and MrsF, one exchange in MrsR2 and four exchanges in MrsK2), with the exception of the N-terminus of MrsG. However, resequencing of the wild type producer indicated that a thymidine residue in position 980 was missing in the original sequence of mrsG. After sequence correction, the MrsG amino acid sequences were identical. In the intergenic region between mrsE and fbaB, a short sequence of 147 bp is inserted in the genome of B. amyloliquefaciens FZB42 (bp 3774591 to 3774738) that is not present in the producer strain of mersacidin and did not yield any hits in the databases. This sequence is flanked by two distinct regions with sequence similarity to the mersacidin gene cluster. The upstream 38 bp region is similar to the sequence found downstream of mrsE, i. e. the sequence upstream of the putative mrsA operator. The 89 bp region found downstream of the 147 bp insert is homologous to the sequence downstream of mrsT, contains the inverted repeat that is thought to delimit the mersacidin biosynthetic gene cluster and is not present in B. amyloliquefaciens DSM 7T. This might indicate that the biosynthetic part of the gene cluster was lost from B. amyloliquefaciens FZB42 during evolution.
Figure 1. The (partial) mersacidin biosynthesis gene clusters of the mersacidin producer B. sp. HIL Y-85,54728, B. amyloliquefaciens FZB42 and its derivatives.
The mersacidin gene cluster of the original producer strain B. sp. HIL Y-85,54728 (A) consists of the immunity genes mrsFGE (green colors), the structural gene mrsA (light blue), the modification enzymes mrsD and mrsM (dark blue colors), the exporter containing a protease domain mrsT (purple) and the regulatory genes mrsR1, mrsR2 and mrsK2 (yellow and orange colors). The genome of B. amyloliquefaciens FZB42 (B) harbors a partial mersacidin gene cluster consisting of the immunity genes mrsFGE and the regulatory genes mrsK2 and mrsR2. The genes are found at the same site as in the original producer strain, i. e. between ycdJ and fbaB. In the mutant strain B. amyloliquefaciens mrs1 (C), a partial completion of the mersacidin gene cluster was reached by competence transformation using genomic DNA of a mersacidin deletion mutant (B. sp. HIL Y-85,54728 Rec1). An erythromycin resistance (ermB) cassette substituting mrsA served as selection marker. mrsR1 is most probably not transcribed in this mutant because of a polar effect. The completion of the mersacidin gene cluster in B. amyloliquefaciens mrs1 pPAR1 (D) was achieved in trans by transformation with the plasmid pPAR1, carrying the structural gene mrsA and mrsR1, yielding B. amyloliquefaciens mrs1 pPAR1.
A previous comparison of minimum inhibitory concentration (MIC) values of the wild type producer and an mrsK2R2 knockout clone, which does not express the mersacidin immunity genes mrsFGE, had demonstrated that expression of mrsK2R2FGE increased the resistance to mersacidin about threefold [4]. MIC determinations of the wild type producer (25 mg/l) and B. amyloliquefaciens FZB42 (25 mg/l) demonstrated that B. amyloliquefaciens FZB42 was at least as resistant to mersacidin as the producer strain and therefore this organism was chosen as amenable to mersacidin production.
Reconstitution of mersacidin production in B. amyloliquefaciens FZB42
In order to transfer the biosynthetic part of the mersacidin biosynthetic gene cluster into B. amyloliquefaciens FZB42, chromosomal DNA of Bacillus sp. HIL Y-85,54728 Rec1 was utilized. This strain harbors the complete mersacidin gene cluster including the operator sequence, apart from mrsA and its promoter that have been replaced by an erythromycin resistance cassette [2]. Therefore, the use of erythromycin as a selection marker for successful integration of the gene cluster was possible during transformation experiments. After a competence transformation, 15 erythromycin resistant colonies were isolated and insertion of the biosynthetic part of the mersacidin biosynthesis gene cluster was confirmed by PCR, employing the primers mrsE681.f and ermB665.r annealing in mrsE and ermB as well as mrsT2251.f and fbaB283.r annealing in mrsT and fabB. A PCR of comK, employing primers located in the intergenic region (FZBfor, FZBrev) that did not match the sequence of the wild type producer strain confirmed that the clones were indeed B. amyloliquefaciens FZB42 transformants and did not derive from spores of the producer strain. Subsequently, one of these clones, B. amyloliquefaciens FZB42 mrs1, was transformed with pPAR1 [3], which harbors mrsA and mrsR1, by competence transformation. The regulator MrsR1 is essential for mersacidin biosynthesis [4] and is transcribed from the mrsA promoter (Bierbaum, unpublished results).
