Abstract
Extensive studies over the last 30 years have demonstrated that vascular smooth muscle cell (SMC) differentiation and phenotypic modulation is controlled by a dynamic array of environmental cues. The identification of the signaling mechanisms by which these environmental cues regulate SMC phenotype has been more difficult because of our incomplete knowledge of the transcription mechanisms that regulate SMC-specific gene expression. However, recent advances in this area have provided significant insight, and the goal of this review is to summarize the signaling mechanisms by which extrinsic cues control SMC differentiation.
Introduction
Smooth muscle cells (SMC) provide structural support to the vasculature and control blood pressure and blood flow through highly regulated contractile mechanisms. Differentiated SMC express a variety of SMC-specific contractile and contractile-associated proteins that contribute to these functions including SM myosin heavy chain (SM MHC), SM22, calponin, and SM α-actin. Importantly, unlike cardiac and skeletal muscle cells, SMC retain significant plasticity even in adult animals. Thus, in response to vessel injury, SMC undergo phenotypic modulation, a process characterized by decreased SMC differentiation marker gene expression and increased proliferatiion, migration, and matrix synthesis. The search for the transcription regulatory mechanisms that ultimately govern the process of SMC differentiation has been complicated by the plasticity of this cell-type and the fact that SMC derive from multiple precursors throughout the embryo 1. The MADS box transcription factor, serum response factor (SRF), regulates most SMC differentiation marker genes by binding to highly conserved CArG cis elements (CC(A/T)6GG) that are present within nearly all of the SMC-specific promoters (see 2 for review). However, SRF cannot be considered a master regulator of SMC differentiation because it is a ubiquitously expressed protein that also regulates cardiac- and skeletal-muscle-specific genes expression and the expression of a variety of early response and structural genes 3. SRF-dependent transcription is controlled to a large extent by SRF’s interaction with additional transcription factors and co-activators. The first SRF co-factors described were the ternary complex factors (TCFs) like Elk-1 that regulate early response gene expression and that interact with SRF when phosphorylated by MAP-kinase (see 4 for review). Some Nkx and Gata family members are expressed in certain SMC sub-types (Nkx3.1, Nkx3.2, and Gata-6) and have been shown interact with SRF to regulate SMC differentiation marker gene expression 5, 6.
The discovery of the cardiac/SMC-selective SRF co-factor, myocardin, was a major advance in our understanding of the transcription mechanisms that regulate SMC-specific gene expression 7. Unlike the Nkx and Gata factors, myocardin does not contain a DNA binding domain but powerfully transactivates cardiac and SMC-specific gene expression by physically interacting with SRF 7. Importantly, expression of myocardin activated SMC differentiation marker gene expression in a variety of non-SMC cells while deletion of myocardin in the mouse resulted in embryonic lethality at E10.5 due, at least in part, to failure of SMC differentiation within the developing dorsal aorta 8, 9. Interestingly, cardiac development did not seem to be affected by deletion of myocardin. Additional analyses of ES cells and chimeric mice indicated that myocardin null cells could differentiate into SMC indicating that other transcription mechanisms could compensate for its loss at least under some conditions 10. Two myocardin-related transcription factors, MRTF-A and MRTF-B, have been identified that have similar transcriptional properties to myocardin 11. Although the MRTFs are expressed more ubiquitously, both are strongly expressed in SMC and have been shown to be required for SMC-specific gene expression in several SMC culture models 12–14. Importantly, separate groups have shown that deletion of MRTF-Bin the mouse resulted in defective SMC differentiation of the cardiac neural crest cells that populate the aortic arch 15, 16. In addition, deletion of MRTF-A prevented the up-regulation of SMC-specific gene expression that normally occurs in myoepithelial cells of the mammary gland during lactation 17, 18. Taken together, these data indicate that the myocardin transcription factors have unique functions that are required for SMC differentiation marker gene expression and vascular development in vitro and in vivo. Unfortunately, the overlapping expression patterns, functional redundancy, and potential heterodimerization between myocardin factors has made it difficult to determine their precise roles in SMC, especially for those aspects of SMC function that do not directly involve specification such as the changes in SMC-specific gene expression that are known to occur during environmental stresses.
Regulation of SRF
A critical step in the activation of SMC-specific gene expression is SRF binding to CArG elements, and several mechanisms regulate this interaction. High SRF expression in all three muscle cell-types likely promotes SRF binding to the relatively low affinity CArG elements present within the muscle-specific promoters. An increase in SRF expression correlates well with the appearance of the SMC during development 19 and can be induced by a number of agonists such as TGF-β that are well-known stimulators of SMC differentiation 20, 21. The transcription mechanisms that regulate SRF expression are not completely clear, but the observation that multiple CArG elements control SRF promoter activity strongly suggests that high SRF expression in muscle cells is facilitated by a positive feedback loop 22, 23. Phosphorylation of Serine103 by a number of kinases increases SRF affinity for CArG elements 24, 25, and results from Garat et al suggested that this mechanism was important for arginine vasopressin-mediated induction of the SM α-actin promoter 26. More recent studies from our lab and Iyer et al demonstrated that phosphorylation within SRF’s core DNA binding domain (at S162 by PKCα and at T159 by PKA) decreased SM α-actin promoter activity by inhibiting SRF binding 27, 28. Importantly, neither of these phosphorylation events affected c-fos promoter activity due to the stabilizing effects of Elk-1 within the ternary complex 27. SRF’s interaction with other transcription factors and co-factors influences SRF binding most likely by steric alterations in SRF conformation. The homeodomain proteins, Nkx3.1, Prx-1, and Barx2b, and the myocardin factors have been shown to enhance SRF binding 5, 29–31 while the homeodomain-only protein (HOP)and YY1 have been shown to inhibit SRF binding 32, 33.
It is extremely important to note that the ability of SRF (or any other factor) to regulate SMC differentiation absolutely requires additional mechanisms that control chromatin structure and transcription factor access to the SMC-specific gene promoters. Consistent with the concept of the “histone code”, binding of SRF to SMC-specific promoters correlates strongly with positive chromatin marks (i.e. H3 acetylation, H3K4 methylation, etc)and a number of labs including our own have shown that the myocardin factors interact with chromatin modifiers 34–39. For a more complete discussion of this important topic we refer the reader to other reviews 40 including the Chen review in this series.
Regulation of myocardin
Clearly, signaling mechanisms that affect myocardin levels and/or activity play a major role in the regulation of SMC differentiation. Hendrix et al demonstrated that myocardin mRNA expression was significantly reduced in rat carotid arteries following wire injury in a time course that closely paralleled the down-regulation of SMC differentiation marker gene expression in this model 41. Although the mechanism for this down regulation is not yet clear, several studies have shown that PDGF-BB inhibits myocardin expression 14, 42 and its release at sites of vascular injury probably contributes to this effect. Interestingly, Doi et al failed to observe a decrease in myocardin protein expression in a rat thoracic aorta injury model perhaps reflecting a differential response to injury governed by SMC origin 43. Several environmental cues that enhance SMC-specific gene expression including angiotensin II and increased calcium influx have been shown to up-regulate myocardin expression 31, 44, 45. Callis et al demonstrated that myocardin expression in cardiomyocytes was up-regulated by BMP-2 46, but a similar effect was not observed in SMC 47. The transcription mechanisms that regulate myocardin expression are starting to be described. Nkx2.5 and NFATc3were shown to bind and activate proximal myocardin promoter fragments in cardiomyocytes, and treatment of myocardial cells with aldosterone or the β-adrenergic agonist, isoproterenol, enhanced NFATc3 binding to the myocardin promoter 48, 49. The relevance of these results is some what unclear given the recent demonstration that myocardin expression in vivo is controlled by an enhancer ~25kb upstream of myocardin’s translational start site 50. This enhancer was activated by MEF2, and several members of the Foxo and TEAD transcription factor families, warranting further studies on the expression and control of these factors in SMC.
The myocardin transactivation domain can be phosphorylated by GSKβ and ERK resulting in decreased SMC differentiation marker gene expression 51–53, and these modifications probably interfere with the transactivation domain’s ability to interact with other cell-selective and/or general transcription factors. Of interest, myocardin has been shown to interact with the estrogen receptor co-activator, SRC3, to enhance SMC-specific expression in human aortic SMC 54. Somewhat surprisingly, treatment of SMC with estrogen had little effect on this mechanism suggesting that the protective effects of estrogen on SMC phenotypic modulation involves additional signals 55. Tang et al demonstrated that NFkB physically interacts with myocardin to inhibit its activity 56. This important observation may explain the down regulation of SMC differentiation marker gene expression observed under inflammatory conditions such as those found in atherosclerosis. Liu et al demonstrated that myocardin activity is also inhibited by its interaction with Foxo4and this mechanism has potential implications on the regulation of SMC phenotype by agonists that stimulate AKT 57. These authors demonstrated that IGF-1/AKT-dependent phosphorylation of Foxo4 induced its translocation from the nucleus to the cytoplasm, thus relieving Foxo4-dependent inhibition of SMC differentiation marker gene expression. The bHLH transcription factors, Msx1 and Msx2, interact with myocardin to inhibit SMC-specific transcription, and the up-regulation of these factors by BMPs may be critical for the modulation of SMC phenotype under conditions that promote vessel calcification 47. Myocardin activity is also inhibited by the bHLH factor, HERP1/HEY2, and the HMG protein, HMG2L1 43, 58. Both of these factors are up-regulated in phenotypically modulated SMC and inhibit myocardin’s interaction with SRF.
The recent demonstration that the myocardin factors are targeted for proteasome-mediated degradation could have significant implication on the regulation of SMC differentiation. We were the first to demonstrate that myocardin factor stability in SMC was enhanced by proteasome inhibition and that MRTF-A was ubiquitylated 59. Xie et al have now shown that the E3 ligase, CHIP (C-terminus of HSC70-intreacting protein), ubiquitylates myocardin in SMC 60. Importantly, these authors demonstrated that siRNA-mediated knockdown of CHIP significantly increased the expression of several SMC differentiation marker genes while overexpression of CHIP in isolated aortic rings decreased SMC contractility. Hu, et. al. identified the E3 ligase, UBR5, as a myocardin interacting protein using a yeast-two-hybrid screen, but surprisingly, UBR5 increased myocardin stability by a mechanism independent of its E3 ligase function 61. Myocardin and MRTF-A can also be modified by the Small Ubiqutin-like MOdifier (SUMO), but because sumolyation activated myocardin, but inhibited MRTF-A 62, 63 the functional significance of this modification in regard to SMC-specific gene expression remains unknown.
