Abstract
Accumulation of damaged guanine nucleobases within genomic DNA, including the imidazole ring opened N6-(2-Deoxy-α,β-D-erythro-pentafuranosyl)-2,6-diamino-4-hydroxy-5-formylamidopyrimidine (Fapy-dG), is associated with progression of age-related diseases and cancer. To evaluate the impact of this mutagenic lesion on DNA structure and energetics, we have developed a novel synthetic strategy to incorporate cognate Fapy-dG site-specifically within any oligodeoxynucleotide sequence. The scheme involves the synthesis of an oligonucleotide precursor containing a 5-nitropyrimidine moiety at the desired lesion site via standard solid-phase procedures. Following deprotection and isolation, the Fapy-dG lesion is generated by catalytic hydrogenation and subsequent formylation. NMR assignment of the Fapy-dG lesion (X) embedded within a TXT trimer reveals the presence of rotameric and anomeric species. The latter have been characterized by synthesizing the tridecamer oligodeoxynucleotide d(GCGTACXCATGCG) harboring Fapy-dG as the central residue and developing a protocol to resolve the isomeric components. Hybridization of the chromatographically isolated fractions with their complementary d(CGCATGCGTACGC) counterpart yields two Fapy-dG·C duplexes that are differentially destabilized relative to the canonical G·C parent. The resultant duplexes exhibit distinct thermal and thermodynamic profiles that are characteristic of α- and β-anomers, the former more destabilizing than the latter. These anomer-specific impacts are discussed in terms of differential repair enzyme recognition, processing and translesion synthesis.
INTRODUCTION
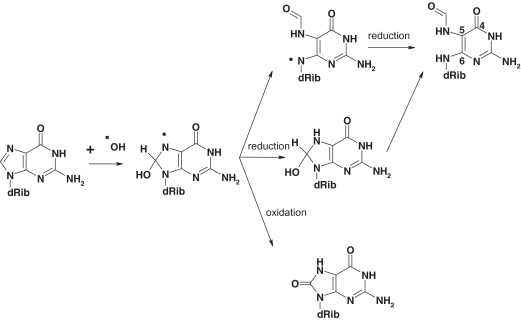
Hydroxyl radicals arising from exposure to environmental oxidants or produced by cell metabolism react with DNA causing thousands of oxidatively damaged nucleotides per diem (1). The reaction of hydroxyl radicals with DNA can generate different base and sugar radicals, including the C8-OH guanine adduct. As illustrated in Scheme 1, oxidation of the C8-OH radical forms 8-oxo-7,8-dihydroxy-2′-deoxyguanosine (8-oxodG), while the competing one-electron reduction that predominates under anoxic conditions generates N6-(2-Deoxy-α,β-D-erythro-pentafuranosyl)-2,6-diamino-4-hydroxy-5-formylamidopyrimidine (Fapy-dG) via imidazole ring opening (2). Formamidopyrimidine (Fapy) lesions are abundant under a variety of experimental conditions. Cell exposure to γ-radiation or treatment with reactive oxygen species generates 8-oxodG and Fapy-dG, the latter forming even in the presence of oxygen (3–6). Mammalian cells possess background levels of both lesions, which rise rapidly following oxidative stress (7–9) and are elevated in a number of circumstances including chronic inflammatory processes and pathological conditions such as Alzheimer’s disease and cancer (10–14).
Scheme 1.
Oxidation pathways from a common precursor that yields 8-oxodG and Fapy-dG.
The base excision repair (BER) pathway efficiently removes Fapy-dG and 8-oxodG lesions from DNA. Pioneering studies using irradiated or photosensitized DNA harboring multiple oxidative lesions have demonstrated that the bacterial Fapy-DNA glycosylase (Fpg, MutM) removes a broad range of oxidative damage, including 8-oxodG, Fapy-dG, methyl-Fapy-dG and Fapy-dA (15–17). These findings have been confirmed in recent investigations of Fapy lesions incorporated site-specifically within DNA constructs to explore the impact of counter-base effects on enzymatic activity (18,19). The specificity constant reported for Fpg excision of Fapy-dG is ∼17-fold higher when it pairs with dC rather than dA. Eukaryotic cells possess several enzymes that can remove Fapy lesions from DNA duplexes including Ogg1, the functional homologue of Fpg. Ogg1 excises both Fapy-dG and 8-oxodG, yet is largely inactive in the presence of Fapy-dA adducts (20–24). Mammalian Nei-like enzymes efficiently remove Fapy-dG and Fapy-dA lesions from damaged duplexes, but do not process 8-oxodG containing substrates, in contrast to Ogg1 and Fpg (25–28).
Despite the impressive number of studies that specifically address the impact of Fapy-dG lesions and their consequence on the host cell, there is a noticeable dearth of parallel synthetic efforts to obtain suitable DNA constructs containing the cognate lesion. The high reactivity of Fapy lesions in standard oligonucleotide synthesis schemes coupled with the challenge of developing efficient synthetic strategies has hampered biochemical and biophysical studies of Fapy-dG embedded within defined DNA duplexes. The existing synthetic approach for preparation of Fapy-containing oligomers requires the synthesis of a phosphoramidite dimer with a standard nucleotide at the lesion 5′-side and its subsequent incorporation into DNA following a modified solid-phase protocol (29,30). Despite these important advances, the dimeric synthesis approach imposes a significant limitation in terms of readily exploring the influence of sequence context on Fapy properties. Specifically, the 5′-neighboring base is generally a thymine as other bases are not easily incorporated within the synthetic scheme to facilitate sequence-dependent studies. This serious deficiency may only be alleviated by a massive synthetic effort that entails preparing all four possible dimeric precursors.
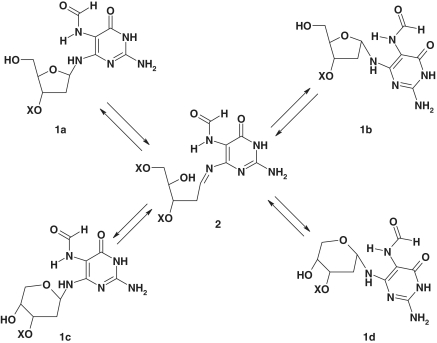
The synthesis and isolation of homogeneous Fapy-dG 2′-oligodeoxynucleotides represent formidable tasks, given that the cognate lesion is chemically labile and exists as a mixture of α- and β-anomers (Scheme 2; 1a and 1b) at the nucleoside level, which re-arrange to the pyranosides 1c and 1d. The impact of lesion anomerization on biological outcomes has only been assessed indirectly via synthesis of chemically stable analogues that do not epimerize. Although proven useful in a number of applications including translesion synthesis assays, such non-hydrolyzable analogues preclude kinetic and thermodynamic characterization of the forces driving Fapy-dG excision/repair. Recent crystallographic studies of Fpg complexes with damaged DNA duplexes employing the carbocyclic analogue of Fapy-dG (i.e. carba-Fapy-dG) (31) have yielded intriguing results in terms of conformational preferences that differ markedly from those reported for 8-oxodG (32). Whereas these differences may be attributed to sequence variations within Fpg (i.e. S77E in Lactococcus lactis), there are legitimate concerns regarding whether non-hydrolyzable analogues (e.g. carba-Fapy-dG) truly mimic their cognate counterparts in lesion recognition processes (33–35). Specifically, analogue binding in the absence of turnover might actually reflect fortuitous interactions that are not necessarily biologically relevant. These findings underscore the need to develop synthetic protocols that yield stable cognate lesions at isomeric resolution for direct assessment and characterization of their biophysical properties and biological outcomes.
Scheme 2.
Proposed mechanism of Fapy isomerization via an open intermediate.
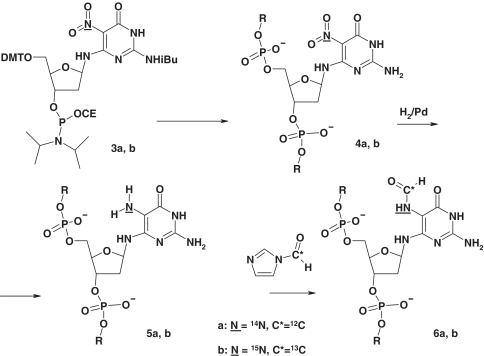
In the present study, we report a novel post-synthetic strategy for the site-specific incorporation of Fapy-dG residues within any oligodeoxynucleotide sequence context. This methodology overcomes inherent limitations of previous approaches by establishing the Fapy moiety as the 5-formamido group in the pyrimidino xenonucleotide (Scheme 3). We discuss the application of this strategy to the preparation of isotopically labeled Fapy-dG incorporated within a single-strand DNA trimer for NMR assignment of the anomeric species. In order to evaluate the impact of Fapy-dG on DNA duplex energetics, we synthesized a tridecameric oligodeoxynucleotide with the lesion embedded as the central residue. Subsequent isolation and hybridization with the corresponding complementary strand generates Fapy-dG·C duplexes exhibiting distinct thermal and thermodynamic properties that are consistent with α- and β-anomers. The combination of structural and thermodynamic analysis provides insights into the properties of Fapy-dG·C duplexes, which sets the stage for a detailed characterization of the respective anomers in terms of lesion formation, translesion replication and BER.
