Abstract
Introns in a wide range of organisms including plants, animals and fungi are able to increase the expression of the gene that they are contained in. This process of intron-mediated enhancement (IME) is most thoroughly studied in Arabidopsis thaliana, where it has been shown that enhancing introns are typically located near the promoter and are compositionally distinct from downstream introns. In this study, we perform a comprehensive comparative analysis of several sequenced plant genomes. We find that enhancing sequences are conserved in the multi-cellular plants but are either absent or unrecognizable in algae. IME signals are preferentially located towards the 5′-end of first introns but also appear to be enriched in 5′-UTRs and coding regions near the transcription start site. Enhancing introns are found most prominently in genes that are highly expressed in a wide range of tissues. Through site-directed mutagenesis in A. thaliana, we show that IME signals can be inserted or removed from introns to increase or decrease gene expression. Although we do not yet know the specific mechanism of IME, the predicted signals appear to be both functional and highly conserved.
INTRODUCTION
Since the discovery of introns in the late 1970s (1), there have been considerable efforts to understand their function and evolution. Initially, it was difficult to understand the role of sequences that are transcribed only to then be spliced out of the mature transcripts. Now, however, we recognize that introns can play important roles in gene regulation via alternative splicing and nonsense-mediated decay. A less well-known role of introns is that they can provide a boost to gene expression. Introns that are known to enhance expression have been observed in diverse organisms including plants, insects, mice and humans (2–5). This positive effect on gene expression has been named intron-mediated enhancement (IME) (6).
In IME, the increase in gene expression coincides with an increase in mRNA accumulation (3,7–10). It is important to note that IME is not due to the presence of intronic enhancers, although some enhancing introns can also contain such enhancer elements. While enhancers may be located upstream or downstream from a gene, introns involved in IME must be located in transcribed sequences in order to increase expression (3,6,11). Furthermore, expression-enhancing introns cause little or no increase in radio—labeled RNA generated in nuclear run-on transcription assays (7,10,12). This argues in general against any mechanism of enhancement that involves transcription initiation.
One critical feature of IME is that not all introns are capable of enhancing expression and many introns have no effect on expression. Splicing, and therefore exon junction complexes, are therefore not sufficient to induce IME. Among enhancing introns, the increase is generally between 2- and 10-fold but can be 100-fold in some cases (13,14). Typically, introns that are located nearer to the 5′-end of a gene have more enhancing power than those at the 3′-end (3,5,6,15,16). A key experiment for understanding the mechanism of IME was changing the position of an enhancing intron in Arabidopsis thaliana (17). In this study, the level of enhancement was seen to decrease as an intron was moved towards the 3′-end of a reporter gene, and the IME effect was abolished when the intron was moved >1 Kb from the start of the transcript.
Molecular experiments designed to identify sequences responsible for IME have had limited success. Chimeras between enhancing and non-enhancing introns demonstrate that multiple regions of an enhancing intron are sufficient for IME. We recently reported a computational approach to identifying IME signals (18). This work featured an algorithm that we developed, called the IMEter, that predicts how much an intron enhances expression. The IMEter is based on k-mer frequency differences between promoter-proximal and promoter-distal introns. Although we have some ability to predict how much any given intron enhances expression, the mechanism behind IME remains mysterious. One model that is consistent with the available data, is that intron sequences mediate a change in the transcription machinery which renders it more processive. In the presence of IME signals, the machinery is more likely to extend through the entirety of the gene and produce a mature transcript complete with a poly-A tail. In the absence of IME signals, RNA polymerase may dissociate more easily and produce immature transcripts (19). To date, experimental studies on IME have been restricted to only very small numbers of genes, and no analysis of IME has been performed at a genome-wide level.
In this study, we explore the sequence contexts of IME in a variety of plant genomes. We show that IME signals are not evenly distributed within introns, but are concentrated towards the 5′-end. Though IME signals are most abundant in introns, they are also enriched in 5′-UTRs and in coding sequences (CDS) that are near to the start of a transcript. This indicates that IME probably occurs at the level of the transcript rather than the intron. Genes with the most powerful IME signals appear to be highly and widely expressed housekeeping genes. IME signals appear to be conserved in the majority of plant genomes sequenced to date including various monocot and dicot species, as well as a lycophyte (Selaginella moellendorfii) and a moss (Physcomitrella patens). However, IME signals are either absent or not recognizable in the two species of algae that were studied. Although we do not yet know the specific mechanism, the fact that IME signals are highly conserved suggests that there is a common mechanism.
MATERIALS AND METHODS
Data sets
Genome sequences and annotations were downloaded for the following species (annotation release details are listed in parentheses): A. thaliana (TAIR7), Oryza sativa var. japonica (v6.0), Vitis vinifera (genoscope v1), Populus trichocarpa (JGI v1.1), Sorghum bicolor (JGI v1.4), Selaginella moellendorffii (JGI v1.0), Physcomitrella patens (JGI v1.1), Chlamydomonas reinhardtii (JGI v3.1) and Volvox carteri (JGI v1.0).
Annotations from each genome were processed to ensure all the genes were complete, non-redundant and of sufficient quality. Genes were required to contain both 5′-UTR and 3′-UTR and genes with unusually short CDS (<50 amino acids), short introns (<60 nt), or long introns (>1000 nt) were removed. When genes possessed alternatively-spliced transcripts, only the first numbered isoform was retained.