Regulation of SMC differentiation by RhoA
Early studies from the Treisman lab demonstrated that SRF-dependent transcription was regulated by the small GTPase, RhoA 64, 65. Based upon these results, we were the first to show that RhoA activity was an important determinant of SMC differentiation marker gene expression 66, but the mechanisms involved were still unknown. A seminal study by Miralles et al demonstrated that RhoA-dependent actin polymerization promoted the nuclear localization of MRTF-A and that G-actin binding to MRTF-A’s N-terminal RPEL domains inhibited this translocation 67. MRTF-B nuclear localization is also controlled by this mechanism, but since the myocardin RPEL domains do not bind G-actin very strongly, myocardin is constitutively nuclear 68. Many groups have now shown that RhoA activity is required for SMC-specific gene expression in a variety of SMC differentiation models 69–74 and for the up-regulation of SMC-specific transcription in response to many environmental cues including thrombin, sphingosine 1-phosphate (S1P), TGF-β, calcium, BMP-2, and cell tension 44, 72, 74–80. In several of these models the activation of SMC-specific gene expression was accompanied by MRTF nuclear accumulation as measured by nuclear fractionation, immunohistochemistry, or localization of GFP-tagged MRTF variants 75, 78–80. In addition, we have shown that RhoA-dependent SMC-specific promoter activity was inhibited by expression of a nuclear localization deficient variant of MRTF-A that traps endogenous MRTFs in the cytoplasm 12. Since this intervention does not affect myocardin activity or MRTF expression, these data provided direct evidence for MRTF nuclear translocation as an important signaling mechanism in the control of SMC-specific gene expression.
The mechanisms that regulate RhoA in SMC have not been completely described and are likely to be complex. Like all small GTPases, RhoA activation is tightly regulated by GTPase Activating Proteins (GAPs) that facilitate RhoA’s intrinsic GTPase activity (inhibiting RhoA) and Guanine Exchange Factors (GEFs) that facilitate exchange of GDP for GTP (activating RhoA) (see 81 for review). In SMC, RhoA activity is mainly regulated by agonists such as angiotensin II (AII), thrombin, and S1P that signal through G-protein coupled receptors (GPCRs). GPCR-dependent activation of RhoA is mediated, at least in part, by Gα12/13-dependent activation of the RGS family of RhoGEFs (LARG, p115RhoGEF, and PDZRhoGEF) 82 and/or Gαq/11-dependent activation of the Trio family of RhoGEFS (Trio, Duet, and p63RhoGEF) 83, 84. SMC typically express multiple receptors for the same agonist and because these receptors have different G-protein coupling properties it has been difficult to predict or determine the relative importance of these proteins to SMC-specific transcription. Using a combination of receptor sub-type-specific agonists for the fiveS1P receptors (S1PR1-5), we and others have demonstrated that S1PR2 is a major activator of RhoA in SMC and is required for S1P-dependent up-regulation of SMC-specific gene expression 85, 86. Our data also suggest that theS1PR2 signals mainly through LARG to activate RhoA in SMC 86. Interestingly, S1PR2deficient mice do not show an overt SMC phenotype, but Shimizu et al have recently demonstrated that they are more susceptible to injury-induced restenosis 87 suggesting that S1P signaling through this pathway helps maintain SMC differentiation in vivo. Angiotensin II-dependent activation of RhoA in SMC is much more complex and has been shown to be mediated byJAK2-dependent activation of p115RhoGEF 88, inhibition of p190RhoGAP 89, up-regulation of LARG 90, and calcium-activated phosphorylation of PDZ-RhoGEF by the tyrosine kinase, PYK2 91.
The signaling mechanisms downstream of RhoA that control MRTF nuclear localization involve multiple RhoA effectors that enhance actin polymerization. Rho-kinase stimulates actin polymerization by inhibiting the disassembly of actin polymers through LIM-kinase-dependent inhibition of cofilin 92. Pharmacologic inhibition of Rho-kinase withY-27632 significantly attenuates SMC-specific gene expression, but does not completely inhibit it 12, 44, 75. The diaphanous-related formins (DRFs), mDia1 and mDia2, are RhoA effectors that directly catalyze actin polymerization through a mechanism that involves profilin 93, 94, and we have shown that both are highly expressed in SMC, and are required for SMC differentiation marker gene expression 95. Another DRF, FHOD1 (formin homology domain containing protein-1), is also highly expressed in SMC 96. Although FHOD1 binds Rac and not RhoA, its activity is regulated by ROCK-dependent phosphorylation 97, and we have shown that this signaling mechanism contributes to SMC-specific transcription 96. The RhoA effector, protein kinase-N, was shown to up-regulate SMC-specific gene expression by a mechanism yet to be described 77.
Taken together these data strongly suggest that RhoA-dependent regulation of MRTF nuclear localization is an important mechanism for the regulation of SMC differentiation marker gene expression and is facilitated in SMC by a combination of parameters including high SRF and myocardin factor expression levels, strong GPCR-dependent activation of RhoA, and relatively high expression of downstream RhoA effectors. Finally, it is important to note that like SRF, RhoA also regulates cell proliferation and several studies have demonstrate that RhoA signaling was required for the stimulation of SMC growth by AII, thrombin, mechanical stretch, and vessel injury 98–102. However, it is also clear that SMC growth and differentiation are not mutually exclusive and that activation of RhoA by itself is not sufficient to stimulate SMC proliferation 75, 95, 103, 104.
Regulation of SMC differentiation by TGF-β
TGF-β is a multifunctional cytokine that regulates vascular development and maintenance by controlling the growth, differentiation, and matrix synthesizing properties of EC, SMC, and lymphocytes. In most cells including SMC, TGF-β signals mainly through a heteromeric complex of two serine/threonine kinase receptors, the type IITGF-β receptor (TβRII)and the type I receptor, ALK5 (see 105 for review). Within the ligand-induced receptor complex, constitutively active TβRII phosphorylates ALK5 resulting in the recruitment and phosphorylation-dependent activation of Smads 2 and 3. The activated Smads complex with Smad4 and then translocate to the nucleus where they stimulate gene expression. The Smads contain DNA binding motifs and have been shown to interact with DNA at a consensus Smad binding element (GTCT). However, because Smads bind these elements with very low affinity it is thought that they regulate transcription in combination with additional transcription factors and co-factors.
Extensive studies have shown that TGF-β strongly stimulates SMC differentiation marker gene expression in a number of cell types including 10T1/2, Monc-1, mesenchymal and embryonic stem cells, lung fibroblasts, and aortic SMC 20, 106–113. This effect requires activation of Smad2 and/or Smad3, and several studies have demonstrated that these Smads interact with the SMC-specific promoters at putative Smad binding elements 107, 108, 114, 115. The effects of TGF-β are also CArG/SRF-dependent 20, and studies by Qui et al have shown that Smad3 physically interacts with SRF to facilitate SMC-specific gene expression 114. Interestingly, this group demonstrated that Smad3 can recruit myocardin to the SM22 promoter by a direct interaction between these transcription factors 115. Nishimura, et. al. have shown that Smad3 also interacts with δEF1, a zinc finger- and homeodomain-containing protein expressed in SMC 112. δEF1 was up-regulated by TGF-β, formed a complex with SRF and Smad3 on the SM α-actin promoter and was required for the full effects of TGF-β on SMC-specific gene expression. TGF–β also up-regulates the expression of many proteins that have secondary effects on SMC phenotype including many matrix and matrix remodeling proteins (see 116 for review). The discovery that the effects of TGF-β on SMC-specific gene expression were mediated at least in part by the NADPH oxidase, Nox4, 117–120 was particularly intriguing and supported previous reports that reactive oxygen species regulate SMC-specific transcription 121, 122. Interestingly, reactive oxygen species have been shown to regulate a number of critical SMC processes 123 but the precise mechanisms involved are currently unknown.
By poorly understood mechanisms, TGF-β directly activates (within minutes) several signaling pathways that affect SMC phenotype (see 124 for review). Activation of p38, most-likely by TGF-β-activated kinase, was shown to required for TGF-β-induced SM α-actin expression in fibroblasts and Pac-1 SMC 77, 125, 126. Activation of p38 also inhibits cell cycle progression and is required for TGF-β’s ability to inhibit SMC proliferation 127. Lien et al have demonstrated that TGF-β activates AKT in 10T1/2 cells and that this signaling pathway was required for TGF-β’s effects on SMC-specific transcription 128. TGF-β also activates RhoA in SMC and SMC precursors, and inhibition of RhoA signaling prevented TGF-β-dependent SMC differentiation marker gene expression in these models 74, 77. In our hands, TGF-β is a relatively weak and slow activator of RhoA and did not stimulate MRTF nuclear localization in SMC at early time-points (< 2h) 12 suggesting a different mechanism for its requirement. Interestingly, inhibition of RhoA prevented Smad2 and Smad3 nuclear localization 74, but this potentially important mechanism will require further investigation. Recent studies have shown that TGF-β up-regulates the expression of multiple RhoA signaling proteins including the Rho GEFs, Net-1 and GEF-H1, and RhoB 129–131 suggesting that TGF-β’s ability to promote long-term activation of RhoA signaling may contribute to its effects on SMC differentiation.
Notch signaling plays a dual role in the regulation of SMC differentiation
Recent studies have demonstrated that Notch signaling plays an important role in vascular development and maintenance (see 132 for reviews). Four single-pass transmembrane Notch receptors (Notch1–4) have been described in mammals with Notch3 most strongly expressed in SMC. Importantly, the Notch receptors interact with ligands that are also transmembrane proteins (Jagged1 and 2 and Delta-like1, 3, and 5) thus limiting Notch signaling to adjacent cells. Activation of Notch by ligand binding results in proteolytic cleavage of the receptor by γ-secretase, release of the Notch intracellular domain (NICD), and translocation of the NICD to the nucleus where it interacts with the multifunctional transcription regulator, RBPJ. In the absence of NICD, RBPJ binds and represses target genes by recruiting HDACs and other transcriptional repressors. Displacement of the repressive factors upon NICD binding and the eventual recruitment of transcriptional activators such as MAML and histone acetyltransferases results in activation of Notch target genes. Major gene targets of Notch are the Hes and Hey/HERP transcription regulators.