Scheme 3.
Post-synthetic generation of Fapy-dG in DNA.
MATERIALS AND METHODS
Reagents and equipment
Chemicals and solvents were purchased from Fluka-Sigma-Aldrich and DNA syntheses were performed on an Applied Biosystems instrument using reagents from Glenn Research. Isotopically labeled compounds were purchased from Cambridge Isotope Laboratories. HPLC analysis and purification were performed on a Gilson system equipped with a Zorbax C18 semi-preparative column (7.5 × 300 mm). Further purification was achieved using a high resolution Mono QTM HR 5/5 (4.6 × 100 mm) pre-packed anion exchange column (Amersham Pharmacia, Piscataway, NJ, USA). NMR spectra were recorded on Varian Inova instruments operating at 400, 500 and 600 MHz, or on a Bruker Avance spectrometer at 700 MHz. Time domain data were processed using Vnmr (Varian, CA, USA), Felix or NMRPipe (36) software. Two dimensional spectra were visualized using Sparky (37). Oligodeoxynucleotides were analyzed by nano-ESI on a Micromass Platform time-of-flight mass spectrometer operated in negative mode. Molecular modeling was performed using HyperChem (Hypercube, Inc.).
Preparation of phosphoramidites for solid-phase DNA synthesis
Details of the synthesis are presented as Supplementary Data.
Synthesis of oligodeoxynucleotides
The NPym-dG phosphoramidite 3 was prepared using the standard protocol (38) (Supplementary Scheme S1). Oligodeoxynucleotide synthesis followed a standard solid-phase protocol using ultra-mild phosphoramidites. Saturated ammonia treatment for 2 h at 50°C removed all protecting groups and the oligomers were purified using a standard solid-phase protocol.
Standard procedure for post-synthetic reduction and formylation of 5-nitropyrimidine oligodeoxynucleotides
Approximately 300 OD260 of oligodeoxynucleotide was dissolved in 1 ml of water, followed by the addition of 500 μl of methanol and 50 μl of triethylamine to maintain a basic medium. A total of 2 mg of Pd black was added to the mixture and shaken under hydrogen (3 atm pressure) for 3–6 h. The reaction was monitored by mass-spectroscopy. After the starting material disappeared completely, 50 ml of mercaptoethanol was added to the mixture followed by 1 ml of a freshly prepared THF solution of formylimidazole (38). The addition was conducted portion-wise via a syringe with 200 μl of solution injected at 30 min time intervals over 2 h. When MS revealed that the aminopyrimidine oligodeoxynucleotide had completely disappeared, the reaction mixture was centrifuged, evaporated to remove organic solvents and the remaining water solution diluted with 0.1 M triethylammonium acetate buffer and purified using ion-pair reverse phase HPLC with a linear gradient of acetonitrile, 0–40%, 30 min in 0.1 M TEAA.
Analytical and preparative ion exchange chromatographic protocol to isolate and identify Fapy-dG oligodeoxynucleotide species
Purification of Fapy-dG fractions was performed via anion-exchange chromatography employing previously published procedures (39) with specific modifications conducive for the isolation of anomeric Fapy-dG species. The anion-exchange column was equilibrated with TA buffer composed of 50 mM Tris–HCl and 15% acetonitrile (pH 8.0) at a flow rate of 1.5 ml min−1. The reverse-phase purified and lyophilized Fapy-dG samples were suspended in de-ionized water (1.5–2.0 ml), injected into the column and the fractions eluted with a linear gradient (typically 20–80%) of TA buffer containing 1.0 M NaCl over a period of 20 min with the outflow monitored at 260 nm. The procedure was repeated by reapplying the enriched isolated fractions following a 3-fold dilution in TA buffer. In the final step, each of the primary components (i.e. Fractions I and II) eluted at 11 and 11.5 min, respectively. The fractions were collected conservatively to avoid peak overlap and immediately combined with an excess of complementary strand in TA buffer. The resultant disproportionate single-strand mixtures were then reapplied into the column to separate the duplex from the excess of single-strand species. The duplex peaks that elute typically at ∼14 min (i.e. ∼0.7 M NaCl) were collected and immediately diluted in cold absolute ethanol and equilibrated at −20°C for a minimum of 30 min. The ethanol precipitated material was collected in a Model 5417R refrigerated centrifuge (Brinkmann Instruments, Westbury, NY, USA) operating at 16 000 rpm for 20 min at 5.0°C. The resultant pellet was dried in a SpeedVac Concentrator (Savant, Hicksville, NY, USA) under vacuum and suspended in cold deionized water followed by freezing in liquid nitrogen. The procedure was repeated and the final pellet suspended in a buffer comprising 10 mM sodium phosphate and 150 mM NaCl (pH 7.0).
Fapy-dG·C duplex concentration determination and standard preparation
An aliquot of the duplex stock in 10 mM sodium phosphate and 150 mM NaCl (pH 7.0) was diluted in the identical buffer system to prepare a Fapy-dG·C standard for concentration determination. The absorbance was measured spectrophotometrically by monitoring the temperature-dependent hyperchromicity at 260 nm via conventional methods. Linear extrapolation of the post-transition baseline to 25°C yields A260 of the dissociated oligodeoxynucleotide that represents the sum of the single-strand extinction coefficients measured via traditional enzyme digestion methods (40,41). Alternatively, an aliquot of the duplex was digested using an excess of nucleases and the resultant absorbance divided by the sum of the extinction coefficients calculated for the nucleotides. The extinction coefficient for the Fapy-dG single strand was corroborated via the method of continuous fractions by hybridizing defined ratios of the complementary strand with known extinction coefficient in isothermal mixing experiments and casting the data in the form of Job plots (42).
Thermodynamic analysis
Temperature-dependent UV melting experiments were performed on an Aviv Model 14 UV/Vis spectrophotometer (Aviv Biomedical, Inc., Lakewood, NJ, USA). Samples in quartz cuvettes of 1.0 cm path length were heated in the thermostatted sample compartment over the temperature range of 0–90.0°C. The absorbance at 260 nm was recorded at 0.5°C increments following integration for 10 s to monitor the hyperchromicity change as a function of temperature. The duplex dissociation temperatures and van’t Hoff thermodynamic parameters have been determined by shape analysis of the temperature-dependent hyperchromicity profiles for a bimolecular two-state dissociation process assuming a zero heat capacity in accordance with traditional protocols (43).
RESULTS AND DISCUSSION
Rationale for characterizing cognate Fapy-dG at anomeric resolution
Fapy lesions have been the subject of considerable interest and inquiry since their discovery over three decades ago as some of the most ubiquitous lesions implicated in DNA mutagenesis. Despite progress in elucidating its promutagenic properties and in vivo processing by the BER pathway, there are no structural or thermodynamic characterizations of DNA duplexes harboring a cognate Fapy-dG. One of the primary obstacles impeding systematic investigations is the fact that a canonical undamaged DNA base in a conventional β-configuration may potentially yield a mixture of α- and β-anomeric species upon conversion to Fapy. Moreover, the equilibrium ratio and sequence context-dependence of the isomerization process within the DNA duplex is not adequately understood. Since epimerization rates are conceivably faster than competing deglycosylation reactions, there is supporting evidence to conclude that isomeric species are present in the cell during biological processes (44), thereby rendering characterization of BER processing specificities a formidable task. A recent study on Fapy-dG promutagenic potential in Escherichia coli suggests that the lesion is 3- to 5-fold less mutagenic than 8-oxodG embedded within an identical sequence context (45). Conversely, Fapy-dG may promote G to T transversions in mammalian (COS) cells at a greater magnitude than 8-oxodG, given that the latter occurs in a highly sequence-context dependent manner (46). The structural and energetic determinants associated with Fapy recognition and the possible stabilization of its mutagenic intermediates remains a matter of inquiry. Since most of these studies employ either synthetic strategies that produce anomeric mixtures or utilize stable bio-isosteric analogues, systematic investigations of the isolated cognate Fapy-dG conformers are warranted to elucidate unequivocally the anomer-dependent replication and/or repair outcomes.
