Abstract
The pandemic (H1N1) 2009 virus is unique in many aspects, especially in its genetics and evolution. In this paper, we examine the molecular mechanisms underlying the evolution of this novel virus through a comprehensive bioinformatics analysis, and present results in the context of a review of the literature. The pandemic virus was found to arise from a reassortment of two swine viruses, each of which ultimately arose from interspecies transmission. It experienced fast evolutionary rates and strong selection pressures, diverging into two different clusters at the early pandemic stage. Cluster I became extinct at the end of 2009 whereas cluster II continued to circulate at much lower rates in 2010. Therefore, on August 10 of 2010 the WHO declared the end of the pandemic. Important mutations associated with host specificity, virulence, and drug resistance were detected in the pandemic virus, indicating effective transmission and increased severity in humans. Much has been learned about the evolutionary dynamics of this pandemic virus; however, it is still impossible to predict when the next pandemic will occur and which virus will be responsible. Improved surveillance at different levels (both national and international) and in different hosts (especially in swine) appears to be crucial for early detection and prevention of future influenza pandemics.
Keywords: influenza, pandemic H1N1, evolution, genetic mutation, genome reassortment, phylodynamics
1. Introduction
Influenza viruses, belonging to the family Orthomyxoviridae (Smith et al., 1933), contain seven to eight segments of single-stranded, negative-sense RNA. They are classified into three types, A, B, and C, according to the antigenic variations in their nucleoprotein (NP) and matrix protein (MP) segments (Webster and Bean, 1978), with relevance to humans for each type. Influenza A can infect multiple mammalian species; its natural hosts, however, are wild aquatic birds. Influenza A is the most common and the most severe human pathogen among the three types (Webster and Bean, 1978; Webster et al., 1992). Influenza B is almost exclusively a human pathogen, whereas influenza C is less common than the other types and usually only causes mild disease in children (Katagiri et al., 1983).
Influenza A viruses are classified into subtypes based upon the antigenic properties of their hemagglutinin (HA) and neuraminidase (NA) glycoproteins (Matsuzaki et al., 2006). So far, 16 HA (H1 - H16) and 9 NA (N1- N9) serotypes have been described (Fouchier et al., 2007), with over 100 identified subtypes found in the NCBI Influenza Virus Resource (Bao et al., 2008). The most common subtypes circulating in human populations are H1N1 and H3N2. Other subtypes of avian origin have also infected humans; currently there are heightened concerns about a pandemic threat from the H5N1 highly pathogenic avian influenza (HPAI) virus (Guan et al., 2004; Pappaioanou, 2009). In the past century, the world experienced three influenza pandemics, the Spanish Flu (1918-1920), caused by H1N1, the Asian Flu (1957-1958), caused by H2N2, and the Hong Kong Flu (1968-1969), caused by H3N2. The first pandemic of this century was caused by the 2009 (H1N1) influenza A virus, also known as H1N1pdm, of the H1N1 subtype (Smith et al., 2009).
A novel swine-origin influenza A virus was initially responsible for outbreaks of influenza-like illness in Mexico, which spread rapidly in the US and then to the rest of the world. On April 21, 2009, the US Centers for Disease Control and Prevention (CDC) confirmed 2 cases of a febrile respiratory illness in children from southern California caused by infection with a novel influenza A (H1N1) virus (CDC, 2009a). By June 11, 2009, nearly 30,000 cases of 2009 H1N1 virus had been confirmed across 74 countries (Figure 1), prompting the WHO to declare the start of the 2009 influenza pandemic. The number of confirmed human cases of H1N1pdm reported to WHO peaked in November, 2009 and decreased significantly in Jan, 2010. In August, 2010, the WHO declared the end of the year-long pandemic (Figure 1).
Figure 1.
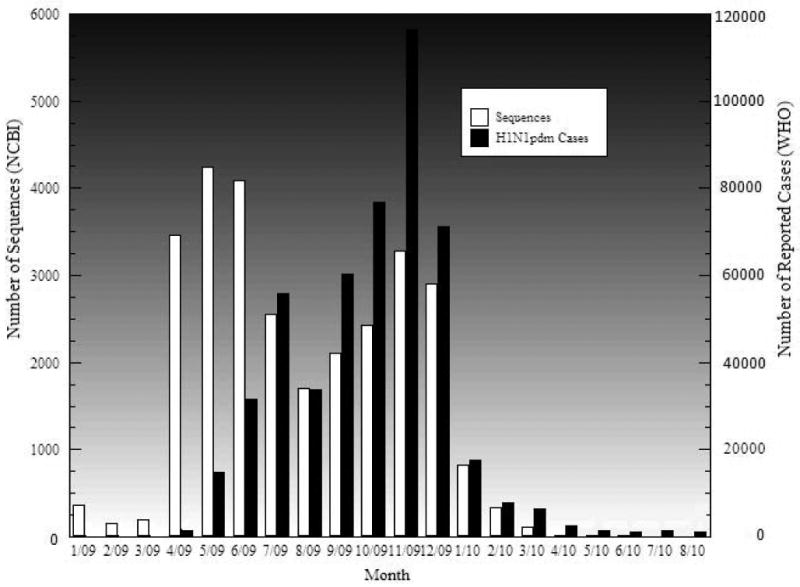
Number of confirmed human cases of pandemic (H1N1) 2009 reported to WHO and the number of sequences submitted to NCBI during the pandemic period.
1.1. Genes and proteins of influenza A viruses
The influenza A viral genome is made up of eight RNA segments, ranging from 890 to 2,341 nucleotides in length, and encoding eleven proteins (Table 1) (Ghedin et al., 2010; Lamb and Choppin, 1983; Steinhauer and Skehel, 2002). Three gene segments encode two proteins each; each of the rest five segments encodes for a single protein (Holmes et al., 2005). The functions of the eleven proteins are given in Table 1. The HA and NA genes encode the major surface virion proteins, hemagglutinin and neuraminidase. Hemagglutinin is responsible for binding to sialic acid at the termini of glycans acting as receptors on the host cell plasma membrane (Huang et al., 1973; Burnett, 1948; Wilson et al., 1981). Neuraminidase is involved in the final step of the replication cycle, the release of mature virions (Frosner et al., 1975). The PB2, PB1, and PA genes, respectively, encode the three subunits of the RNA polymerase. The NP gene encodes the nucleoprotein, which is associated with viral genomic RNA binding to support transcription and replication (Klumpp et al., 1997). Both the M gene and the NS gene, respectively, encode two different proteins using different reading frames of the same RNA.