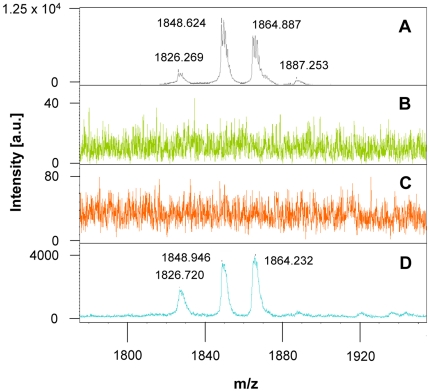
In order to demonstrate the expression of mersacidin, MALDI-TOF analysis of the culture supernatant of B. amyloliquefaciens mrs1 pPAR1 was performed and spectra were compared to those of B. amyloliquefaciens FZB42 (wild type) as well as the mrs1 strain, missing mrsA (Fig. 2). The typical mersacidin masses [1826 Da: mersacidin + H, 1848 Da: mersacidin + Na and 1864 Da: mersacidin + K] were detected in the culture supernatant of the strain harboring pPAR1, indicating expression of fully modified mersacidin. In contrast, the spectra of B. amyloliquefaciens FZB42 and of B. amyloliquefaciens mrs1 did not show any mersacidin-related mass peaks.
Figure 2. MALDI-TOF mass spectra of pure mersacidin and culture supernatants of B. amyloliquefaciens clones.
Panel (A) shows pure mersacidin (control). The spectra of culture supernatants of B. amyloliquefaciens FZB42 (B) as well as of B. amyloliquefaciens mrs1 (C), which harbor only a part of the mersacidin gene cluster or miss the structural gene mrsA, respectively, show no mersacidin production. In contrast, the culture supernatant of B. amyloliquefaciens mrs1 pPAR1 (D) is characterized by the presence of the typical mersacidin-related mass signals [1826 Da: mersacidin + H, 1848 Da: mersacidin + Na and 1864 Da: mersacidin + K] which demonstrate the production of mersacidin by this strain.
On the other hand, in agar well diffusion assays with supernatants of cultures incubated in the absence of chloramphenicol, the inhibition zones of B. amyloliquefaciens mrs1 pPAR1 against M. luteus, S. aureus SG511 and Bacillus megaterium were not significantly larger than those produced by B. amyloliquefaciens FZB42 or B. amyloliquefaciens mrs1. The reason for this observation was that B. amyloliquefaciens FZB42 is able to produce an array of antimicrobial and antifungal substances including polyketides (bacillaene, difficidin, macrolactin), lipopeptides (surfactin, fengycin, bacillomycin D), two siderophores (bacillibactin, product of nrs cluster), the antimicrobial dipeptide bacilysin as well as the thiazole/oxazole containing antibiotic plantazolicin [14], [15]. Comparative MALDI-TOF spectra of B. amyloliquefaciens FZB42 and its mutant strains indeed indicated the presence of the lipopeptide surfactin and the antifungal compounds fengycin and bacillomycin D in culture supernatants of all tested B. amyloliquefaciens strains (data not shown). The activity of surfactin was also detected by hemolysis on Columbia blood agar plates. In conclusion, the antimicrobial activity of mersacidin was probably masked by the activity of surfactin in the agar well diffusion assays. The production of several antibacterial products by Bacillus strains is far from unusual. For example, secondary antibacterial compounds are also detected in the culture supernatants of the producer strain of mersacidin (Bierbaum, unpublished results) and even B. subtilis 168 - in spite of the mutation in sfp that inhibits production of the lipopeptides encoded in the genome [16] (surfactin [17] and plipastatin/fengycin [18]) - is still able to excrete at least four other antibacterial compounds, i. e. sublancin 168 [19], subtilosin A [20], bacilysocin [21] and bacilysin [22].
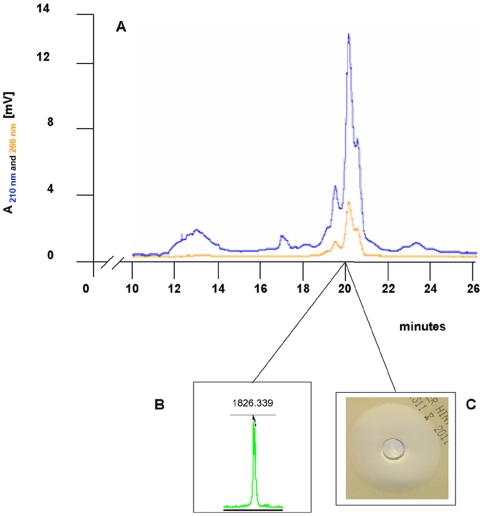
In order to demonstrate that the mersacidin produced by B. amyloliquefaciens mrs1 pPAR1 was correctly modified and showed antimicrobial activity, it was partially purified by two consecutive HPLC runs from cultures grown in the presence of chloramphenicol. The HPLC fractions that were active against M. luteus and that showed the typical adsorption spectrum of mersacidin were further analyzed by MALDI-TOF and showed the presence of the typical mass signals (Fig. 3). These results indicated B. amyloliquefaciens mrs1 pPAR1 had produced active and fully modified mersacidin.