Synthesis of nitropyrimidine oligodeoxynucleotides
The low chemical stability of the Fapy moiety coupled with the ability of the Fapy-dG nucleoside to isomerize represents a major challenge for solid-phase DNA synthesis. Previous studies have demonstrated that the Fapy nucleosides readily isomerize to yield a mixture of anomeric 2′-deoxyribofuranosides (Scheme 2; 1a and 1b) (47). Moreover, deprotection of the 5′-hydroxyl in Fapy furanosides leads by rearrangement to an anomeric mixture of corresponding pyranosides (i.e. 1c and 1d). The latter renders a standard solid-phase protocol unsatisfactory for the synthesis of Fapy-containing DNA. A solution to this problem has been provided by the Greenberg group, who initially reported the synthesis of Fapy-dG containing DNA using a modified 5′- to 3′-solid-phase protocol (29), and subsequently prepared the dinucleotide phosphoramidite with protected Fapy-dG at its 3′-end (48). Since these approaches require substantial modification of the standard solid-phase protocol using intermediates that are not trivial to synthesize, we decided to develop a universal post-synthetic method for the generation of Fapy lesions in DNA. Our strategy is based on reduction of a nitropyrimidine residue present in oligomeric DNA followed by formylation of the resulting amino group in aqueous media. Initially, we surmised that the problem of furanoside isomerization to the pyranoside with the 5′-hydroxyl group unprotected might be due to an increase in the acid-sensitivity of the system given the basic nature of the associated pyrimidine. We hypothesized that this might be solved by retaining the 5-nitro group, the progenitor of the formamido group, during oligomer synthesis. This reasoning is based on the idea that the strongly electron-withdrawing nitro group would deplete the electrons on the 6-amino group, thus reducing its ability to facilitate acid-catalyzed opening of the ribosyl ring. This concept subsequently proved to be correct. Following reduction of the nitro group, formylation of the 5-amino group in the precursor Fapy lesion is expected to be highly selective since all other amino and hydroxy functions in the deprotected oligomers are much less basic in character. To verify the feasibility of this synthesis strategy, we prepared a model TpNPympT trimer 7 (Figure 1) using NPym-dG amidite 3 as a building block. For preparation of the starting nucleoside, we utilized a scheme similar to that reported by the Greenberg group (48) implementing specific modifications that allowed us to obtain the amidite as a pure β-anomer (Supplementary Scheme S1).
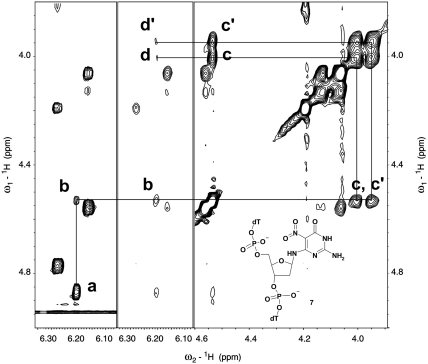
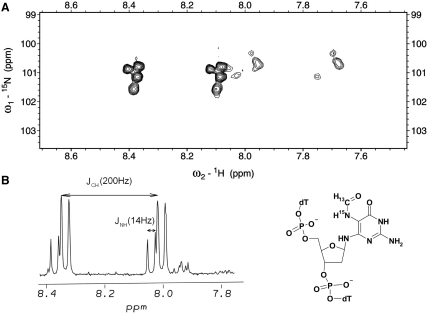
Figure 1.
Expanded region of 1H-TOCSY (left panel) and NOESY (central and right panels) spectra of the major isomer of the TpNPypT trimer 7. Cross-peaks between the sugar protons of the nitropyrimidine nucleotide are assigned as follows: a: 1′3′, b: 1′–4′, c, c′: 4′–5′, d, d′: 1′–5′.
Generation of the TpNPympT trimer
We employed a standard solid-phase protocol to synthesize the TpNPympT trimer with the slight modification of increasing the coupling time of NPym-dG amidite to 5 min, which resulted in a 97% coupling efficiency approximating that of standard amidites. Deprotection in concentrated ammonia followed by trityl-on HPLC purification yields trimer 7, which is detritylated via standard treatment with acetic acid. Subsequent HPLC analysis of the resulting trimer reveals only two components, one of which is the predominant species reflective of a non-equilibrium mixture. Examination of 1H-NMR spectra confirms that the major fraction is a single compound, and comparison with the crude trimer reveals that the second component is also a single compound (Supplementary Figure S5). 2D-NOESY and TOCSY spectra of the major component (Figure 2) corroborated the furanoside structure of trimer 7, indicating that isomerization of NPym-dG to pyranosides does not occur during the monomer synthesis deprotection and coupling steps. Such isomerization requires significant time for completion and all four isomers would be present in the mixture during the reaction course (49). The spectral evidence allows us to conclude that the minor component is the α-furanoside isomer, which arises during deprotection and chromatographic separation, thus eliminating the possibility of pyranoside isomer formation. Our results are therefore consistent with previous studies, indicating that pyranoside rearrangement is unattainable when Fapy-dG is successfully incorporated into oligodeoxynucleotides, whereas anomerization can potentially occur within the DNA strand (29,49,50). As reported in subsequent sections, we have utilized the identical synthetic scheme to prepare the tridecameric d(GCGTACXCATGCG) oligodeoxynucleotides, where X denotes a nitropyrimidine residue. In order to minimize possible degradation of the nitropyrimidine residue, we employed ultra-mild protective groups on the dC, dA and dG phosphoramidites used in the solid-phase synthesis, followed by standard deprotection and purification protocols.
Figure 2.
(A) Expanded region of a [13C-coupled] 1H-15N-HSQC spectrum (700 MHz) of the doubly labeled Fapy-dG containing trimer. (B) Expanded region of a [13C, 15N-coupled] 1H-NMR spectrum (600 MHz) illustrating the formyl proton resonances.
Post-synthetic treatment and structural characterization of the TpNPympT trimer
We dissolved the TpNPympT trimer in 30% methanol and reduced the nitro group by hydrogenation over Pd black in the presence of triethylamine, employing ESI-MS to monitor reaction completion (i.e. 3–6 h). After reduction, the formylation reaction is performed directly without removal of the catalyst or purification of the aminopyrimidine to minimize possible oxidation of the reduced oligonucleotide. We selected N-formylimidazole as the formylating agent since it does not decompose immediately in the presence of water. This mild formylating agent is prepared from N,N′-carbonyldiimidazole and formic acid as described in (51) and used as a THF solution. The reaction normally requires 2–6 h for completion and is monitored by ESI-MS. The Fapy-containing trimer is purified by reverse-phase HPLC with an approximate yield of 60% based on the nitropyrimidine precursor.
Structural characterization of the trimer confirms the presence of a single formyl susbtitution attached exclusively to the N5 amino group. Specifically, we prepared the TpNPympT trimer with the nitrogroup 15N labeled, and formylated the reduction product with 13C-formyl imidazole, obtaining a trimer containing a double labeled (13C,15N) Fapy moiety (Figure 2). The 1H-NMR spectrum of this trimer (Figure 2) reveals the formyl proton with one-bond 13C–H (∼200 Hz) and two-bond 15N–H (15 Hz) couplings, indicating that the formamido group is situated at N5 as opposed to alternate positions (i.e. N2 or N6). Since both post-synthetic steps (i.e. catalytic hydrogenation and treatment with formyl imidazole in aqueous media) do not affect natural nucleotides, and incorporation of the nitropyrimidylamino residue into DNA follows standard solid-phase protocols, the proposed synthetic scheme represents the first universal method for the preparation of DNA containing a Fapy lesion in any sequence context. The versatility of the synthetic strategy can therefore be exploited to incorporate either Fapy-dG or Fapy-dA within defined oligodeoxynucleotide sequences.
