Abstract
The deposition of the histones H3/H4 onto DNA to give the tetrasome intermediate and the displacement of H3/H4 from DNA are thought to be the first and the last steps in nucleosome assembly and disassembly, respectively. Anti-silencing function 1 (Asf1) is a chaperone of the H3/H4 dimer that functions in both of these processes. However, little is known about the thermodynamics of chaperone–histone interactions or the direct role of Asf1 in the formation or disassembly of histone–DNA complexes. Here, we show that Saccharomyces cerevisiae Asf1 shields H3/H4 from unfavorable DNA interactions and aids the formation of favorable histone–DNA interactions through the formation of disomes. However, Asf1 was unable to disengage histones from DNA for tetrasomes formed with H3/H4 and strong nucleosome positioning DNA sequences or tetrasomes weakened by mutant (H3K56Q/H4) histones or non-positioning DNA sequences. Furthermore, Asf1 did not associate with preformed tetrasomes. These results are consistent with the measured affinity of Asf1 for H3/H4 dimers of 2.5 nM, which is weaker than the association of H3/H4 for DNA. These studies support a mechanism by which Asf1 aids H3/H4 deposition onto DNA but suggest that additional factors or post-translational modifications are required for Asf1 to remove H3/H4 from tetrasome intermediates in chromatin.
INTRODUCTION
The nucleosome is the fundamental repeating unit of the chromatin structure found in eukaryotes, in which the DNA wraps around a core histone octamer formed by one H3/H4 heterotetramer flanked by two H2A/H2B heterodimers (1–3). It is now well established that the processes of assembling the nucleosome from newly synthesized and recycled histones as well as disassembling chromatin for all DNA-dependent processes such as DNA replication, DNA repair and transcription are orchestrated by a variety of ATP-dependent remodeling complexes and histone chaperones (4,5). Histone chaperones are acidic proteins that facilitate histone deposition, exchange and eviction during nucleosome assembly and disassembly (5–9).
As a chaperone of histones H3/H4, Anti-silencing Function 1 (Asf1) has an extensive range of functions in chromatin regulation. Asf1 was initially found to be a suppressor of gene silencing when over-expressed in Saccharomyces cerevisiae (10,11). Deletion of S. cerevisiae Asf1 (yAsf1) leads to defects in DNA replication, repair, transcription and histone modification (12–20). Although there are species-specific variations in the isoforms of Asf1, it is conserved across eukaryotic species as a central H3/H4 chaperone (21–25). The crystal structures of Asf1 bound to the H3/H4 dimer show that Asf1 binds to H3 at the H3/H4 dimerization surface, physically blocking formation of the H3/H4 heterotetramer and importantly, interactions with both H3 and H4 are required for Asf1 histone chaperone function (26–28). The C-terminus of histone H4 adopts a dramatically different conformation when bound to Asf1 relative to its position in the nucleosome (3), which led to our suggestion that the H4 tail could facilitate chromatin disassembly/ assembly via a ‘strand capture mechanism’ (26). Furthermore, the findings that yAsf1 mediates chromatin disassembly from promoters during transcriptional activation and chromatin disassembly and reassembly during transcriptional elongation (14,19,29–31) leaves open the possibility that Asf1 might directly participate in the nucleosome disassembly process.
Whereas the detailed interactions of H3/H4 within a nucleosome (3) and bound to Asf1 (26,28) are known, the dynamic transition between them remains a mystery. Quantitative studies of histone interactions with another histone chaperone, Nap1, reveal that the binding affinity of histones for Nap1 is weaker than the affinity of histones for DNA (32,33). Furthermore, the histone affinity for DNA is quite sensitive to the ability of the DNA sequence to favorably position the nucleosome (32,34). Interestingly, histone modifications such as the acetylation of H3 at K56 alter the dynamic behavior of the nucleosome and weaken the affinity of the histones for DNA (32,35). The presentation of H3/H4 by Asf1 to acetyltransferases in vivo is critical for the acetylation of H3 at K56 (17,36). However, it has been difficult to dissociate the potential direct role of Asf1 in nucleosome disassembly via a strand capture or any other mechanism from the indirect effects of weaker histone–DNA contacts generated by H3 K56 acetylation, which are also mediated by Asf1 (17,37).
Knowledge about the thermodynamics of chaperone–histone interactions in general, and of Asf1–histone interactions in particular is quite limited. The earliest step in nucleosome assembly pathway involves the deposition of two H3/H4 dimers on the DNA resulting in the intermediate known as the tetrasome (38,39). Here, we focused on the action of Asf1 in tetrasome assembly and disassembly in vitro under conditions of physiological ionic strength, which provide much clearer insights into the histone chaperone capabilities of Asf1.
