Abstract
Human RNA polymerase (Pol) III-transcribed genes are thought to share a simple termination signal constituted by four or more consecutive thymidine residues in the coding DNA strand, just downstream of the RNA 3′-end sequence. We found that a large set of human tRNA genes (tDNAs) do not display any T≥4 stretch within 50 bp of 3′-flanking region. In vitro analysis of tDNAs with a distanced T≥4 revealed the existence of non-canonical terminators resembling degenerate T≥5 elements, which ensure significant termination but at the same time allow for the production of Pol III read-through pre-tRNAs with unusually long 3′ trailers. A panel of such non-canonical signals was found to direct transcription termination of unusual Pol III-synthesized viral pre-miRNA transcripts in gammaherpesvirus 68-infected cells. Genome-wide location analysis revealed that human Pol III tends to trespass into the 3′-flanking regions of tDNAs, as expected from extensive terminator read-through. The widespread occurrence of partial termination suggests that the Pol III primary transcriptome in mammals is unexpectedly enriched in 3′-trailer sequences with the potential to contribute novel functional ncRNAs.
INTRODUCTION
In eukaryotes, multiple specialized RNA polymerases (Pols) ensure proper transcription of the genome. In yeast and metazoans, three Pols share the task of transcribing rRNAs and tRNAs (Pols I and III), mRNAs (Pol II), snRNAs, snoRNAs and other non-protein-coding RNAs (ncRNAs) participating in a variety of cellular processes (Pols II and III). To date, most RNAs that intervene in regulatory processes such as RNA interference are thought to be produced by Pol II, but there is increasing evidence for the involvement of Pol III in the transcription of regulatory RNAs (1,2).
As the final step in the transcription cycle, proper termination is critical for successful gene expression. Primarily, transcription termination not only allows for transcript release and facilitates the recycling of RNA polymerases for further rounds of transcription (3), but it also ensures that other promoters are not perturbed by read-through polymerases from upstream genes. In all eukaryotes, the three nuclear RNA polymerases use different strategies to fulfill these requirements (4). Among these strategies, the one adopted by Pol III is undoubtedly the simplest, in spite of the fact that Pol III is the largest and the most complex among the three nuclear Pols (5). Pol III is unique in recognizing a simple run of thymidine residues on the coding DNA strand as a termination signal (6). Moreover, a facilitated reinitiation pathway, relying on proper termination, operates in the case of Pol III to ensure high transcript supply (7–9). The minimal signal thought to be sufficient for Pol III termination varies among different eukaryotes. T4 suffices for termination and is the most frequent signal in mammals, while yeast Pol III termination minimally requires five consecutive Ts, with T6 and T7 being the most frequent terminators (10–12).
Termination signal recognition is an intrinsic property of Pol III, even though high efficiency of termination (and reinitiation) may require the presence of accessory factors (4). In yeast, terminator recognition by Pol III requires the C37–C53 subunit subassembly, whose presence limits the processivity of Pol III elongation thus facilitating RNA release at poly(dT) tracts (13). Even though the Pol III terminator is one of the first transcriptional regulatory elements identified in eukaryotes, how such a simple signal can suffice for transcription termination is not known, and several exceptions to the general Pol III termination rule have been reported. In particular, read-through at canonical termination signals has been observed for several Pol III templates (14–16), and a few alternative elements, such as various combinations of two to three consecutive dT residues (17–20), long stretches of dAs (21) and potential hairpins at the 3′-end of transcribed regions (18), have sparsely been described in the literature as serving as Pol III termination signals. The sequence context in which the T-run is embedded was also shown to play a significant role in its recognition as a termination signal (6,12,20,22,23).
The tRNA 3′-ends are not generated by Pol III termination, but require cleavage of pre-tRNA 3′ trailing sequences, a process that might involve multiple endoribonucleases, including tRNase Z (24,25). The length of the 3′-trailing sequence of Pol III transcripts is dictated by the location and the strength of the termination signal downstream of the mature 3′-end. Distantly located terminators, or weak terminators allowing for substantial Pol III read-through, have the potential to generate pre-tRNAs with long 3′ trailers, whose processing to generate mature tRNAs could in principle couple tRNA maturation to the production of downstream encoded, functional RNA species. This strategy has been documented, for example, in the case of the Arabidopsis thaliana tRNA-snoRNA dicistronic coding units (26) and, more recently, for the tRNA-like-miRNA dicistronic units in the murine gammaherpesvirus genome, where Pol III recognition of strong terminators also proved to prevent the Drosha-independent biogenesis of downstream encoded viral miRNA. Collectively, these results suggested that strong or relaxed constraints at the termination level might actually play subtle roles in global small RNA biogenesis (27–29).
That at least four consecutive Ts are required for human Pol III termination is a widely employed rule for the prediction of Pol III transcript ends. However, the validity of such rule has never been addressed experimentally in a comprehensive way, and the termination signals naturally occurring downstream of Pol III-transcribed genes in the human genome have not yet been analyzed systematically. Based on previous analysis, T4 appears to be the most frequent tRNA gene terminator in mammalian genomes (12). We wondered whether other previously unrecognized signals, characterized by the presence of just T3 and/or T2 runs, can also regulate human Pol III termination. We found evidence for the widespread utilization of such signals, and for the frequent occurrence of Pol III terminator read-through potentially leading to the synthesis of a large set of novel ncRNAs.
MATERIALS AND METHODS
Computational identification of human tRNA genes
The 274 Saccharomyces cerevisiae tRNA genes were previously identified in ref. (30). The 675 human tRNA genes with a canonical T≥4 terminator within a 1-kb flanking region downstream of the tRNA coding sequence were identified with the software Pol3Scan (30,31), which was run on the human release 36 of the EMBL database. Based on the statistical analysis of tRNA promoter regions, and making use of weight matrices and weight vectors for scoring, the program discriminates between tRNA genes and related class III elements on the basis of the presence of a transcriptional terminator signal and of the base-pairing within the aminoacyl stem. To include in the analysis tRNA genes with distant terminators (up to 1 kb), looser constraints were imposed for the distance of a canonical termination site. Together with 675 tRNA gene candidates, the software identified more than 13 000 tRNA-like sequences (e.g. tRNA derived SINEs), which were discarded for having a combined A- and B-box score below the cut-off or insufficient base pairing in the aminoacyl stem. Among the tRNA gene candidates, 463 were also identified as tDNAs by tRNAscan-SE (32). All the tRNA genes selected for the in vitro transcription assay were validated as tDNAs by both tRNAscan-SE and RepeatMasker analysis (http://repeatmasker.org).
Plasmid construction
Using oligonucleotides listed in Supplementary Table S2, eight human tRNA genes and their 3′ trailers were PCR amplified from buccal cell genomic DNA with recombinant Taq DNA polymerase (Fermentas) and cloned into pGEM-T-Easy (Promega). The 7SLUAS-γHV68 tRNA4-miR-M1-5-like transcription unit for the experiment in Figure 4 (sequence reported in Supplementary Table S3) was purchased from Mr Gene GmbH (Regensburg, Germany) and subcloned into the NotI/XhoI sites of an AmpR → KanR version of pSuper (Oligoengine, Seattle, USA). All the mutants described in the text were generated by site-specific mutagenesis with either Pfu DNA polymerase (Promega) or Phusion DNA polymerase (Finnzymes), following the Dpn I-based QuickChangeTM protocol (Stratagene).
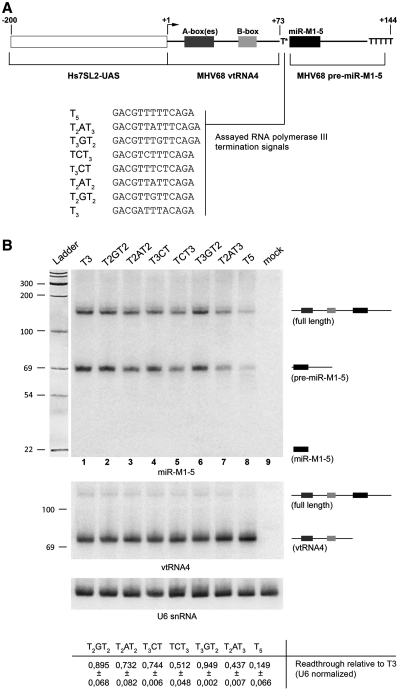
Figure 4.