Notch signaling has been shown to promote SMC differentiation marker gene expression, but its effects are likely depend upon cell context. Over-expression of NICD in 10T1/2 or human SMC cells stimulated SMC differentiation marker gene expression, and RBPJ was shown to interact with the SM α-actin promoter by ChIP assays 133, 134. Endothelial-specific deletion of Jagged1 resulted in embryonic lethality and severe defects in SMC investment of vessels 135. In addition, neural crest cell-specific expression of a dominant negative MAML that inhibits all Notch family members disrupted aortic arch development and reduced SMC differentiation 136. The decrease in artery maturation observed in Notch3 deficient mice 137 and the requirement for Notch3 in EC-dependent mural cell differentiation 138 provide additional evidence that Notch activation is important for SMC identity. Interestingly, a mutation in Notch3 is causal for CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephaly), a neurovascular disorder associated with SMC abnormalities 139.
A number of studies have demonstrated that expression of the NICD inhibited SMC differentiation marker gene expression significantly complicating our understanding of this pathway 140, 141. These inhibitory effects are due, at least in part, to up regulation of the canonical Notch target genes of the HERP/HEY family which have been shown to inhibit SMC differentiation marker gene expression by interfering with SRF/myocardin binding to CArG elements and by directly inhibiting the function of the NICD/RBPJ complex 43, 142. HERP1/HEY2 is up-regulated following vascular injury and HERP1/HEY2 knockout mice have reduced neointimas suggesting that that this mechanism plays a role during SMC phenotypic modulation 43, 143. Further supporting this idea, Notch3 deficient mice do not develop pulmonary hypertension in response to hypoxia and knockdown of HES-5 in human pulmonary artery SMC decreased proliferation and increased SMC differentiation marker gene expression 144. Thus, in regard to the regulation of SMC differentiation, the eventual outcome of Notch activation seems to depend upon the efficacy and timing of these competing transcriptional events. Notch receptor and ligand expression levels are known to change during development and following injury 145 and it will be important to determine whether alterations in these parameters have significant effects on the dynamics of Notch activation. It will also be critical to determine how the Notch pathway in SMC is influenced by additional signaling and transcription mechanisms especially those that regulate chromatin structure.
Inhibition of SMC differentiation by PDGF-BB
The growth factor, PDGF-BB, is a critical regulator of early vascular development. It is highly expressed by EC and is required for the initial recruitment and subsequent proliferation of pericytes and SMC within the maturing vasculature 146. However, treatment of differentiated SMC with PDGF-BB strongly stimulates SMC phenotypic modulation by enhancing SMC proliferation and migration and down-regulating SMC differentiation marker gene expression (see 147 for review). The observations that PDGF-BB is released upon vessel injury 148 and that inhibition of PDGF-BB signaling inhibits neointimal growth 149 strongly suggest that it is a major regulator of SMC phenotype in vivo. It is clear that PDGF-BB regulates SMC growth and differentiation by multiple, but partially overlapping mechanisms. Activation of tyrosine kinase receptors such as PDGFRβ triggers the Ras/Raf/MEK/ERK kinase cascade leading to Elk-1 phosphorylation and the SRF-dependent up-regulation of multiple early response growth genes. Importantly, myocardin and Elk-1 bind to the same region of SRF and compete for SRF binding 150. Thus, in PDGF-BB treated SMC, phosphorylated Elk-1 inhibited SMC differentiation marker gene by displacing myocardin (and likely the MRTFs) from the SMC-specific promoters 150, 151. ERK-dependent phosphorylation of MRTF-A at Ser454 was also shown to inhibit MRTF-A nuclear accumulation in HeLa cells 152. Results from the Owens laboratory suggest that the negative effects of PDGF-BB are at least partially mediated by the pluripotency transcription factor, KLF4. These authors demonstrated that KLF4was strongly induced by PDGF-BB and vascular injury, and inhibited SMC differentiation marker gene expression by decreasing myocardin expression, interfering with SRF/myocardin factor binding to the SMC-specific promoters, and modulating the chromatin environment near the SMC-specific promoters 14, 153, 154. Although this group has also identified a G/C repressor within the SM22 promoter that is required for its down-regulation upon injury or treatment with PDGF-BB 155, 156, it is somewhat unclear whether the inhibitory effects of KLF4 are mediated by this cis element.
Integrin-matrix signaling
Early studies demonstrated that SMC differentiation during development correlated strongly with a change in basement membrane composition from fibronectin, which supports SMC proliferation, to collagen IV and laminin, which promote SMC differentiation 157–159. Matrix degradation and the re-expression of fibronectin and other growth promoting matrix components such as collagen I also occur following vessel injury and likely contribute to SMC phenotypic modulation 160. Matrix components signal through heterodimeric integrin receptors composed of α and β subunits and the mechanisms by which integrins regulate cell growth are fairly well described (see 161 for review). In brief, integrin activation leads to the formation of multiprotein focal adhesion signaling complexes and the activation of the non-receptor tyrosine kinases, FAK and c-src. Tyrosine phosphorylation of FAK, Shc, and other focal adhesion scaffolding proteins such paxillin a p130Cas, leads to activation of the Raf/Ras/MEK/ERK kinase cascade through Grb2/Sos-dependent mechanisms. As discussed above, up- or down-regulation of ERK signaling may explain the effects of integrin-matrix signaling on SMC differentiation marker gene expression, but other mechanisms are likely to be involved. Orr et al recently demonstrated that plating SMC on collagen IV, but not on collagen I, enhanced myocardin expression, although the mechanism for this effect was not clear 162. Integrin signaling is also a major regulator of the actin cytoskeleton and is required for a cell’s ability to sense and respond to mechanical forces (see 163 for review). Although the effects of these cues on SMC-specific gene expression are mediated, at least in part, by RhoA/MRTF-dependent mechanisms 76, 80, 164, 165, the signaling pathways by which integrins, FAK, and mechanical forces regulate RhoA in cells are only partially understood 166. Interestingly, Kogata et al have shown that integrin-linked kinase (ILK), a very weak serine/threonine kinase that interacts with the β1 integrin receptor, negatively regulates RhoA activity in SMC and that deletion of ILK in PDGFRβ expressing cells in vivo resulted in defects in SMC investment and hypercontractility 167. Although Wu, et al have demonstrated that ILK negatively regulates SMC differentiation marker gene expression 168, additional studies will be required to confirm that these effects were due to alterations in RhoA signaling.
Interestingly, several SMC-selective integrin and/or signaling molecules have been described that regulate SMC differentiation marker gene expression. First, in close collaboration with the Taylor lab, we have shown that FAK activity in SMC is regulated by a dominant negative FAK variant termed FRNK (FAK-related non-kinase) that is selectively expressed in large arteries 169. FRNK expression is up-regulated during early post-natal development and following vascular injury when SMC are transitioning from the proliferative to contractile phenotype, and FRNK promotes this transition by inhibiting SMC growth and migration and by stimulating SMC differentiation marker gene expression 170, 171. In a second collaborative effort with the Taylor lab, we demonstrated that the SMC-selective LIM protein, leupaxin, interacts with SRF to activate SMC-specific gene expression most likely by scaffolding additional transcription co-activators through its multiple LIM domains 172. Importantly, leupaxin localizes to focal adhesions and the nucleus, and we demonstrated that nuclear translocation of leupaxin was inhibited by plating cells on fibronectin or by expression of an active FAK variant. Chang et al demonstrated that the SMC-specific LIM proteins, CRP1 and CRP2 enhanced SMC-specific gene expression by facilitating SRFs interaction with Gata–6 173. The CRPs have been shown to bind α-actinin and to localize to actin stress fibers within the cytoplasm 174. Whether CRP nuclear/cytoplasmic shuttling is regulated by integrin-matrix signaling is unclear but seems likely. The four and a half LIM domain-containing protein, FHL2, is a CArG-dependent cardiac/SMC-selective protein that also interacts with SRF 175. In contrast to the effects of leupaxin and the CRPs, FHL2 was shown to inhibit SMC-specific transcription by competitively interfering with MRTF binding to SRF. Like leupaxin, FHL2 localizes to focal adhesions and its nuclear accumulation was enhanced by activation of RhoA 176 suggesting that FHL2 may act as a feedback inhibitor of RhoA/SRF-dependent gene expression.
Summary and Unanswered Questions
The discovery of the myocardin factors has provided a regulatory framework for our understanding of the control of SMC phenotype. Figures 1 summarizes many of the signaling mechanisms that stimulate and/or maintain SMC differentiation while figure 2 summarizes those that promote SMC phenotypic modulation. It is important to emphasize that significant cross-talk exists between these signaling mechanisms and that SMC phenotype likely reflects the integrated sum of these pathways. It is clear that many important questions still remain. What are the epigenetic mechanisms that allow transcription factor access to the SMC-specific genes? What are the mechanisms that regulate myocardin factor expression? To what extent does RhoA-dependent MRTF nuclear localization regulate SMC differentiation in vivo? How do the direct and indirect actions of Notch affect SMC differentiation? Can we define the cis regulatory elements and trans-acting factors that mediate Smad interactions with the SMC-specific promoters. What are the mechansims by which integrin-matrix signaling regulate RhoA and the localization of the LIM protein SRF co-factors in SMC? Do signaling pathways have differential effects on SMC phenotype based upon developmental origin? Can we identify novel signaling targets for the treatment of cardiovascular diseases that involve SMC phenotypic modulation? The recent development of tools for studying signaling, transcription, and chromatin networks on a genome-wide basis will certainly facilitate these efforts and will hopefully increase our understanding of the complex signaling and transcription mechanisms that regulate SMC differentiation.
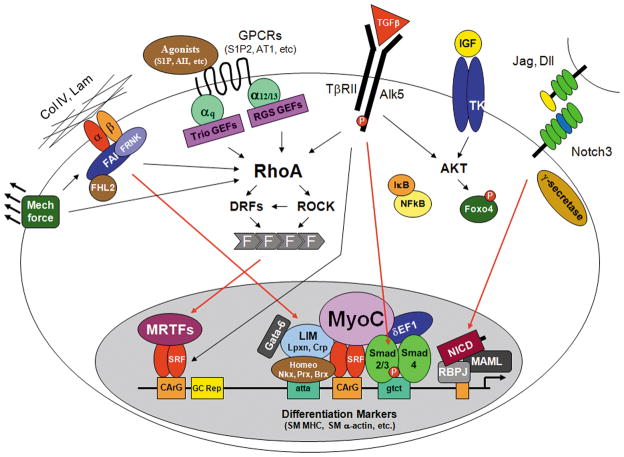
Figure 1. Signaling pathways that stimulate and/or maintain SMC differentiation.
The red arrows denote nuclear translocation events.