The IMEter
We have previously described the IMEter algorithm elsewhere (18), though a brief summary will be presented here. The IMEter takes an input sequence and reports a log-odds score based on the frequencies of pentamers in that sequence. A positive IMEter score indicates that the input sequence is similar to the nucleotide content of proximal introns in Arabidopsis [i.e. those introns that are closer to the transcription start site (TSS)]. Conversely, a negative IMEter score indicates similarity to introns that are distal to the TSS. High scoring introns are inferred to be those that contain IME signals and which are capable of enhancing expression. For all the studies in this article, the IMEter pentamers were derived from A. thaliana introns.
In addition to calculating an IMEter score for an entire sequence, one can also calculate the score throughout a sequence using a sliding window approach. In this article we use a window size of 50 nt with a step size of 25 nt. This allows one to visualize which regions of a sequence are contributing most to the overall IMEter score. We also extend this approach to calculate the average IMEter score of a window of sequence from many different sequences. That is, we can extract a window of sequence from all introns that are the same distance from the TSS, and then calculate an average IMEter score for that window.
The IMEter v2.0 is similar to the IMEter 1.0 in many respects. The parameter estimation is the same as in the previous version and uses the same pentamer frequencies from proximal and distal introns. The scoring mechanism has changed in order to identify and score only the high scoring regions (HSRs) within any intron sequence. The procedure is (i) compute the score for each pentamer, (ii) smooth the scores in a 6 nt window (iii) identify HSRs as positive scoring regions over a threshold and (iv) weight the HSRs by their distance from the start of the intron using a geometric distribution. The final score is the sum of the weighted HSRs.
Modified introns
Introns with the desired sequence were synthesized (Epoch Biolabs, Sugar Land, TX, USA), verified by sequencing and inserted as PstI restriction fragments into a TRP1:GUS reporter gene, whose expression in single-copy transgenic lines was measured as described previously (18).
Detecting orthologs of A. thaliana genes
We identified orthologs of A. thaliana genes in several other plant species by using the BLASTP algorithm. Protein sequences for each Arabidopsis gene were searched against the proteomes of eight other plant species (using an expected cutoff value of 10−6). We then retained proteins that were the best reciprocal matches, and we only kept putative orthologs when the gene structure and number of exons/introns were comparable to that in A. thaliana.
Expression analysis
Information on gene expression from 14 Arabidopsis cDNA libraries was obtained from the Arabidopsis MPSS Plus database (http://mpss.udel.edu/at/). These libraries were constructed using mRNA from diverse tissues, and from various treatments of A. thaliana. Expression levels and the maximal rate of change in transcript levels (Rmax) from (20), were provided by Daniel C. Jeffares.
KEGG pathways and GO term analysis
GENECODIS (21) is a web server application (http://genecodis.dacya.ucm.es/analysis/) that generates a functional analysis from a user-specified list of genes. A statistical test is applied (hypergeometric distribution) to identify functional categories, and their combinations, that are significantly enriched in the specified list of genes relative to the set of all genes for that species. Arabidopsis genes from the IME+ category were analyzed for Gene Ontology (GO) terms and KEGG categories that were overrepresented. The GO term analysis was performed with the ‘level 3’ option. The result of the GENECODIS analysis consists of a list of A. thaliana annotations (or combinations of annotations), the functional categories that are overrepresented, and the corresponding P-values.
RESULTS
The highest IMEter scores are found in the 5′-ends of A. thaliana transcripts
To begin a whole-genome analysis of A. thaliana we split ‘confirmed’ genes (those required to have a full length cDNA) into two classes: IME+ and IME−. Genes in the IME+ class (2580 genes, 17 000 introns) have at least one intron with an IMEter score >20 (Supplementary Figure S1A). The IME− class contains all other genes (8083 genes, 42 260 introns). This threshold score of 20 corresponds to a predicted 5 × increase in expression (18). The majority (90%) of introns in the IME+ set are first introns.
The G + C content of all A. thaliana introns lies in a range of ∼20–50% G + C (Supplementary Figure S1B); IME+ introns display a slightly higher G + C content (mean G + C = 33.6%) than all introns (mean G + C = 32.4%). However, not all introns with a high G + C content exhibit a high IMEter score and there is no clear correlation between IMEter score and G + C content (Supplementary Figure S1C).