Figure 3. Purification of mersacidin from culture supernatant of B. amyloliquefaciens mrs1 pPAR1.
The culture supernatant of B. amyloliquefaciens mrs1 pPAR1 was applied to a Poros RP-HPLC column and eluted in a gradient of 30 % to 42 % acetonitrile (containing 0.1 % TFA). Active fractions were pooled and lyophilized. The resulting lyophilizate was resuspended in 5 % acetonitrile and applied to a Nucleosil RP-C18 column (A). The antimicrobial activity of the fractions was assayed in agar well diffusion tests against M. luteus (diameter 3.2 cm) (C) and analyzed by MALDI-TOF (B). Active fractions eluted after 20 min in a gradient of 50 – 65 % acetonitrile (0.1 % TFA) and were characterized by the presence of a peak with 1826.339 Da representing mersacidin.
The producer strain of mersacidin belongs to the species B. amyloliquefaciens
Upon comparison of various DNA sequences (yvnB, czcO, hpr, baeD, hemE, and comK) obtained from the producer strain of mersacidin to the sequences of the B. amyloliquefaciens FZB42 genome in NCBI, a high similarity between both strains became obvious. Table 1 demonstrates that all sequences, including intergenic regions, showed about 98.5 % nucleotide sequence identity to those of B. amyloliquefaciens FZB42 and about 93.5 % identity to the sequences of the type strain B. amyloliquefaciens DSM 7T, whereas the identity to B. subtilis 168 was considerably lower (77.4 %).
Table 1. Results of discontiguous megablasts employing nucleotide sequences of Bacillus sp. HIL Y-85,54728.
| gene | B. amyloliquefaciens FZB42 | B. amyloliquefaciens DSM 7T | B. subtilis 168 | |||
| identical bases/total bases | percent identity | identical bases/total bases | percent identity | identical bases/total bases | percent identity | |
| yvnB | 628/638 | 98.4 | 595/638 | 93.2 | 464/638 | 72.7 |
| czcO | 747/758 | 98.5 | 164/192 | 85.4 | 651/788 | 82.6 |
| baeD | 337/343 | 98.2 | 319/343 | 93.0 | 174/240 | 72.5 |
| yhaI, hpr | 768/779 | 98.5 | 746/779 | 95.7 | 632/782 | 80.8 |
| hemE | 217/220 | 98.6 | 216/220 | 98.1 | 167/212 | 78.7 |
| comK | 802/814 | 98.5 | 780/814 | 95.8 | 559/720 | 77.6 |
| mean value | 98.5 | 93.5 | 77.4 | |||
During BLAST searches, the genome sequence of B. amyloliquefaciens FZB42 always showed the highest similarity to the sequence of the producer strain. B. amyloliquefaciens DSM 7 scored second, with the exception of the czcO region which seems to be partially missing in this strain, followed by B. subtilis 168 in third position (with the single exception of hemE, here B. subtilis W23 scored third with 168/212 identical bases).
The presence of baeD showed that Bacillus sp. HIL Y-85,54728 carries at least parts of the bacillaene gene cluster that was described for B. amyloliquefaciens FZB42 [13] and B. amyloliquefaciens DSM 7T [23]. A biochemical identification test demonstrated that, in contrast to B. subtilis 168, B. amyloliquefaciens FZB42 and Bacillus sp. HIL Y-85,54728 both were able to metabolize xylose, lactose and starch and did not grow at 50°C nor in the presence of 10 % NaCl. Furthermore, both strains did not produce acid from trehalose and mannitol. The only difference between B. amyloliquefaciens FZB42 and Bacillus sp. HIL Y-85,54728 was the absence of gelatinase in the latter strain. 16S rRNA sequencing was performed with a PCR product that had been obtained using chromosomal DNA of Bacillus sp. HIL Y-85,54728 as a template. An NCBI BLAST search yielded a close similarity to B. amyloliquefaciens FZB42 16S rRNA (Table 2), indicating that Bacillus sp. HIL Y-85,54728 belongs to the species B. amyloliquefaciens. This was confirmed by the sequence of the gyrase gene gyrA (Fig. 4) and, therefore, we propose that the producer strain of mersacidin should be renamed “B. amyloliquefaciens HIL Y-85,54728”.