Table 1. The eight segments of Influenza A, coding eleven proteins, their lengths and functions.
| Segment | Size(nt) | Protein | Function |
|---|---|---|---|
| 1 (PB2) | 2,341 | Polymerase Basic 2 | Transcriptase: cap binding |
| 2 (PB1) | 2,341 | Polymerase Basic 1 | Transcriptase: elongation |
| PB1-F2 (Frame 2) | Virulence | ||
| 3 (PA) | 2,233 | Polymerase acidic | Transcriptase: protease activity |
| 4 (HA) | 1,778 | Hemaggluttinin | Binding the virus to the host cell |
| 5 (NP) | 1,565 | Nucleoprotein | RNA binding; part of transcriptase complex |
| 6 (NA) | 1,413 | Neuraminidase | Release of mature virions from host cell |
| 7 (M or MP) | 1,027 | M1, Matrix protein 1 | Component of viral envelope |
| M2, Matrix protein 2 | Integral membrane protein: ion channel | ||
| 8 (NS) | 890 | NS 1(Non-structural 1) | Counters interferon, a product of the host immune system |
| NS2/NEP (Non-structural 2/Nuclear Export Protein) | Effects on cellular RNA transport from the host cell nucleus to the cytoplasm, splicing, translation |
1.2. Molecular mechanisms of influenza A virus evolution
To survive as a successful pathogen, the influenza A virus relies on two evolutionary mechanisms, mutation and genome reassortment, to continuously evade the surveillance of host immune systems and adapt to novel hosts (Webster and Bean, 1978). The lack of a proofreading ability of the influenza A RNA-dependent RNA polymerase facilitates rapid evolution (Drake, 1993; Drake and Holland, 1999). Mutations can cause small changes, often called genetic drift. Mutations at critical positions on HA and NA can alter their antigenic characteristics, resulting in antigenic drift. Viruses whose HA and/or NA genes have drifted can eventually achieve higher fitness, become dominant, rapidly sweep through the human population—causing an epidemic.
The segmented nature of the influenza viral genome allows for segment exchange (termed reassortment) when distinct viruses co-infect the same host cell and generate progeny with a mixed genome. Reassortment can involve any of the eight segments. In the case of the surface proteins, i.e., HA and NA, this reassortment process is termed antigenic shift (Desselberger et al., 1978; Webster et al., 1974). Genome reassortment establishes a high probability for the creation of pandemic viral strains, which can evade the human immune system and eventually cause widespread infection (Chen et al., 2008). The human influenza A viruses of the 1957 and 1968 pandemics, which killed millions of people, are believed to have arisen through reassortment between human and avian viruses (Webster, 2002; Webster et al., 1992). In the past 250 years, there have likely been 10 to 20 pandemics, presumably because of genome reassortments (Webster, 1998). Thus, early detection of genome reassortments is important in global influenza surveillance.
2. From whence did pandemic (H1N1) 2009 virus come?
Most influenza researchers agree that the pandemic (H1N1) 2009 (H1N1pdm) virus arose from a reassortment of two swine influenza viruses, namely, a North American H1N2 and a Eurasian H1N1, each of which themselves arose from reassortments (Figure 2) (Garten et al., 2009; Gibbs et al., 2009; Smith et al., 2009; Trifonov et al., 2009). The North American swine virus was itself formed by at least two previous reassortments in swine, contributing six segments: PB2, PB1, PA, HA, NP, and NS. The known triple-reassortant swine H3N2 was first detected in 1998; it originated from genome reassortment of classical swine H1N1 (contributing NS, NP and MP), avian H1N1 (PB2 and PA) and human H3N2 (HA, NA and PB1). Subsequently, the immediate North American swine progenitor of H1N1pdm, i.e., H1N2, was first detected in 1999. It is a reassortant of triple reassortant H3N2 (PB2, PB1, PA, NP, NA, M and NS) and classical swine H1N1 (HA).
Figure 2.
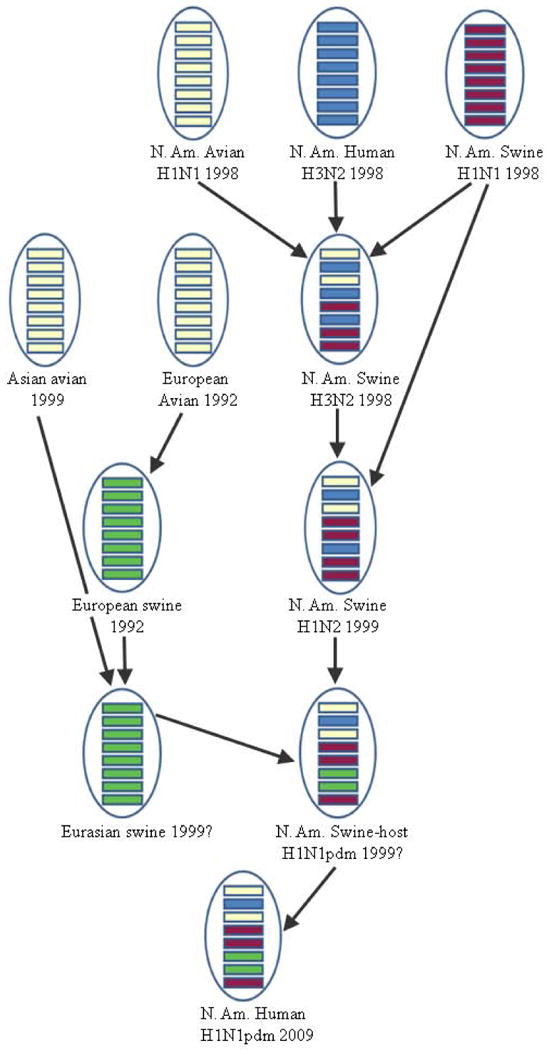
Multiple reassortments contributed to the pandemic (H1N1) 2009 virus.
The Eurasian swine H1N1 contributing to the pandemic H1N1 virus was also formed through at least two reassortments, with both NA and MP being transferred from host avian, but at different times. Genomic analyses by Smith et al. (2009) revealed that the estimated duration of unsampled diversity between the pandemic strains and the closest swine strains is around 17 years (i.e., 1992) for the NA gene and 12 years (i.e., 1997) for the MP gene. Using bioinformatics tools SWeBLAST and PATRISTIC, Gibbs et al. (2010) found the pandemic NA gene is closest to those of swine H1N1 viruses isolated in Europe in 1991-1993, and the pandemic MP gene is closest to those of H3N2 viruses isolated in Asia in 1999-2000. Both studies show strong agreement in the emergence time of the avian-like NA and MP genes in Eurasian swine.
Using FluGenome, a Web tool developed by us for influenza virus lineage and genotype prediction (Lu et al., 2007), the H1N1pdm reference strain A/California/04/2009 was classified as genotype [C,D,E,1A,A,1F,F,1A], with a lineage assigned sequentially to each of the eight genomic segments. All swine viruses from Europe and Asia from 1991 to 1999, inclusively, with the lineage 1F for NA and the lineage F for MP were identified. Pairwise BLASTs of those identified sequences against A/California/04/2009 were performed. The two isolates with the highest sequence similarity values compared to the H1N1pdm reference strain were found to be A/swine/England/WVL7/1992(H1N1) for the MP segment and A/swine/Hong Kong/5190/99(H3N2) for the NA segment. This provides evidence of the earlier contribution of the NA segment, likely coming from Europe, and the later contribution of the MP segment, likely coming from Asia. Regarding the other six segments coming from North American swine, a summary of a comprehensive BLAST analysis of all eight segments confirms the predicted host of origin, available as Supplemental File 1, with the raw data in Supplemental File 2.
3. How has the virus evolved during the pandemic period?
Molecular evolutionary analyses of pandemic sequences available as of September 2009 revealed two major clusters, Cluster I originating from Mexico, Texas and California and Cluster II originating from New York (Fereidouni et al., 2009; Shiino et al., 2010). However, the authors caution that further study is needed to determine if the differences between cluster I and cluster II viruses are due to time, geography, or other factors (Fereidouni et al., 2009). Large scale phylogenetic analyses conducted with 1,850 concatenated genome sequences of the pandemic 2009 viruses show that the resulting tree has a topology (i.e., two clusters) similar to that described in Fereidouni et al. (2009) (Figure 3). The two clusters were observed in individual trees constructed with sequences of PB2, HA, NP, NA, MP, and NS, but were not found in the PA and PB1 trees (Supplemental File 4). In the NA tree, sequences from Wisconsin were found in Cluster II rather than in Cluster I, as in the trees of concatinated genome sequences and sequences of the other five gene segments. However, our analyses revealed that the boostrap values in support of the two clusters were all relatively low, no matter which phylogenetic method (i.e., Neighbor-Joining or Maximum Likelihood) was used.