Conformational properties and isomerism of Fapy-dG in single-strand DNA
One of the problematic issues in characterizing the biophysical properties of Fapy-dG lesions and their impact on nucleic acid structure and energetics is the feasibility of accurately determining the overall degree of isomerism within single strand and duplex DNA. Concurrently, one must understand the factors that play an integral role in modulating Fapy-dG equilibrium anomerization following incorporation of the cognate lesion within the DNA chain. The presence of a doubly labeled 13C,15N Fapy-oligodeoxynucleotide within the TXT model system provides us with a valuable tool for studying the conformational behavior of the formamido group and Fapy-dG isomerism. The [13C-coupled] 1H-15N-HSQC spectrum of the TpFapyGpT trimer exhibits two groups of resonances with one-bond 1H-13C-splitting as evidenced in Figure 2. This observation is in full accordance with earlier reports of Cadet and co-workers (49) who determined that Fapy-dG adopts two primary conformations corresponding to the cis- and trans-isomers around the formamido bond. The cis-isomer is the predominant species and is characterized by a CHO resonance that is shifted downfield. Since the formyl chemical shifts in both isomers and their populations correspond approximately to those observed for the nucleoside in prior investigations (49,52), we conclude that the formamido group of Fapy-dG exists predominantly as the cis-isomer in single-stranded oligodeoxynucleotide.
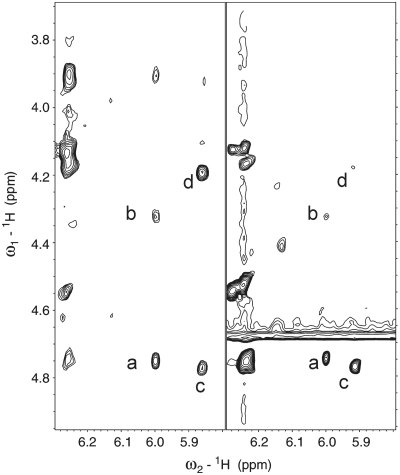
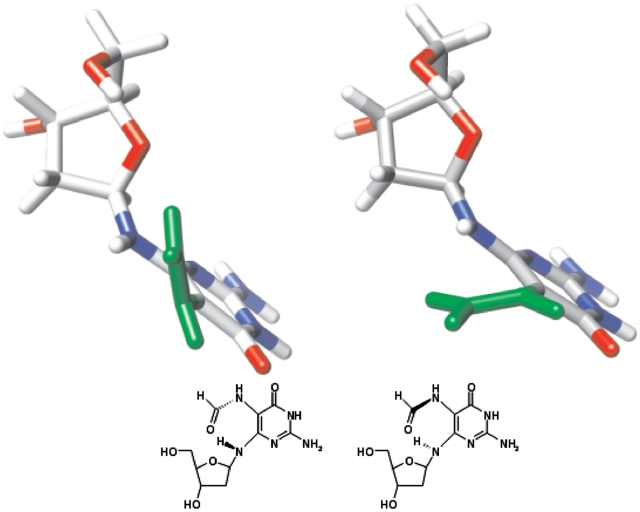
Inspection of the [13C-coupled] 1H-15N-HSQC spectrum for the TXT trimer (Figure 2) reveals that in addition to cis–trans isomerism of the amido group, the Fapy-dG moiety exists in four different states. NMR spectroscopy provides partial insight regarding the identity of these conformers. Specifically, NOESY and TOCSY spectra reveal that the 1′-proton of one Fapy-dG isomer is closer to its 4′- than the 3′-sugar proton, whereas the opposite situation is encountered for the second isomer (Figure 3). Based on such characteristics, we assign the anomeric configuration of these isomers as β for the former and α for the latter. Due to the absence of spin–spin correlation and NOE interactions between the formyl and sugar protons, the NMR spectra do not furnish additional information regarding the nature of the second set of resonances. Nevertheless, the observed double set of signals might conceivably arise from the non-planar Fapy aglycone (52) that can exist in two conformations with no mirror symmetry (Figure 4). Molecular modeling reveals that the transition between these two conformations is restricted by the steric clash between H-N6 and H-N5, and the pyrimidine O-4 and formyl oxygen. Consequently, the resultant conformers exhibit traits of atropisomerism. Whereas the presence of the atropisomers is not detected in the 1D NMR spectra of the Fapy nucleoside (52), the highly unsymmetrical environment of an oligodeoxynucleotide reveals the magnetic non-equivalence of the formyl proton and 5-nitrogen. The latter are clearly visible in the 1H-15N-HSQC spectrum, primarily along the nitrogen axis (Figure 2). Our detailed structural analysis of the Fapy trimer including possible conformations of the lesion within the TXT context sets the stage for characterization of longer oligodeoxynucleotide sequences harboring Fapy-dG. Such studies enable further investigation of the biophysical and biological properties of the corresponding DNA duplexes and modulation of lesion properties due to Fapy isomerization.
Figure 3.
Expanded region of a NOESY (1 s mixing time, 277 K) (left) and a TOCSY (70 ms isotropic mixing, 298 K) (right) spectra of the Fapy-dG trimer. Interactions between the sugar protons of the Fapy-dG residue identify the anomeric forms of the lesion. Labeled peaks are assigned as follows: a: 1′–3′ (α), b: 1′–4′ (α), c: 1′–3′ (β)-5′, d: 1′–4′ (β).
Figure 4.
Atropisomeric states of Fapy-dG residue. These forms were minimized with HyperChem using the Amber force field. The formyl group is depicted in green.
Characterization of Fapy-dG incorporated within a tridecameric oligodeoxynucleotide
Our primary objective is to elucidate the energetic and structural signatures of homogeneous Fapy-dG species. Given the degree of isomerism evident in NMR spectra of the TXT trimer, the resultant equilibrium mixture may effectively preclude isolation and characterization of the respective Fapy anomers. We utilized our novel synthetic strategy to embed Fapy-dG (represented as X) as the central residue within the d(GCGTACXCATGCG) oligodeoxynucleotide and employed a standard purification protocol. Mass spectroscopic analysis of the reverse phase purified Fapy-dG strand reveals an apparently homogeneous species with an expected mass of 3992.7 Da. Following hybridization with its complementary counterpart d(CGCATGCGTACGC) to form the tridecameric Fapy-dG oligodeoxynucleotide duplex, thermal dissociation profiles exhibit non-characteristic biphasic melting behavior (data not shown). Analysis of the transitions suggests the presence of two Fapy-dG species with distinct thermal stabilities. Previous studies have reported similar thermal dissociation profiles for an N5-alkylated Fapy adduct in which the corresponding Me-Fapy-dG duplexes display biphasic melting behavior (53). These findings have been attributed to the presence of an α/β-anomeric mixture with differential thermal stabilities. The bulky aflatoxin B Fapy-dG lesion exhibits comparable behavior (54) with the notable exception that its adduct intercalating properties shift the α/β-anomeric equilibrium in favor of the β-conformation. Remarkably, favorable stacking interactions actually stabilize the β-anomeric-intercalated conformer relative to its parent duplex (54). In order to separate and characterize the respective components within our Fapy-dG isomeric mixture, we have introduced a series of chromatographic fractionation steps into the standard purification protocol. In the following sections, we describe the isolation, purification and characterization of the two predominant isomers of natural Fapy-dG and provide compelling evidence that these isomers represent the α- and β-anomers.
Chromatographic separation of the isomeric Fapy-dG·C oligodeoxynucleotides
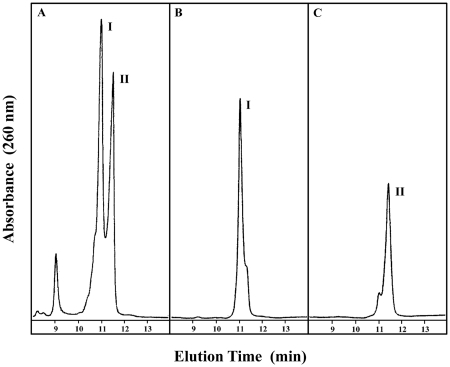
Conventional reverse-phase chromatography of oligodeoxynucleotides harboring cognate Fapy-dG yields a pseudo-homogeneous deprotected species as reflected in the equilibrium mixture of isomers with distinct thermal stabilities (data not shown). Isolation of the natural Fapy anomers necessitates incorporation of a multi-step ion exchange chromatographic protocol to fractionate the heterogeneous mixture. Although ion exchange chromatography is normally employed for separations based on size and/or charge, application of this technique allows resolution of two major fractions eluting at lower (Fraction I) and higher (Fraction II) ionic strengths as depicted in Figure 5A. Successive re-chromatographic isolation of the respective fractions (Figure 5B and C) yields separable species exhibiting indistinguishable mass spectra that are characterized by identical molecular weights (i.e. Fraction I = 3992.71 ± 0.85 Da; Fraction II = 3993.01 ± 0.89 Da) consistent with the predicted value of 3992.7 Da for the Fapy-dG oligodeoxynucleotides (Supplementary Figures S7 and S8). Although the origins for this unique chromatographic behavior warrants further exploration, our results suggest that subtle local changes at the lesion site and/or within neighboring bases provide alternate interactions with the separating chromatographic medium that may result in the distinct elution times/volumes. In fact, recent studies have reported that oligodeoxynucleotides of identical base composition can be discriminated on the basis of their relative nucleotide sequence (55), suggesting that the differential retention ability of the ion exchange resin may resolve sequence context interactions and/or local conformational changes under specific solution conditions. Following exchange on a G-25 sepharose gel filtration column to triethylammonium acetate buffer (pH 6.0), each of the fractions interconvert rapidly yielding a mixture of Fractions I and II. The finding that each oligodeoxynucleotide fraction isomerizes to the starting mixture as a function of time and solution conditions provides additional corroboration of their anomeric nature. The exchanged fractions maintain their original mass and may be re-chromatographed to isolate the Fapy-dG anomers. Each of the fractions comprising the single-strand sequence d(GCGTACXCATGCG) harboring Fapy-dG as the central residue is rapidly hybridized with its complementary counterpart d(CGCATGCGTACGC) to circumvent the unavoidable re-equilibration of isomeric species. The isolated Fapy-dG·C duplexes are subsequently re-chromatographed and buffer exchanged to ensure preparation of homogeneous solutions for temperature-dependent studies.