Table 2. Comparison of the sequences of the Bacillus sp. HIL Y-85,54728 16S rRNA genes with all rrn paralogs of Bacillus amyloliquefaciens DSM 7T, Bacillus amyloliquefaciens FZB42 and Bacillus subtilis 168.
| strain/nucleotide positiona | 181 | 186 | 203 | 286 | 466 | 473 | 484 |
| B. sp. HIL Y-85,54728 | C/Gb | C/T | G | A/G | G | A | C |
| B. amyloliquefaciens FZB42 | G | C/T | G | A/G | G | A | C |
| B. amyloliquefaciens DSM 7T | C | T | A/G | G | G | A | C |
| B. subtilis 168 | G | A | A | A | A | G | T |
The nucleotide positions correspond to [29]. All other bases are conserved between the four strains.
The PCR product was directly sequenced, therefore, it contained all rrn paralogs and gave three mixed positions. An alignment of all seven paralogs of the B. amyloliquefaciens FZB42 16S rRNA with the sequence of the producer strain showed that two of the three mixed positions of Bacillus sp. HIL Y-85,54728 vary in B. amyloliquefaciens FZB42 in the same manner. The third mixed position (181) contained the C that is present in all rrn paralogs of B. amyloliquefaciens DSM 7T and the G that is found in all paralogs of B. amyloliquefaciens FZB42.
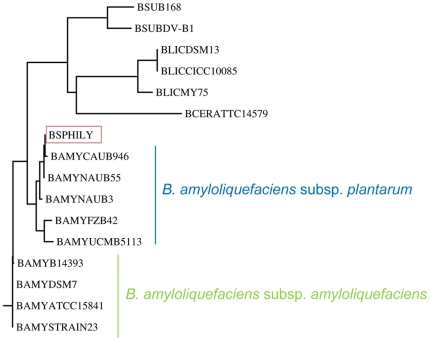
Figure 4. Phylogenetic tree based on the partial nucleotide sequence of the gyrA gene.
The tree was calculated based on the gyrA nucleotide sequences of the mersacidin producer (BHILY, marked by a red box) and different members of the genus Bacillus (NCBI accession numbers in brackets) [B. amyloliquefaciens = BAMY strains: FZB42 (CP000560), CAUB946 (FN652789), S23 (FN652780), ATCC15841 (FN662838), DSM7 (FN597644), NAUB3 (FN652783), NAUB55 (FN652801), UCMB5113 (AY212974); B. licheniformis = BLIC strains: MY75 (EU073420), DSM13 (BLi00007), CICC10085 (GQ355995); B. subtilis = BSUB strains: 168 (BSU00070), DV1-B1 (EF134416) and B. cereus = BCER strain: ATCC14579 (BC0006)]. The mersacidin wild type producer is placed among the members of the subspecies B. amyloliquefaciens subsp. plantarum; it does not belong to the subspecies amyloliquefaciens that consists of strains closely related to the type strain B. amyloliquefaciens DSM 7T. It is also clearly not a member of the species B. subtilis, B. cereus or B. licheniformis.
B. amyloliquefaciens FZB42 was isolated from the plant rhizosphere and has recently been defined as the type strain of a group of growth-promoting plant associated B. amyloliquefaciens strains (B. amyloliquefaciens subsp. plantarum) [24]. In addition to their ability to colonize plant roots, the members of the plantarum subspecies are discriminated from the subspecies B. amyloliquefaciens subsp. amyloliquefaciens by differences in the gyrA and cheA nucleotide sequence, hydrolysis of cellulose and an increased ability to produce nonribosomal secondary metabolites like fengycin and difficin [24]. In a taxonomic tree that was calculated from the gyrA nucleotide sequences of the mersacidin producer and different members of the genus Bacillus, the producer strain is found in the cluster formed by the members of the plantarum subspecies, suggesting a close association between the mersacidin wild type producer and these strains (Fig. 4). The similarity was not confined to the gyrA gene but was also reflected by the higher overall nucleotide sequence identity (98.5 %) of the mersacidin wild type producer and B. amyloliquefaciens subsp. plantarum FZB42 in comparison to the value reached by B. amyloliquefaciens subsp. amyloliquefaciens DSM 7T (93.5 %) (Table 1).
A distinguishing feature of the subspecies B. amyloliquefaciens subsp. plantarum is their ability to colonize Arabidopsis roots [24]. In fact, nearly all members of this subspecies were isolated from plants, plant roots or like B. amyloliquefaciens FZB42 from infested soil. The only exception is represented by strain UCMB5113 that was isolated from soil. In contrast, the producer strain of mersacidin originates from soil of a salt pan in Mulund, India, and unfortunately a plant association of the original isolate was not mentioned in the first report [1]. However, the strain did not grow in the presence of 10 % salt in the laboratory, indicating that the salt pan might not be its natural biotope and that its presence in the sample might rather be due to its ability to form long-lived spores.