Figure 3.
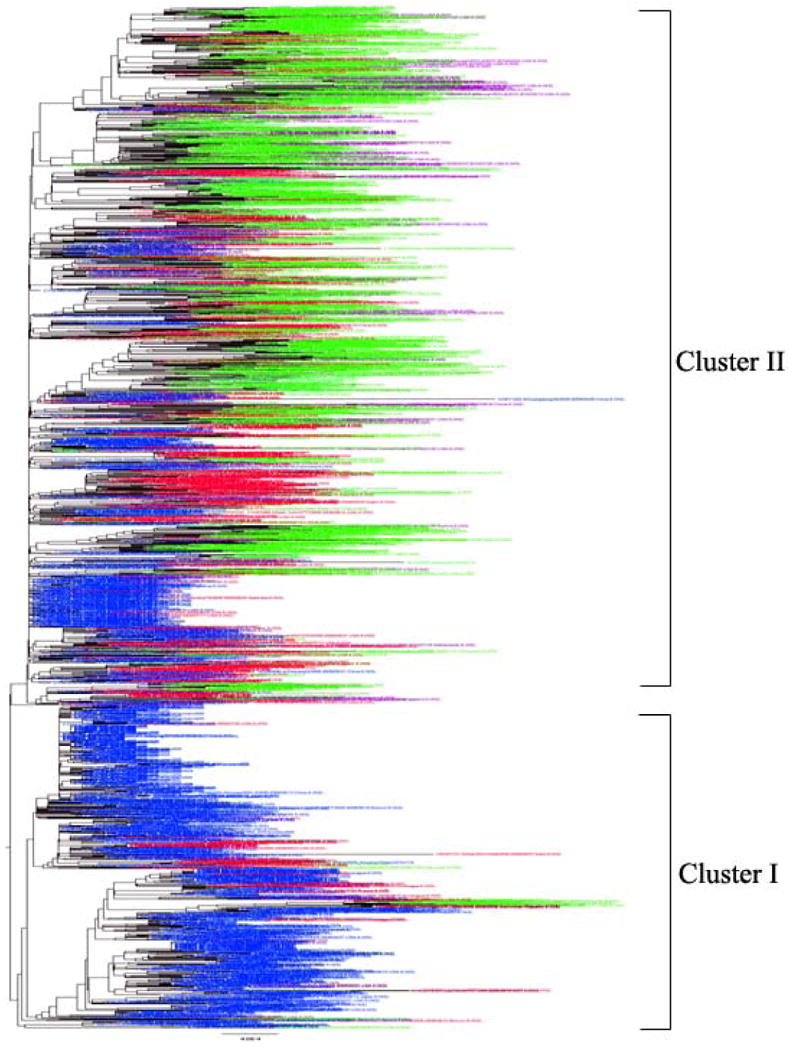
Maximum likelihood tree using concatenated genome sequences of all pandemic (H1N1) 2009 viral strains available in NCBI. Viral strains isolated from April-June, 2009 are colored in blue; July- September, 2009 in red; October-December 2009 in green; January-March, 2010 in purple.
Previous analyses found that Cluster I occurred earlier than Cluster II, which is in agreement with the results of our analysis (Figure 3). Among over 1800 sequences, the earliest viral strains in Cluster I and Cluster II are A/California/2009 and A/New York/2009, which were detected on April 1 and April 24, 2009, respectively. In addition, our analyses showed that Cluster I had died off by the end of 2009, whereas Cluster II has been circulating continuously since April 2009. The virus became less active in 2010, as shown by the reported cases of pandemic human infection and viral sequences isolated and submitted to NCBI (Figure 1). This observation provides evidence to support the WHO's declaration of the end of the pandemic on August 10, 2010.
4. Genome reassortment among pandemic (H1N1) 2009 viruses and between pandemic and other viruses
The current state of knowledge of reassortment among pandemic (H1N1) 2009 viruses comes primarily from phylogenetic analyses of existing strains. Fereidouni et al. (2009) found one subcluster (comprised of Japanese viruses) has seven of eight segments like Cluster I viruses, but has an NP segment most like cluster II viruses, which points to the possibility of reassortment between the two main clusters. Similar observations were found by Shiino et al. (2010), where one subcluster consists of the MP and NS segments coming from Cluster I, with the HA, NP and NA segments coming from Cluster II. Our phylogenetic analyses also revealed a likely reassortment in the pandemic May to June Wisconsin viruses (Supplemental File 4). The NA segments of Wisconsin viruses groups with Cluster II, whereas the remaining segments (except PA and PB1, which do not separate into a Cluster I and Cluster II) group with Cluster I.
There is evidence that human pandemic (H1N1) 2009 virus has re-infected swine herds in several countries (Nagarajan et al., 2010; Song et al., 2010; Poon et al., 2010; Vijaykrishna et al., 2010). According to a study in Korea (Song et al., 2010) and one in Australia (Deng et al., 2010), there is no evidence yet that the re-infected H1N1pdm virus has itself reassorted with swine-origin viruses. However, to the contrary, Vijaykrishna et al. (2010) did find reassortment of pandemic (H1N1) 2009 virus with swine viral strains in a Hong Kong abattoir (Vijaykrishna et al., 2010).
Most of these swine reassortants do not have high pathogenicity; however, there has been one recent report of a highly pathogenic-to-swine reassortant virus, with a mortality rate of 10% (Maurer-Stroh et al., 2009). An in vitro study of reassortments between seasonal human H1N1 / H3N2 and pandemic (H1N1) 2009 by Shih et al. (2009) showed an increased pathogenicity of reassortants, specifically ones with the PB2, PB1 and NP of seasonal H1N1 and the PA of H1N1pdm. There had been previous intermittent reports of swine workers infected with swine viruses without subsequent sustained human-to-human transmission, e.g., A/Iowa/CEID23/2005 (H1N1) (Gray et al., 2007). All this argues for at least increased surveillance (and possibly increased vaccination) in swine herds, both for the economic impact to swine producers and ultimately to the consumer, and for the possible eventual adverse health impact on humans.
5. Evolutionary rates and selection
High evolutionary rates were found in the influenza A H1N1pdm virus. The mean rate varies from 2.34 × 10-3 to 3.67 × 10-3 substitutions per site per year during the early outbreak period of the pandemic (Smith et al., 2009). The evolutionary rate for each segment sequence during the whole pandemic period varies from 3.65 × 10-3 to 6.17 × 10-3 substitutions per site per year, which are relatively higher than the rates found in the early outbreak period (Table 2). Analyses of non-synonymous to synonymous ratios (dN/dS) in all eight segments during the whole pandemic period showed that all the genes of human pandemic (H1N1) 2009 had higher dN/dS ratios than the corresponding swine genes (Table 2). The ratios are largely comparable when using sequences from the early pandemic period and from the whole period, except for the NS segment.