Figure 5.
Anion exchange chromatographic profiles depicting fractionation of reverse-phase-purified Fapy-dG-containing deoxyoligonucleotide. Panel A illustrates the resolution of two major species eluting at lower (Fraction I) and higher (Fraction II) ionic strength. Successive re-chromatographic isolation of each fraction (as visualized in panels B and C) yields homogeneous species that retain their respective elution times and exhibit indistinguishable mass spectra (Supplementary Figures S7 and S8).
Impact of Fapy-dG isomerization on DNA duplex energetics
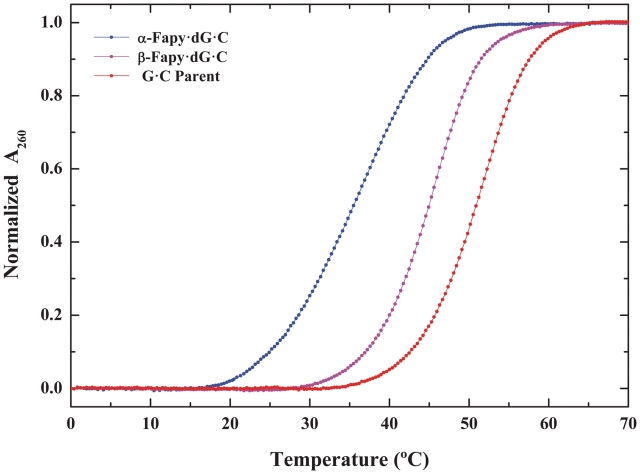
The thermal and thermodynamic stability of the isolated Fapy-dG·C duplexes has been evaluated by temperature-dependent UV spectroscopy. Thermal dissociation profiles of the Fraction I- and Fraction II-derived Fapy-dG·C duplexes and the canonical G·C parent are presented in Figure 6. Inspection of the profiles reveals that the duplexes harboring each of the chromatographically resolvable Fapy-dG isomers exhibit monophasic transitions characteristic of a single anomeric species. The transition temperatures of the differentially destabilized Fapy-dG·C duplexes are 35.1 and 44.7°C. Evaluation of the duplex energetics via van’t Hoff analysis of the denaturation profiles reveals that both Fapy-dG·C duplexes are destabilized relative to the parent G·C tridecamer. The energetic profiles summarized in Table 1 delineate the magnitudes of the observed thermal (ΔTm ∼ −15.4 and −6.0°C) and thermodynamic (ΔΔGvH ∼ −5.5 and −1.5 kcal·mol−1) destabilizations for Fapy-dG·C duplexes I and II, respectively. Significantly, the transition temperatures of the isolated Fapy-dG·C duplexes are in qualitative agreement with the midpoints of a biphasic dissociation profile previously reported for Me-Fapy-dG·C (53). Moreover, one observes comparable differential thermal destabilization of Fapy-dG·C (i.e. ΔTm ∼ −15.4 and −6.0°C) and Me-Fapy-dG·C (i.e. ΔTm ∼ −20 and −8°C) relative to the corresponding parent duplex. Considering the diversity of solution conditions and sequence contexts employed, as well as the distinct nature of the lesions studied, the similarities observed between Fapy-dG·C and Me-Fapy-dG·C are remarkable. In subsequent sections, we provide evidence for our assignment of the isolated oligomeric Fapy-dG·C isomers.
Figure 6.
Thermally-induced dissociation profiles of α-Fapy-dG·C (blue), β-Fapy-dG·C (magenta) and canonical G·C (red) duplexes expressed as normalized absorbance at 260 nm.
Table 1.
Thermal and thermodynamic dissociation parameters of α- and β-Fapy-dG·C duplexes relative to the parent G·C tridecamer
| Duplex acronym | Tm (°C) | ΔTm (°C) | ΔGvH (kcal mol−1) | ΔΔGvH (kcal mol−1) |
|---|---|---|---|---|
| Parent G·C | 50.8 | 15.9 | ||
| α-Fapy-dG·C | 35.4 | −15.4 | 10.6 | −5.3 |
| β-Fapy-dG·C | 44.7 | −6.0 | 14.4 | −1.5 |
Isolated fractions I and II correspond to the Fapy-dG α- and β-anomers
Synthetic approaches designed to incorporate the Fapy-dG lesion within DNA duplexes have historically utilized isomeric mixtures comprised of varying relative ratios. The resultant single-strand species are hybridized with their corresponding complementary strands to form a heterogeneous mixture of Fapy-dG duplexes (29,30,48,50). Consequently, data accumulated on the cognate lesion to date does not specifically resolve the unique properties of each isomer populated within the DNA host duplex. The dearth of investigations aimed at resolving isomeric species may be attributed to the practical notion that once incorporated within DNA, sequence environment plays a significant role on the overall anomeric distribution (50) regardless of initial isomeric ratios in the free adduct. In the present study, we find that under specific solution and chromatographic conditions, single-strand Fapy-dG oligonucleotides may be resolved into two primary species that exhibit unique thermal dissociation profiles upon duplex formation. Our findings infer that the Fapy-dG conformation within each of the isolated isomeric single strands may be preserved upon association to form the DNA host duplex, provided the isolation and hybridization processes are conducted promptly and under optimized solution conditions. Our data therefore challenges the conventional wisdom that anomeric properties can only be evaluated via synthesis of stable bio-isosteric analogues. Nevertheless, the wealth of data accumulated on such analogues has proven insightful in terms of assessing the biological and biophysical impacts of Fapy-dG and their modulation by epimerization. In the following section, we discuss our data in view of recent advances employing stable bio-isosteric analogues to characterize lesion anomerization properties.
Conformationally stable analogues for assessment of Fapy-dG α- and β-anomer properties
In order to alleviate the need to resolve potential anomer-specific properties of Fapy lesions, a number of studies have reported the synthesis of stable bio-isosteric analogues mimicking each of the Fapy-dG anomers (30,56–58). The latter do not undergo anomerization or processing by the cellular repair machinery and may be incorporated into template-primer duplexes for in vitro and/or in vivo mutagenesis studies. A specific example is the conformationally stable β-anomer of Fapy-dG (i.e. β-C-Fapy-dG) in which N6 is replaced by a methylene group (56). The latter has been synthesized and its biochemical and biophysical properties characterized extensively (18,19,30,59). Carbocyclic derivatives designed to resemble cognate damage in the absence of turnover have been employed to study a number of lesions (33,60–63), as these analogues mimic the structural and biochemical properties of the cognate lesion (61). Conformationally stable Fapy-dG isomers have been prepared as carba-analogues to mimic both the β- (57,64) and α- (58) anomeric conformations. NMR analysis of the α-carba-Fapy·dG reveals that in spite of local perturbations at the lesion site, the overall duplex structure is maintained (62). Comparison of these carba-analogues within identical sequence contexts reveals that the Fapy-dG α-anomer is thermally destabilized to a greater extent than its β-anomeric counterpart (58). The finding that the Fapy-dG α-anomer is more destabilizing than the β-anomer is not unprecedented and is generally supported by studies comparing the β-conformation of canonical nucleobases with their α-counterparts. The latter are relatively rare yet readily repaired in vivo by specific endonucleases. One such example is α-adenosine (α-dA) that thermally destabilizes the parent duplex and is processed by endonuclease IV (65,66). Recent evidence reveals that endo IV processes α-anomers of Fapy lesions including those of Fapy-dG (50) and Me-Fapy-dG (67). Collectively, the existing database of Fapy-dG analogues conforms to the consensus regarding the relative energetic impacts of α- and β-isomers. Our observations on the cognate Fapy-dG anomeric species provide further corroboration of such anomer-specific impacts that may assume a critical role in modulating lesion-mediated cytotoxicity and/or mutagenicity.