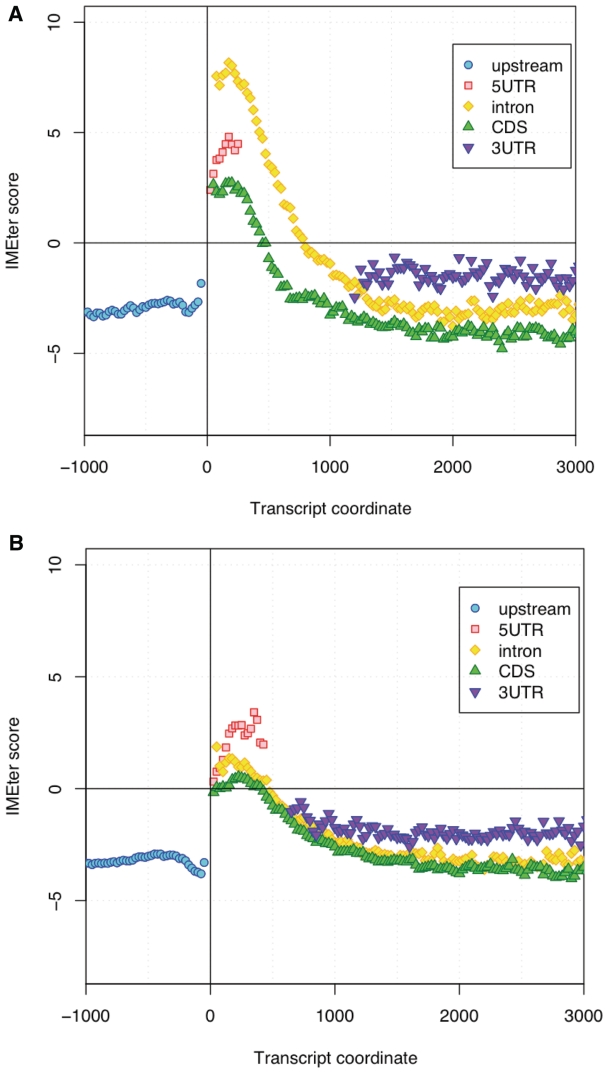
To investigate how IMEter scores are distributed across the length of each gene we computed IMEter scores in sliding windows moving both upstream and downstream of the TSS. This allowed us to observe IMEter score variation not just within introns, but also within intergenic sequences, untranslated regions and coding exons. Within the IME+ set of genes we find that regions of introns near to the TSS have the highest IMEter scores (Figure 1). These high scoring intronic regions are chiefly due to sequences present in first introns; second introns that are located at the same distance from the TSS as first introns have lower IMEter scores (Supplementary Figure S2). IMEter scores peak at a distance of ∼200 nt from the TSS and decrease to negative values at distances of 800–900 nt. These results support the previous observations that for an intron to enhance it has to be closer than 1000 nt to the TSS (17), and that average IMEter scores for introns decline with distance from the TSS (18).
Figure 1.
IMEter scores are highest at the 5′-end of transcribed regions. (A) IME+ data set (B) IME− data set.
Sequences from 5′-UTRs also have a relatively high IMEter score and these scores peak at the same distance from the TSS as introns (Figure 1). CDS likewise show an increase in IMEter scores in the same region, though to a much lesser extent. Upstream regions and sequences from 3′-UTRs appear uniform with respect to their IMEter score and remain negative on average. In the IME− set, IMEter scores in introns are much lower, but the same pattern of enrichment is still observable. The highest IMEter scores in the IME− set belong to 5′-UTRs and not introns, though the IMEter scores are lower than 5′-UTR sequences in the IME+ set. One reason for why IME− introns exhibited much lower scores is because any high scoring introns had already been partitioned into the IME+ set. Beyond distances of 1000 nt from the TSS, both data sets are indistinguishable on the basis of their IMEter scores. Overall, IME signals appear to be enriched in the 5′-region of transcripts and most abundant in first introns.
Reanalysis of experimental A. thaliana data reveal a punctate IME signal
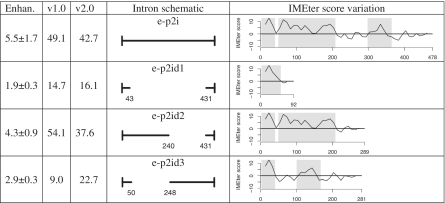
Several previous studies in A. thaliana have attempted to identify the IME signal by analysis of deletion- and hybrid introns. We reanalyzed these studies to determine if the IMEter could provide some insight into the results. Specifically, we used a new version of the IMEter which calculates scores using a sliding window approach. This can be used to reveal local variations in IMEter score in addition to calculating the IMEter score for the entire intron. In (22), the authors create several deletion constructs in introns from the profilin PRF2 gene. Compared to their deletion constructs which produce moderate increases in expression (1.9× to 4.3×), it is the wild-type intron with no deletions that produces the highest increase in expression (5.5× increase, Figure 2). However, the highest IMEter score occurs in the second deletion construct and not the wild-type intron (54.1 versus 49.1). Conversely, the first deletion intron, which removes the most sequence of any of the deletion constructs, showed the lowest increase in expression (1.9×) but did not have the lowest IMEter score. Running the IMEter with a sliding window approach reveals that the highest scoring IME region is located in the 5′-end of this intron (Supplementary Figure S3). The IMEter score is calculated from the entire intron sequence and so any non-enhancing regions (which might be expected at the 3′-end of long introns) can substantially lower the total IMEter score.
Figure 2.
IMEter analysis of PRF2 deletions. First column lists expression enhancement relative to an intronless control. Second and third columns show IMEter scores (using version 1.0 and 2.0) for each intron. Fourth column shows a schematic representation of the hybrid structure, with the original construct names provided. The final column shows IMEter score density computed in 50 nt windows, gray regions correspond to peaks of high IMEter score predicted by IMEter v2.0.
A similar situation exists in studies of the first intron of the petunia (Petunia x hybrida) actin-depolymerizing factor 1 (PhADF1) gene. This intron has been shown to induce strong and constitutive expression of that gene in vegetative tissues of transgenic A. thaliana. Three independent transgenic plants harboring single copies of each construct were analyzed along with various deletion constructs (23). The wild-type PhADF1 intron strongly enhances gene expression (7.1 × increase) but the IMEter produces a very negative score (−44.7) for this sequence (Supplementary Figure S4A). Only one of the deletion constructs produces a positive IMEter score, and in this construct approximately the last two-thirds of the wild-type intron sequence is removed. The wild-type intron is relatively long (>1500 nt) and most of the regions of this intron that produce positive IMEter scores are located in the first half of the sequence.