Table 2. Evolutionary rates and dN/dS ratios of eight H1N1pdm viral segments.
| Gene | Mean evolutionary rate* | dN/dS ratio | |||
|---|---|---|---|---|---|
| 04/09- 03/10 | 04-05/09 | 04/09- 03/10 | 04-05/09 | Reference swine virus | |
| PB2 | 4.56 | 2.60 | 0.18 | 0.15 | 0.11 |
| PB1 | 4.38 | 2.34 | 0.15 | 0.21 | 0.06 |
| PA | 3.65 | 2.45 | 0.25 | 0.11 | 0.06 |
| HA | 6.17 | 3.67 | 0.42 | 0.32 | 0.21 |
| NP | 3.69 | 2.59 | 0.10 | 0.18 | 0.05 |
| NA | 3.76 | 3.65 | 0.23 | 0.26 | 0.18 |
| MP | 3.87 | 2.55 | 0.19 | 0.19 | 0.05 |
| NS | 4.34 | 2.62 | 0.83 | 2.15 | 0.23 |
× 10-3 substitutions per site per year
6. Genetic mutations
The pandemic H1N1 2009 virus evolved rapidly, with new mutations that are likely to provide the viral strain with more opportunities to survive and spread (Fitch et al., 1991). Although many mutations are found in H1N1pdm viruses, only a few mutations were reported to benefit the new viruses in terms of host receptor binding, virulence, and drug resistance (Table 3), which will be discussed below.
Table 3. Mutations of the pandemic (H1N1) virus and their associated functions.
| Segment | Mutation | Function |
|---|---|---|
| 1(PB2) | D701N | Increased host range, polymerase activity, and pathogenicity |
| E627K | Increased virulence | |
| 4(HA) | D190E | Both involved in host receptor specificity for the α2-3 and |
| D222G/E | α2-6 sialic acid receptors | |
| 5(NP) | V100I | Increased transmissibility or infectivity |
| 6(NA) | I223V | Decreased neuraminidase inhibitor sensitivities |
| H274Y | Oseltamivir resistance | |
| 7(M2) | S31N | Adamantine resistance |
The hemagglutinin (HA) segment plays an essential role in viral cell entry. It has been found that viral strains with residues D190/D225 are human-specific, D190/G225 swine-specific, and E190/G225 avian-specific. The H1N1pdm HA has D190/D225, supporting the efficient transmissibility of these viruses among humans (Potdar et al., 2010). A mutation (D225G/E) was found in the HA protein in the pandemic virus; this may allow the virus to have dual hosts, humans and swine (Chen et al., 2010). Our structural analyses of the H1N1pdm HA reveals that this virus is closer to the 1918 H1N1 pandemic viruses than to seasonal H1N1 influenza viruses, suggesting the preexisting immunity against the 2009 H1N1pdm viruses in elderly persons (Figure 4) (Xu et al., 2010). Significant amino acid differences occur in the four antigenic regions (Sa, Sb, Ca and Cb) of HA when comparing the pandemic and seasonal influenza viruses. High sequence similarity (∼80%) was found between the two pandemic viruses (1918 and 2009), with the differences restricted mainly to the Ca region of HA. This agrees with the findings of Booy et al. (2009), Morens et al. (2010), and Hancock et al. (2009) that cross-reactive antibodies to H1N1pdm do exist in older people.
Figure 4.
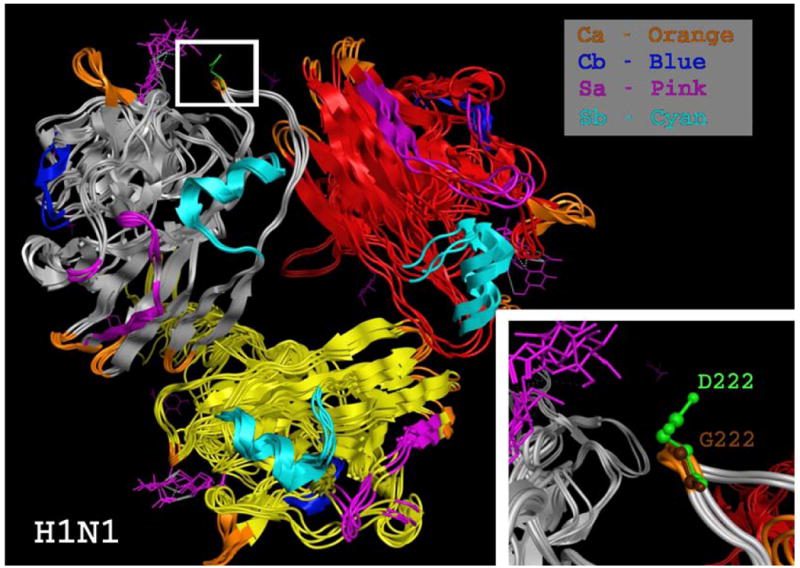
Structural alignment of the HA segment of three different influenza A H1N1 viruses showing a mutation at position D222G. The different chains: red, grey, and yellow, show the different subunits of the HA trimer. Antigenic regions of HA are also color coded (Ca, Orange; Cb, Blue; Sa, Pink; Sb, Cyan).The H1N1 HA segments of the viruses used for this alignment are A/Darwin/2001/2009 (3M6S), A/Swine/Iowa/15/1930(1RUY), and A/SouthCarolina/1/1918(1RUZ). The smaller region at the lower right corner zooms in on the Ca region to show the D222G mutation.
Hancock et al. (2009) used microneutralization assays to test for level of antibody response to H1N1pdm in serum samples from various age groups, both before and after vaccination with seasonal flu vaccine. They did not find any cross-protective pre-existing immunity against H1N1pdm in pediatric or younger-adult age groups, but for those aged 60 years or older, even before seasonal flu vaccination, they did find a significant cross-protective immunity. Seasonal flu vaccination produced no measurable change in antibody titers against H1N1pdm.
A single residue in the PB2 subunit of the influenza polymerase, amino acid 627, regulates polymerase activity in a species-specific fashion, with a lysine(K) mainly found in human viruses and a glutamic acid (E) in avian viruses (Subbarao et al., 1993). PB2 K627 also correlates with enhanced polymerase activity and mortality in mammals (Steel et al., 2009), and moderately enhanced replication in pigs, consistent with pigs serving as an intermediary viral reservoir between birds and humans (Manzoor et al., 2009). The current pandemic (H1N1) 2009 virus was found to exhibit mainly K627, resulting in high levels of viral replication in human cells. Other changes within PB2, such as the D701N mutation, are associated with increased host range, polymerase activity, and pathogenicity in mammalian systems and humans (Doudnaa and Mehle, 2009). An in vitro experiment using cell cultures in a mouse model does not support that PB2-E627K or D701N mutations are responsible for enhanced virulence of the pandemic virus (Jagger et al., 2010). Additionally, a mutation has been linked to increased transmissibility or infectivity, namely, the valine to isoleucine V100I mutation in the NP segment, which was found to occur during the short period of time between pandemic alert levels phase 4 and phase 6 (Pan et al., 2010).