In conclusion, we could show here that it is possible to produce mersacidin in B. amyloliquefaciens FZB42. The successful production of fully modified and active mersacidin by this strain provides an appropriate in vivo expression system for the construction and expression of mersacidin analogs. The vast array of antibacterial and antifungal compounds that is already excreted by this organism is thought to provide competitive advantage in the rhizosphere [25]. It also harbors genes which are nearly identical to the immunity genes of mersacidin and which will afford additional protection against competing strains that excrete this lantibiotic. The strategy employed here, i. e. to use an organism that already possesses the immunity genes of a lantibiotic for production of the same substance, proved successful and led to the production of active and fully modified mersacidin.
Materials and Methods
Strains, plasmids, culture conditions and media
All bacterial strains and plasmids used in this study are listed in table 3. Strains were stored as 50 % glycerol stocks at −80°C. Bacillus strains were cultured in tryptic soy broth (TSB, Oxoid, Wesel, Germany) or on tryptic soy agar. Escherichia coli strains were cultivated in LB. All cultures were maintained at 37°C. For genetically manipulated strains, antibiotics were added to the growth media (ampicillin, 40 µg/ml; erythromycin, 25 µg/ml; chloramphenicol, 20 µg/ml).
Table 3. Strains and plasmids used in this study.
| Microorganism/plasmid | Function | Source/reference |
| E. coli K12 strains | Cloning hosts | |
| B. subtilis 168 | Type strain of B. subtilis | ATCC 23857[30] |
| B. sp. HIL Y-85,54728 | Wild type mersacidin producer strain, strain collection of Sanofi-Aventis (Frankfurt, Germany), no. FH 1658 | [1] |
| B. sp. HIL Y-85,54728 Rec1 | Mersacidin wild type producer deletion mutant: mrsA replaced by ermB, no mersacidin production | [2] |
| B. amyloliquefaciens FZB42 | Wild type strain, carrying the 5′ part of the mersacidin gene cluster (mrsKR2FGE) | [13] |
| B. amyloliquefaciens mrs1 | Mutant strain carrying the mersacidin gene cluster; mrsA is replaced by ermB | this study |
| M. luteus ATCC 4698 | Indicator strain | ATCC 4698 |
| S. aureus SG511 | Indicator strain | [31] |
| B. megaterium KM | Indicator strain | ATCC 13632 |
| pCU1 | Shuttle vector | [32] |
| pPAR1 | pCU1 harboring the promoter of mrsA, mrsA and the regulator gene mrsR1 | [3] |
Purification of nucleic acids and sequencing
Genomic DNA was prepared using the PrestoSpinD Bug Kit according to the recommendations of the supplier (Molzym, Bremen, Germany). Plasmid DNA was isolated using the Gene-Jet™ Plasmid Miniprep Kit (Fermentas, St. Leon-Rot, Germany). All nucleic acids were analyzed by agarose gel electrophoresis and by spectrophotometry (Nanodrop Technologies, Wilmington, USA).
DNA sequencing was performed by Sequiserve (Vaterstetten, Germany) or Seqlab (Göttingen, Germany). Plasmid DNA and PCR products were dissolved in EB buffer (Qiagen, Hilden, Germany). Primers used for PCR analyses are listed in table 4. Sequencing primers were designed using the Primer3: WWW primer tool (http://biotools.umassmed.edu/bioapps/primer3_www.cgi).
Table 4. Primers used in this study.
| Primer | Gene | Sequence | Source |
| 16s1550.r | 16sRNA genes (rrn), B. amyloliquefaciens | AAGGAGGTGATCCAGCCG | [27] modified |
| 16s9.f | 16sRNA genes (rrn), B. amyloliquefaciens | AGAGTT TGATCCTGGCTCAG | [27] modified |
| ermB665.r | ermB | CAATTTAAGTACCGTTACTTATGAGC | this study |
| fbaB283.r | fbaB, B. amyloliquefaciens FZB42 | CTCCCGCATGACTGATATTCCTC | this study |
| gyrA_F | gyrA, B. amyloliquefaciens FZB42 | CAGTCAGGAAATGCGTACGTCCTT | [28] |
| gyrA_R | gyrA, B. amyloliquefaciens FZB42 | CAAGGTAATGCTCCAGGCATTGCT | [28] |
| FZBfor | comK, B. amyloliquefaciens FZB42 | ATGGGGTCGAAGGTCATTGAG | this study |
| FZBrev | comK, B. amyloliquefaciens FZB42 | CAGCTCCCGCAAAATAAAGTCG | this study |
| mrsE681.f | mrsE, mersacidin gene cluster | TGTCTCGGTCTCCTGGTTTACG | this study |
| mrsT2251.f | mrsT, mersacidin gene cluster | GGATAGACAGAAAGCTACGCTGC | this study |
Construction of the mersacidin producing B. amyloliquefaciens FZB42 mutants
The biosynthetic part of the mersacidin gene cluster was transferred to B. amyloliquefaciens FZB42 by competence transformation [26] using genomic DNA of the strain B. sp. HIL Y-85,54728 Rec1, which harbors a selection marker (ermB resistance cassette) instead of mrsA. Transformants were selected on TSA (0.3 µg/ml erythromycin) and were subsequently cultured on TSA containing erythromycin at a final concentration of 25 µg/ml. The transfer of the gene cluster was confirmed by PCR using primer combinations that anneal within ermB (ermB665.r), mrsE (mrsE681.f), mrsT (mrsT2251.f) and the downstream coding ORF fbaB/iolJ (fbaB283.r) (see Fig. 1). To exclude the possibility that these colonies might derive from germinated spores of Bacillus sp. HIL Y-85,54728 Rec1 that had not been eliminated during the gDNA preparation, differential PCRs were performed using the comK primers FZBfor and FZBrev. The 3′ ends of these primers anneal to single nucleotide polymorphisms of the sequence of B. amyloliquefaciens FZB42, but not to those of B. sp. HIL Y-85,54728. The resulting clone was named B. amyloliquefaciens mrs1.