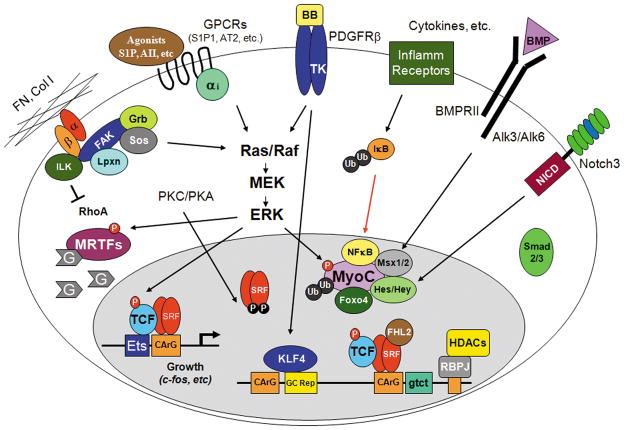
Figure 2. Signaling pathways that promote the phenotypic modulation of SMC.
Note that myocardin levels are significantly reduced. The red arrows denote nuclear translocation events.
Acknowledgments
I would like to thank all of my colleagues for the pleasure of discussing their work. More importantly, I apologize for the many important pathways and studies that have been omitted due to space constraints.
Sources of Funding -This work was supported by NIH grantsHL-070953and HL-082883
Footnotes
Disclosures -none
References
- 1.Majesky MW. Developmental basis of vascular smooth muscle diversity. Arterioscler Thromb Vasc Biol. 2007;27:1248–1258. doi: 10.1161/ATVBAHA.107.141069. [DOI] [PubMed] [Google Scholar]
- 2.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 3.Sun Q, Chen G, Streb JW, Long X, Yang Y, Stoeckert CJ, Jr, Miano JM. Defining the mammalian CArGome. Genome Res. 2005 doi: 10.1101/gr.4108706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Carson JA, Fillmore RA, Schwartz RJ, Zimmer WE. The smooth muscle gamma-actin gene promoter is a molecular target for the mouse bagpipe homologue, mNkx3-1, and serum response factor. J Biol Chem. 2000;275:39061–39072. doi: 10.1074/jbc.M006532200. [DOI] [PubMed] [Google Scholar]
- 6.Nishida W, Nakamura M, Mori S, Takahashi M, Ohkawa Y, Tadokoro S, Yoshida K, Hiwada K, Hayashi K, Sobue K. A triad of serum response factor and the GATA and NK families governs the transcription of smooth and cardiac muscle genes. J Biol Chem. 2002;277:7308–7317. doi: 10.1074/jbc.M111824200. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 8.Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc Natl Acad Sci U S A. 2003;100:9366–9370. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci U S A. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pipes GC, Sinha S, Qi X, Zhu CH, Gallardo TD, Shelton J, Creemers EE, Sutherland L, Richardson JA, Garry DJ, Wright WE, Owens GK, Olson EN. Stem cells and their derivatives can bypass the requirement of myocardin for smooth muscle gene expression. Dev Biol. 2005;288:502–513. doi: 10.1016/j.ydbio.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Wang DZ, Li S, Hockemeyer D, Sutherland L, Wang Z, Schratt G, Richardson JA, Nordheim A, Olson EN. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc Natl Acad Sci U S A. 2002;99:14855–14860. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinson JS, Medlin MD, Lockman K, Taylor JM, Mack CP. Smooth muscle cell-specific transcription is regulated by nuclear localization of the myocardin-related transcription factors. Am J Physiol Heart Circ Physiol. 2007;292:H1170–1180. doi: 10.1152/ajpheart.00864.2006. [DOI] [PubMed] [Google Scholar]
- 13.Du KL, Chen M, Li J, Lepore JJ, Mericko P, Parmacek MS. Megakaryoblastic leukemia factor-1 transduces cytoskeletal signals and induces smooth muscle cell differentiation from undifferentiated embryonic stem cells. J Biol Chem. 2004;279:17578–17586. doi: 10.1074/jbc.M400961200. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida T, Gan Q, Shang Y, Owens GK. Platelet-derived growth factor-BB represses smooth muscle cell marker genes via changes in binding of MKL factors and histone deacetylases to their promoters. Am J Physiol Cell Physiol. 2007;292:C886–895. doi: 10.1152/ajpcell.00449.2006. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Zhu X, Chen M, Cheng L, Zhou D, Lu MM, Du K, Epstein JA, Parmacek MS. Myocardin-related transcription factor B is required in cardiac neural crest for smooth muscle differentiation and cardiovascular development. Proc Natl Acad Sci U S A. 2005;102:8916–8921. doi: 10.1073/pnas.0503741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh J, Richardson JA, Olson EN. Requirement of myocardin-related transcription factor-B for remodeling of branchial arch arteries and smooth muscle differentiation. Proc Natl Acad Sci U S A. 2005;102:15122–15127. doi: 10.1073/pnas.0507346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Chang S, Qi X, Richardson JA, Olson EN. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol Cell Biol. 2006;26:5797–5808. doi: 10.1128/MCB.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, Boyd K, Xu W, Ma J, Jackson CW, Fu A, Shillingford JM, Robinson GW, Hennighausen L, Hitzler JK, Ma Z, Morris SW. Acute myeloid leukemia-associated Mkl1 (Mrtf-a) is a key regulator of mammary gland function. Mol Cell Biol. 2006;26:5809–5826. doi: 10.1128/MCB.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Browning CL, Culberson DE, Aragon IV, Fillmore RA, Croissant JD, Schwartz RJ, Zimmer WE. The developmentally regulated expression of serum response factor plays a key role in the control of smooth muscle-specific genes. Dev Biol. 1998;194:18–37. doi: 10.1006/dbio.1997.8808. [DOI] [PubMed] [Google Scholar]
- 20.Hautmann MB, Madsen CS, Owens GK. A transforming growth factor beta (TGFbeta) control element drives TGFbeta-induced stimulation of smooth muscle alpha-actin gene expression in concert with two CArG elements. J Biol Chem. 1997;272:10948–10956. doi: 10.1074/jbc.272.16.10948. [DOI] [PubMed] [Google Scholar]
- 21.Hirschi KK, Lai L, Belaguli NS, Dean DA, Schwartz RJ, Zimmer WE. Transforming growth factor-beta induction of smooth muscle cell phenotpye requires transcriptional and post-transcriptional control of serum response factor. J Biol Chem. 2002;277:6287–6295. doi: 10.1074/jbc.M106649200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spencer JA, Misra RP. Expression of the serum response factor gene is regulated by serum response factor binding sites. J Biol Chem. 1996;271:16535–16543. doi: 10.1074/jbc.271.28.16535. [DOI] [PubMed] [Google Scholar]
- 23.Belaguli NS, Schildmeyer LA, Schwartz RJ. Organization and myogenic restricted expression of the murine serum response factor gene. A role for autoregulation. J Biol Chem. 1997;272:18222–18231. doi: 10.1074/jbc.272.29.18222. [DOI] [PubMed] [Google Scholar]
- 24.Miranti CK, Ginty DD, Huang G, Chatila T, Greenberg ME. Calcium activates serum response factor-dependent transcription by a Ras-and Elk-1-independent mechanism that involves a Ca2+/calmodulin-dependent kinase. Mol Cell Biol. 1995;15:3672–3684. doi: 10.1128/mcb.15.7.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manak JR, Prywes R. Phosphorylation of serum response factor by casein kinase II: evidence against a role in growth factor regulation of fos expression. Oncogene. 1993;8:703–711. [PubMed] [Google Scholar]
- 26.Garat C, Van Putten V, Refaat ZA, Dessev C, Han SY, Nemenoff RA. Induction of smooth muscle alpha-actin in vascular smooth muscle cells by arginine vasopressin is mediated by c-Jun amino-terminal kinases and p38 mitogen-activated protein kinase. J Biol Chem. 2000;275:22537–22543. doi: 10.1074/jbc.M003000200. [DOI] [PubMed] [Google Scholar]
- 27.Iyer D, Chang D, Marx J, Wei L, Olson EN, Parmacek MS, Balasubramanyam A, Schwartz RJ. Serum response factor MADS box serine-162 phosphorylation switches proliferation and myogenic gene programs. Proc Natl Acad Sci U S A. 2006;103:4516–4521. doi: 10.1073/pnas.0505338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blaker AL, Taylor JM, Mack CP. PKA-dependent phosphorylation of serum response factor inhibits smooth muscle-specific gene expression. Arterioscler Thromb Vasc Biol. 2009;29:2153–2160. doi: 10.1161/ATVBAHA.109.197285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hautmann MB, Thompson MM, Swartz EA, Olson EN, Owens GK. Angiotensin II-induced stimulation of smooth muscle alpha-actin expression by serum response factor and the homeodomain transcription factor MHox. Circ Res. 1997;81:600–610. doi: 10.1161/01.res.81.4.600. [DOI] [PubMed] [Google Scholar]
- 30.Herring BP, Kriegel AM, Hoggatt AM. Identification of Barx2b, a serum response factor-associated homeodomain protein. J Biol Chem. 2001;276:14482–14489. doi: 10.1074/jbc.M011585200. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida T, Hoofnagle MH, Owens GK. Myocardin and Prx1 contribute to angiotensin II-induced expression of smooth muscle alpha-actin. Circ Res. 2004;94:1075–1082. doi: 10.1161/01.RES.0000125622.46280.95. [DOI] [PubMed] [Google Scholar]
- 32.Chen F, Kook H, Milewski R, Gitler AD, Lu MM, Li J, Nazarian R, Schnepp R, Jen K, Biben C, Runke G, Mackay JP, Novotny J, Schwartz RJ, Harvey RP, Mullins MC, Epstein JA. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110:713–723. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- 33.Martin KA, Gualberto A, Kolman MF, Lowry J, Walsh K. A competitive mechanism of CArG element regulation by YY1 and SRF: implications for assessment of Phox1/MHox transcription factor interactions at CArG elements. DNA Cell Biol. 1997;16:653–661. doi: 10.1089/dna.1997.16.653. [DOI] [PubMed] [Google Scholar]
- 34.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao D, Wang Z, Zhang CL, Oh J, Xing W, Li S, Richardson JA, Wang DZ, Olson EN. Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Mol Cell Biol. 2005;25:364–376. doi: 10.1128/MCB.25.1.364-376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis CA, Haberland M, Arnold MA, Sutherland LB, McDonald OG, Richardson JA, Childs G, Harris S, Owens GK, Olson EN. PRISM/PRDM6, a transcriptional repressor that promotes the proliferative gene program in smooth muscle cells. Mol Cell Biol. 2006;26:2626–2636. doi: 10.1128/MCB.26.7.2626-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lockman K, Taylor JM, Mack CP. The histone demethylase, Jmjd1a, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ Res. 2007;101:e115–123. doi: 10.1161/CIRCRESAHA.107.164178. [DOI] [PubMed] [Google Scholar]