Concerning drug resistance, nearly all of the 2009 pandemic H1N1 viruses were sensitive to neuraminidase inhibitors; only sporadic Oseltamivir-resistant viruses with the H275Y mutation in the NA segment were reported. On the contrary, Oseltamivir was found more effective for pandemic (H1N1) 2009 than for seasonal H1N1 influenza with or without the H275Y mutation (Ikematsu et al., 2010). Additionally, the mutation I222V that has been shown to affect susceptibility to neuraminidase inhibitors (Deyde et al., 2010; Monto et al, 2006) was also detected in a North Carolina pandemic specimen (A/North Carolina/15/2009). The pandemic virus contains the adamantine resistance-conferring change S31N in the M2 protein (Dawood et al., 2009; Deyde et al., 2010; Garten et al., 2009).
7. Discussion
7.1. Evolutionary history of pandemic (H1N1) 2009 virus
It appears clear that the pandemic (H1N1) 2009 virus arose from a reassortment between North American and Eurasian swine viruses (Garten et al., 2009; Smith et al., 2009); both the North American and the Eurasian immediate progenitors of the pandemic virus itself originated from at least two reassortments. One salient feature of this paper is its comprehensive information regarding the subtypes, contributing gene segments, and the time of emergence of the previous reassortants, which are provided through a careful review of published analyses and additional analyses of sequences collected during the entire pandemic period, available in GenBank (Figure 1) (Brockwell-Staats et al., 2009; Garten et al., 2009; Gibbs et al., 2009; Smith et al., 2009; Trifonov et al., 2009). In addition, our analyses further confirmed that the precursors of the pandemic H1N1 virus went unnoticed or unsampled in swine herds for 9 – 12 years for the six North American swine influenza viral genes and 12 – 17 years for the two Eurasian genes (NA and MP) (Gibbs et al., 2009; Smith et al., 2009; Trifonov et al., 2009); this strongly suggests that increased surveillance is critical in swine herds in order to prevent a future event like the 2009 H1N1 pandemic.
Regarding where the Eurasian swine and North American reassortant swine viruses reassorted, we speculate that the likely place of origin for H1N1pdm was North America, with the most likely place of all being Mexico. The overwhelming majority of the earliest cases of infections were reported from Mexico first and then the United States. Additionally, the two pandemic viral genes originating from Eurasian swine viruses were detected much earlier in North American swine viruses (e.g., A/Swine/Virginia/670/1987) than were the six North American genes detected in Eurasian swine viruses (e.g., A/Swine/Korea/ASAN04/2006) (Trifonov et al., 2009). Similarly, Smith et al. (2009) found a sister lineage to H1N1pdm with seven out of its eight segments in an H1N2 2004 isolate (A/Swine/Hong Kong/915/2004).
Since 1994 when the North American Free Trade Agreement (NAFTA) was signed into law, US swine have been exported in increasing numbers to Mexico. According to USDA statistics, Mexican pork consumption of US pork was 31% in 2005, in comparison to only 6% in 1996 (Zahniser, 2006). Since the known triple-reassortant H3N2 virus arose in 1998, a number of reassortants have been detected in both swine and humans, including the latest pandemic H1N1 (Brockwell-Staats et al., 2009; Zhou et al., 1999). Although we find piquant the theory of Gibbs et al. (2009) that H1N1pdm was a lab mixture of several strains that escaped, we think it more likely that movement of live pigs between Eurasia and North America facilitated the mixing of diverse swine influenza viruses; their reassortment has lead to the novel H1N1pdm virus. Solovyov et al. (2009) did a comparative analysis of evolutionary rates of several subtypes of influenza viruses in various hosts, which they conclude provides strong support for the unsampled pig herd hypothesized origin of H1N1pdm, and contradicts Gibbs et al.'s escaped lab mixture hypothesis. Domestic pigs have been considered a mixing vessel of avian, human and swine viruses, producing new influenza viruses that cause pandemics in both swine and humans (Webster et al., 1992). Therefore, increased surveillance in swine herds is important, both for the economic impact to swine producers and ultimately to the consumer, and for the possible eventual adverse health impact on humans.
7.2. Evolutionary dynamics of pandemic H1N1 virus
Previous studies using sequences available as of September 2009 revealed the pandemic virus diverged into two clusters, with Cluster I originating from Mexico, Texas and California, and Cluster II originating from New York (Fereidouni et al., 2009; Shiino et al., 2009). Our large scale phylogenetic analyses using all complete genome sequences available as of August 2010 showed a tree topology similar to those reported in previous studies, but the bootstrap support for the two clusters is low. This may be due to small genetic differences among pandemic viral sequences. Based upon 1,850 available whole genome sequences, the average p-distances (portion of nucleotide sites different between sequences) were estimated to be 0.002 for the MP, NA, NP, NS, PA, PB1 and PB2 segments and 0.003 for HA. According to Shiino et al. (2009) only nine nucleotides / amino acid residues were found to be cluster-specific (Shiino et al., 2009). Most mutations (around 98%) occurred randomly, which might diminish the phylogenetic signals of the two clusters, resulting in low bootstrap values, when a large number of sequences collected worldwide and the entire pandemic period were analyzed. However, micro-evolution could be observed using samples collected from local regions and within restricted time periods as demonstrated by Shiino et al. (2009).
Cluster I viruses were found to occur earlier than Cluster II viruses (Fereidouni et al. 2009; Shiino et al. 2009), with which our analysis of large datasets agree. This finding makes sense for the following reasons. First, the earliest outbreak of the pandemic virus was reported in Mexico in March 2009, and the first case was reported in California on April 14, 2009, whereas the cases of infection in New York was first reported on April 24 (CDC, 2009b; Lessler et al., 2009). Secondly, based upon tMRCA, Cluster I viruses appeared two months earlier than Cluster II viruses (Shiino et al., 2010). Thirdly, our study shows the earliest sequences found in Cluster I and Cluster II occur on April 1 and April 24, 2009, respectively. One important finding with large scale genome sequence analyses is that Cluster I viruses died off in the end of 2009; Cluster II viruses had been circulating, but they were less active on a global scale in 2010 (Figure 1), which supports the WHO's declaration that the pandemic ended in August 2010.
Reassortment was found between different clusters of the pandemic viruses in humans and between the pandemic human H1N1 viruses and swine viruses in pigs (Fereidouni et al., 2009; Shiino et al., 2009; Vijaykrishna et al., 2010). Swine infection by human viruses was reported previously in several cases, including swine infected by human H1N2 in the US and Canada in 2003, and by human H1N1 in 2005 (Gramer, 2009; Noble et al., 1993). The ongoing isolation of human-like H1N1 and H1N2 influenza viruses in swine suggests that the viruses have adapted to swine and are capable of causing respiratory disease in swine. It has been known that the respiratory tract of swine possesses both avian and human receptors for influenza viruses. Swine act as a mixing vessel for novel reassorted viruses (Webster et al., 1992).
The pandemic H1N1 viruses evolved much faster than their progenitor swine viruses (Table 2). Consequently, a large number of mutations were generated, but most mutations were found to be random. Mutations known to be associated with host specificity and virulence were detected in the pandemic viruses, indicating they can be transmitted effectively in humans and cause increased severity. Different patient populations were affected by H1N1pdm than by H1N1/H3N2 seasonal influenza. Structural comparison revealed that the virus should be less severe in older people (Xu.et al., 2009). The structure of the pandemic 2009 HA is more similar to that of the pandemic 1918 HA compared to seasonal influenza's HA. The 1918 H1N1 virus continued to circulate in humans prior to 1958. Older people might have been infected by the 1918-57 H1N1 and acquired a certain amount of immune system protection against 2009 H1N1pdm. Harvard epidemiologists, analyzing data from seven regions around the globe, found the highest hospitalization rates for H1N1pdm were among infants, with much lower rates among those over 50 and even lower rates among those over 60 (Jacobs et al., 2009).