In vivo transcription analysis of non-canonical terminators. (A) Schematic representation of the reporter constructs used to evaluate termination efficiencies in vivo. The upstream region of a human 7SL RNA gene (Hs7SL2-UAS) was fused to a viral tDNA-like region (γHV68 vtRNA4) separated from a pre-miRNA coding region of the same viral transcription unit (γHV68 pre-miR-M1-5) by a canonical or non-canonical termination signal (T*; tested sequence reported below). The positions of vtRNA4 internal promoter elements (B-box and multiple putative A-boxes) are approximately indicated. A backup T5 terminator was placed ∼70 bp downstream of T*. (B) Plasmid constructs carrying the indicated sequences as a T* terminator were transfected into HeLa cells, RNAs were extracted after 36 h and analyzed by northern blot using different probes, specific for miR-M1-5 (top panel), the upstream tRNA-like region (middle panel) and U6 snRNA as an internal standard (bottom panel). The migration positions of the various transcript species are diagrammed on the right, while those of RNA size markers run in parallel are reported on the left. Termination efficiency with each of the constructs was estimated from the total levels of the RNA species (primary transcript and pre miR-M1-5) whose synthesis requires read-through of the first terminator, normalized for the levels of U6 snRNA, and reported (below the images) as a fraction of the levels of read-through RNAs observed with the T3 element (assumed to be a totally ineffective terminator, thus allowing for maximal read-through). Reported values are the mean of two independent experiments, with the corresponding deviation. Mature miR-M1-5 RNA was not detected (even though the same procedure allowed to detect other miRNAs—data not shown) probably because it accumulates at very low levels (27).
For the construction of 3′-overhanged linear templates, equimolar amounts of the two oligonucleotides TRNAV18FF_for (5′-AGA CGA GCT CTG GGG GTG TAG CTC AGT GG) and TRNAV18FF_rev (5′-AAA AGA CCG GAA CTA AGT ATT C) were used to PCR-amplify (with Phusion DNA polymerase) from pGEM T-Easy-TRNAV18 (WT or containing the different non-canonical terminators), a linear fragment containing the SacI restriction site at its 5′-end followed by the TRNAV18 coding sequence. Amplicons were digested with Sac I, yielding 146-bp 3′-overhanged fragments that were gel-purified with QIAquick gel extraction kit (Qiagen) and used for transcription experiments.
In vitro transcription and processing reactions
All plasmids for in vitro transcription reactions were purified with the Qiagen Plasmid Midi kit (Qiagen). For promoter-dependent in vitro transcription, reaction mixtures (25 μl) contained 500 ng of template DNA, 70 mM KCl, 1 mM MgCl2, 1.5 mM DTT, 11.5% glycerol, 20 mM Tris–HCl pH 7.9, 5 mM phosphocreatine, 2 μg/ml alpha-amanitin, 0.4 U/μl SUPERase-In (Ambion), 40 μg of HeLa cell nuclear extract (33), 0.5 mM ATP, CTP and GTP, 10 μCi of [α-32P]UTP and either 0.025 or 0.25 mM UTP. Reactions were allowed to proceed for 25 min at 30°C before being stopped by addition of 75 μl H2O and 100 μl of phenol:chloroform (1:1). Purified labeled RNA products were resolved on a 6% polyacrylamide, 7 M urea gel and visualized and quantified with a Personal Imager FX and the Quantity One software (Bio-Rad).
For factor-free transcription assays, reaction mixtures (25 μl) contained 50 ng of linear DNA template, 60 mM KC1, 2 mM MgCl2, 1.5 mM DTT, 10% glycerol, 10 mM Tris–HCl pH 7.9, 50 μg/ml BSA, 0.32 U/μl SUPERase-In, 0.5 mM ATP, CTP and GTP, 0.025 mM UTP, 0.3 mM CpU dinucleotide primer, 10 μCi of [α-32P]UTP and 10 ng of human RNA polymerase III purified from a stable HeLa cell line expressing an FHM-tagged version of subunit RPC32, either alpha or beta isoforms, as described (34). Reactions were pre-incubated for 15 min at 30°C before addition of CpU and NTPs, then allowed to proceed for 25 min at 30°C before separating and visualizing the labeled RNAs.
Northern analysis
HeLa cells were maintained in Dulbecco’s modified Eagle medium (DMEM, Invitrogen) containing 5% fetal bovine serum and propagated in 5% CO2 at 37°C. Cells (3 × 106 on 150 mm plates) were transiently transfected with 12 μg of plasmid containing the tRNA4-miR-M1-5 expression cassette and 500 ng of the EGFP expressing vector pEGFP-C1, using the jetPEI™ kit (Polyplus Transfection) as described by the manufacturer. Thirty-six hours after transfection, total RNA extraction was performed with Trizol (Invitrogen) and RNA quality was verified by Agilent analysis. Northern analysis was performed as described (35), except that 20 μg of total RNA were resolved on a 11% denaturing polyacrylamide gel in TBE 1× and transferred onto Hybond-N+ membrane (GE Healthcare). Pre-hybridization and hybridization were performed at 30°C with 5× Denhardt reagent, 5× SSC, 0.5% SDS and 0.1 mg/ml salmon sperm DNA. The membrane was probed with 5′ 32P-radiolabeled oligodeoxynucleotides complementary to the γHV68 miR-M1-5, γHV68 tRNA4 and human U6 snRNA sequences (Supplementary Table S2). Data were recovered by phosphorimager exposition of the filters.
Analysis of viral transcription units
The mouse fibroblast cell line 3T3 (ATCC CRL-1658) was infected with the γHV68 strain WUMS (ATCC VR-1465), whose stocks were passaged, grown, and titer determined as previously described (36). The procedures for RNA extraction and 3′-end analysis, whose detailed description can be found in the Supplementary Data section, involved selection for 5′ triphosphate-containing molecules, that were then circularized and used as templates for inverse RT-PCR reactions with primer sets specific to either γHV68 pol III-1, pol III-4 or pol III-5.
Generation and analysis of Pol III ChIP-seq data
During analysis of Pol III ChIP-Seq data (37), we noticed frequent trailing of peaks in the 3′ flanking regions of tRNA genes. To analyze this phenomenon further, we first checked that the accumulation of tags within the tRNA 3′ flanking regions was above the general background observed in the genome. For this purpose, the 3 080 000 000 bp of the human genome assembly (NCBI36/hg18) were split into roughly 61 600 000 bins of 50 bp. All the bins corresponding to peaks detected by sissrs within the RNA coding sequence of annotated (or new) Pol III genes (37), as well as bins in simple repeats, SINE and LINE repeats, and satellite regions, were then eliminated from the analysis, which left 38 661 remaining bins. Of those, 207 had a tag score higher than 5, and 57 of them were at a maximum distance of 150 bp from a tRNA gene, 49 in the 3′ flanking region and eight in the 5′ flanking region. We concluded that the tag accumulation observed up- and downstream of tRNA genes was clearly above the general genome-wide background. Figure 5 was generated as detailed in the corresponding legend. It may be useful to compare the Pol III profile in Figure 5A with the one reported in Figure 3C of the previous ChIP-seq study (37). There, the pol III profile refers to peak maxima distribution relative to the transcription start site (TSS), and only one point was plotted for each gene. In contrast, Figure 5A in this work is based on cumulated tag number: the plot represents the cumulated number of plus and minus tags at each position around the TSS of 622 tRNA genes, giving a much more precise representation of the pol III profile in the region immediately downstream of the CDS.