A series of six hybrid introns containing fragments of an enhancing intron (UBQ10 intron 1) within the context of an otherwise non-enhancing intron (COR15a intron 2) (18) revealed that while the enhancing sequences are distributed throughout the UBQ10 intron, the IMEter scores of the hybrid introns do not always accurately reflect their known level of enhancement. For instance, the hybrid introns CCUU and UCCC produce about the same level of enhancement in expression (5.3× and 5.1×, respectively), but their IMEter scores are very different (36.3 and 3.1, respectively). The sliding window approach reveals a good correlation between regions of high IMEter score and the presence of UBQ10 intron sequence (Supplementary Figure S4B).
We have also produced a series of systematic deletions of the UBQ10 intron; each of these deletions removes one of four separate regions of the intron (these regions are depicted in Supplementary Figure S4C). Additionally, we engineered a deletion construct that removes the middle two regions of this intron. The results from these deletions suggest that the IME effect is weakly additive. The wild-type intron strongly enhances (13.3× increase) but deletions to the 2nd, 3rd, 4th or middle regions of this intron all result in a drop in expression (10.1×, 8.2×, 11.4× and 5.9×, respectively). Interestingly, it is only the deletion of the 1st region of the UBQ10 intron that does not significantly alter expression (13.1×). This deletion construct has a lower IMEter score relative to the wild-type intron (75.1 versus 90.0).
IMEter 2.0
The results from the hybrid and deletion studies provide several examples where the IMEter score of an intron does not agree with the intron’s observed level of enhancement. One of the shortcomings of the IMEter is that the score is calculated from the entire intron. However, we expect sequences that are far away from the promoter to have less influence on expression compared with those that are near the 5′-end of the intron. Furthermore, the experimental results suggest that introns either enhance expression or they have no effect; we have not found any evidence that a spliceable intron can reduce expression below that of an intronless control. So the meaning of a negative IMEter score is questionable.
Based on these observations, we have developed a new version of the IMEter (v2.0) that addresses all of these issues. The new IMEter score for an intron is based only on the positive scoring regions (negative scoring regions are now ignored). Additionally, the contribution of positive scoring regions is now weighted depending on the distance of the region from the promoter (‘Materials and Methods’ section). This means that very high scoring regions towards the end of a very long intron will count far less towards the overall IMEter score than similar regions that occur near the start of an intron.
Overall, the new version of the IMEter is a much better predictor of how well any intron will enhance expression. For the introns depicted in Figure 2 and Supplementary Figure S4, there is now a much stronger correlation between their known level of enhancement and their IMEter score (r = 0.31 and r = 0.67; v1.0 and v2.0, respectively; Supplementary Figure S5). For the profilin deletion data, IMEter v2.0 now correctly awards the lowest IMEter score to the lowest enhancing intron and the highest IMEter score to the most enhancing intron (Figure 2). Similar improvements are also seen in UBQ10/COR15a hybrids (Supplementary Figure S4B).
Adding or removing specific sequences to introns can enhance or abolish the IME effect
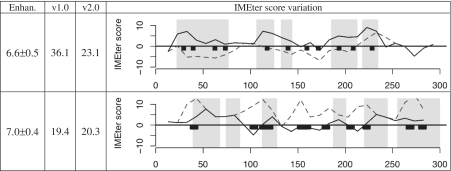
In order to determine if the IME signals were functional, we performed site-directed mutagenesis. The sequences of the enhancing UBQ10 intron and the non-enhancing COR15a intron were modified to create large alterations in IMEter score via minimal nucleotide changes. The sliding window version of the IMEter was used to reveal the location of the individual sequences within the UBQ10 intron that contribute most to its overall IMEter score (Figure 3). A total of 46 nt underlying the highest peaks were rearranged to reduce the IMEter score without changing the nucleotide composition of the intron. The six highest peaks in the UBQ10 intron have an overrepresentation of the pentamer CGATT, and these sequences were all converted to TACTG. The resulting intron had an IMEter score (v.2.0) of 20.3 and produced a 7.0 × increase in mRNA accumulation; this was substantially less than the wild-type version of the intron (IMEter score = 47.7, expression increase = 13.1×). We then proceeded to see whether this CGATT sequence could be used to raise the IMEter score of the poorly-enhancing COR15a intron. Modifications were made to 42 nt in order to add eleven new copies of this sequence. These changes led to an increase in mRNA accumulation from 1.7 × (wild-type intron) to 6.6×, and raised the IMEter score from 6.1 to 20.3 (Figure 3).
Figure 3.
IMEter analysis of introns with site-directed mutations. Top COR15a. Bottom UBQ10. Solid lines show IMEter score variation across the modified intron sequences, whereas dashed lines show IMEter score variation of the original introns. Black boxes indicate mutagenized regions. Gray regions denote peaks of high IMEter score predicted by IMEter v2.0. Enhancement values and IMEter scores refer to the modified intron sequences.