7.3. Influenza surveillance and prevention
Regarding pork production since the late 1990s: In the US and globally prior to that time, there were numerous small family farms, raising both crops and livestock, including hogs. Who can say what drove market forces, but the profit margin in hog production became increasingly slim, driving the small hog producer out of business (D. Christman, personal communication). This led to the rise, in the US (which agrees with a global trend) of 70% of hog production being controlled by only 50 producers by 2001, with the largest operations having over 700,000 hogs (Moore, 2004). This in turn led to increased crowding, which led to increased stress on the hogs, as measured by decreased feed consumption. As is widely known in complementary medicine and is being investigated in the emerging cross-disciplinary field of science known as psychoneuroimmunology, increased stress leads to decreased immune functioning (Mercola, 2003; Yang and Glaser, 2002). So the situation is greatly increased mingling of hogs (with also an increase in global trade of swine) at the very time when their immune systems are least likely to be able to cope. Is it any wonder there has been a proliferation of reassortment among swine viruses since 1998, after decades of only one subtype in swine, namely, classical H1N1, and the increased transmission of swine viruses to humans, and back again to the swine, and so on? This again argues increased surveillance in swine herds is essential for monitoring genetic changes of influenza viruses.
Although the cross-protective immunity that the oldest members of the population have is the likely explanation for the lower morbidity among the elderly for H1N1pdm, most likely there is another cause at work in the case of the increased hospital admissions in the young aged 0-4, namely a young immune system's propensity to overreact with a cytokine storm, a potentially fatal immune reaction in which the immune system gets stuck in a feedback loop between immune cells and one of their regulatory products, cytokines(Leon et al., 2009; Vogel et al., 2009). The highest risk groups for 2009 H1N1pdm are children (Olsen et al., 2009) and pregnant women. In Canada, the odds ratio for hospital admission for pregnant women versus non-pregnant women of child-bearing age was calculated as 8.4, with a 95% confidence interval of 6.5 to 10.9 (Helferty et al., 2009). In other words, you were 6 to 11 times more likely to be hospitalized for H1N1pdm if you were pregnant than if you were a non-pregnant woman of child-bearing age.
Researchers at St. Jude Children's Research Hospital established another link between a risk factor and H1N1pdm that had not previously been established with H1N1/H3N2 seasonal flu, namely the risk factor of obesity. It was found that a staggering 80% to 90% of obese mice inoculated with strain A/California/07/09 succumbed, compared to approximately 20% of non-obese mice. They found this high mortality rate in both genetically obese and diet-induced obese mice (O'Brien et al., 2009). This finding agrees with a case-cohort design study done by researchers at the CDC, who compared cases of hospitalizations and deaths from 2009 H1N1pdm from April to July, 2009 to a cohort of hospitalized patients taken from the 2003-2006 National Health and Nutrition Examination Survey (Morgan et al., 2009). The study examined whether or not obesity alone was an independent risk factor for hospitalization and death from H1N1pdm. They found that morbid obesity alone without an ACIP-recognized chronic medical condition, such as asthma or diabetes, was associated with a greater risk of hospitalization from H1N1pdm (odds ratio = 4.7, with a 95% confidence interval of 2.4-9.9).
The CDC recommends annual flu vaccination for all but the youngest infants (an infant less than 6 months old cannot mount an immune reaction to a vaccine), with an even stronger recommendation for high-risk groups, i.e., children, pregnant women, patients with comorbidities, morbid obesity, and the elderly (CDC, 2010). With the near-global consolidation of swine production into larger facilities and the near-global rise in obesity, we expect the next flu pandemic will arrive probably sooner than the roughly 40-year interval since the last.
Supplementary Material
Acknowledgments
This publication was made possible by NIH grant numbers R01 LM009985-01A1. The authors also acknowledge the UCRCA, the University of Nebraska at Omaha, for continuous funding support to this research program. The authors are grateful to Andy Zhong and Todd Herpy for their help with structural analysis. We would also like to thank Thaine Rowley for a Perl script to concatenate the eight segments of the viral sequences for phylogenetic analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
MC Christman, Email: mchristman@unomaha.edu.
A Kedwaii, Email: akedwaii@unomaha.edu.
J Xu, Email: jianpenxu@unomaha.edu.
RO Donis, Email: rvd6@CDC.GOV.
References
- Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. The influenza virus resource at the National Center for Biotechnology Information. J Virol. 2008;82(2):596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy R, Khandaker G, Heron L, Yin J, Doyle B, Tudo K, Hueston L, Gilbert GL, MacIntyre CR, Dwyer DE. Options for the Control of Influenza VII. Hong Kong SAR, China: 2009. Cross-reacting antibodies against pandemic influenza A (H1N1) 2009 virus in elderly Australians. In Press. [DOI] [PubMed] [Google Scholar]
- Brockwell-Staats C, Webster RG, Webby RJ. Diversity of Influenza Viruses in Swine and the Emergence of a Novel Human Pandemic Influenza A (H1N1) Influenza Other Respi Viruses. 2009;3(5):207–213. doi: 10.1111/j.1750-2659.2009.00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett F. Mucins and mucoids in relation to influenza virus action. III. Inhibition of virus haemagglutination by glandular mucins. Aust J Exp Biol Med Sci. 1948;26:403–411. doi: 10.1038/icb.1948.38. [DOI] [PubMed] [Google Scholar]
- CDC. Swine Influenza A (H1N1) Infection in Two Children - Southern California. 2009a 2009 March-April; http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5815a5.htm. [PubMed]
- CDC. Morbidity and Mortality Weekly Report. Atlanta, Georgia: The Centers for Disease Control and Prevention; 2009b. March-April, 2009. Swine Influenza A (H1N1) Infection in Two Children - Southern California. [PubMed] [Google Scholar]
- CDC. Key Facts about Influenza (Flu) & Flu Vaccine. The Centers for Disease Control and Prevention; 2010. [Google Scholar]
- Chen H, Wen X, To KK, Wang P, Tse H, Chan JF, Tsoi HW, Fung KS, Tse CW, Lee RA, Chan KH, Yuen KY. Quasispecies of the D225G substitution in the hemagglutinin of pandemic influenza A(H1N1) 2009 virus from patients with severe disease in Hong Kong, China. J Infect Dis. 2010;201(10):1517–21. doi: 10.1086/652661. [DOI] [PubMed] [Google Scholar]
- Chen LM, Davis CT, Zhou H, Cox NJ, Donis RO. Genetic compatibility and virulence of reassortants derived from contemporary avian H5N1 and human H3N2 influenza A viruses. PLoS Pathog. 2008;4(5):e1000072. doi: 10.1371/journal.ppat.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng YM, Iannello P, Smith I, Watson J, Daniels P, Komadina N, Barr I, Harrower B, Wong F. No reassortment in pandemic (H1N1) 2009 viruses isolated from swine in Australia In Options for the Control of Influenza VII. Hong Kong SAR, China: 2010. In Press. [Google Scholar]