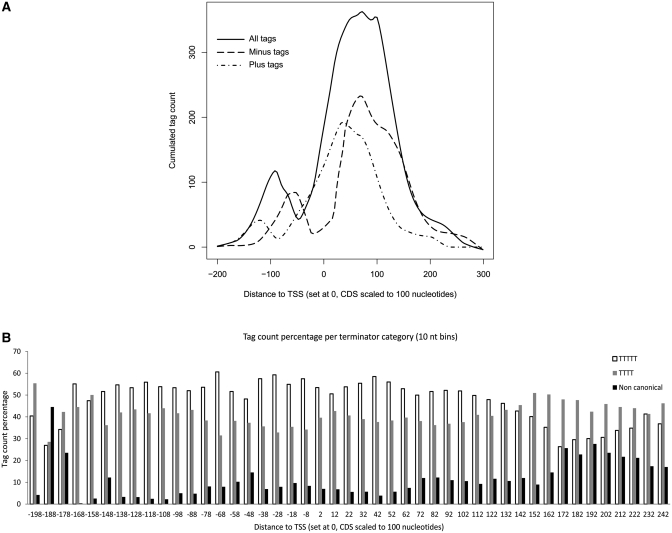
Figure 5.
Genome-wide analysis of Pol III association to the 3′-flanking regions of tRNA genes. (A) Tag accumulation over tRNA genes. The 622 tRNA genes listed in Supplementary Table S3 in ref. (37) were considered for the analysis. The transcription start site (TSS) was set at 0, and the RNA coding sequence of all tRNA genes was scaled to 100 nt. The tag count accumulation profiles were plotted using the lowess function in R (54). To generate the solid line (all tags), we first virtually generated the 60-nt fragments corresponding to each 36-nt tag and cumulated the central position for each of these fragments relative to the tRNA gene TSS (position 0). To generate the two different dashed lines, we cumulated the first positions of the plus tags and the last positions of the minus tags, respectively, relative to the TSS. (B) Tag accumulation over tRNA genes with different terminators. We considered three groups of tRNA genes containing various kinds of terminators (T≥5, T4 and non-canonical terminators) within 50 nt past the 3′ end of the RNA coding region. The graph displays total tag (as represented by the central position of the corresponding 60-nt fragments) counts per 10-nt bins relative to the position of the TSS.
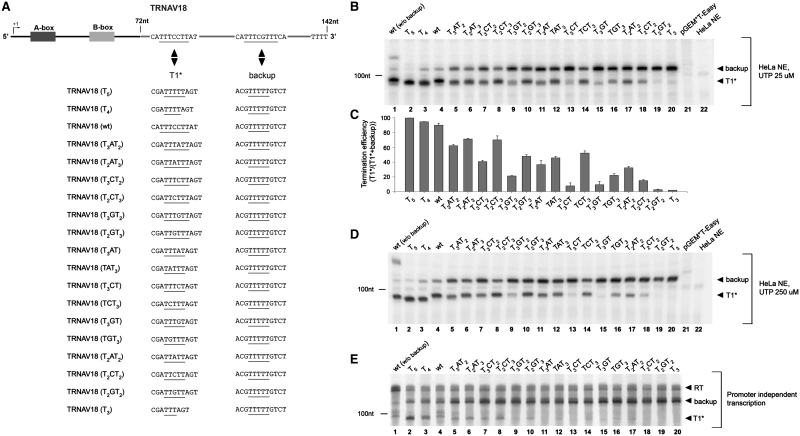
Figure 3.
Mutational analysis of non-canonical terminators. (A) Schematic representation of TRNAV18-derived templates. The first two termination signals in the 3′-flanking region of TRNAV18 were replaced by the non-canonical termination sequences under analysis (T1*) and a backup terminator, respectively. Termination signals are underlined. (B) TRNAV18-derived templates, carrying the indicated sequences at the T1* terminator position, were transcribed in vitro using a HeLa cell nuclear extract in the presence of a 25 µM UTP concentration. The migration positions of TRNAV18-derived transcripts ending either at the first non-canonical terminator (T1*) or the backup terminator (backup) are indicated on the right. The migration position of a 100-nt RNA size marker run in parallel is indicated on the left. (C) Evaluation of termination efficiencies observed in the experiment in (B), conducted in triplicate. Termination efficiency with each construct is expressed as the ratio between the amount of T1*-terminated transcript versus total (T1* + backup) transcripts. Error bars refer to the standard error of three independent measurements. (D) In vitro transcription reactions were conducted as in (A), but in the presence of a 250 µM UTP concentration. (E) Transcription was initiated by immunopurified Pol III containing the beta isoform of RPC32 subunit (34), in the absence of other transcription proteins, by using TRNAV18-derived, 3′-overhanged linear templates containing the same T1* and backup termination sequences reported in (A). The migration positions of transcripts terminated at either T1* or the backup terminator, and run-off transcripts (RT), are indicated on the right. The migration position of a 100-nt RNA size marker run in parallel is indicated on the left. Very similar results were obtained using immunopurified Pol III containing the alpha isoform of RPC32.
RESULTS
RNA polymerase III canonical terminators for human tRNA genes are unexpectedly distant from the discriminator nucleotide
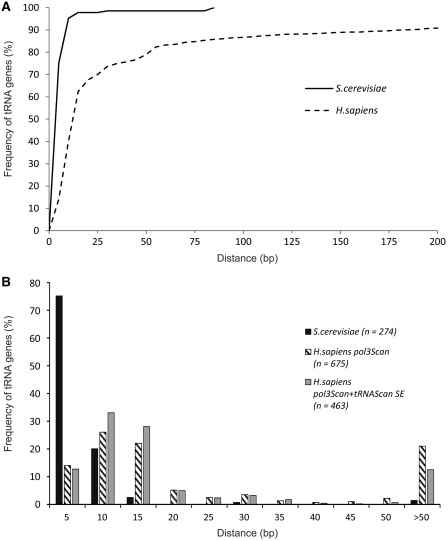
Based on previous statistical analyses, the yeast tRNA gene (tDNA) terminator can be described as a run of five or more T residues always starting within the first 40-bp downstream of the end of the mature tRNA coding sequence, with the most frequent signal being T6–7 (12). The same analysis revealed that, in contrast, a run of only four Ts is by far the most frequent terminator in mammals and that, unexpectedly, a number of tDNAs lack a recognizable terminator within 40 bp of 3′-flanking region. This point was more directly assessed by analyzing the distribution of the positions of Pol III canonical terminators in the 3′-flanking regions of S. cerevisiae and human tDNAs. To this end, T≥5 (for yeast tDNAs) or T≥4 (for human tDNAs) canonical termination signals were sorted in 5-bp bins covering a 1-kb flanking region downstream of each tDNA. For each bin, the percentage of tDNAs containing a correspondingly located canonical terminator was plotted as a function of the distance from the last nucleotide of the mature tRNA sequence and represented with a cumulative distribution function. As shown in Figure 1A, >97% of S. cerevisiae tDNAs have a canonical terminator within the first 15 bp of 3′-flanking region, thus pointing to strict termination requirements immediately downstream of the tRNA coding region. The same analysis was then conducted on an inventory of 675 tDNAs identified in the human genome by Pol3Scan (30,31). This search algorithm was initially employed, instead of the more widely used tRNAscan-SE (32), because Pol3Scan makes explicit predictions about the presence and position of Pol III terminators. The search algorithm could easily be adapted to our purpose by removing any requirement for a maximal distance of the first T≥4 stretch. In the human tDNA set, the percentage of genes with a canonical terminator within the first 15 bp amounts to 50–60% only, with the curve approximating a plateau much more slowly than for yeast tDNAs. As further evidenced by the histogram plot in Figure 1B, and in contrast to S. cerevisiae, human tDNAs are surprisingly enriched in canonical terminators located at ≥50 bp from the 3′ end of tRNA coding sequence (>20% of the set, corresponding to ∼120 individual transcription units), suggesting that human Pol III transcription extends into a broader region downstream of tRNA genes. The Pol3scan-based sample was further screened with tRNA-scan SE, producing a set of 463 bona fide tRNA genes. The analysis of this set still revealed at least 60 genuine tRNA genes (∼12% of total) with a remote terminator (Figure 1B).
Figure 1.