IME signals are conserved in rice
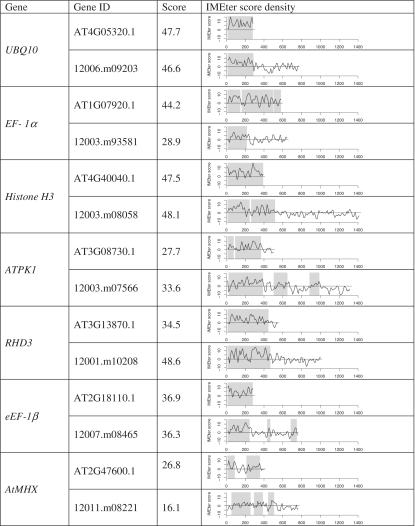
We have previously shown that an IMEter trained from rice (O. sativa) introns is effective in predicting the enhancement level of Arabidopsis introns (18). This is a little surprising because the 5′-ends of rice introns are much more GC-rich than Arabidopsis introns (Supplementary Figure S6). Rice introns are also much longer on average (418 nt compared to 167 nt). Although orthologous rice and Arabidopsis introns are generally too diverged to align to each other, we wanted to determine if there were any similarities that could be detected by IMEter v2.0. In our previous work, we described a set of 21 Arabidopsis introns that had been shown to enhance expression. We were able to unambiguously identify orthologs for nine of these introns in rice. Although most of the rice introns are much longer than their Arabidopsis counterparts, they contain very similar amounts of sequence with positive IMEter scores (Figure 4). On average, the pairs of introns differ in length by 437 nt, but the length of positive scoring regions differs by only 87 nt. Despite the differences in sequence composition and length, IMEter signals appear to be highly conserved at the 5′-ends of these introns.
Figure 4.
Comparison of IMEter scores in a set of orthologous O. sativa and A. thaliana introns. First two columns list the Arabidopsis gene name and gene identifiers for each pair of Arabidopsis (top) and rice (bottom) orthologs. Third column shows the v2.0 IMEter score for the whole intron. The final column shows the IMEter score density computed in 50 nt windows, gray regions correspond to peaks of high IMEter score predicted by IMEter v2.0.
IMEter signals are conserved across a wide range of plants
IME has been reported in at least 18 plant species and one study has reported that introns from a dicot species can still elevate gene expression if inserted into a monocot species (24). This suggests that IME signals might be conserved across different plant species. The availability of a number of sequenced plant genomes affords the opportunity to study the conservation of IMEter scores across a wide phylogenetic range. In addition to A. thaliana and O. sativa, other plant species with suitable genome data include grape (Vitis vinifera), a tree (Populus trichocarpa), a cereal (Sorghum bicolor), a lycophyte (Selaginella moellendorffii), a moss (Physcomitrella patens) and two algae (Chlamydomonas reinhardtii and Volvox carteri).
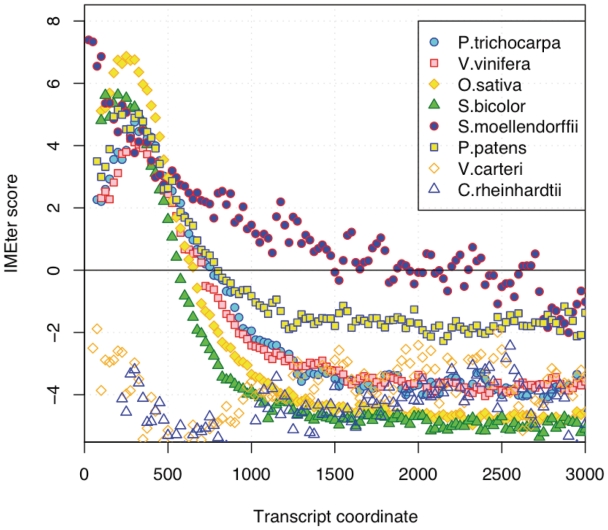
To explore IME conservation, we calculated IMEter scores in sliding windows for each intron of each genome. Scores were then averaged for windows at each increasing distance from the TSS. There is a striking correlation in the distribution of IMEter scores across the introns in nearly all of the species that were studied (Figure 5). For six of the eight species, we find that intronic IMEter scores all peak in the same region of the transcript and then decline to negative values further downstream. The two algal species are the main exceptions to this pattern with negative IMEter scores throughout the length of their transcripts. The other outlier is the lycophyte S. moellendorfii, the introns of which show the highest IMEter scores in the windows of sequence that are immediately adjacent to the TSS. They also continue to produce high IMEter scores over a much longer distance than in any other species.
Figure 5.
IMEter score distribution in introns from a range of plant species. Each point represents the average IMEter score (y-axis) for introns that start at a specified distance from the TSS (x-axis).
Since transcribed regions can have a biased nucleotide composition due to mutations in the DNA repair process (25), it could be argued that the observed distribution of IMEter scores is the result of mutation bias. Given the mostly uniform pattern of IMEter scores across the set of species, it might be expected that any biases in nucleotide composition might also be similar between species. However, we observe that there is considerable variation in nucleotide composition along the transcripts of the different species (Supplementary Figure S6). This would suggest that transcription biased mutation is not responsible for the similarities in the distribution of IMEter scores.