- Desselberger U, Nakajima K, Alfino P, Pedersen FS, Haseltine WA, Hannoun C, Palese P. Biochemical evidence that “new” influenza virus strains in nature may arise by recombination (reassortment) Proc Natl Acad Sci U S A. 1978;75(7):3341–5. doi: 10.1073/pnas.75.7.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyde VM, Sheu TG, Trujillo AA, Okomo-Adhiambo M, Garten R, Klimov AI, Gubareva LV. Detection of molecular markers of drug resistance in 2009 pandemic influenza A (H1N1) viruses by pyrosequencing. Antimicrob Agents Chemother. 2010;54(3):1102–10. doi: 10.1128/AAC.01417-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudnaa JA, Mehle A. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc Natl Acad Sci U S A. 2009;106(50):21312–21316. doi: 10.1073/pnas.0911915106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW. Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci U S A. 1993;90(9):4171–5. doi: 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc Natl Acad Sci U S A. 1999;96(24):13910–3. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fereidouni SR, Beer M, Vahlenkamp T, Starick E. Differentiation of two distinct clusters among currently circulating influenza A(H1N1)v viruses, March-September2009. Euro Surveill. 2009;14(46) [PubMed] [Google Scholar]
- Fitch WM, Leiter JM, Li XQ, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc Natl Acad Sci U S A. 1991;88(10):4270–4. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RAM, Munster VJ, Keawcharoen J, Osterhaus ADME, Kuiken T. Virology of avian influenza in relations to wild birds. Journal of wildlife diseases. 2007;43(3_Supplement):7–14. [Google Scholar]
- Frosner G, Gerth HJ, Flehmig B, Vallbracht A. Functional significance of neuraminidase in the replication cycle of influenza viruses. Zentralbl Bakteriol [Orig A] 1975;232:178–188. [PubMed] [Google Scholar]
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, López-Gatell H, Olivera H, López I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD, Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325(5937):197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E, Wentworth DE, Halpin RA, Lin X, Bera J, DePasse J, Fitch A, Griesemer S, Hine E, Katzel DA, Overton L, Proudfoot K, Sitz J, Szczypinski B, St George K, Spiro DJ, Holmes EC. Unseasonal transmission of H3N2 influenza A virus during the swine-origin H1N1 pandemic. J Virol. 2010;84(11):5715–8. doi: 10.1128/JVI.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs AJ, Armstrong JS, Downie JC. From where did the 2009 ‘swine-origin’ influenza A virus (H1N1) emerge? Virol J. 2009 Nov 24;6:207. doi: 10.1186/1743-422X-6-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramer MR. Reassortant human/swine H1N1 and H1N2 influenza virus infections in US swine. Journal of swine health and production. 2009:463–464. [Google Scholar]
- Gray GC, McCarthy T, Capuano AW, Setterquist SF, Olsen CW, Alavanja MC. Swine workers and swine influenza virus infections. Emerg Infect Dis. 2007;13(12):1871–8. doi: 10.3201/eid1312.061323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Poon LL, Cheung CY, Ellis TM, Lim W, Lipatov AS, Chan KH, Sturm-Ramirez KM, Cheung CL, Leung YH, Yuen KY, Webster RG, Peiris JS. H5N1 influenza: a protean pandemic threat. Proc Natl Acad Sci U S A. 2004;101(21):8156–61. doi: 10.1073/pnas.0402443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, Brammer TL, Cox NJ, Tumpey TM, Katz JM. Cross-Reactive Antibody Responses to the 2009 Pandemic H1N1 Influenza Virus. N Engl J Med. 2009;361:1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- Helferty M, Vachon J, Tarasuk J, Rodin R, Spika J, Pelletier L. Options for the Control of Influenza VII. Hong Kong SAR, China: 2009. Epidemiology of hospitalized pandemic H1N1 2009 cases in Canada: a comparison of the first and second pandemic waves. In Press. [Google Scholar]
- Holmes EC, Ghedin E, Miller N, Taylor J, Bao Y, St George K, Grenfell BT, Salzberg SL, Fraser CM, Lipman DJ, Taubenberger JK. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 2005;3(9):e300. doi: 10.1371/journal.pbio.0030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RT, Rott R, Klenk HD. On the receptor of influenza viruses. 1. Artificial receptor for influenza virus. Z Naturforsch [C] 1973;28:342–345. [PubMed] [Google Scholar]
- Ikematsu H, Kawai N, Iwaki N, Hirotsu N. Options for the Control of Influenza VII. Hong Kong, China: 2010. Clinical symptoms and the effectiveness of neuraminidase inhibitor for patients with pandemic influenza H1N1 2009 in Japan: Comparison with seasonal H1N1 influenza in the 2007-2008 and 2008-2009 seasons. In Press. [DOI] [PubMed] [Google Scholar]
- Jacobs J, Archer B, Baker M, Cowling B, Heffernan R, Mercer G, Uez O, Tchetgen E, Lipsitch M. Options for the Control of Influenza VII. Hong Kong SAR, China: 2009. Epidemiologic evidence on age-related immunity to 2009 H1N1 influenza. In Press. [Google Scholar]
- Jagger BW, Memoli MJ, Sheng ZM, Qi L, Hrabal RJ, Allen GL, Dugan VG. The PB2-E627K Mutation Attenuates Viruses Containing the 2009 H1N1 Influenza Pandemic Polymerase. mBio. 2010:1–10. doi: 10.1128/mBio.00067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri S, Ohizumi A, Homma M. An outbreak of type C influenza in a children's home. J Infect Dis. 1983;148(1):51–6. doi: 10.1093/infdis/148.1.51. [DOI] [PubMed] [Google Scholar]
- Klumpp K, Ruigrok RW, Baudin F. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. Embo J. 1997;16:1248–1257. doi: 10.1093/emboj/16.6.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RA, Choppin PW. The gene structure and replication of influenza virus. Annu Rev Biochem. 1983;52:467–506. doi: 10.1146/annurev.bi.52.070183.002343. [DOI] [PubMed] [Google Scholar]
- Leon A, Lei Y, Wang P, Huang J, Lin J, Farooqui A, Guan Y, Kelvin D. Options for the Control of Influenza VII. Hong Kong SAR, China: 2009. Isolation and characterization of three 2009-H1N1 influenza strains from Nanchang, China. In Press. [Google Scholar]
- Lessler J, Reich NG, Cummings DA, New York City Department of Health and Mental Hygiene Swine Influenza Investigation Team. Nair HP, Jordan HT, Thompson N. Outbreak of 2009 pandemic influenza A (H1N1) at a New York City school. N Engl J Med. 2009;361(27):2628–36. doi: 10.1056/NEJMoa0906089. [DOI] [PubMed] [Google Scholar]
- Lu G, Rowley T, Garten R, Donis R. FluGenome: a web tool for genotyping influenza A virus. Nucleic Acids Res. 2007;35:W275–9. doi: 10.1093/nar/gkm365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoor R, Sakoda Y, Nomura N, Tsuda Y, Ozaki H, Okamatsu M, Kida H. PB2 protein of a highly pathogenic avian influenza virus strain A/chicken/Yamaguchi/7/2004 (H5N1) determines its replication potential in pigs. J Virol. 2009;83(4):1572–8. doi: 10.1128/JVI.01879-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Katsushima N, Nagai Y, Shoji M, Itagaki T, Sakamoto M, Kitaoka S, Mizuta K, Nishimura H. Clinical features of influenza C virus infection in children. J Infect Dis. 2006;193(9):1229–35. doi: 10.1086/502973. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Ma J, Lee RT, Sirota FL, Eisenhaber F. Mapping the sequence mutations of the 2009 H1N1 influenza A virus neuraminidase relative to drug and antibody binding sites. Biol Direct. 2009;4:18. doi: 10.1186/1745-6150-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercola J. More evidence that stress is major factor for infections. [Last accessed: January 13, 2011];2003 http://articles.mercola.com/sites/articles/archive/2003/07/16/stress-infections.aspx.