For reconstitution of mersacidin production, the mersacidin structural gene (mrsA) was introduced in trans on the plasmid pPAR1 by competence transformation, yielding B. amyloliquefaciens mrs1 pPAR1. The presence of the plasmids as well as the plasmid integrity was analyzed by plasmid isolation and gel electrophoresis.
Producer self-protection against mersacidin
To test the susceptibility of B. amyloliquefaciens FZB42, B. amyloliquefaciens mrs1, and B. amyloliquefaciens mrs1 pPAR1 to mersacidin, the minimal inhibitory concentration (MIC) of mersacidin was determined by arithmetic broth microdilution. Serial twofold dilutions of mersacidin were prepared in polystyrene round bottom microtiter plates (Greiner, Frickenhausen, Germany) using half concentrated Mueller Hinton II broth (Difco, Detroit, USA) containing 1 mM CaCl2. An inoculum of 5×105 CFU/ml was employed in a final volume of 200 µl. The MICs were calculated from the lowest concentration of mersacidin resulting in the complete inhibition of visible bacterial growth after 16 hours of incubation at 37°C and compared to the MIC of the mersacidin producer B. sp. HIL Y-85,54728.
Mersacidin production by Bacillus amyloliquefaciens mrs1 pPAR1
The production of mersacidin by B. amyloliquefaciens mrs1 harboring pPAR1 was assayed in 50 ml synthetic medium (2 x BPM) [2] in the presence of chloramphenicol. The cells were grown for 24 hours at 37°C with agitation (180 rpm). For further analysis the culture supernatant was sterilized by filtration and stored at −20°C. The detection of antimicrobial activity in 50 µl of culture supernatant and HPLC fractions was performed by agar well diffusion assays on Mueller-Hinton agar II plates (Difco, Detroit, USA) seeded with the indicator strains Micrococcus luteus ATCC 4698, Bacillus megaterium KM and Staphylococcus aureus SG511 in wells with a diameter of 7 mm. After incubation at 37°C overnight, the growth inhibition zones were measured.
In order to remove the chloramphenicol and other antibiotics excreted by B. amyloliquefaciens FZB42, 5 ml of the culture supernatant containing 0.1 % trifluoroacetic acid (TFA, Sigma-Aldrich, Taufkirchen, Germany) were applied to a Poros RP-HPLC-column (10R2, 10064.6 mm Perseptive Biosystems, Freiburg, Germany) and eluted in a gradient of 30 % to 42 % acetonitrile (containing 0.1 % TFA). The peaks were detected measuring the absorbance at 210 or 220 and 266 nm. The fractions were collected and assayed for the antimicrobial activity against M. luteus ATCC 4698 in agar well diffusion assays. The active fractions of 85 ml of culture supernatant were lyophilized and purified further using an RP C-18 column (Nucleosil-100-C18, 250×4.5 mm; Schambeck SFD GmbH, Bad Honnef, Germany) with a gradient of 50 to 65 % acetonitrile (containing 0.1 % TFA). For MALDI-TOF analysis (Bruker Biflex, Bruker Daltonics, Bremen, Germany) of culture supernatant and active HPLC fractions, 20 µl of each fraction were concentrated 1∶10 using a rotational Vacuum Concentrator (RVC 2–18, Christ, Osterode, Germany). Then, a 1 µl sample was mixed with 2 µl matrix (alpha-cyano-4-hydroxycinnamic acid in acetonitrile: 0.1 % TFA in water, 1∶3). The mixtures were spotted onto the MALDI target and air-dried. Mass spectra were measured in positive ion mode in the range of 500 to 4000 Da and analyzed by Flexanalysis 2.0 (Bruker Daltonics).