Computational analysis of tDNA terminator position. (A) Cumulative distribution of canonical terminators in the 3′-flanking regions of 274 S. cerevisiae and 675 H. sapiens tRNA genes, identified with the software Pol3Scan (30,31). Numbers on the x-axis indicate the distance (in bp) of the first T of the terminator element from the end of the tRNA coding sequence. Distances were clustered in 5-bp bins. Canonical terminators are defined as a T≥4 or T≥5 stretch for H. sapiens and S. cerevisiae, respectively. The chart does not take into account terminators located at >200 bp, which represent ∼9% of the total number (the original Pol3Scan analysis included terminators located up to 1 kb from the reported 3′-end of the tRNA coding sequence). (B) The same data used for the plot in (A) were chartered in a histogram which depicts the frequency distribution of canonical terminators in each 5-bp bin. In the last bin (>50), all termination sites located at a distance >50 bp (and <1 kb) are subsumed. The analysis was also conducted on a subset of 463 tDNAs that were recovered with both Pol3scan and tRNAscan-SE search algorithms.
Human RNA polymerase III recognizes novel non-canonical terminators in vitro
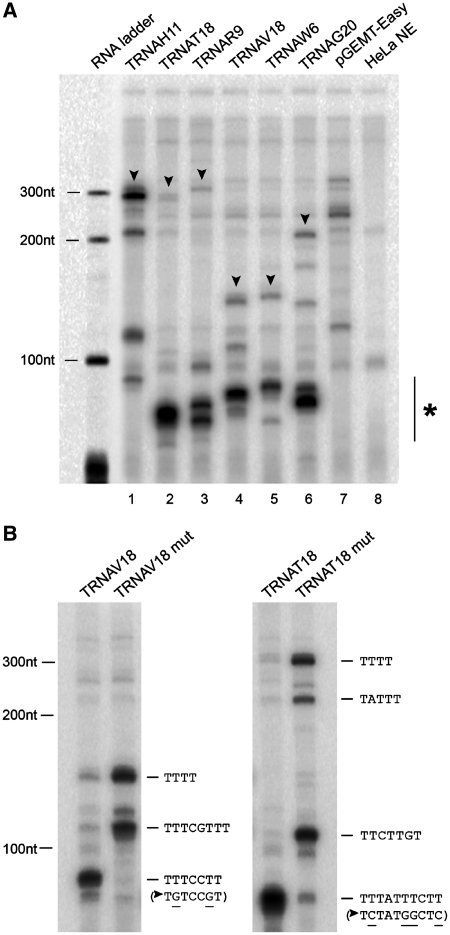
Six representative human tRNA genes, displaying the closest T≥4 at different (>50 bp) distances from the 3′ end of tRNA coding sequence, were cloned and transcribed in vitro using a HeLa cell nuclear extract (the sequences of these templates and of the corresponding 3′-flanking regions are reported in Supplementary Table S1). As shown in Figure 2A (lanes 1–6) these templates were efficiently transcribed by Pol III, but in no case was a single RNA product observed, as would be expected for full and exclusive termination at the canonical T≥4 sequence. Rather, the observed transcripts patterns could be explained by assuming that termination can occur in vitro at some of the many T-rich elements occurring upstream of the canonical terminator in all tDNA 3′-flanks (sequences underlined in Supplementary Table S1). In particular, it seemed possible that some of these putative signals are very active as terminators, thus leading to transcript patterns dominated by a single, tRNA-sized transcript (e.g. lanes 2 and 6), while others allow for more consistent read-through and thus for heterogeneous transcript patterns with a significant representation of longer RNAs (lanes 1 and 3–5). Alternatively, tRNA-sized RNAs could derive from processing of longer pre-tRNAs; pre-tRNA processing or degradation, however, were practically absent under our in vitro transcription conditions (see below and Supplementary Figure S1 for more details). To directly test the hypothesis of premature termination as the main source of tRNA-sized transcripts, we introduced point mutations into the first putative termination element of the TRNAV18 and TRNAT18 transcription units, and analyzed their effects on in vitro transcription patterns. As shown in Figure 2B, when the putative non-canonical terminators were mutated to T-poor elements, the shorter predominant transcripts disappeared, and transcription produced several long read-through transcripts terminated at the downstream predicted (non-canonical or canonical) termination sites, thus suggesting that the smaller than expected RNAs in Figure 2A derive from early termination rather than pre-tRNA processing. Altogether, the above data show that: (i) natural human tRNA genes can be transcribed into pre-tRNAs with unusually long 3′-trailers; (ii) several types of dT-rich sequences downstream of human tDNAs can act as non-canonical termination signals for Pol III.
Figure 2.
In vitro transcription analysis of tRNA genes with a distanced canonical terminator. (A) In vitro transcription reactions were performed in HeLa nuclear extract using 0.5 µg of the indicated tDNA templates (lanes 1–6), the empty vector (lane 7) or no DNA (lane 8). The size of the transcripts in each lane is generally consistent with termination at the putative non-canonical sites underlined in Supplementary Table S1. The vertical line with asterisk on the right indicates the migration position range of RNA products whose deduced size corresponds to pre-tRNAs with no or very short 3′ trailer (referred to as ‘tRNA-sized’ in the text). The bands whose migration positions correspond to the ones predicted for transcripts terminated at the canonical signals (see Supplementary Table S1) are indicated by arrowheads on the gel image. An end-labeled RNA ladder was run in parallel. (B) In vitro transcription of TRNAV18 and TRNAT18 wild-type templates (left lane in each panel) and their respective mutants in the closest non-canonical termination element (right lane in each panel). Indicated on the right of each panel are the migration positions of transcripts terminated at the different non-canonical or canonical sites (predicted from the sequences of the 3′-flanking regions reported in Supplementary Table S1), together with the sequence of the termination signals (and of their mutant variants, denoted by arrowheads).
Biochemical analysis of non-canonical terminators
The T-rich elements that appear to induce termination in the above examples are T3CCT2 (TRNAV18, in which this element is further followed by the T-rich stretch ATATCTAT) and T3AT3CT2 (TRNAT18). These are only two out of an enormous number of possible dT-rich sequences that could function as Pol III terminators. Since an exhaustive investigation of the sequence requirements for human Pol III termination appeared too arduous a task, we chose to select a significant, yet easily testable set of putative non-canonical terminators for further biochemical analysis. More specifically, we decided to concentrate on all possible combinations of the following types: T3VT2, T2VT3, T3VT, TVT3 (V = A or C or G), characterized by the presence of three consecutive T residues separated from one or two T residues by any non-T nucleotide, and T2VT2, which we considered as the most degenerated signal for termination based on previous literature (19,38). Our choice was supported by the observation that at least one of these putative termination signals occurred upstream of T≥4, and within the first 50 bp of 3′-flanking region, in the majority of tRNA genes with a distant canonical terminator. To analyze termination efficiency, we first engineered a series of expression plasmids containing mutated versions of TRNAV18 in which the TTTCCTT sequence, inducing termination in the native context, is replaced by the sequences to be analyzed for their termination capacity, including T4/5 and T3 as positive and negative controls for termination, respectively (Figure 3A). To avoid as much as possible any contribution to termination efficiency from neighboring sequences, we embedded the putative terminator elements within a sequence context in which no T residues are encountered in the first two positions either upstream or downstream of the tested sequence. We also introduced a strong back-up terminator (T5) in place of the native TTTCGTTT sequence in the 3′-flanking region of TRNAT18, to have downstream terminated transcripts in case of read-through at the non-canonical sites. All these constructs were tested as Pol III templates in vitro in the presence of either 25 or 250 µM UTP (Figure 3B and D). As expected, Pol III termination at the canonical T4 and T5 sites (lanes 2 and 3) was very efficient, with read-through products being absent (T5) or very poorly represented (T4), while an isolated T3 motif (lacking any other dT residue in the first two upstream or downstream positions) was almost completely inactive as a terminator (Figure 3B and D, lane 20). The absence of shorter than expected RNA products with the T3 template indicates that no significant processing of the 3′’-trailers of primary transcripts occurs under the employed reaction conditions. This point was further verified by pulse-chase in vitro transcription experiments and by northern hybridization analysis of in vitro transcription products with probes specific for downstream sequences (Supplementary Figure S1).