- 38.Zhou J, Zhang M, Fang H, El-Mounayri O, Rodenberg JM, Imbalzano AN, Herring BP. The SWI/SNF chromatin remodeling complex regulates myocardin-induced smooth muscle-specific gene expression. Arterioscler Thromb Vasc Biol. 2009;29:921–928. doi: 10.1161/ATVBAHA.109.187229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang M, Fang H, Zhou J, Herring BP. A novel role of Brg1 in the regulation of SRF/MRTFA-dependent smooth muscle-specific gene expression. J Biol Chem. 2007;282:25708–25716. doi: 10.1074/jbc.M701925200. [DOI] [PubMed] [Google Scholar]
- 40.McDonald OG, Owens GK. Programming smooth muscle plasticity with chromatin dynamics. Circ Res. 2007;100:1428–1441. doi: 10.1161/01.RES.0000266448.30370.a0. [DOI] [PubMed] [Google Scholar]
- 41.Hendrix JA, Wamhoff BR, McDonald OG, Sinha S, Yoshida T, Owens GK. 5′ CArG degeneracy in smooth muscle alpha-actin is required for injury-induced gene suppression in vivo. J Clin Invest. 2005;115:418–427. doi: 10.1172/JCI22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jie W, Guo J, Shen Z, Wang X, Zheng S, Wang G, Ao Q. Contribution of myocardin in the hypoxia-induced phenotypic switching of rat pulmonary arterial smooth muscle cells. Exp Mol Pathol. 2010;89:301–306. doi: 10.1016/j.yexmp.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 43.Doi H, Iso T, Yamazaki M, Akiyama H, Kanai H, Sato H, Kawai-Kowase K, Tanaka T, Maeno T, Okamoto E, Arai M, Kedes L, Kurabayashi M. HERP1 inhibits myocardin-induced vascular smooth muscle cell differentiation by interfering with SRF binding to CArG box. Arterioscler Thromb Vasc Biol. 2005;25:2328–2334. doi: 10.1161/01.ATV.0000185829.47163.32. [DOI] [PubMed] [Google Scholar]
- 44.Wamhoff BR, Bowles DK, McDonald OG, Sinha S, Somlyo AP, Somlyo AV, Owens GK. L-type voltage-gated Ca2+ channels modulate expression of smooth muscle differentiation marker genes via a rho kinase/myocardin/SRF-dependent mechanism. Circ Res. 2004;95:406–414. doi: 10.1161/01.RES.0000138582.36921.9e. [DOI] [PubMed] [Google Scholar]
- 45.Tharp DL, Wamhoff BR, Turk JR, Bowles DK. Upregulation of intermediate-conductance Ca2+-activated K+ channel (IKCa1) mediates phenotypic modulation of coronary smooth muscle. Am J Physiol Heart Circ Physiol. 2006;291:H2493–2503. doi: 10.1152/ajpheart.01254.2005. [DOI] [PubMed] [Google Scholar]
- 46.Callis TE, Cao D, Wang DZ. Bone Morphogenetic Protein Signaling Modulates Myocardin Transactivation of Cardiac Genes. Circ Res. 2005 doi: 10.1161/01.RES.0000190670.92879.7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayashi K, Nakamura S, Nishida W, Sobue K. Bone morphogenetic protein-induced MSX1 and MSX2 inhibit myocardin-dependent smooth muscle gene transcription. Mol Cell Biol. 2006;26:9456–9470. doi: 10.1128/MCB.00759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ueyama T, Kasahara H, Ishiwata T, Nie Q, Izumo S. Myocardin expression is regulated by Nkx2.5, and its function is required for cardiomyogenesis. Mol Cell Biol. 2003;23:9222–9232. doi: 10.1128/MCB.23.24.9222-9232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang K, Long B, Zhou J, Li PF. miR-9 and NFATc3 regulate myocardin in cardiac hypertrophy. J Biol Chem. 2010;285:11903–11912. doi: 10.1074/jbc.M109.098004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Creemers EE, Sutherland LB, McAnally J, Richardson JA, Olson EN. Myocardin is a direct transcriptional target of Mef2, Tead and Foxo proteins during cardiovascular development. Development. 2006;133:4245–4256. doi: 10.1242/dev.02610. [DOI] [PubMed] [Google Scholar]
- 51.Badorff C, Seeger FH, Zeiher AM, Dimmeler S. Glycogen synthase kinase 3beta inhibits myocardin-dependent transcription and hypertrophy induction through site-specific phosphorylation. Circ Res. 2005;97:645–654. doi: 10.1161/01.RES.0000184684.88750.FE. [DOI] [PubMed] [Google Scholar]
- 52.Deng H, Dokshin GA, Lei J, Goldsmith AM, Bitar KN, Fingar DC, Hershenson MB, Bentley JK. Inhibition of glycogen synthase kinase-3beta is sufficient for airway smooth muscle hypertrophy. J Biol Chem. 2008;283:10198–10207. doi: 10.1074/jbc.M800624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taurin S, Sandbo N, Yau DM, Sethakorn N, Kach J, Dulin NO. Phosphorylation of myocardin by extracellular signal-regulated kinase. J Biol Chem. 2009;284:33789–33794. doi: 10.1074/jbc.M109.048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li HJ, Haque Z, Lu Q, Li L, Karas R, Mendelsohn M. Steroid receptor coactivator 3 is a coactivator for myocardin, the regulator of smooth muscle transcription and differentiation. Proc Natl Acad Sci U S A. 2007;104:4065–4070. doi: 10.1073/pnas.0611639104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pare G, Krust A, Karas RH, Dupont S, Aronovitz M, Chambon P, Mendelsohn ME. Estrogen receptor-alpha mediates the protective effects of estrogen against vascular injury. Circ Res. 2002;90:1087–1092. doi: 10.1161/01.res.0000021114.92282.fa. [DOI] [PubMed] [Google Scholar]
- 56.Tang RH, Zheng XL, Callis TE, Stansfield WE, He J, Baldwin AS, Wang DZ, Selzman CH. Myocardin inhibits cellular proliferation by inhibiting NF-kappaB(p65)-dependent cell cycle progression. Proc Natl Acad Sci U S A. 2008;105:3362–3367. doi: 10.1073/pnas.0705842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu ZP, Wang Z, Yanagisawa H, Olson EN. Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev Cell. 2005;9:261–270. doi: 10.1016/j.devcel.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 58.Zhou J, Hu G, Wang X. Repression of smooth muscle differentiation by a novel high mobility group box-containing protein, HMG2L1. J Biol Chem. 2010;285:23177–23185. doi: 10.1074/jbc.M110.109868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hinson JS, Medlin MD, Taylor JM, Mack CP. Regulation of myocardin factor protein stability by the LIM-only protein FHL2. Am J Physiol Heart Circ Physiol. 2008;295:H1067–H1075. doi: 10.1152/ajpheart.91421.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie P, Fan Y, Zhang H, Zhang Y, She M, Gu D, Patterson C, Li H. CHIP represses myocardin-induced smooth muscle cell differentiation via ubiquitin-mediated proteasomal degradation. Mol Cell Biol. 2009;29:2398–2408. doi: 10.1128/MCB.01737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu G, Wang X, Saunders DN, Henderson M, Russell AJ, Herring BP, Zhou J. Modulation of myocardin function by the ubiquitin E3 ligase UBR5. J Biol Chem. 2010;285:11800–11809. doi: 10.1074/jbc.M109.079384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Li A, Wang Z, Feng X, Olson EN, Schwartz RJ. Myocardin sumoylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol Cell Biol. 2007;27:622–632. doi: 10.1128/MCB.01160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakagawa K, Kuzumaki N. Transcriptional activity of megakaryoblastic leukemia 1 (MKL1) is repressed by SUMO modification. Genes Cells. 2005;10:835–850. doi: 10.1111/j.1365-2443.2005.00880.x. [DOI] [PubMed] [Google Scholar]
- 64.Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 65.Gineitis D, Treisman R. Differential usage of signal transduction pathways defines two types of serum response factor target gene. J Biol Chem. 2001;276:24531–24539. doi: 10.1074/jbc.M102678200. [DOI] [PubMed] [Google Scholar]
- 66.Mack CP, Somlyo AV, Hautmann M, Somlyo AP, Owens GK. Smooth muscle differentiation marker gene expression is regulated by RhoA-mediated actin polymerization. J Biol Chem. 2001;276:341–347. doi: 10.1074/jbc.M005505200. [DOI] [PubMed] [Google Scholar]
- 67.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 68.Guettler S, Vartiainen MK, Miralles F, Larijani B, Treisman R. RPEL motifs link the serum response factor cofactor MAL but not myocardin to Rho signaling via actin binding. Mol Cell Biol. 2008;28:732–742. doi: 10.1128/MCB.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu J, Landerholm TE, Wei JS, Dong XR, Wu SP, Liu X, Nagata Ki K, Inagaki M, Majesky MW. Coronary Smooth Muscle Differentiation from Proepicardial Cells Requires RhoA-Mediated Actin Reorganization and p160 Rho-Kinase Activity. Dev Biol. 2001;240:404–418. doi: 10.1006/dbio.2001.0403. [DOI] [PubMed] [Google Scholar]
- 70.Liu HW, Halayko AJ, Fernandes DJ, Harmon GS, McCauley JA, Kocieniewski P, McConville J, Fu Y, Forsythe SM, Kogut P, Bellam S, Dowell M, Churchill J, Lesso H, Kassiri K, Mitchell RW, Hershenson MB, Camoretti-Mercado B, Solway J. The RhoA/Rho kinase pathway regulates nuclear localization of serum response factor. Am J Respir Cell Mol Biol. 2003;29:39–47. doi: 10.1165/rcmb.2002-0206OC. [DOI] [PubMed] [Google Scholar]
- 71.Gorenne I, Jin L, Yoshida T, Sanders JM, Sarembock IJ, Owens GK, Somlyo AP, Somlyo AV. LPP expression during in vitro smooth muscle differentiation and stent-induced vascular injury. Circ Res. 2006;98:378–385. doi: 10.1161/01.RES.0000202802.34727.fd. [DOI] [PubMed] [Google Scholar]