Species identification of B. sp. HIL-Y-85,54728
The abilities of the wild type mersacidin producer strain, B. amyloliquefaciens FZB42 and B. subtilis 168 to ferment dextrose, maltose, lactose, sucrose, xylose, trehalose, mannitol and starch, to reduce sulfur and nitrate and to hydrolyze tryptophan and gelatin were compared using a conventional biochemical identification test. All inoculated tubes were incubated at 37°C for 24 h. Furthermore, growth at high salt concentrations (10 %) and high temperature (50°C) as well as motility were tested. The PCR product of the 16S rRNA genes was sequenced using the primers 16s9.f and 16s1550.r [27]. The gene coding for the gyrase subunit A (gyrA) was partially sequenced employing the primers gyrA_F and gyrA_R [28].
Bioinformatic tools and nucleotide sequence accession numbers
The sequences of the chromosome of B. subtilis 168 and B. amyloliquefaciens FZB42 are deposited in the NCBI database CoreNucleotide under the accession numbers NC_000964 and NC_009725, respectively. The sequence of the mersacidin gene cluster has the accession number AJ250862. Further sequences of the mersacidin producer strain have been deposited at NCBI under the following accession numbers: yvn, JF519627; czcO, JF519628; hpr, JF519629; baeD, JF519630; hemE, JF519631; comK, JF519632; gyrA, JF519633.
Blasts were performed at the NCBI nucleotide website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and ClustalW 2.0.12 (http://mobyle.pasteur.fr/cgi-bin/portal.py#forms::clustalw-multialign) were employed for multi sequence alignments of gyrA and calculation of the phylogenetic tree.
Acknowledgments
Mersacidin was kindly provided by Sanofi-Aventis (Frankfurt, Germany). The authors want to thank R. Borriss for the gift of Bacillus amyloliquefaciens FZB42.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by a grant of the DFG to G. Bierbaum (Deutsche Forschungsgemeinschaft Bi504-9/1, FOR 854) and the Bonfor program of the Medical Faculty of the University of Bonn. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chatterjee S, Lad SJ, Phansalkar MS, Rupp RH, Ganguli BN, et al. J Antibiot. Vol. 45. Tokyo: 1992. Mersacidin, a new antibiotic from Bacillus. Fermentation, isolation, purification and chemical characterization. pp. 832–838. [DOI] [PubMed] [Google Scholar]
- 2.Altena K, Guder A, Cramer C, Bierbaum G. Biosynthesis of the lantibiotic mersacidin: organization of a type B lantibiotic gene cluster. Appl Environ Microbiol. 2000;66:2565–2571. doi: 10.1128/aem.66.6.2565-2571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitz S, Hoffmann A, Szekat C, Rudd B, Bierbaum G. The lantibiotic mersacidin is an autoinducing peptide. Appl Environ Microbiol. 2006;72:7270–7277. doi: 10.1128/AEM.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guder A, Schmitter T, Wiedemann I, Sahl HG, Bierbaum G. Role of the single regulator MrsR1 and the two-component system MrsR2/K2 in the regulation of mersacidin production and immunity. Appl Environ Microbiol. 2002;68:106–113. doi: 10.1128/AEM.68.1.106-113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brötz H, Bierbaum G, Leopold K, Reynolds PE, Sahl HG. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob Agents Chemother. 1998;42:154–160. doi: 10.1128/aac.42.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Appleyard AN, Choi S, Read DM, Lightfoot A, Boakes S, et al. Dissecting structural and functional diversity of the lantibiotic mersacidin. Chem Biol. 2009;16:490–498. doi: 10.1016/j.chembiol.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szekat C, Jack RW, Skutlarek D, Färber H, Bierbaum G. Construction of an expression system for site-directed mutagenesis of the lantibiotic mersacidin. Appl Environ Microbiol. 2003;69:3777–3783. doi: 10.1128/AEM.69.7.3777-3783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuksel S, Hansen JN. Transfer of nisin gene cluster from Lactococcus lactis ATCC 11454 into the chromosome of Bacillus subtilis 168. Appl Microbiol Biotechnol. 2007;74:640–649. doi: 10.1007/s00253-006-0713-y. [DOI] [PubMed] [Google Scholar]
- 9.Liu W, Hansen JN. Conversion of Bacillus subtilis 168 to a subtilin producer by competence transformation. J Bacteriol. 1991;173:7387–7390. doi: 10.1128/jb.173.22.7387-7390.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan MP, McAuliffe O, Ross RP, Hill C. Heterologous expression of lacticin 3147 in Enterococcus faecalis: comparison of biological activity with cytolysin. Lett Appl Microbiol. 2001;32:71–77. doi: 10.1046/j.1472-765x.2001.00864.x. [DOI] [PubMed] [Google Scholar]