The termination efficiency at each of the T3VT2, T2VT3, T3VT, TVT3 and T2VT2 motifs was lower than at the canonical T4/5, but higher than at isolated T3 [with the exception of T2GT2 (lane 19) that like T3 was almost completely inactive]. These sequence elements thus all behaved as sub-optimal terminators. The actual termination efficiency, however, was found to vary greatly among the different variants, according to two general principles. First, motifs of the type T3VT2/T2VT3 (which can be viewed as interrupted T5 elements) tend to be more efficient terminators than motifs of the type T3VT/TVT3 (interrupted T4 elements maintaining a T3), that in turn tend to be more efficient terminators than T2VT2 motifs (lacking any T3). Second, within each of the asymmetric types of motifs (T3VT2/T2VT3 and T3VT/TVT3), the variant in which T3 comes at the end of the motif is invariably a much better terminator than the variant having the same base composition but opposite orientation, with T3 at the start of the motif. Compare, for example, lane 9 (T3GT2) with lane 10 (T2GT3), or lane 13 (T3CT) with lane 14 (TCT3) in Figure 3B and D. About 50% termination efficiency was observed with TCT3, but termination dropped to <10% with the oppositely oriented T3CT. Another interesting observation emerging from the data in Figure 3 is that significant termination can also take place at two of the ‘minimal’ T2VT2 motifs, T2CT2 (15% termination) and, even more clearly, T2AT2 (32% termination). It should be pointed out that the tested terminators represent a minimal set, and that the effect of the sequence context on termination efficiency was not explored. We anticipate that many more combinations of T2/T3 runs, especially if contiguous to generally T-rich sequences, will act as Pol III terminators (as exemplified by the T3CCT2 and T3AGT2 downstream of TRNAV18).
The efficiency of Pol III termination is known to be affected by UTP concentration, with terminator read-through being favored at higher UTP concentrations (12,39). When the UTP concentration in the in vitro termination assays was raised from 25 to 250 µM, closer to the estimated intracellular concentrations of UTP in mammalian cells [0.567 ± 0.460 mM; (40)], only a slight general decrease in the absolute termination efficiencies was observed. Under these conditions, however, the differences between oppositely oriented asymmetric motifs turned out to be accentuated: for example, substantially no termination was observed with T3GT, while the corresponding oppositely oriented variant gave ∼30% termination (cf. lanes 15 and 16 in Figure 3D).
Strictly speaking, what was assessed by the above analyses should be referred to as terminator site recognition, rather than termination, as the termination process also includes transcript release, a step that we did not directly address. It thus remains possible that the RNA species which we consider as the products of non-canonical termination actually represent non-released transcripts that are part of stalled complexes induced by the T-rich non-canonical signals. If that was the case, however, the comparable levels of transcripts deriving from canonical and non-canonical signal recognition could only be explained by assuming that no more than a single round of transcription occurs during in vitro transcription reactions. Such an assumption contrasts with the high extent of transcription reinitiation typical of the Pol III system (8,9).
To address whether human RNA polymerase III itself is able to discriminate among the different non-canonical terminators, we carried out promoter- and factor-independent transcription assays on 3′-overhanged templates containing the entire set of non-canonical terminators. As expected on the basis of previous analyses of Pol III initiation on tailed templates (23,41), overall termination efficiency at both canonical and non-canonical signals was seriously reduced under these conditions (in particular, terminated transcripts were almost undetectable at terminators of the T3VT, TVT3 and T2VT2 types). Mechanistically, the lower termination efficiency observed in this experiment may reflect, at least in part, an unknown level of hybrid formation due to reduced RNA strand displacement on tailed templates (42). Despite these limitations, the relative termination efficiency profile by purified Pol III appeared to be maintained in the absence of other nuclear proteins (Figure 3E), in agreement with the early observation that Pol III itself is able to discriminate between weak and strong terminators (22).
The experiments in Figure 3 are based on the introduction of putative terminator elements in the same sequence context, the TRNAV18 3′-flanking region. To verify that the non-canonical terminators work similarly when they occur in their native tDNA context, we tested by in vitro transcription two human tDNAs (TRNAH3 and TRNAL12) displaying in their 3′-flanking regions supposedly weak (T3CT) and strong (T3AT2) non-canonical terminators, respectively (Supplementary Figure S2). Pol III terminated at these natural sites with relative efficiencies comparable to the ones observed with engineered TRNAV18 templates in Figure 3.
Non-canonical Pol III termination in mammalian cells
To assess whether non-canonical Pol III termination also occurs in vivo, we constructed a series of dicistronic units closely resembling the pol III-4 locus of murine gammaherpesvirus 68 (γHV68) (27,29) (Figure 4A). The wild-type viral transcription unit was modified by insertion of a strong termination signal (T5) immediately downstream of the pre-miR-M1-5 sequence and replacement of the 5′-flanking region of tRNA4 (a tDNA-like element including a functional internal type-2 promoter) with the upstream region of a human 7SL RNA gene which was shown to enhance expression of type-2-promoter class III genes in vivo (43,44). The T5, T2AT3, T3GT2, TCT3, T3CT, T2AT2, T2GT2 and T3 sequences were inserted into this termination reporter plasmid, between the tRNA-like and the pre-miRNA coding moieties of the template. If termination occurs at this position, no synthesis of the full-length primary transcript, nor of the pre-miRNA, should be observed. The constructs were transfected into HeLa cells, and transcription products were analyzed by northern blot using probes specific for either the upstream tRNA-like or the downstream pre-miRNA portion. As shown in Figure 4B, the maximal level of readthrough transcripts (represented by the ∼150-nt primary transcript and by the ∼70-nt pre-miRNA derived from it) were observed with the T3 construct (lane 1), for which no termination is expected to occur between the upstream tRNA-like portion and the downstream pre-miRNA moiety of the reporter gene. Quite unexpectedly, detectable levels of readthrough transcripts were also produced from the T5 template (lane 8), suggesting that significant readthrough can also occur at canonical terminators in vivo, at least within the sequence context of this construct. Intermediate termination efficiencies were observed at the non-canonical signals, with the exception of T2GT2 (lane 2) that, similar to its behavior in vitro, produced very poor termination, and, unexpectedly, T3GT2 (lane 6). In general, the differences in termination efficiencies among the non-canonical signals closely matched the ones observed in vitro, with T2AT3 terminating better than T3GT2, and TCT3 imposing significantly stronger termination that the inversely oriented T3CT (cf. lanes 4 and 5). These data demonstrate that termination at non-canonical signals, as well as read-through at signals predicted to be strong terminators, can occur in vivo.
To address how widespread non-canonical Pol III termination is, we took advantage of high-resolution Pol III ChIP-seq datasets, which allowed us to measure the extent of Pol III association with tDNA 3′ trailer regions on a genome-wide scale. In particular, we exploited a dataset of sequence tags generated by ultra high-throughput sequencing of human genomic DNA co-immunoprecipitated with Pol III-specific antibodies directed against the POLR3D subunit (37). These data consist of 36-nt sequence tags corresponding to the end of fragments which were on an average 60-nt long. For each tag, we first virtually generated the corresponding 60-nt fragment and cumulated the central position for each of these fragments relative to the tRNA gene TSS (position 0) to generate the solid line shown in Figure 5A. Note that in this graph the length of the tRNA coding sequences of all tRNA genes was normalized to 100. We observed a major peak over the tRNA genes, as well as a minor peak centered at about position −100, which is discussed further below. Strikingly, the major peak was not centered on the tRNA coding region, but displayed a 3′ tail extending up to 180-nt past the end of the tRNA coding sequence, suggesting Pol III occupancy far downstream of the region corresponding to the end of mature tRNA. To analyze this phenomenon further, we considered the upper strand (or plus) and lower strand (or minus) tags, relative to the direction of transcription, of the various tRNA genes separately. Thus, the plus tags correspond to the fragment ends located upstream of tRNA-transcribing Pol III, whereas the minus tags correspond to the fragment ends located downstream of tRNA-transcribing Pol III. As shown in Figure 5A, which shows the cumulated first positions of the plus tags and last positions of the minus tags relative to the TSS, the plus tags are enriched upstream and around the TSS, as expected, whereas the minus tags are enriched in downstream regions, as far as 180-nt downstream of the end of the tRNA coding sequence. Significantly, however, plus tags were found past position +100 downstream of the end of the tRNA coding sequence, indicating that polymerases were present as far downstream as 160-nt downstream of the tRNA coding region (considering that the average size of sequenced fragments was 60 nt).