Relevance of our findings in terms of Fapy-dG mutagenic potential
Lesion outcomes in vivo are likely to be dependent on cell type, polymerase properties, replication/repair proficiencies and sequence-context effects. Parallel in vitro energetic characterizations yield insights regarding the propensity of a particular lesion to exhibit mutagenic or replication blocking properties. As an empirical probe of mutagenicity/cytotoxicity, we already have demonstrated that energy data can assist in predicting a lesion outcome during polymerase-mediated replication (68; C.A.S.A. Minetti., D.P. Remeta, C.R. Iden, F. Johnson and K.J. Breslauer, submitted for publication). The present study provides compelling evidence that β-anomeric species are significantly more stable than their α-counterparts, a finding that is generally consistent with previous observations employing synthetic analogues. The use of stable carbocyclic analogues in template-primer extension analysis reveals unique anomer-dependent consequences on polymerase-mediated replication. Such damage-induced biological outcomes may be correlated with differential energetic impacts as we have proposed for a number of oxidative lesions and exocyclic adducts (68; C.A.S.A. Minetti., D.P. Remeta, C.R. Iden, F. Johnson and K.J. Breslauer, submitted for publication).
Our finding that cognate α-Fapy-dG significantly destabilizes the host duplex is consistent with studies demonstrating that α-carba-Fapy-dG blocks polymerase-mediated DNA synthesis. Conversely, the lower degree of destabilization observed for the cognate β-Fapy-dG corroborates previous assertions that the β-anomer is solely responsible for the mutagenic potential of this lesion. The latter is confirmed by the observation that polymerases may bypass β-Fapy-dG analogues in an error-free or error-prone manner depending on cell type and sequence context (58). Ongoing studies to characterize the replication and repair of cognate Fapy-dG duplexes should provide significant insight regarding the modulatory impact of anomerization on biological processes at the cellular level. Our novel large-scale post-synthetic strategy and anomeric separation protocol will be exploited to furnish a complete structural and thermodynamic characterization of duplexes containing the Fapy-dG α- and β-anomers within varying sequence contexts and as a function of the counterbase.
Lesion analogues and their cognate counterparts are not necessarily isoenergetic
Despite the absence of a detailed structure for the cognate Fapy-dG lesion, a number of reports suggest that conformationally stable Fapy-dG analogues may fulfill this void. Preliminary studies on the solution conformation of β-carba-Fapy-dG reveals that the lesion adopts conventional Watson–Crick interactions yet significantly disrupts the duplex structure to an extent that exceeds the promutagenic 8-oxodG lesion (62). While bio-isosteric analogues are designed to mimic nascent damage, there is evidence that Fapy analogues might not always resemble the cognate lesion given the greater thermal/thermodynamic destabilization observed for the latter in specific cases. Representative examples include the β-C-Fapy-dG analogue in which N6 is replaced by the methylene group (18,56), and the carbocyclic β-carba-Fapy-dG analogue in which O4′ is replaced by a CH2 moiety (57,58). Interestingly, a study employing β-carba-Fapy-dG reveals aberrant temperature-dependent profiles that are not typical of a homogeneous species (64). Under the premise that bio-isosteric analogues are resistant to anomerization, the anomalous biphasic behavior reported for β-carba-Fapy-dG is intriguing and may actually arise from the choice of sequence context and solution conditions. Our energy data on the cognate isomeric species assigned as β-Fapy·dG reveals that lesion-induced thermal and thermodynamic destabilizations are generally less dramatic than those reported for conformationally stable β-analogues. Inasmuch as studies of Fapy-analogues have furnished significant insight on lesion-induced perturbations, caution must be exercised when attempting to extrapolate these findings to elucidate energetic/structural properties of the cognate lesion within genomic DNA and underlying biological consequences at the molecular level.
Although valuable information has been gleaned from studies employing surrogate Fapy analogues, there are reports suggesting that specific interactions with glycosylases might differ from those of the cognate lesion. The basis for such concerns are the distinct lesion-residue interactions observed when comparing crystal structures of the Fpg glycosylase complexed with the carba-Fapy-dG analogue (31) versus a mutant form bound to the cognate 8-oxodG lesion (32). Recent molecular mechanics studies reveal that lesion-substrate interactions with Fpg occur via a syn-conformation (34), yet crystal structure analysis suggests that the carba-Fapy-dG analogue adopts a non-planar anti-conformation in its complex with Fpg (31). These disparities have been attributed to a number of factors, including the participation of a non-conserved amino acid residue in Fpg:carba-Fapy-dG interactions (i.e. S77E in L. lactis Fpg). Considering that the carba-analogue interacts with a residue that is not conserved within the Fpg-family of glycosylases, it has been hypothesized that the crystallographic structure of the Fpg:carba-Fapy-dG complex might not accurately reflect biologically relevant Fpg interactions that mediate cognate lesion recognition and processing. Future studies employing Fpg mutants that lack catalytic activity yet retain wild-type binding ability are required to probe cognate Fapy-dG lesion interactions, the energy landscape of BER recognition, and the impact of lesion anomerization on the overall process.
CONCLUSION
The experimental strategy outlined herein provides a facile and robust synthetic scheme for the large-scale production of Fapy-dG embedded within any oligodeoxynucleotide sequence. The overall utility of the methodology has been enhanced by optimizing a multistep purification protocol that permits chromatographic isolation of tridecameric deoxynucleotides containing cognate Fapy-dG at anomeric resolution. Hybridization of the isomeric species with their complementary counterpart yields two Fapy-dG·C duplexes that exhibit unique thermal and thermodynamic properties. The resultant duplexes are differentially destabilized relative to the canonical G·C parent and thereby reflect two primary conformations of the cognate Fapy-dG lesion that are consistent with α- and β-anomers, respectively. Comparison of the respective energetic profiles reveals that α-Fapy-dG·C is more destabilizing than β-Fapy-dG·C. Our findings underscore the importance of characterizing anomer-specific impacts in terms of biological outcomes, particularly given the fact that anomerization is likely to occur within genomic DNA and pose significant challenges to the cellular replication/repair machinery.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Institutes of Health (CA047995 and ES017368).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Adelman R, Saul R, Ames B. Oxidative damage to DNA: relation to species metabolic rate and life span. Proc. Natl Acad. Sci. USA. 1988;85:2706–2708. doi: 10.1073/pnas.85.8.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dizdaroglu M, Kirkali G, Jaruga P. Formamidopyrimidines in DNA: mechanisms of formation, repair, and biological effects. Free Radical Biol. Med. 2008;45:1610–1621. doi: 10.1016/j.freeradbiomed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Aruoma OI, Halliwell B, Dizdaroglu M. Iron ion-dependent modification of bases in DNA by the superoxide radical-generating system hypoxanthine/xanthine oxidase. J. Biol. Chem. 1989;264:13024–13028. [PubMed] [Google Scholar]