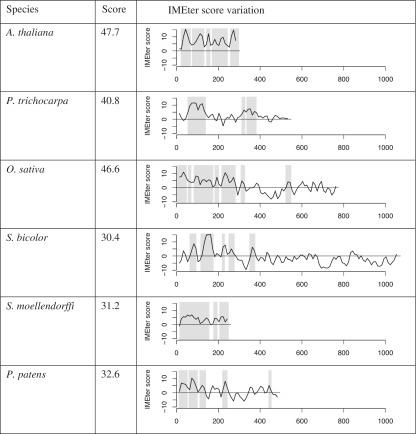
The results shown in Figure 5 indicate that IME signals are generally conserved among higher plants. To explore the conservation in greater depth, we examined orthologs of the first intron of the A. thaliana UBQ10 gene. We chose UBQ10 because it a well characterized and highly conserved gene. We were able to unambiguously identify the orthologs of this intron in six other plant species. We find that all of these orthologous introns produce a high IMEter score (>30) and all contain many peaks of high IMEter score (Figure 6). The species with short introns (A. thaliana and S. moellendorffi) have high IMEter scores throughout the intron, while species with longer introns tend to have high-scoring regions preferentially located at the 5′-end.
Figure 6.
IMEter scores of the first intron of UBQ10 orthologs in various plant species. Second column lists IMEter v2.0 scores for the whole intron. Third column shows IMEter score density. Gray regions denote peaks of high IMEter score predicted by IMEter v2.0.
Enhancing introns tend to be longer
We compared genes from the IME+ and IME− data sets to see if we could discern any useful properties of genes that have enhancing introns. One notable difference is that first introns appear much longer in the IME+ data set (38% longer on average). However, other metrics such as the lengths of transcripts, CDSs, exons and UTRs appear broadly similar between both data sets (Supplementary Table S1). More generally, there is a slight positive correlation between intron length and intron IMEter score (r = 0.463, n = 59 260). We expected that very long transcripts might require more IME signal. However, we did not find any correlation between the IMEter score of an intron and the length of the total transcript (Supplementary Figure S7).
Expression studies reveal the genes with strong IME signals are expressed in a variety of tissues at consistently high levels
To see whether the expression patterns of genes differ between the IME+ and IME− data sets, we used massively parallel signature sequencing (MPSS) experiments taken from seventeen A. thaliana mRNA libraries (26). The MPSS expression data reveals that genes from the IME+ data set tend to be expressed in more libraries than genes from the IME− data set (Supplementary Figure S8). The proportion of genes that were expressed in all seventeen mRNA libraries was twice as high in the high IMEter set than in the low IMEter set (31 versus 15%).
We also made use of Arabidopsis expression data that has been collected from a set of individual microarray experiments (20). These authors pooled data from separate time-course experiments that each measured the change in gene expression levels in response to various environmental stresses The pooled data records both the overall expression level of each gene as well as the maximal rate of change in transcript levels (as recorded by the ‘Rmax’ statistic). We extracted the list of genes from their data and then took the highest IMEter-scoring intron from each gene. We then divided this set of introns into four even-sized categories based on their IMEter score. There is a good correlation between expression level and IMEter score, and genes that have introns with the highest IMEter scores have the highest levels of expression (Supplementary Figure S9A). Introns with high IMEter scores also have significantly lower Rmax values than introns with low IMEter scores (Supplementary Figure S9B). This suggests that introns with high IMEter scores tend to belong to genes that have relatively constant levels of expression, even when being exposed to stress conditions. In contrast, introns with low IMEter scores belong to genes that are much more variable in their expression.
KEGG pathways and GO term analysis reveal that genes with strong IME signals tend to be housekeeping genes
To understand the functional aspects of genes affected by IME, we attempted to detect functional descriptors that were significantly overrepresented in the set of IME+ genes.
The GENECODIS tool (‘Materials and Methods’ section) identified several KEGG pathways (27) and Gene Ontology (GO) terms that were significantly enriched in the IME+ data set (Supplementary Table S2). The KEGG pathways ontology shows an enrichment in ubiquitin mediated proteolysis, oxidative phosphorylation, ribosome and proteasome and glycolysis. GO term analysis revealed that genes in the IME+ data set are enriched in binding, catalytic activity, structural molecule activity, transporter activity, translation regulator activity and molecular transducer activity. These data fit well with all the previous results in this section. House keeping genes tend to be widely, highly and constantly expressed.
DISCUSSION
Our previous work in A. thaliana focused primarily on the UBQ10 intron. Hybrid introns identified multiple regions that were sufficient for enhancing expression; it was therefore assumed that IME signals were dispersed. Several experiments in this study show that the previous interpretation was only partially correct. IME signals are discrete and also somewhat additive (Supplementary Figure S4C). Powerful enhancing introns therefore tend to have many IME signals. Short introns, such as in A. thaliana UBQ10, are densely packed with enhancing signals along their entire length. Genes with long introns, including orthologs of UQB10, show that IME signals are concentrated at the 5′-end of introns. After examining such introns, it was obvious that the IMEter 1.0 calculation, which based the score on the entire intron, was flawed. The improvements in the 2.0 version take into account both the discrete nature of the signals and their distance from the TSS. The IMEter 2.0 is available online from http://korflab.ucdavis.edu/.
IME+ introns are longer than IME− introns (Supplementary Table S1), and this mirrors the general trend that first introns are longer in the majority of species (28). It may be that first introns need to be longer in order to accommodate IME signals. Although little is known about IME signals outside of several plant species, it may be that animals and fungi also embed enhancing signals in first introns. Surprisingly, IME signals can also occur in the 5′-UTR or the CDS. Given the model that IME signals increase RNA polymerase processivity, it seems obvious in retrospect that enhancing signals could occur anywhere in the 5′-end of a transcript. The descriptive phrase ‘intron-mediated enhancement’ may therefore require modification some day. But since the majority of signals are in introns, and since the function of such signals outside of introns have yet to be verified experimentally, ‘IME’ is still an appropriate initialism.