- 72.Martin K, Weiss S, Metharom P, Schmeckpeper J, Hynes B, O’Sullivan J, Caplice N. Thrombin stimulates smooth muscle cell differentiation from peripheral blood mononuclear cells via protease-activated receptor-1, RhoA, and myocardin. Circ Res. 2009;105:214–218. doi: 10.1161/CIRCRESAHA.109.199984. [DOI] [PubMed] [Google Scholar]
- 73.Jin L, Yoshida T, Ho R, Owens GK, Somlyo AV. The actin-associated protein Palladin is required for development of normal contractile properties of smooth muscle cells derived from embryoid bodies. J Biol Chem. 2009;284:2121–2130. doi: 10.1074/jbc.M806095200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen S, Crawford M, Day RM, Briones VR, Leader JE, Jose PA, Lechleider RJ. RhoA modulates Smad signaling during transforming growth factor-beta-induced smooth muscle differentiation. J Biol Chem. 2006;281:1765–1770. doi: 10.1074/jbc.M507771200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lockman K, Hinson JS, Medlin MD, Morris D, Taylor JM, Mack CP. Sphingosine 1-Phosphate Stimulates Smooth Muscle Cell Differentiation and Proliferation by Activating Separate Serum Response Factor Co-factors. J Biol Chem. 2004;279:42422–42430. doi: 10.1074/jbc.M405432200. [DOI] [PubMed] [Google Scholar]
- 76.Zeidan A, Nordstrom I, Albinsson S, Malmqvist U, Sward K, Hellstrand P. Stretch-induced contractile differentiation of vascular smooth muscle: sensitivity to actin polymerization inhibitors. Am J Physiol Cell Physiol. 2003;284:C1387–1396. doi: 10.1152/ajpcell.00508.2002. [DOI] [PubMed] [Google Scholar]
- 77.Deaton RA, Su C, Valencia TG, Grant SR. Transforming growth factor-beta1-induced expression of smooth muscle marker genes involves activation of PKN and p38 MAPK. J Biol Chem. 2005;280:31172–31181. doi: 10.1074/jbc.M504774200. [DOI] [PubMed] [Google Scholar]
- 78.Lagna G, Ku MM, Nguyen PH, Neuman NA, Davis BN, Hata A. Control of phenotypic plasticity of smooth muscle cells by bone morphogenetic protein signaling through the myocardin-related transcription factors. J Biol Chem. 2007;282:37244–37255. doi: 10.1074/jbc.M708137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jeon ES, Park WS, Lee MJ, Kim YM, Han J, Kim JH. A Rho kinase/myocardin-related transcription factor-A-dependent mechanism underlies the sphingosylphosphorylcholine-induced differentiation of mesenchymal stem cells into contractile smooth muscle cells. Circ Res. 2008;103:635–642. doi: 10.1161/CIRCRESAHA.108.180885. [DOI] [PubMed] [Google Scholar]
- 80.Zhao XH, Laschinger C, Arora P, Szaszi K, Kapus A, McCulloch CA. Force activates smooth muscle alpha-actin promoter activity through the Rho signaling pathway. J Cell Sci. 2007;120:1801–1809. doi: 10.1242/jcs.001586. [DOI] [PubMed] [Google Scholar]
- 81.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 82.Suzuki N, Nakamura S, Mano H, Kozasa T. Galpha 12 activates Rho GTPase through tyrosine-phosphorylated leukemia-associated RhoGEF. Proc Natl Acad Sci U S A. 2003;100:733–738. doi: 10.1073/pnas.0234057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rojas RJ, Yohe ME, Gershburg S, Kawano T, Kozasa T, Sondek J. Galphaq directly activates p63RhoGEF and Trio via a conserved extension of the Dbl homology-associated pleckstrin homology domain. J Biol Chem. 2007;282:29201–29210. doi: 10.1074/jbc.M703458200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lutz S, Shankaranarayanan A, Coco C, Ridilla M, Nance MR, Vettel C, Baltus D, Evelyn CR, Neubig RR, Wieland T, Tesmer JJ. Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs. Science. 2007;318:1923–1927. doi: 10.1126/science.1147554. [DOI] [PubMed] [Google Scholar]
- 85.Wamhoff BR, Lynch KR, Macdonald TL, Owens GK. Sphingosine-1-phosphate receptor subtypes differentially regulate smooth muscle cell phenotype. Arterioscler Thromb Vasc Biol. 2008;28:1454–1461. doi: 10.1161/ATVBAHA.107.159392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Medlin MD, Staus DP, Dubash AD, Taylor JM, Mack CP. Sphingosine 1-phosphate receptor 2 signals through leukemia-associated RhoGEF (LARG), to promote smooth muscle cell differentiation. Arterioscler Thromb Vasc Biol. 2010;30:1779–1786. doi: 10.1161/ATVBAHA.110.209395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shimizu T, Nakazawa T, Cho A, Dastvan F, Shilling D, Daum G, Reidy MA. Sphingosine 1-phosphate receptor 2 negatively regulates neointimal formation in mouse arteries. Circ Res. 2007;101:995–1000. doi: 10.1161/CIRCRESAHA.107.159228. [DOI] [PubMed] [Google Scholar]
- 88.Guilluy C, Bregeon J, Toumaniantz G, Rolli-Derkinderen M, Retailleau K, Loufrani L, Henrion D, Scalbert E, Bril A, Torres RM, Offermanns S, Pacaud P, Loirand G. The Rho exchange factor Arhgef1 mediates the effects of angiotensin II on vascular tone and blood pressure. Nat Med. 2010;16:183–190. doi: 10.1038/nm.2079. [DOI] [PubMed] [Google Scholar]
- 89.Bregeon J, Loirand G, Pacaud P, Rolli-Derkinderen M. Angiotensin II induces RhoA activation through SHP2-dependent dephosphorylation of the RhoGAP p190A in vascular smooth muscle cells. Am J Physiol Cell Physiol. 2009;297:C1062–1070. doi: 10.1152/ajpcell.00174.2009. [DOI] [PubMed] [Google Scholar]
- 90.Ying Z, Jin L, Palmer T, Webb RC. Angiotensin II up-regulates the leukemia-associated Rho guanine nucleotide exchange factor (RhoGEF), a regulator of G protein signaling domain-containing RhoGEF, in vascular smooth muscle cells. Mol Pharmacol. 2006;69:932–940. doi: 10.1124/mol.105.017830. [DOI] [PubMed] [Google Scholar]
- 91.Ying Z, Giachini FR, Tostes RC, Webb RC. PYK2/PDZ-RhoGEF links Ca2+ signaling to RhoA. Arterioscler Thromb Vasc Biol. 2009;29:1657–1663. doi: 10.1161/ATVBAHA.109.190892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science. 1999;285:895–898. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- 93.Tominaga T, Sahai E, Chardin P, McCormick F, Courtneidge SA, Alberts AS. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol Cell. 2000;5:13–25. doi: 10.1016/s1097-2765(00)80399-8. [DOI] [PubMed] [Google Scholar]
- 94.Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. Embo J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Staus DP, Blaker AL, Taylor JM, Mack CP. Diaphanous 1 and 2 regulate smooth muscle cell differentiation by activating the myocardin-related transcription factors. Arterioscler Thromb Vasc Biol. 2007;27:478–486. doi: 10.1161/01.ATV.0000255559.77687.c1. [DOI] [PubMed] [Google Scholar]
- 96.Staus DP, Blaker AL, Medlin MD, Taylor JM, Mack CP. Formin homology domain-containing protein 1 regulates smooth muscle cell phenotype. Arterioscler Thromb Vasc Biol. 2011;31:360–367. doi: 10.1161/ATVBAHA.110.212993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Takeya R, Taniguchi K, Narumiya S, Sumimoto H. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. Embo J. 2008;27:618–628. doi: 10.1038/emboj.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martin CB, Mahon GM, Klinger MB, Kay RJ, Symons M, Der CJ, Whitehead IP. The thrombin receptor, PAR-1, causes transformation by activation of Rho-mediated signaling pathways. Oncogene. 2001;20:1953–1963. doi: 10.1038/sj.onc.1204281. [DOI] [PubMed] [Google Scholar]
- 99.Numaguchi K, Eguchi S, Yamakawa T, Motley ED, Inagami T. Mechanotransduction of rat aortic vascular smooth muscle cells requires RhoA and intact actin filaments. Circ Res. 1999;85:5–11. doi: 10.1161/01.res.85.1.5. [DOI] [PubMed] [Google Scholar]
- 100.Yamakawa T, Tanaka S, Numaguchi K, Yamakawa Y, Motley ED, Ichihara S, Inagami T. Involvement of Rho-kinase in angiotensin II-induced hypertrophy of rat vascular smooth muscle cells. Hypertension. 2000;35:313–318. doi: 10.1161/01.hyp.35.1.313. [DOI] [PubMed] [Google Scholar]
- 101.Morishige K, Shimokawa H, Eto Y, Kandabashi T, Miyata K, Matsumoto Y, Hoshijima M, Kaibuchi K, Takeshita A. Adenovirus-mediated transfer of dominant-negative rho-kinase induces a regression of coronary arteriosclerosis in pigs in vivo. Arterioscler Thromb Vasc Biol. 2001;21:548–554. doi: 10.1161/01.atv.21.4.548. [DOI] [PubMed] [Google Scholar]
- 102.Sawada N, Itoh H, Ueyama K, Yamashita J, Doi K, Chun TH, Inoue M, Masatsugu K, Saito T, Fukunaga Y, Sakaguchi S, Arai H, Ohno N, Komeda M, Nakao K. Inhibition of rho-associated kinase results in suppression of neointimal formation of balloon-injured arteries. Circulation. 2000;101:2030–2033. doi: 10.1161/01.cir.101.17.2030. [DOI] [PubMed] [Google Scholar]
- 103.Seasholtz TM, Majumdar M, Kaplan DD, Brown JH. Rho and Rho kinase mediate thrombin-stimulated vascular smooth muscle cell DNA synthesis and migration. Circ Res. 1999;84:1186–1193. doi: 10.1161/01.res.84.10.1186. [DOI] [PubMed] [Google Scholar]
- 104.Alberts AS, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- 105.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 106.Kurpinski K, Lam H, Chu J, Wang A, Kim A, Tsay E, Agrawal S, Schaffer DV, Li S. Transforming growth factor-beta and notch signaling mediate stem cell differentiation into smooth muscle cells. Stem Cells. 2010;28:734–742. doi: 10.1002/stem.319. [DOI] [PubMed] [Google Scholar]