- 4.Aruoma OI, Halliwell B, Gajewski E, Dizdaroglu M. Damage to the bases in DNA induced by hydrogen-peroxide and ferric ion chelates. J. Biol. Chem. 1989;264:20509–20512. [PubMed] [Google Scholar]
- 5.Gajewski E, Rao G, Nackerdien Z, Dizdaroglu M. Modification of DNA bases in mammalian chromatin by radiation-generated free-radicals. Biochemistry. 1990;29:7876–7882. doi: 10.1021/bi00486a014. [DOI] [PubMed] [Google Scholar]
- 6.Dizdaroglu M, Rao G, Halliwell B, Gajewski E. Damage to the DNA bases in mammalian chromatin by hydrogen-peroxide in the presence of ferric and cupric ions. Arch. Biochem. Biophys. 1991;285:317–324. doi: 10.1016/0003-9861(91)90366-q. [DOI] [PubMed] [Google Scholar]
- 7.Nackerdien Z, Olinski R, Dizdaroglu M. DNA-base damage in chromatin of gamma-irradiated cultured human-cells. Free Radical Res. Com. 1992;16:259–273. doi: 10.3109/10715769209049179. [DOI] [PubMed] [Google Scholar]
- 8.Pouget JP, Douki T, Richard MJ, Cadet J. DNA damage induced in cells by gamma and UVA radiation as measured by HPLC/GC-MS and HPLC-EC and comet assay. Chem. Res. Toxicol. 2000;13:541–549. doi: 10.1021/tx000020e. [DOI] [PubMed] [Google Scholar]
- 9.Nyaga SG, Jaruga P, Lohani A, Dizdaroglu M, Evans MK. Accumulation of oxidatively induced DNA damage in human breast cancer cell lines following treatment with hydrogen peroxide. Cell Cycle. 2007;6:1472–1478. [PubMed] [Google Scholar]
- 10.Malins DC, Haimanot R. Major alterations in the nucleotide structure of DNA in cancer of the female breast. Cancer Res. 1991;51:5430–5432. [PubMed] [Google Scholar]
- 11.Malins DC, Polissar NL, Gunselman SJ. Progression of human breast cancers to the metastatic state is linked to hydroxyl radical-induced DNA damage. Proc. Natl Acad. Sci. USA. 1996;93:2557–2563. doi: 10.1073/pnas.93.6.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang JQ, Markesbery WR, Lovell MA. Increased oxidative damage in nuclear and mitochondrial DNA in mild cognitive impairment. J. Neurochem. 2006;96:825–832. doi: 10.1111/j.1471-4159.2005.03615.x. [DOI] [PubMed] [Google Scholar]
- 13.Lovell MA, Markesbery WR. Oxidative DNA damage in mild cognitive impairment and late-stage Alzheimers disease. Nucleic Acids Res. 2007;35:7497–7504. doi: 10.1093/nar/gkm821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirkali G, Tunca M, Genc S, Jaruga P, Dizdaroglu M. Oxidative DNA damage in polymorphonuclear leukocytes of patients with familial Mediterranean fever. Free Radic. Biol. Med. 2008;44:386–393. doi: 10.1016/j.freeradbiomed.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 15.Boiteux S, O’Connor TR, Laval J. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987;6:3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tchou J, Kasai H, Shibutani S, Chung MH, Laval J, Grollman AP, Nishimura S. 8-Oxoguanine (8-Hydroxyguanine) DNA glycosylase and its substrate-specificity. Proc. Natl Acad. Sci. USA. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Substrate-specificity of the Escherichia-coli Fpg protein (formamidopyrimidine DNA glycosylase) - excision of purine lesions in DNA produced by ionizing-radiation or photosensitization. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 18.Wiederholt CJ, Delaney MO, Greenberg MM. Interaction of DNA containing Fapy · dA or its C-nucleoside analogues with base excision repair enzymes. Implications for mutagenesis and enzyme inhibition. Biochemistry. 2002;41:15838–15844. doi: 10.1021/bi025903e. [DOI] [PubMed] [Google Scholar]
- 19.Wiederholt CJ, Delaney MO, Pope MA, David SS, Greenberg MM. Repair of DNA containing Fapy center dot dG and its beta-C-nucteoside analogue by formamidopyrimidine DNA glycosylase and MutY. Biochemistry. 2003;42:9755–9760. doi: 10.1021/bi034844h. [DOI] [PubMed] [Google Scholar]
- 20.Karahalil B, Girard PM, Boiteux S, Dizdaroglu M. Substrate specificity of the Ogg1 protein of Saccharomyces cerevisiae: excision of guanine lesions produced in DNA by ionizing radiation or hydrogen peroxide metal ion generated free radicals. Nucleic Acids Res. 1998;26:1228–1232. doi: 10.1093/nar/26.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dherin C, Radicella JP, Dizdaroglu M, Boiteux S. Excision of oxidatively damaged DNA bases by the human alpha-hOgg1 protein and the polymorphic alpha-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 1999;27:4001–4007. doi: 10.1093/nar/27.20.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dherin C, Dizdaroglu M, Doerflinger H, Boiteux S, Radicella JP. Repair of oxidative DNA damage in Drosophila melanogaster: identification and characterization of dOgg1, a second DNA glycosylase activity for 8-hydroxyguanine and formamidopyrimidines. Nucleic Acids Res. 2000;28:4583–4592. doi: 10.1093/nar/28.23.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morales-Ruiz T, Birincioglu M, Jaruga P, Rodriguez H, Roldan-Arjona T, Dizdaroglu M. Arabidopsis thaliana Ogg1 protein excises 8-hydroxyguanine and 2,6-diamino-4-hydroxy-5-formamidopyrimidine from oxidatively damaged DNA containing multiple lesions. Biochemistry. 2003;42:3089–3095. doi: 10.1021/bi027226u. [DOI] [PubMed] [Google Scholar]
- 24.Krishnamurthy N, Haraguchi K, Greenberg MM, David SS. Efficient removal of formamidopyrimidines by 8-oxoguanine glycosylases. Biochemistry. 2008;47:1043–1050. doi: 10.1021/bi701919u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl Acad. Sci. USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaruga P, Birincioglu M, Rosenquist TA, Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 2004;43:15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- 27.Roy LM, Jaruga P, Wood TG, McCullough AK, Dizdaroglu M, Lloyd RS. Human polymorphic variants of the NEIL1 DNA glycosylase. J. Biol. Chem. 2007;282:15790–15798. doi: 10.1074/jbc.M610626200. [DOI] [PubMed] [Google Scholar]
- 28.Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc. Natl Acad. Sci. USA. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haraguchi K, Greenberg MM. Synthesis of oligonucleotides containing Fapy · dG(N6-(2-deoxy-alpha,beta-D-erythro-pentofuranosyl)-2,6-diamino-4-hydroxy-5-formamidopyrimidine) J. Am. Chem. Soc. 2001;123:8636–8637. doi: 10.1021/ja0160952. [DOI] [PubMed] [Google Scholar]
- 30.Haraguchi K, Delaney MO, Wiederholt CJ, Sambandam A, Hantosi Z, Greenberg MM. Synthesis and characterization of oligodeoxynucleotides containing formamidopyrimidine lesions and nonhydrolyzable analogues. J. Am. Chem. Soc. 2002;124:3263–3269. doi: 10.1021/ja012135q. [DOI] [PubMed] [Google Scholar]
- 31.Coste F, Ober M, Carell T, Boiteux S, Zelwer C, Castaing B. Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase. J. Biol. Chem. 2004;279:44074–44083. doi: 10.1074/jbc.M405928200. [DOI] [PubMed] [Google Scholar]
- 32.Fromme JC, Verdine GL. DNA lesion recognition by the bacterial repair enzyme MutM. J. Biol. Chem. 2003;278:51543–51548. doi: 10.1074/jbc.M307768200. [DOI] [PubMed] [Google Scholar]
- 33.Minetti CASA, Remeta DP, Zharkov DO, Plum GE, Johnson F, Grollman AP, Breslauer KJ. Energetics of lesion recognition by a DNA repair protein: Thermodynamic characterization of formamidopyrimidine-glycosylase (Fpg) interactions with damaged DNA duplexes. J. Mol. Biol. 2003;328:1047–1060. doi: 10.1016/s0022-2836(03)00365-6. [DOI] [PubMed] [Google Scholar]
- 34.Song K, Kelso C, de los Santos C, Grollman AP, Simmerling C. Molecular simulations reveal a common binding mode for glycosylase binding of oxidatively damaged DNA lesions. J. Am. Chem. Soc. 2007;129:14536–14537. doi: 10.1021/ja075128w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song K, Hornak V, De los Santos C, Grollman AP, Simmerling C. Molecular mechanics parameters for the FapydG DNA lesion. J. Comput. Chem. 2008;29:17–23. doi: 10.1002/jcc.20625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. MRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Goddard TD, Kneller DG. San Francisco: University of California; 2008. http://www.cgl.ucsf.edu/home/sparky/ [Google Scholar]