The minor peak in the 5′ flanking region seems to correspond to either a paused enzyme or a very short transcription unit located just upstream of many tRNA genes. We have not pursued this information further, but we note that such short transcription units could be divergent relative to the tRNA genes (since the tags were considered plus or minus tags relative to tRNA genes, whose direction of transcription is known). This possibility is reminiscent of the observation that DNA-bound yeast TFIIIB is capable of directing Pol III transcription in both directions, at least in vitro (45). Although we have no data suggesting that the independent binding of TFIIIB to the TATA box plays a role in this phenomenon as shown in yeast, we speculate that bi-directional transcription might also occur at human tRNA genes.
To evaluate the contribution of non-canonical terminator read-through to Pol III 3′ trailing, all tRNA genes were divided into three subsets characterized, respectively, by having T≥5, T4 and non-canonical termination signals within the first 50 nt following the tRNA coding sequence. The tag count percentage contributed by each subset at the different positions around the tRNA coding sequence (10-nt bins) was then computed and plotted (Figure 5B). Strikingly, the contribution of the ‘non-canonical terminator’ tDNA set to cumulated tag counts was much more pronounced for the most downstream positions (from ∼170 downwards, corresponding to distances higher than ∼70 nt from the 3′ end of tRNA coding sequence), thus suggesting that Pol III 3′-trail signals in the ChIP-seq data might indeed be due to widespread terminator read-through by Pol III, which is more prominent at non-canonical terminators but, quite unexpectedly, also relevant at canonical termination signals. The data in Supplementary Figure S3 illustrate partial 3′ trailing and transcription past non-canonical terminators at two individual tRNA genes, TRNAQ22 and TRNAH3.
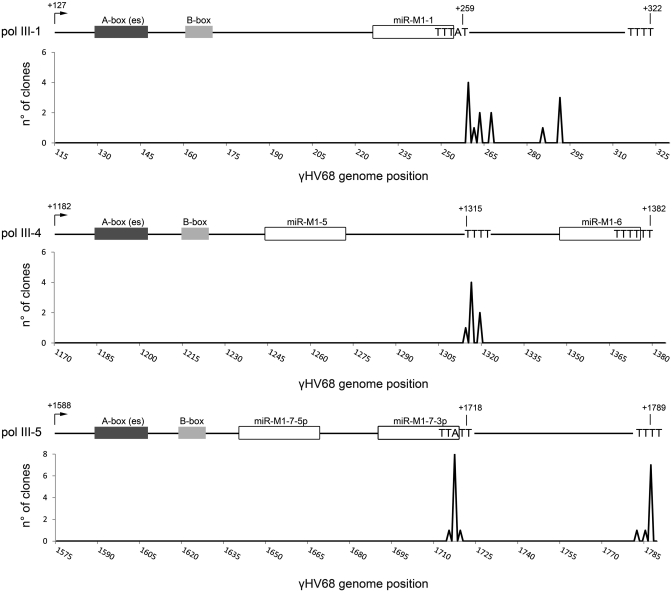
Finally, to gain further evidence for the occurrence of non-canonical Pol III termination in mammalian cells, we concentrated on some of the Pol III-dependent miRNA transcription units recently described in the genome of murine virus γHV68 (27–29). In particular, non-canonical terminators are predicted in the pol III-1 and pol III-5 transcription units, just downstream of the miR-M1-1 and miR-M1-7-3p, respectively, while a canonical T4 terminator is present downstream of miR-M1-5 in the pol III-4 transcription unit (Figure 6). We and others previously analyzed the γHV68 pol III transcripts by northern blotting RNA from infected cells. These analyses demonstrated the substantial use of an alternate terminator at the base of the first stem loop (band of ∼135 nt), as well as frequent read-through of the full-length transcript to the canonical terminator (band of ∼200 nt) [cf. Figure 1C in ref. (27) and Figure 1B in ref. (28)]. These dual products resulting from alternative termination (bands of ∼135 and 200 nt) were also detected by northern analysis of the viral pol III-4 and pol III-5 transcripts during virus infection (Supplementary Figure S4). To more accurately define the termination sites within these transcripts revealed by northern blot, we used ligation mediated RT-PCR and cloning. To this end, total RNA was extracted from cultured mouse 3T3 cells 36 h after infection with γHV68, and then subjected to biochemical modification of the 5′-end and circularization of RNA templates, followed by RT-PCR product cloning and sequencing. As illustrated in Figure 6, sequences indicate that the non-canonical transcription termination signals in pol III-1 and pol III-5 are recognized by mouse Pol III, as is the canonical T4 in the Pol III-4 transcription unit. As the 3′-end of the transcripts were mapped exactly in correspondence of the predicted termination sites, it is highly unlikely that these ends were generated by maturation of longer transcripts. In vitro and in vivo expression analysis of another γHV68 Pol III-miRNA transcription unit, pol III-2, further suggested that a TCT3 non-canonical terminator, located between the miR-M1-2 and miR-M1-3 coding sequences, is partially active and thus has the potential to modulate the synthesis of a downstream encoded miRNA (Supplementary Figure S5).
Figure 6.
Mapping the 3′-end of pol III-1, pol III-4 and pol III-5 transcripts of γHV68. Total RNA from γHV68-infected cells was treated to select for 5′ triphosphate containing molecules that were then circularized and used as templates for inverse RT–PCR reactions with primer sets specific to either γHV68 pol III-1, pol III-4, or pol III-5. Amplified RT–PCR products were cloned into the pCR4-TOPO vector. Individual colonies were then grown for plasmid DNA recovery and sequencing. The mapped transcriptional stop sites of the triphosphate containing RNA molecules of γHV68 pol III-1, pol III-4 and pol III-5 are shown. Histograms demonstrate the number of sequences ending at each genome position indicated. Above each histogram is a linear representation of each γHV68 pol III transcript with grey boxes representing the locations of Pol III promoter elements, white boxes representing the locations of the viral miRNA coding regions, and right-pointing arrows marking the TSS.
Altogether, the results of in vivo analyses suggest that two types of non-canonical Pol III termination occur in mammalian cells: efficient termination at non-canonical signals and incomplete termination at canonical signals.
DISCUSSION
This work provides two major contributions to our knowledge of mammalian genome transcription by RNA polymerase III. First, it redefines the transcription termination signal for RNA polymerase III, by demonstrating that human tRNA gene transcription is frequently terminated at elements whose sequence does not conform to the widely accepted T≥4 rule for Pol III terminators. Second, it shows that in mammals Pol III termination tends to be leaky, leading to read-through transcription behind both canonical and non-canonical terminator sequences into the 3′-flanking regions of known Pol III-transcribed genes. These two findings have interesting implications both for our basic, mechanistic view of the Pol III transcription cycle and for our understanding of the human Pol III transcriptome, whose contribution to human ncRNA biogenesis has still not been fully defined (1,46).
Even though examples of Pol III termination at non-T≥4 signals had previously been reported, the finding that a large variety of non-canonical signals are of general use for human tRNA gene transcription termination came quite unexpected. The impetus for non-canonical terminator sequence search came from the observation that a substantial number of human tRNA genes lack any recognizable terminator in the first 50 bp of 3′-flanking region (in which all previously characterized Pol III termination signals do occur). There are two immediate implications of this observation: either Pol III termination for these genes takes place at unusually distant positions (thus generating unusually long pre-tRNAs) or termination occurs at usual distances, but due to unusual terminators. By characterizing the termination properties of a subset of such apparently ‘terminator-less’ tDNAs, we found evidence for both types of irregular behavior.