- Monto AS, McKimm-Breschkin JL, Macken C, Hampson AW, Hay A, Klimov A, Tashiro M, Webster RG, Aymard M, Hayden FG, Zambon M. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob Agents Chemother. 2006;50(7):2395–402. doi: 10.1128/AAC.01339-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JR. Swine production: a global perspective. Nicholasville, Kentucky, USA: Alltech Inc; 2004. [Google Scholar]
- Morens DM, Taubenberger JK, Fauci AS. The 2009 H1N1 Pandemic Influenza Virus: What Next? MBio. 2010;1(4):e00211–10. doi: 10.1128/mBio.00211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan OW, Bramley A, Fowlkes A, Freedman DS, Taylor TH, Gargiullo P, Belay B, Jain S, Cox C, Kamimoto L, Fiore A, Finelli L, Olsen SJ, Fry AM. Morbid obesity as a risk factor for hospitalization and death due to 2009 pandemic influenza A(H1N1) disease. PLoS One. 2009;5(3):e9694. doi: 10.1371/journal.pone.0009694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S, McGregor MS, Wentworth DE, Hinshaw VS. Antigenic and genetic conservation of the haemagglutinin in H1N1 swine influenza viruses. J Gen Virol. 1993;74(Pt 6):1197–200. doi: 10.1099/0022-1317-74-6-1197. [DOI] [PubMed] [Google Scholar]
- Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360(25):2605–15. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- O'Brien KB, Govorkova EA, Webby RJ, McCullers JA, Schultz-Cherry S. Options for the Control of Influenza VII. Hong Kong SAR, China: 2009. Obesity is a risk factor for developing severe influenza infection. In Press. [Google Scholar]
- Olsen SJ, Baggett H, Chittaganpitch M, Thamthitiwat S, Prapasiri P, Naorat S, Ditsungnoen D, Simmerman JM, Chantra S, Sawanpanyalert P, Maloney SA, Akarasewi P. Options for the Control of Influenza VII. Hong Kong SAR, China: 2009. Clinical characteristics of hospitalized influenza patients by subtype in Thailand, 2009. In Press. [Google Scholar]
- Pan C, Cheung B, Tan S, Li C, Li L, Liu S, Jiang S. Genomic signature and mutation trend analysis of pandemic (H1N1) 2009 influenza A virus. PLoS One. 2010;5(3):e9549. doi: 10.1371/journal.pone.0009549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappaioanou M. Highly pathogenic H5N1 avian influenza virus: cause of the next pandemic? ILAR J. 2009;51(3):268–80. doi: 10.1016/j.cimid.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Potdar VA, Chadha MS, Jadhav SM, Mullick J, Cherian SS, Mishra AC. Genetic characterization of the influenza A pandemic (H1N1) 2009 virus isolates from India. PLoS One. 2010;5(3):e9693. doi: 10.1371/journal.pone.0009693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiino T, Okabe N, Yasui Y, Sunagawa T, Ujike M, Obuchi M, Kishida N, Xu H, Takashita E, Anraku A, Ito R, Doi T, Ejima M, Sugawara H, Horikawa H, Yamazaki S, Kato Y, Oguchi A, Fujita N, Odagiri T, Tashiro M, Watanabe H. Molecular evolutionary analysis of the influenza A(H1N1)pdm, May-September, 2009: temporal and spatial spreading profile of the viruses in Japan. PLoS One. 2010;5(6):e11057. doi: 10.1371/journal.pone.0011057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459(7250):1122–5. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- Smith GJ, Bahl J, Vijaykrishna D, Zhang J, Poon LL, Chen H, Webster RG, Peris JS, Guan Y. Dating the emergence of pandemic influenza viruses. Proc Natl Acad Sci U S A. 2009;106(28):11709–12. doi: 10.1073/pnas.0904991106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W, Andrewes CH, Laidlaw PP, Timbury MC. A virus obtained from influenza patients. Lancet. 1933;(225):66–68. [Google Scholar]
- Solovyov A, Greenbaum B, Palacios G, Lipkin WI, Rabadan R. Host dependent evolutionary patterns and the origin of 2009 H1N1 pandemic influenza. PLoS Curr. 2010;2:RRN1147. doi: 10.1371/currents.RRN1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Lee J, Pascua P, Baek Y, Kwon H, Park K, Choi H, Shin Y, Song J, Kim C, Choi Y. Options for the Control of Influenza VII. Hong Kong SAR, China: 2010. Occurrence of human-to-swine transmission of the pandemic (H1N1) 2009 influenza virus in South Korea. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel J, Lowen AC, Mubareka S, Palese P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009;5(1):e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer DA, Skehel JJ. Genetics of influenza viruses. Annu Rev Genet. 2002;36:305–32. doi: 10.1146/annurev.genet.36.052402.152757. [DOI] [PubMed] [Google Scholar]
- Subbarao EK, London W, Murphy BR. A Single Amino Acid in the PB2 Gene of Influenza A Virus Is determinant of host range. J virol. 1993;67(4):1761–4. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov V, Khiabanian H, Rabadan R. Geographic dependence, surveillance, and origins of the 2009 influenza A (H1N1) virus. N Engl J Med. 2009;361(2):115–9. doi: 10.1056/NEJMp0904572. [DOI] [PubMed] [Google Scholar]
- Vogel GE, Back T, Blasini R, Doerr HW, Hofler H, Komm C, Manych M, Moeller R, Schaefer B, Schoettler M, Tsokos M, Wutzler P. Options for the Control of Influenza VII. Hong Kong SAR, China: 2009. Influenza, MRSA, cytokines: diagnosis, treatment, prevention - a possible strategy for outpatient care. In Press. [Google Scholar]
- Webster RG. Textbook of influenza. Oxford: Blackwell Science; 1998. Evolution and ecology of influenza viruses: interspecies transmission; pp. 109–119. [Google Scholar]
- Webster RG. The importance of animal influenza for human disease. Vaccine. 2002;20 2:S16–20. doi: 10.1016/s0264-410x(02)00123-8. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ. Genetics of Influenza Virus. Annual Review of Genetics. 1978;12(1):415–431. doi: 10.1146/annurev.ge.12.120178.002215. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56(1):152–79. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Isachenko VA, Carter M. A new avian influenza virus from feral birds in the USSR: recombination in nature? Bull World Health Organ. 1974;51(4):325–32. [PMC free article] [PubMed] [Google Scholar]
- Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A° resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Jr, Wilson IA. Structural Basis of Preexisting Immunity to the 2009 H1N1 Pandemic Influenza Virus. Science. 2010;328(5976):357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang EV, Glaser R. Stress-induced immunomodulation and the implications for health Review Article. International Immunopharmacology. 2002;2(2-3):315–324. doi: 10.1016/s1567-5769(01)00182-5. [DOI] [PubMed] [Google Scholar]
- Zahniser S. Doha, NAFTA, and California Agriculture. USDA, ed. Sacramento, California: 2006. U.S.-Mexico Agricultural Trade during the NAFTA Era. [Google Scholar]
- Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon K, Krauss S, Webster RG. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73(10):8851–6. doi: 10.1128/jvi.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