- 38.Beaucage SL. Oligodeoxyribonucleotide synthesis: phosphoramidite approach, In Protocols for Oligonucleotides and Analogs. In: Agrawal S, editor. Methods in Molecular Biology Series. Vol. 20. Totowa, NJ: Human Press; 1993. pp. 33–61. [DOI] [PubMed] [Google Scholar]
- 39.Gelfand CA, Plum GE, Mielewczyk S, Remeta DP, Breslauer KJ. A quantitative method for evaluating the stabilities of nucleic acids. Proc. Natl Acad. Sci. USA. 1999;96:6113–6118. doi: 10.1073/pnas.96.11.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griswold BL, Humoller FL, McIntyre AR. Inorganic phosphates and phosphate esters in tissue extracts. Anal. Chemistry. 1951;23:192–194. [Google Scholar]
- 41.Minetti CA, Remeta DP, Dickstein R, Breslauer KJ. Energetic signatures of single base bulges: thermodynamic consequences and biological implications. Nucleic Acids Res. 2010;38:97–116. doi: 10.1093/nar/gkp1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Job P. Formation and stability of inorganic complexes in solution. Ann. Chim. 1928;9:113–203. [Google Scholar]
- 43.Marky LA, Breslauer KJ. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 44.Greenberg MM, Hantosi Z, Wiederholt CJ, Rithner CD. Studies on N4-(2-deoxy-D-pentofuranosyl)-4,6-diamino-5-formamidopyrimidine (Fapy·dA) and N6-(2-deoxy-D-pentofuranosyl)-6-diamino-5-formamido-4-hydroxypyrimidine (Fapy·dG) Biochemistry. 2001;40:15856–15861. doi: 10.1021/bi011490q. [DOI] [PubMed] [Google Scholar]
- 45.Patro JN, Wiederholt CJ, Jiang YL, Delaney JC, Essigmann JM, Greenberg MM. Studies on the replication of the ring opened formamidopyrimidine, Fapy·dG in Escherichia coli. Biochemistry. 2007;46:10202–10212. doi: 10.1021/bi700628c. [DOI] [PubMed] [Google Scholar]
- 46.Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK. Genetic effects of oxidative DNA damages: comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boiteux S, Belleney J, Roques BP, Laval J. Two rotameric forms of open ring 7-methylguanine are present in alkylated polynucleotides. Nucleic Acids Res. 1984;12:5429–5439. doi: 10.1093/nar/12.13.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang YL, Wiederholt CJ, Patro JN, Haraguchi K, Greenberg MM. Synthesis of oligonucleotides containing Fapy · dG (N-6-(2-deoxy-alpha,beta-D-erythropentofuranosyl)-2,6-diamino-4-hydroxy-5-formamidopyrimidine) using a 5'-dimethoxytrityl dinucleotide phosphoramidite. J. Org. Chem. 2005;70:141–149. doi: 10.1021/jo048253o. [DOI] [PubMed] [Google Scholar]
- 49.Raoul S, Bardet M, Cadet J. Gamma irradiation of 2'-deoxyadenosine in oxygen-free aqueous solutions: identification and conformational features of formamidopyrimidine nucleoside derivatives. Chem. Res. Toxicol. 1995;8:924–933. doi: 10.1021/tx00049a005. [DOI] [PubMed] [Google Scholar]
- 50.Patro JN, Haraguchi K, Delaney MO, Greenberg MM. Probing the configurations of formamidopyrimidine lesions Fapy· dA and Fapy·dG in DNA using endonuclease IV. Biochemistry. 2004;43:13397–13403. doi: 10.1021/bi049035s. [DOI] [PubMed] [Google Scholar]
- 51.Lown WJ, Krowicki K. Efficient total synthesis of the oligopeptide antibiotic netropsin and distamycin. J. Org. Chem. 1985;50:3774–3779. [Google Scholar]
- 52.Burgdorf LT, Carell T. Synthesis, stability, and conformation of the formamidopyrimidine G DNA lesion. Chem. Eur. J. 2002;8:293–301. doi: 10.1002/1521-3765(20020104)8:1<293::aid-chem293>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 53.Christov PP, Brown KL, Kozekov ID, Stone MP, Harris TM, Rizzo CJ. Site-specific synthesis and characterization of oligonucleotides containing an N-6-(2-deoxy-d-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-methylformamidopyrimidine lesion, the ring-opened product from N7-methylation of deoxyguanosine. Chem. Res. Toxicol. 2008;21:2324–2333. doi: 10.1021/tx800352a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mao H, Deng ZW, Wang F, Harris TM, Stone MP. An intercalated and thermally stable FAPY adduct of aflatoxin B-1 in a DNA duplex: Structural refinement from H-1 NMR. Biochemistry. 1998;37:4374–4387. doi: 10.1021/bi9718292. [DOI] [PubMed] [Google Scholar]
- 55.Buncek M, Backovska V, Holasova S, Radilova H, Safarova M, Kunc F, Haluza R. Unusual chromatographic behavior of oligonucleotide sequence isomers on two different anion exchange HPLC columns. Anal. Biochem. 2006;348:300–306. doi: 10.1016/j.ab.2005.10.047. [DOI] [PubMed] [Google Scholar]
- 56.Delaney MO, Greenberg MM. Synthesis of oligonucleotides and thermal stability of duplexes containing the beta-C-nucleoside analogue of Fapy· dG. Chem. Res. Toxicol. 2002;15:1460–1465. doi: 10.1021/tx025588x. [DOI] [PubMed] [Google Scholar]
- 57.Ober M, Muller H, Pieck C, Gierlich J, Carell T. Base pairing and replicative processing of the formamidopyrimidine-dG DNA lesion. J. Am. Chem. Soc. 2005;127:18143–18149. doi: 10.1021/ja0549188. [DOI] [PubMed] [Google Scholar]
- 58.Buesch F, Pieck JC, Ober M, Gierlich J, Hsu GW, Beese LS, Carell T. Dissecting the differences between the alpha and beta anomers of the oxidative DNA lesion FaPydG. Chem. Eur. J. 2008;14:2125–2132. doi: 10.1002/chem.200701373. [DOI] [PubMed] [Google Scholar]
- 59.Wiederholt CJ, Greenberg MM. Fapy-dG instructs Klenow exo(-) to misincorporate deoxyadenosine. J. Am. Chem. Soc. 2002;124:7278–7279. doi: 10.1021/ja026522r. [DOI] [PubMed] [Google Scholar]
- 60.Johnson F, Dorman G, Rieger RA, Marumoto R, Iden CR, Bonala R. Synthesis of enzymatically noncleavable carbocyclic nucleosides for DNA-N-glycosylase studies. Chem. Res. Toxicol. 1998;11:193–202. doi: 10.1021/tx9701539. [DOI] [PubMed] [Google Scholar]
- 61.de los Santos C, El-khateeb M, Rege P, Tian K, Johnson F. Impact of the Cl' configuration of abasic sites on DNA duplex structure. Biochemistry. 2004;43:15349–15357. doi: 10.1021/bi048400c. [DOI] [PubMed] [Google Scholar]
- 62.Lukin M, Zaliznyak T, Attaluri S, Johnson F, de los Santos C. NMR characterization of DNA with a single carba-Fapy. J. Biomol. Struct. Dyn. 2007;24:613. [Google Scholar]
- 63.Minetti C, Remeta DP, Breslauer KJ. A continuous hyperchromicity assay to characterize the kinetics and thermodynamics of DNA lesion recognition and base excision. Proc. Natl Acad. Sci. USA. 2008;105:70–75. doi: 10.1073/pnas.0710363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ober M, Linne U, Gierlich J, Carell T. The two main DNA lesions 8-oxo-7,8-dihydroguanine and 2,6-diamino-5-formamido-4-hydroxypyrimidine exhibit strongly different pairing properties. Angewandte Chemie-International Edition. 2003;42:4947–4951. doi: 10.1002/anie.200351287. [DOI] [PubMed] [Google Scholar]
- 65.Ide H, Shimizu H, Kimura Y, Sakamoto S, Makino K, Glackin M, Wallace SS, Nakamuta H, Sasaki M, Sugimoto N. Influence of α-deoxyadenosine on the stability and structure of DNA. Thermodynamic and molecular mechanics studies. Biochemistry. 1995;34:6947–6955. doi: 10.1021/bi00021a006. [DOI] [PubMed] [Google Scholar]
- 66.Aramini JM, Cleaver SH, Pon RT, Cunningham RP, Germann MW. Solution structure of a DNA duplex containing an alpha-anomeric adenosine: insights into substrate recognition by endonuclease IV. J. Mol. Biol. 2004;338:77–91. doi: 10.1016/j.jmb.2004.02.035. [DOI] [PubMed] [Google Scholar]
- 67.Christov P, Banerjee S, Stone M, Rizzo C. Selective incision of the -N5-Methyl-formamidopyrimidine anomer by Escherichia coli endonuclease IV. J. Nucleic Acids. 2010 doi: 10.4061/2010/850234. doi:10.4061/2010/850234 [Epub ahead of print, 25 July 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Minetti CA, Remeta DP, Johnson F, Iden CR, Breslauer KJ. Impact of alpha-hydroxy-propanodeoxyguanine adducts on DNA duplex energetics: opposite base modulation and implications for mutagenicity and genotoxicity. Biopolymers. 2010;93:370–382. doi: 10.1002/bip.21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.