- 107.Chen S, Kulik M, Lechleider RJ. Smad proteins regulate transcriptional induction of the SM22alpha gene by TGF-beta. Nucleic Acids Res. 2003;31:1302–1310. doi: 10.1093/nar/gkg224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hu B, Wu Z, Phan SH. Smad3 mediates transforming growth factor-beta-induced alpha-smooth muscle actin expression. Am J Respir Cell Mol Biol. 2003;29:397–404. doi: 10.1165/rcmb.2003-0063OC. [DOI] [PubMed] [Google Scholar]
- 109.Hirschi KK, Rohovsky SA, D’Amore PA. PDGF, TGF-beta, and heterotypic cell-cell interactions mediate endothelial cell-induced recruitment of 10T1/2 cells and their differentiation to a smooth muscle fate. J Cell Biol. 1998;141:805–814. doi: 10.1083/jcb.141.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sinha S, Hoofnagle MH, Kingston PA, McCanna ME, Owens GK. Transforming growth factor-beta1 signaling contributes to development of smooth muscle cells from embryonic stem cells. Am J Physiol Cell Physiol. 2004;287:C1560–1568. doi: 10.1152/ajpcell.00221.2004. [DOI] [PubMed] [Google Scholar]
- 111.Chen S, Lechleider RJ. Transforming growth factor-beta-induced differentiation of smooth muscle from a neural crest stem cell line. Circ Res. 2004;94:1195–1202. doi: 10.1161/01.RES.0000126897.41658.81. [DOI] [PubMed] [Google Scholar]
- 112.Nishimura G, Manabe I, Tsushima K, Fujiu K, Oishi Y, Imai Y, Maemura K, Miyagishi M, Higashi Y, Kondoh H, Nagai R. DeltaEF1 mediates TGF-beta signaling in vascular smooth muscle cell differentiation. Dev Cell. 2006;11:93–104. doi: 10.1016/j.devcel.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 113.Jeon ES, Moon HJ, Lee MJ, Song HY, Kim YM, Bae YC, Jung JS, Kim JH. Sphingosylphosphorylcholine induces differentiation of human mesenchymal stem cells into smooth-muscle-like cells through a TGF-beta-dependent mechanism. J Cell Sci. 2006;119:4994–5005. doi: 10.1242/jcs.03281. [DOI] [PubMed] [Google Scholar]
- 114.Qiu P, Feng XH, Li L. Interaction of Smad3 and SRF-associated complex mediates TGF-beta1 signals to regulate SM22 transcription during myofibroblast differentiation. J Mol Cell Cardiol. 2003;35:1407–1420. doi: 10.1016/j.yjmcc.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 115.Qiu P, Ritchie R, Fu Z, Cao D, Cumming J, Miano JM, Wang DZ, Li HJ, Li L. Myocardin Enhances Smad3-Mediated Transforming Growth Factor-{beta}1 Signaling in a CArG Box-Independent Manner. Smad-Binding Element Is a Critical cis Element for SM22{alpha} Transcription In Vivo. Circ Res. 2005 doi: 10.1161/01.RES.0000190604.90049.71. [DOI] [PubMed] [Google Scholar]
- 116.Ruiz-Ortega M, Rodriguez-Vita J, Sanchez-Lopez E, Carvajal G, Egido J. TGF-beta signaling in vascular fibrosis. Cardiovasc Res. 2007;74:196–206. doi: 10.1016/j.cardiores.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 117.Cucoranu I, Clempus R, Dikalova A, Phelan PJ, Ariyan S, Dikalov S, Sorescu D. NAD(P)H oxidase 4 mediates transforming growth factor-beta1-induced differentiation of cardiac fibroblasts into myofibroblasts. Circ Res. 2005;97:900–907. doi: 10.1161/01.RES.0000187457.24338.3D. [DOI] [PubMed] [Google Scholar]
- 118.Martin-Garrido A, Brown DI, Lyle AN, Dikalova A, Seidel-Rogol B, Lassegue B, Martin AS, Griendling KK. NADPH oxidase 4 mediates TGF-beta-induced smooth muscle alpha-actin via p38MAPK and serum response factor. Free Radic Biol Med. 2010;50:354–362. doi: 10.1016/j.freeradbiomed.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Clempus RE, Sorescu D, Dikalova AE, Pounkova L, Jo P, Sorescu GP, Schmidt HH, Lassegue B, Griendling KK. Nox4 is required for maintenance of the differentiated vascular smooth muscle cell phenotype. Arterioscler Thromb Vasc Biol. 2007;27:42–48. doi: 10.1161/01.ATV.0000251500.94478.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xiao Q, Luo Z, Pepe AE, Margariti A, Zeng L, Xu Q. Embryonic stem cell differentiation into smooth muscle cells is mediated by Nox4-produced H2O2. Am J Physiol Cell Physiol. 2009;296:C711–723. doi: 10.1152/ajpcell.00442.2008. [DOI] [PubMed] [Google Scholar]
- 121.Su B, Mitra S, Gregg H, Flavahan S, Chotani MA, Clark KR, Goldschmidt-Clermont PJ, Flavahan NA. Redox regulation of vascular smooth muscle cell differentiation. Circ Res. 2001;89:39–46. doi: 10.1161/hh1301.093615. [DOI] [PubMed] [Google Scholar]
- 122.Sato M, Kawai-Kowase K, Sato H, Oyama Y, Kanai H, Ohyama Y, Suga T, Maeno T, Aoki Y, Tamura J, Sakamoto H, Nagai R, Kurabayashi M. c-Src and hydrogen peroxide mediate transforming growth factor-beta1-induced smooth muscle cell-gene expression in 10T1/2 cells. Arterioscler Thromb Vasc Biol. 2005;25:341–347. doi: 10.1161/01.ATV.0000152608.29351.8f. [DOI] [PubMed] [Google Scholar]
- 123.Clempus RE, Griendling KK. Reactive oxygen species signaling in vascular smooth muscle cells. Cardiovasc Res. 2006;71:216–225. doi: 10.1016/j.cardiores.2006.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shi-wen X, Parapuram SK, Pala D, Chen Y, Carter DE, Eastwood M, Denton CP, Abraham DJ, Leask A. Requirement of transforming growth factor beta-activated kinase 1 for transforming growth factor beta-induced alpha-smooth muscle actin expression and extracellular matrix contraction in fibroblasts. Arthritis Rheum. 2009;60:234–241. doi: 10.1002/art.24223. [DOI] [PubMed] [Google Scholar]
- 126.Sousa AM, Liu T, Guevara O, Stevens J, Fanburg BL, Gaestel M, Toksoz D, Kayyali US. Smooth muscle alpha-actin expression and myofibroblast differentiation by TGFbeta are dependent upon MK2. J Cell Biochem. 2007;100:1581–1592. doi: 10.1002/jcb.21154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Seay U, Sedding D, Krick S, Hecker M, Seeger W, Eickelberg O. Transforming growth factor-beta-dependent growth inhibition in primary vascular smooth muscle cells is p38-dependent. J Pharmacol Exp Ther. 2005;315:1005–1012. doi: 10.1124/jpet.105.091249. [DOI] [PubMed] [Google Scholar]
- 128.Lien SC, Usami S, Chien S, Chiu JJ. Phosphatidylinositol 3-kinase/Akt pathway is involved in transforming growth factor-beta1-induced phenotypic modulation of 10T1/2 cells to smooth muscle cells. Cell Signal. 2006;18:1270–1278. doi: 10.1016/j.cellsig.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 129.Lee J, Moon HJ, Lee JM, Joo CK. Smad3 regulates Rho signaling via NET1 in the transforming growth factor-beta-induced epithelial-mesenchymal transition of human retinal pigment epithelial cells. J Biol Chem. 2010;285:26618–26627. doi: 10.1074/jbc.M109.073155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vardouli L, Vasilaki E, Papadimitriou E, Kardassis D, Stournaras C. A novel mechanism of TGFbeta-induced actin reorganization mediated by Smad proteins and Rho GTPases. Febs J. 2008;275:4074–4087. doi: 10.1111/j.1742-4658.2008.06549.x. [DOI] [PubMed] [Google Scholar]
- 131.Tsapara A, Luthert P, Greenwood J, Hill CS, Matter K, Balda MS. The RhoA activator GEF-H1/Lfc is a transforming growth factor-beta target gene and effector that regulates alpha-smooth muscle actin expression and cell migration. Mol Biol Cell. 2010;21:860–870. doi: 10.1091/mbc.E09-07-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gridley T. Notch signaling in the vasculature. Curr Top Dev Biol. 2010;92:277–309. doi: 10.1016/S0070-2153(10)92009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Doi H, Iso T, Sato H, Yamazaki M, Matsui H, Tanaka T, Manabe I, Arai M, Nagai R, Kurabayashi M. Jagged1-selective notch signaling induces smooth muscle differentiation via a RBP-Jkappa-dependent pathway. J Biol Chem. 2006;281:28555–28564. doi: 10.1074/jbc.M602749200. [DOI] [PubMed] [Google Scholar]
- 134.Noseda M, Fu Y, Niessen K, Wong F, Chang L, McLean G, Karsan A. Smooth Muscle alpha-actin is a direct target of Notch/CSL. Circ Res. 2006;98:1468–1470. doi: 10.1161/01.RES.0000229683.81357.26. [DOI] [PubMed] [Google Scholar]
- 135.High FA, Lu MM, Pear WS, Loomes KM, Kaestner KH, Epstein JA. Endothelial expression of the Notch ligand Jagged1 is required for vascular smooth muscle development. Proc Natl Acad Sci U S A. 2008;105:1955–1959. doi: 10.1073/pnas.0709663105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.High FA, Zhang M, Proweller A, Tu L, Parmacek MS, Pear WS, Epstein JA. An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation. J Clin Invest. 2007;117:353–363. doi: 10.1172/JCI30070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Domenga V, Fardoux P, Lacombe P, Monet M, Maciazek J, Krebs LT, Klonjkowski B, Berrou E, Mericskay M, Li Z, Tournier-Lasserve E, Gridley T, Joutel A. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004;18:2730–2735. doi: 10.1101/gad.308904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu H, Kennard S, Lilly B. NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed JAGGED1. Circ Res. 2009;104:466–475. doi: 10.1161/CIRCRESAHA.108.184846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cecillion M, Marechal E, Maciazek J, Vayssiere C, Cruaud C, Cabanis EA, Ruchoux MM, Weissenbach J, Bach JF, Bousser MG, Tournier-Lasserve E. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 140.Proweller A, Pear WS, Parmacek MS. Notch signaling represses myocardin-induced smooth muscle cell differentiation. J Biol Chem. 2005;280:8994–9004. doi: 10.1074/jbc.M413316200. [DOI] [PubMed] [Google Scholar]
- 141.Morrow D, Scheller A, Birney YA, Sweeney C, Guha S, Cummins PM, Murphy R, Walls D, Redmond EM, Cahill PA. Notch-mediated CBF-1/RBP-J{kappa}-dependent regulation of human vascular smooth muscle cell phenotype in vitro. Am J Physiol Cell Physiol. 2005;289:C1188–1196. doi: 10.1152/ajpcell.00198.2005. [DOI] [PubMed] [Google Scholar]