- 11.Heidrich C, Pag U, Josten M, Metzger J, Jack RW, et al. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl Environ Microbiol. 1998;64:3140–3146. doi: 10.1128/aem.64.9.3140-3146.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caetano T, Krawczyk JM, Mosker E, Süßmuth RD, Mendo S. Heterologous expression, biosynthesis, and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli. Chem Biol. 2011;18:90–100. doi: 10.1016/j.chembiol.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Chen XH, Koumoutsi A, Scholz R, Eisenreich A, Schneider K, et al. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol. 2007;25:1007–1014. doi: 10.1038/nbt1325. [DOI] [PubMed] [Google Scholar]
- 14.Chen XH, Koumoutsi A, Scholz R, Schneider K, Vater J, et al. Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J Biotechnol. 2009;140:27–37. doi: 10.1016/j.jbiotec.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 15.Scholz R, Molohon KJ, Nachtigall J, Vater J, Markley AL, et al. Plantazolicin, a novel microcin B17/streptolysin S-like natural product from Bacillus amyloliquefaciens FZB42. J Bacteriol. 2010;193:215–24. doi: 10.1128/JB.00784-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 17.Julkowska D, Obuchowski M, Holland IB, Seror SJ. Comparative analysis of the development of swarming communities of Bacillus subtilis 168 and a natural wild type: critical effects of surfactin and the composition of the medium. J Bacteriol. 2005;187:65–76. doi: 10.1128/JB.187.1.65-76.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuge K, Ano T, Hirai M, Nakamura Y, Shoda M. The genes degQ, pps, and lpa-8 (sfp) are responsible for conversion of Bacillus subtilis 168 to plipastatin production. Antimicrob Agents Chemother. 1999;43:2183–2192. doi: 10.1128/aac.43.9.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paik SH, Chakicherla A, Hansen JN. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J Biol Chem. 1998;273:23134–23142. doi: 10.1074/jbc.273.36.23134. [DOI] [PubMed] [Google Scholar]
- 20.Babasaki K, Takao T, Shimonishi Y, Kurahashi K. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J Biochem. 1985;98:585–603. doi: 10.1093/oxfordjournals.jbchem.a135315. [DOI] [PubMed] [Google Scholar]
- 21.Tamehiro N, Okamoto-Hosoya Y, Okamoto S, Ubukata M, Hamada M, et al. Bacilysocin, a novel phospholipid antibiotic produced by Bacillus subtilis 168. Antimicrob Agents Chemother. 2002;46:315–320. doi: 10.1128/AAC.46.2.315-320.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hilton MD, Alaeddinoglu NG, Demain AL. Synthesis of bacilysin by Bacillus subtilis branches from prephenate of the aromatic amino acid pathway. J Bacteriol. 1988;170:482–484. doi: 10.1128/jb.170.1.482-484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rückert C, Blom J, Chen X, Reva O, Borriss R. J Biotechnol. In press; 2011. Genome sequence of B. amyloliquefaciens type strain DSM7(T) reveals differences to plant-associated B. amyloliquefaciens FZB42. [DOI] [PubMed] [Google Scholar]
- 24.Borriss R, Chen X, Rueckert C, Blom J, Becker A, et al. Relationship of Bacillus amyloliquefaciens clades associated with strains DSM7T and FZB42: a proposal for Bacillus amyloliquefaciens subsp. 2010. amyloliquefaciens subsp. nov. and Bacillus amyloliquefaciens subsp. plantarum subsp. nov. based on their discriminating complete genome sequences. Int J Syst Evol Microbiol. In press. [DOI] [PubMed]
- 25.Koumoutsi A, Chen XH, Henne A, Liesegang H, Hitzeroth G, et al. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol. 2004;186:1084–1096. doi: 10.1128/JB.186.4.1084-1096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reva ON, Dixelius C, Meijer J, Priest FG. Taxonomic characterization and plant colonizing abilities of some bacteria related to Bacillus amyloliquefaciens and Bacillus subtilis. FEMS Microbiol Ecol. 2004;48:249–259. doi: 10.1016/j.femsec.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Idriss EE, Makarewicz O, Farouk A, Rosner K, Greiner R, et al. Extracellular phytase activity of Bacillus amyloliquefaciens FZB45 contributes to its plant-growth-promoting effect. Microbiology. 2002;148:2097–2109. doi: 10.1099/00221287-148-7-2097. [DOI] [PubMed] [Google Scholar]
- 30.Zeigler DR, Pragai Z, Rodriguez S, Chevreux B, Muffler A, et al. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol. 2008;190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sass P, Bierbaum G. Native graS mutation supports the susceptibility of Staphylococcus aureus strain SG511 to antimicrobial peptides. Int J Med Microbiol. 2009;299:313–322. doi: 10.1016/j.ijmm.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Augustin J, Rosenstein R, Wieland B, Schneider U, Schnell N, et al. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur J Biochem. 1992;204:1149–1154. doi: 10.1111/j.1432-1033.1992.tb16740.x. [DOI] [PubMed] [Google Scholar]