Our biochemical analysis of non-canonical terminators confirms, on the one hand, the ability of Pol III to recognize DNA sequence signals that are among the simplest known modulatory elements in transcription; on the other hand, it underlines the exquisite selectivity of Pol III in distinguishing among very similar sequence element for termination. The finding that T3 (and even T2) elements, if embedded in particular sequence contexts, can act as remarkably efficient terminators was not completely unexpected, given previous reports of similar observations [see for example (19) and references therein; (20)]. However, the systematic analysis of non-canonical terminator variants revealed unexpected features of the Pol III termination reaction that helps to shed light on its mechanism. In particular, we found that sequences having the same composition, and almost identical predicted stability in terms of RNA/DNA hybrid duplex (47), differed significantly in their ability to induce termination. This was especially evident with the oppositely oriented sequence pairs TCT3/T3CT and TGT3/T3GT. With both pairs, termination was much more efficient when the run of 3 Ts was at the 3′ end of the motif. A possible molecular explanation of such a ‘discriminating taste’ is that there could be a requirement for a few consecutive Us at the 3′-end of RNA in order for Pol III to terminate. Relevant to this hypothesis, it has been proposed recently that the binding of C53 subunit to RNA might contribute to Pol III termination (48). We speculate that an RNA end with three consecutive Us could be a better binding substrate for C53 (or, more generally, for Pol III) in a Pol III-RNA interaction required for transcription termination. As an additional remark, we note that in the S. cerevisiae Pol III system, the CT dinucleotide was shown to act as a terminator-weakening element when placed immediately downstream to a minimal T5 termination signal (12). By analogy, the poor termination signal TTTCT can be seen as composed by a T3 element followed by the CT dinucleotide that would be responsible for terminator weakness. Even though Pol III seems to intrinsically distinguish among different non-canonical terminators (Figure 3), our data do not rule out the possibility that accessory proteins (such as La, that discriminates the number of Us at RNA 3′ ends) (41,49) and RNA structural features could also influence the selectivity of Pol III termination at non-canonical signals.
We generally conclude from our study that the human and mouse Pol III transcription systems display reduced transcription termination stringency, at least as regards the sequence and position of cis-acting signals, in comparison to what is known in yeast, where tRNA gene terminators tend to be clear-cut (T≥5 with no exceptions) and very close to the 3′ end of the coding sequence. Even though evidence of in vivo Pol III termination leakiness has not been directly searched for in S. cerevisiae as we did in the human system, the available evidence suggests that leakiness must be strongly disfavored in yeast, where 85% of the termination signals are runs of six or more T residues, that tend to impose full termination in vitro, and Pol III readthrough possibly occurring at T5 signals is counteracted by closely juxtaposed, longer T runs (11,12). The tight yeast configuration is expected to result in more rapid and efficient tRNA synthesis, especially because efficient termination is required for facilitated Pol III reinitiation to occur (7,13). One appealing interpretation of the difference between yeast and human Pol III terminators is that, in the yeast genome, tRNA gene templates are under selection for high-level production of tRNAs required for sustained protein synthesis in these rapidly growing unicellular organisms, while in humans (and, in principle, in all multicellular organisms for which rapid cell growth is not the predominant task), tDNA transcription units need not to be optimized for termination/reinitiation efficiency.
The assumption of a higher than expected leakiness in human Pol III termination is supported by the results of a genome-wide location analysis of Pol III by ChIP-seq. At tRNA transcription units, we found unexpectedly high levels of Pol III association to the 3′-flanking regions, whose simplest explanation is terminator read-through and trailer RNA synthesis by a fraction of polymerases. Such a behavior is more pronounced where canonical termination signals are lacking, but it could also occur at classical terminators. Pol III readthrough past canonical stop signals is consistent with our observation that even T5 display some leakiness in vivo in specifically devised, termination reporter constructs (Figure 4), and with the observation that the γHV68 transcription unit pol III-4 contains a canonical T4 upstream of miR-M1-6 coding sequence, yet this miRNA is readily detected in infected cells (Figure 6) (27). The general tendency of Pol III to trespass on the 3′-flanking regions of its target genes has two relevant implications. The first one is that the tRNA 3′ end maturation process and its regulation are expected to be more complex in humans than in yeast, where most pre-tRNA trailers are expected not to be longer than 15 nt. The existence of a large and heterogeneous set of trailer sequences that need to be removed to generate mature tRNA species offers the possibility of novel levels of post-transcriptional regulation of tRNA biogenesis. A second, even more intriguing implication of widespread Pol III read-through is the possibility to generate tRNA-independent functional RNA species by transcription of tDNA flanking regions. A few recent reports provide strong support for this idea, by showing that small RNAs derived from tRNA 3′ trailers accumulate in the cytoplasm of human cells, where they can play regulatory roles (50–52). From an evolutionary perspective, tDNAs and, more generally, Pol III transcription units appear to be ideally suited to favor the emergence of novel small RNA species. Pol III promoters are strong, and ensure high-level transcription of any sequence that gets incorporated into the transcription units. Loss or weakening of the termination signal downstream of tRNA and other class III genes is the simplest way to incorporate downstream sequences, giving them the opportunity of being expressed at high efficiency. Such kind of strategy has been found to operate, for example, in the biogenesis of snoRNAs and 5S rRNA (16,26,53), as well as of viral miRNAs (27–29), and could be responsible for the generation of a largely unexplored section of the Pol III transcriptome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fondazione Cariparma (2010), Italian Ministry of Education, University and Research (PRIN Program), AICCRE—Regione Emilia Romagna (to G.D.); from Conseil Régional d’Aquitaine, European Regional Development Fund, Agence Nationale de la Recherche (ANR, ‘REGPOLSTRESS’) and Ligue Contre le Cancer-Comités Gironde et Dordogne (to M.T.); the National Institutes of Health (CA103632 and P30AI054907 to L.v.D.); the Burroughs Wellcome Investigator in Infectious Diseases award and a University of Colorado Technical Transfer Office award (to K.W.D. and L.v.D.). C.P. was supported by a doctoral fellowship from the ‘Università Italo-Francese/Université Franco-Italienne’. V.P., D.R. and N.H. were supported by the University of Lausanne and Swiss National Science Foundation (NSF grant 3100A0-109941 to N.H.). Funding for open access charge: Fondazione Cariparma, Parma, Italy and Conseil Régional d'Aquitaine, France.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Dumay-Odelot H, Durrieu-Gaillard S, Da Silva D, Roeder RG, Teichmann M. Cell growth- and differentiation-dependent regulation of RNA polymerase III transcription. Cell Cycle. 2010;9:3687–3699. doi: 10.4161/cc.9.18.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dieci G, Sentenac A. Detours and shortcuts to transcription reinitiation. Trends Biochem. Sci. 2003;28:202–209. doi: 10.1016/S0968-0004(03)00054-9. [DOI] [PubMed] [Google Scholar]
- 4.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, et al. Structure of eukaryotic RNA polymerases. Annu. Rev. Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- 6.Bogenhagen DF, Brown DD. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24:261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- 7.Dieci G, Sentenac A. Facilitated recycling pathway for RNA polymerase III. Cell. 1996;84:245–252. doi: 10.1016/s0092-8674(00)80979-4. [DOI] [PubMed] [Google Scholar]