The pentamer CGATT appears to be an important part of the IME signal. Experimentally manipulating an intron sequence to contain more of this sequence can turn a poorly-enhancing intron into an highly-enhancing intron (Figure 3). This pentamer is one of many pentamers used by the IMEter to score introns and it is the pentamer which shows the biggest difference in frequency between a set of promoter-proximal and promoter-distal introns. However, other sequences must be playing a role as the enhancing intron of the PRF2 gene does not contain any CGATT pentamers and yet it still enhances gene expression. Likewise, even though all CGATT pentamers were removed from the UBQ10 intron, it still enhances (7.0× compared to 13.1× in the wild-type intron). The CGATT pentamer appears similar to a potential IME-related motif that we previously identified (18), but other signals must also be present.
The IMEter was trained from intron sequences in A. thaliana and yet it appears to be useful when calculating IMEter scores in other species. For example, it can detect enhancing regions in a Petunia hybrida intron sequence (Supplementary Figure S4A), suggesting that the signals responsible for IME must be conserved to some level across different angiosperm species. The comparison between A. thaliana and O. sativa orthologs (Figure 4), and between orthologs of UBQ10 (Figure 6), reveals that although there is little conservation at the sequence level, the scores and locations of the high scoring IMEter regions are highly conserved. More striking evidence of the conservation of IME signals comes from the comparison of IMEter scores in the transcripts of eight different plant species (Figure 5). The pattern of IMEter scores in the introns of a moss (P. patens) appears similar to patterns in the introns of six different monocot and dicot species. This would imply that similar IME signals were present in the ancestor of mosses and vascular plants.
Introns with high IMEter scores tend to be found in genes that are expressed in many different tissues (Supplementary Figure S8). Analyses of expression data, KEGG pathways and GO terms suggest that genes associated with IME signals also tend to be highly-expressed housekeeping genes (Supplementary Figure S9 and Table S2). These results also agree with recent research that suggests that rapidly regulated genes are intron poor (20). The authors of this paper show that genes that undergo rapidly changing expression levels in response to external stresses contain significantly fewer introns. This suggests that introns can either delay regulatory responses, or that they contain signals to stabilize transcription. This latter possibility is further supported by data which show that highly expressed plant genes are typically longer and contain more introns than poorly-expressed genes (29). A link between intron number and expression has also been seen in a yeast species. The 3.8% of Saccharomyces cerevisiae genes that have introns account for 27% of all of the mRNAs (30). This suggests that many genes with introns tend to be highly transcribed and that introns play an important regulatory mechanism in gene expression.
Chromatin modifications that have been correlated with gene expression and whose localization within genes is largely restricted to or excluded from the first 1 Kb downstream of the TSS in Arabidopsis include methylation of histone H3 on lysine 4 or lysine 36 and DNA CpG methylation (31,32). The similarity in the distribution of these marks, IMEter scores and the positions from which introns can stimulate expression suggest that IME may affect, or be affected by, chromatin state.
The reason why enhancing signals occur predominantly in introns is probably because there is less functional constraint; introns are spliced in the nucleus and therefore do not impact the sequence of the mature mRNA. Although we do not yet know the specific mechanism of IME, the fact that the predicted signals are highly conserved and functional suggests that there is a common mechanism. Therefore, studying one system in detail should improve our understanding in all plants. Future work should focus on identifying the molecular players interacting with the signals. For example, although we assume that RNA polymerase is involved at some point, it is not yet known whether IME is mediated by DNA or RNA. While little is known about which specific macromolecules are responsible for IME, we are gaining insights into its ‘language’.
FUNDING
Cooperative State Research, Education and Extension Service, US Department of Agriculture, under Agreement No. 2006-35301-17072 (to A.R.); National Human Genome Research Institute 1RO1HG004348 (to I.K.). Funding for open access charge: National Institutes of Health grant; Institutional accounts.
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors will like to thank Kim Blahnik and Shahram Emami for useful suggestions concerning the design of the experiments.