- 142.Tang Y, Urs S, Liaw L. Hairy-related transcription factors inhibit Notch-induced smooth muscle alpha-actin expression by interfering with Notch intracellular domain/CBF-1 complex interaction with the CBF-1-binding site. Circ Res. 2008;102:661–668. doi: 10.1161/CIRCRESAHA.107.165134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sakata Y, Xiang F, Chen Z, Kiriyama Y, Kamei CN, Simon DI, Chin MT. Transcription factor CHF1/Hey2 regulates neointimal formation in vivo and vascular smooth muscle proliferation and migration in vitro. Arterioscler Thromb Vasc Biol. 2004;24:2069–2074. doi: 10.1161/01.ATV.0000143936.77094.a4. [DOI] [PubMed] [Google Scholar]
- 144.Li X, Zhang X, Leathers R, Makino A, Huang C, Parsa P, Macias J, Yuan JX, Jamieson SW, Thistlethwaite PA. Notch3 signaling promotes the development of pulmonary arterial hypertension. Nat Med. 2009;15:1289–1297. doi: 10.1038/nm.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Morrow D, Guha S, Sweeney C, Birney Y, Walshe T, O’Brien C, Walls D, Redmond EM, Cahill PA. Notch and vascular smooth muscle cell phenotype. Circ Res. 2008;103:1370–1382. doi: 10.1161/CIRCRESAHA.108.187534. [DOI] [PubMed] [Google Scholar]
- 146.Betsholtz C, Lindblom P, Bjarnegard M, Enge M, Gerhardt H, Lindahl P. Role of platelet-derived growth factor in mesangium development and vasculopathies: lessons from platelet-derived growth factor and platelet-derived growth factor receptor mutations in mice. Curr Opin Nephrol Hypertens. 2004;13:45–52. doi: 10.1097/00041552-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 147.Kawai-Kowase K, Owens GK. Multiple repressor pathways contribute to phenotypic switching of vascular smooth muscle cells. Am J Physiol Cell Physiol. 2007;292:C59–69. doi: 10.1152/ajpcell.00394.2006. [DOI] [PubMed] [Google Scholar]
- 148.Uchida K, Sasahara M, Morigami N, Hazama F, Kinoshita M. Expression of platelet-derived growth factor B-chain in neointimal smooth muscle cells of balloon injured rabbit femoral arteries. Atherosclerosis. 1996;124:9–23. doi: 10.1016/0021-9150(95)05742-0. [DOI] [PubMed] [Google Scholar]
- 149.Ferns GA, Raines EW, Sprugel KH, Motani AS, Reidy MA, Ross R. Inhibition of neointimal smooth muscle accumulation after angioplasty by an antibody to PDGF. Science. 1991;253:1129–1132. doi: 10.1126/science.1653454. [DOI] [PubMed] [Google Scholar]
- 150.Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 151.Zhou J, Hu G, Herring BP. Smooth muscle-specific genes are differentially sensitive to inhibition by Elk-1. Mol Cell Biol. 2005;25:9874–9885. doi: 10.1128/MCB.25.22.9874-9885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Muehlich S, Wang R, Lee SM, Lewis TC, Dai C, Prywes R. Serum-induced phosphorylation of the serum response factor coactivator MKL1 by the extracellular signal-regulated kinase 1/2 pathway inhibits its nuclear localization. Mol Cell Biol. 2008;28:6302–6313. doi: 10.1128/MCB.00427-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH, Owens GK. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J Biol Chem. 2005;280:9719–9727. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 154.Deaton RA, Gan Q, Owens GK. Sp1-dependent activation of KLF4 is required for PDGF-BB-induced phenotypic modulation of smooth muscle. Am J Physiol Heart Circ Physiol. 2009;296:H1027–1037. doi: 10.1152/ajpheart.01230.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Regan CP, Adam PJ, Madsen CS, Owens GK. Molecular mechanisms of decreased smooth muscle differentiation marker expression after vascular injury. J Clin Invest. 2000;106:1139–1147. doi: 10.1172/JCI10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wamhoff BR, Hoofnagle MH, Burns A, Sinha S, McDonald OG, Owens GK. A G/C element mediates repression of the SM22alpha promoter within phenotypically modulated smooth muscle cells in experimental atherosclerosis. Circ Res. 2004;95:981–988. doi: 10.1161/01.RES.0000147961.09840.fb. [DOI] [PubMed] [Google Scholar]
- 157.Hedin U, Bottger BA, Forsberg E, Johansson S, Thyberg J. Diverse effects of fibronectin and laminin on phenotypic properties of cultured arterial smooth muscle cells. J Cell Biol. 1988;107:307–319. doi: 10.1083/jcb.107.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Thyberg J, Hultgardh-Nilsson A. Fibronectin and the basement membrane components laminin and collagen type IV influence the phenotypic properties of subcultured rat aortic smooth muscle cells differently. Cell Tissue Res. 1994;276:263–271. doi: 10.1007/BF00306112. [DOI] [PubMed] [Google Scholar]
- 159.Hirst SJ, Twort CH, Lee TH. Differential effects of extracellular matrix proteins on human airway smooth muscle cell proliferation and phenotype. Am J Respir Cell Mol Biol. 2000;23:335–344. doi: 10.1165/ajrcmb.23.3.3990. [DOI] [PubMed] [Google Scholar]
- 160.Raines EW. The extracellular matrix can regulate vascular cell migration, proliferation, and survival: relationships to vascular disease. Int J Exp Pathol. 2000;81:173–182. doi: 10.1046/j.1365-2613.2000.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Luo M, Guan JL. Focal adhesion kinase: a prominent determinant in breast cancer initiation, progression and metastasis. Cancer Lett. 2010;289:127–139. doi: 10.1016/j.canlet.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Orr AW, Lee MY, Lemmon JA, Yurdagul A, Jr, Gomez MF, Bortz PD, Wamhoff BR. Molecular mechanisms of collagen isotype-specific modulation of smooth muscle cell phenotype. Arterioscler Thromb Vasc Biol. 2009;29:225–231. doi: 10.1161/ATVBAHA.108.178749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chan MW, Chaudary F, Lee W, Copeland JW, McCulloch CA. Force-induced myofibroblast differentiation through collagen receptors is dependent on mammalian diaphanous (mDia) J Biol Chem. 2010;285:9273–9281. doi: 10.1074/jbc.M109.075218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Albinsson S, Nordstrom I, Hellstrand P. Stretch of the vascular wall induces smooth muscle differentiation by promoting actin polymerization. J Biol Chem. 2004;279:34849–34855. doi: 10.1074/jbc.M403370200. [DOI] [PubMed] [Google Scholar]
- 166.Assoian RK, Klein EA. Growth control by intracellular tension and extracellular stiffness. Trends Cell Biol. 2008;18:347–352. doi: 10.1016/j.tcb.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kogata N, Tribe RM, Fassler R, Way M, Adams RH. Integrin-linked kinase controls vascular wall formation by negatively regulating Rho/ROCK-mediated vascular smooth muscle cell contraction. Genes Dev. 2009;23:2278–2283. doi: 10.1101/gad.535409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wu Y, Huang Y, Herring BP, Gunst SJ. Integrin-linked kinase regulates smooth muscle differentiation marker gene expression in airway tissue. Am J Physiol Lung Cell Mol Physiol. 2008;295:L988–997. doi: 10.1152/ajplung.90202.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Taylor JM, Mack CP, Nolan K, Regan CP, Owens GK, Parsons JT. Selective expression of an endogenous inhibitor of FAK regulates proliferation and migration of vascular smooth muscle cells. Mol Cell Biol. 2001;21:1565–1572. doi: 10.1128/MCB.21.5.1565-1572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sayers RL, Sundberg-Smith LJ, Rojas M, Hayasaka H, Parsons JT, Mack CP, Taylor JM. FRNK expression promotes smooth muscle cell maturation during vascular development and after vascular injury. Arterioscler Thromb Vasc Biol. 2008;28:2115–2122. doi: 10.1161/ATVBAHA.108.175455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sundberg LJ, Galante LM, Bill HM, Mack CP, Taylor JM. An endogenous inhibitor of focal adhesion kinase blocks Rac1/JNK but not Ras/ERK-dependent signaling in vascular smooth muscle cells. J Biol Chem. 2003;278:29783–29791. doi: 10.1074/jbc.M303771200. [DOI] [PubMed] [Google Scholar]
- 172.Sundberg-Smith LJ, DiMichele LA, Sayers RL, Mack CP, Taylor JM. The LIM protein leupaxin is enriched in smooth muscle and functions as an serum response factor cofactor to induce smooth muscle cell gene transcription. Circ Res. 2008;102:1502–1511. doi: 10.1161/CIRCRESAHA.107.170357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chang DF, Belaguli NS, Iyer D, Roberts WB, Wu SP, Dong XR, Marx JG, Moore MS, Beckerle MC, Majesky MW, Schwartz RJ. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev Cell. 2003;4:107–118. doi: 10.1016/s1534-5807(02)00396-9. [DOI] [PubMed] [Google Scholar]
- 174.Grubinger M, Gimona M. CRP2 is an autonomous actin-binding protein. FEBS Lett. 2004;557:88–92. doi: 10.1016/s0014-5793(03)01451-0. [DOI] [PubMed] [Google Scholar]
- 175.Philippar U, Schratt G, Dieterich C, Muller JM, Galgoczy P, Engel FB, Keating MT, Gertler F, Schule R, Vingron M, Nordheim A. The SRF Target Gene Fhl2 Antagonizes RhoA/MAL-Dependent Activation of SRF. Mol Cell. 2004;16:867–880. doi: 10.1016/j.molcel.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 176.Muller JM, Metzger E, Greschik H, Bosserhoff AK, Mercep L, Buettner R, Schule R. The transcriptional coactivator FHL2 transmits Rho signals from the cell membrane into the nucleus. Embo J. 2002;21:736–748. doi: 10.1093/emboj/21.4.736. [DOI] [PMC free article] [PubMed] [Google Scholar]