- 8.Ferrari R, Rivetti C, Acker J, Dieci G. Distinct roles of transcription factors TFIIIB and TFIIIC in RNA polymerase III transcription reinitiation. Proc. Natl Acad. Sci. USA. 2004;101:13442–13447. doi: 10.1073/pnas.0403851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabart P, Lee J, Willis IM. Facilitated recycling protects human RNA polymerase III from repression by Maf1 in vitro. J. Biol. Chem. 2008;283:36108–36117. doi: 10.1074/jbc.M807538200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allison DS, Hall BD. Effects of alterations in the 3′ flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J. 1985;4:2657–2664. doi: 10.1002/j.1460-2075.1985.tb03984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamada M, Sakulich AL, Koduru SB, Maraia RJ. Transcription termination by RNA polymerase III in fission yeast. A genetic and biochemically tractable model system. J. Biol. Chem. 2000;275:29076–29081. doi: 10.1074/jbc.M003980200. [DOI] [PubMed] [Google Scholar]
- 12.Braglia P, Percudani R, Dieci G. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J. Biol. Chem. 2005;280:19551–19562. doi: 10.1074/jbc.M412238200. [DOI] [PubMed] [Google Scholar]
- 13.Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 2006;25:118–128. doi: 10.1038/sj.emboj.7600915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinmann R, Brendler TG, Raskas HJ, Roeder RG. Low molecular weight viral RNAs transcribed by RNA polymerase III during adenovirus 2 infection. Cell. 1976;7:557–566. doi: 10.1016/0092-8674(76)90206-3. [DOI] [PubMed] [Google Scholar]
- 15.Mazabraud A, Scherly D, Muller F, Rungger D, Clarkson SG. Structure and transcription termination of a lysine tRNA gene from Xenopus laevis. J. Mol. Biol. 1987;195:835–845. doi: 10.1016/0022-2836(87)90488-8. [DOI] [PubMed] [Google Scholar]
- 16.Guffanti E, Ferrari R, Preti M, Forloni M, Harismendy O, Lefebvre O, Dieci G. A minimal promoter for TFIIIC-dependent in vitro transcription of snoRNA and tRNA genes by RNA polymerase III. J. Biol. Chem. 2006;281:23945–23957. doi: 10.1074/jbc.M513814200. [DOI] [PubMed] [Google Scholar]
- 17.Vnencak-Jones CL, Wahab SZ, Zehner ZE, Holmes WM. A human tRNA(iMet) gene produces multiple transcripts. Mol. Cell. Biol. 1987;7:4134–4138. doi: 10.1128/mcb.7.11.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hess J, Perez-Stable C, Wu GJ, Weir B, Tinoco I, Jr, Shen CK. End-to-end transcription of an Alu family repeat. A new type of polymerase-III-dependent terminator and its evolutionary implication. J. Mol. Biol. 1985;184:7–21. doi: 10.1016/0022-2836(85)90039-7. [DOI] [PubMed] [Google Scholar]
- 19.Thomann HU, Schmutzler C, Hudepohl U, Blow M, Gross HJ. Genes, variant genes and pseudogenes of the human tRNA(Val) gene family. Expression and pre-tRNA maturation in vitro. J. Mol. Biol. 1989;209:505–523. doi: 10.1016/0022-2836(89)90590-1. [DOI] [PubMed] [Google Scholar]
- 20.Gunnery S, Ma Y, Mathews MB. Termination sequence requirements vary among genes transcribed by RNA polymerase III. J. Mol. Biol. 1999;286:745–757. doi: 10.1006/jmbi.1998.2518. [DOI] [PubMed] [Google Scholar]
- 21.Emerson BM, Roeder RG. DNA sequences and transcription factor interactions of active and inactive forms of mammalian 5S RNA genes. J. Biol. Chem. 1984;259:7926–7935. [PubMed] [Google Scholar]
- 22.Cozzarelli NR, Gerrard SP, Schlissel M, Brown DD, Bogenhagen DF. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983;34:829–835. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Bai L, Hsieh YJ, Roeder RG. Nuclear factor 1 (NF1) affects accurate termination and multiple-round transcription by human RNA polymerase III. EMBO J. 2000;19:6823–6832. doi: 10.1093/emboj/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopper AK, Pai DA, Engelke DR. Cellular dynamics of tRNAs and their genes. FEBS Lett. 2010;584:310–317. doi: 10.1016/j.febslet.2009.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takaku H, Minagawa A, Takagi M, Nashimoto M. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res. 2003;31:2272–2278. doi: 10.1093/nar/gkg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kruszka K, Barneche F, Guyot R, Ailhas J, Meneau I, Schiffer S, Marchfelder A, Echeverria M. Plant dicistronic tRNA-snoRNA genes: a new mode of expression of the small nucleolar RNAs processed by RNase Z. EMBO J. 2003;22:621–632. doi: 10.1093/emboj/cdg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diebel KW, Smith AL, van Dyk LF. Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters. RNA. 2010;16:170–185. doi: 10.1261/rna.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol. Cell. 2010;37:135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 30.Percudani R, Pavesi A, Ottonello S. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 1997;268:322–330. doi: 10.1006/jmbi.1997.0942. [DOI] [PubMed] [Google Scholar]
- 31.Pavesi A, Conterio F, Bolchi A, Dieci G, Ottonello S. Identification of new eukaryotic tRNA genes in genomic DNA databases by a multistep weight matrix analysis of transcriptional control regions. Nucleic Acids Res. 1994;22:1247–1256. doi: 10.1093/nar/22.7.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haurie V, Durrieu-Gaillard S, Dumay-Odelot H, Da Silva D, Rey C, Prochazkova M, Roeder RG, Besser D, Teichmann M. Two isoforms of human RNA polymerase III with specific functions in cell growth and transformation. Proc. Natl Acad. Sci. USA. 2010;107:4176–4181. doi: 10.1073/pnas.0914980107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat. Protocols. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 36.Virgin HWt, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Canella D, Praz V, Reina JH, Cousin P, Hernandez N. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010;20:710–721. doi: 10.1101/gr.101337.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaack J, Sharp S, Dingermann T, Burke DJ, Cooley L, Soll D. The extent of a eukaryotic tRNA gene. 5′- and 3′-flanking sequence dependence for transcription and stable complex formation. J. Biol. Chem. 1984;259:1461–1467. [PubMed] [Google Scholar]
- 39.Matsuzaki H, Kassavetis GA, Geiduschek EP. Analysis of RNA chain elongation and termination by Saccharomyces cerevisiae RNA polymerase III. J. Mol. Biol. 1994;235:1173–1192. doi: 10.1006/jmbi.1994.1072. [DOI] [PubMed] [Google Scholar]
- 40.Traut TW. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z, Roeder RG. DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol. Cell. 1998;1:749–757. doi: 10.1016/s1097-2765(00)80074-x. [DOI] [PubMed] [Google Scholar]
- 42.Campbell FE, Jr, Setzer DR. Transcription termination by RNA polymerase III: uncoupling of polymerase release from termination signal recognition. Mol. Cell. Biol. 1992;12:2260–2272. doi: 10.1128/mcb.12.5.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Englert M, Felis M, Junker V, Beier H. Novel upstream and intragenic control elements for the RNA polymerase III-dependent transcription of human 7SL RNA genes. Biochimie. 2004;86:867–874. doi: 10.1016/j.biochi.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 44.Roy AM, West NC, Rao A, Adhikari P, Aleman C, Barnes AP, Deininger PL. Upstream flanking sequences and transcription of SINEs. J. Mol. Biol. 2000;302:17–25. doi: 10.1006/jmbi.2000.4027. [DOI] [PubMed] [Google Scholar]
- 45.Whitehall K, Kassavetis A, Geiduschek P. The symmetry of the yeast U6 RNA gene’s TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev. 1995;9:2974–2985. doi: 10.1101/gad.9.23.2974. [DOI] [PubMed] [Google Scholar]
- 46.Pagano A, Castelnuovo M, Tortelli F, Ferrari R, Dieci G, Cancedda R. New snRNA gene-like transcriptional units as sources of regulatory transcripts. PLoS Genet. 2007;3:e1. doi: 10.1371/journal.pgen.0030001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 48.Kassavetis GA, Prakash P, Shim E. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J. Biol. Chem. 2010;285:2695–2706. doi: 10.1074/jbc.M109.074013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y, Intine RV, Mozlin A, Hasson S, Maraia RJ. Mutations in the RNA polymerase III subunit Rpc11p that decrease RNA 3′ Cleavage activity increase 3′-terminal oligo(U) length and La-dependent tRNA processing. Mol. Cell. Biol. 2005;25:621–636. doi: 10.1128/MCB.25.2.621-636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Acker J, Ozanne C, Kachouri-Lafond R, Gaillardin C, Neuveglise C, Marck C. Dicistronic tRNA-5S rRNA genes in Yarrowia lipolytica: an alternative TFIIIA-independent way for expression of 5S rRNA genes. Nucleic Acids Res. 2008;36:5832–5844. doi: 10.1093/nar/gkn549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cleveland WS. LOWESS: A program for smoothing scatterplots by robust locally weighted regression. Am. Stat. 1981;35:54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.