REFERENCES
- 1.Sambrook J. Adenovirus amazes at Cold Spring Harbor. Nature. 1977;268:101–104. doi: 10.1038/268101a0. [DOI] [PubMed] [Google Scholar]
- 2.Buchman AR, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol. Cell. Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callis J, Fromm M, Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- 4.Duncker BP, Davies PL, Walker VK. Introns boost transgene expression in Drosophila melanogaster. Mol. Gen. Genet. 1997;254:291–296. doi: 10.1007/s004380050418. [DOI] [PubMed] [Google Scholar]
- 5.Palmiter RD, Sandgren EP, Avarbock MR, Allen DD, Brinster RL. Heterologous introns can enhance expression of transgenes in mice. Proc. Natl Acad. Sci. USA. 1991;88:478–482. doi: 10.1073/pnas.88.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mascarenhas D, Mettler IJ, Pierce DA, Lowe HW. Intron-mediated enhancement of heterologous gene expression in maize. Plant Mol. Biol. 1990;15:913–920. doi: 10.1007/BF00039430. [DOI] [PubMed] [Google Scholar]
- 7.Dean C, Favreau M, Bond-Nutter D, Bedbrook J, Dunsmuir P. Sequences downstream of translation start regulate quantitative expression of two petunia rbcS genes. Plant Cell. 1989;1:201–208. doi: 10.1105/tpc.1.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607–617. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rethmeier N, Seurinck J, Van Montagu M, Cornelissen M. Intron-mediated enhancement of transgene expression in maize is a nuclear, gene-dependent process. Plant J. 1997;12:895–899. doi: 10.1046/j.1365-313x.1997.12040895.x. [DOI] [PubMed] [Google Scholar]
- 10.Rose AB, Last RL. Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J. 1997;11:455–464. doi: 10.1046/j.1365-313x.1997.11030455.x. [DOI] [PubMed] [Google Scholar]
- 11.Clancy M, Vasil V, Hannah LC, Vasil IK. Maize Shrunken-1 intron and exon regions increase gene expression in maize protoplasts. Plant Sci. 1994;98:151–161. [Google Scholar]
- 12.Rose AB, Beliakoff JA. Intron-mediated enhancement of gene expression independent of unique intron sequences and splicing. Plant Physiol. 2000;122:535–542. doi: 10.1104/pp.122.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maas C, Laufs J, Grant S, Korfhage C, Werr W. The combination of a novel stimulatory element in the first exon of the maize Shrunken-1 gene with the following intron 1 enhances reporter gene expression up to 1000-fold. Plant Mol. Biol. 1991;16:199–207. doi: 10.1007/BF00020552. [DOI] [PubMed] [Google Scholar]
- 14.Zhang SH, Lawton MA, Hunter T, Lamb CJ. atpk1, a novel ribosomal protein kinase gene from Arabidopsis. I. Isolation, characterization, and expression. J. Biol. Chem. 1994;269:17586–17592. [PubMed] [Google Scholar]
- 15.Donath M, Mendel R, Cerff R, Martin W. Intron-dependent transient expression of the maize GapA1 gene. Plant Mol. Biol. 1995;28:667–676. doi: 10.1007/BF00021192. [DOI] [PubMed] [Google Scholar]
- 16.Ho SH, So GM, Chow KL. Postembryonic expression of Caenorhabditis elegans mab-21 and its requirement in sensory ray differentiation. Dev. Dyn. 2001;221:422–430. doi: 10.1002/dvdy.1161. [DOI] [PubMed] [Google Scholar]
- 17.Rose AB. The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J. 2004;40:744–751. doi: 10.1111/j.1365-313X.2004.02247.x. [DOI] [PubMed] [Google Scholar]
- 18.Rose AB, Elfersi T, Parra G, Korf I. Promoter-proximal introns in Arabidopsis thaliana are enriched in dispersed signals that elevate gene expression. Plant Cell. 2008;20:543–551. doi: 10.1105/tpc.107.057190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose AB. Intron-mediated regulation of gene expression. Curr. Top Microbiol. Immunol. 2008;326:277–290. doi: 10.1007/978-3-540-76776-3_15. [DOI] [PubMed] [Google Scholar]
- 20.Jeffares DC, Penkett CJ, Bahler J. Rapidly regulated genes are intron poor. Trends Genet. 2008;24:375–378. doi: 10.1016/j.tig.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317–W322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong YM, Mun JH, Lee I, Woo JC, Hong CB, Kim SG. Distinct roles of the first introns on the expression of Arabidopsis profilin gene family members. Plant Physiol. 2006;140:196–209. doi: 10.1104/pp.105.071316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong YM, Mun JH, Kim H, Lee SY, Kim SG. An upstream region in the first intron of petunia actin-depolymerizing factor 1 affects tissue-specific expression in transgenic Arabidopsis (Arabidopsis thaliana) Plant J. 2007;50:230–239. doi: 10.1111/j.1365-313X.2007.03053.x. [DOI] [PubMed] [Google Scholar]
- 24.Vain P, Finer K, Engler D, Pratt RC, Finer JJ. Intron-mediated enhancement of gene expression in maize (Zea mays L.) and bluegrass (Poa pratensis L.) Plant Cell Rep. 1996;15:489–494. doi: 10.1007/BF00232980. [DOI] [PubMed] [Google Scholar]
- 25.Touchon M, Arneodo A, d'Aubenton-Carafa Y, Thermes C. Transcription-coupled and splicing-coupled strand asymmetries in eukaryotic genomes. Nucleic Acids Res. 2004;32:4969–4978. doi: 10.1093/nar/gkh823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S. The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res. 2004;14:1641–1653. doi: 10.1101/gr.2275604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradnam KR, Korf I. Longer first introns are a general property of eukaryotic gene structure. PLoS ONE. 2008;3:e3093. doi: 10.1371/journal.pone.0003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren XY, Vorst O, Fiers MW, Stiekema WJ, Nap JP. In plants, highly expressed genes are the least compact. Trends Genet. 2006;22:528–532. doi: 10.1016/j.tig.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Ares M, Jr, Grate L, Pauling MH. A handful of intron-containing genes produces the lion's share of yeast mRNA. RNA. 1999;5:1138–1139. doi: 10.1017/s1355838299991379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nature Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 32.Luo C, Lam E. ANCORP: a high resolution approach that generates distinct chromatin state models from multiple genome-wide datasets. Plant J. 2010;63:339–351. doi: 10.1111/j.1365-313X.2010.04236.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.