MATERIALS AND METHODS
Preparation of proteins
Plasmid pET60yAsf1FL was produced by cloning the full-length yAsf1 sequence for amino acids 1–279 into the vector Nova pET-60-DEST (Gateway) between the GST-tag coding sequence and the His6-tag coding sequence. All yAsf1 constructs used had the −1 position proline substituted to a cysteine (Quikchange, Stratagene) for fluorophore attachment. The pGST-V94R vector was derived from the pGST-Asf1tr overexpression vector (27) using the QuikChange mutagenesis kit (Stratagene) and purified as described in (27). Expression of the N-terminal GST-tag and C-terminal His6-tag fusion protein was induced by addition of 0.8 mM IPTG to exponentially growing Escherichia coli Rosetta (pLysS) cells (Novagen) containing plasmid pET60yAsf1FL for 4 h at 27°C. The purification protocol was identical to (27), with the exception of the subsequent purification by nickel affinity chromatography. For this, the released protein was bound to a HisTrap HP column (GE). The column was washed with three column volumes of Buffer A (20 mM Tris–HCl pH 7.9, 1 M NaCl, 5 mM imidazole, 20 mM BME, 10% glycerol, 0.05% Brij-35). The full-length protein with C-terminal His-tag was eluted via a gradient to Buffer B (10 mM Tris–HCl pH 7.9, 0.5 M NaCl, 250 mM imidazole, 10% glycerol, 0.05% Brij-35, 0.5 mM TCEP, 1 mM EDTA).
Xenopus laevis histones H3 and H4 with amino acid residue substitutions H3 C110A and H4 T71C, were expressed and purified as previously described, with modifications that are detailed in the Supplementary material (40,41). Purification of HMGB1 was carried out according to (42).
Preparation of DNA and tetrasomes
DNA fragments of an 80 bp X. laevis 5S rDNA sequence were prepared by PCR using the plasmid pMM and 5SprimerI (CCC AGA AGG CAG CAC AAG G) and 5SprimerII (AGA CGA TAT CGG GCA CTT TCA GG). DNA fragments of 80 bp containing a very strong positioning sequence (‘Widom 601’) from the Widom 601 plasmid (34,43) were prepared by PCR using the 601primerI (GTC GTA GAC AGC TCT AGC A) and 601primerII (TAG GGA GTA ATC CCC TTG). DNA fragments of length 146 bp were produced in the same way, but with primers 601146 F (CTG GAG AAT CCC GGT GC) and 601146 R (CAG GAT GTA TAT ATC TGA CAC GTG C). DNA fragments of 80 bp were produced from a portion of the coding region of the yAsf1 gene that was predicted to be a weak or non-positioning sequence (NPS). The NPS 80 bp fragments were prepared by PCR using the NPSprimerI (TGC TCT TCC GCT TCC T) and NPSprimerII (CGC CTT TGA GTG AGC TGA TAC). The PCR products were purified by ion-exchange chromatography using a DEAE column (Tyopearl MD-G DEAE SPW-1; Tosoh Bioscience LLC) followed by ethanol precipitation. The supercoiled 5SDNA and 601 plasmids were purified by agarose gel electrophoresis from mini-prepped DNA. The corresponding linear DNA fragments were produced by digesting the pMM plasmid with HindIII and the Widom 601 plasmid with EcoRI, respectively.
Purified 80 bp DNA fragments were reconstituted with H3/H4 to form tetrasomes using a continuous salt-gradient dialysis method (41). Tetrasomes were formed at a final molar ratio of tetramer to DNA of 1:1 with a final DNA concentration of 0.15 mg/ml. The mixture was transferred into 3500 MWCO Slide-A-Lyzer Dialysis Cassette (Thermo Scientific) and dialyzed against 300 ml of 10 mM Tris–HCl (pH 7.5), 2 M NaCl, 1 mM EDTA and 1 mM TCEP. A total of 3700 ml of 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, and 1 mM TCEP was added to the 300 ml at a rate of 1 ml/min to give a final salt concentration of 150 mM. Tetrasomes were alternatively formed by direct addition of H3/H4 to the DNA in the presence or absence of Asf1, producing two bands confirmed by quantitation of fluorescence to be histone:DNA ratio twice as large for the top band compared to the bottom band (Figure S3).
Electrophoretic analyses
Mixtures of yAsf1 and/or H3/H4 and DNA were prepared in assembly buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5 mM TCEP). The samples were incubated at 4°C for 1 h prior to electrophoresis unless another temperature or time is specifically stated. Seven percent polyacrylamide gel (59:1 acrylamide:bis-acrylamide) containing 0.2×TBE were pre-run for 60 min in 0.2×TBE at 4°C at ∼5 V/cm. The reactions were electrophoresed at 4°C and 70 V for 2–3 h. Bands were visualized by recording the signal of the histone H4 fluorescein (FM) label, the Alexa Fluor 532 signal of yAsf* and SYBR Green I (Invitrogen) nucleic acid stain fluorescence with a Typhoon 9400 Variable mode imager (GE Healthcare).
Analytical ultracentrifugation
Analytical ultracentrifugation (AUC) experiments were conducted at 20°C on a Beckman Optima XL-A analytical ultracentrifuge equipped with absorbance optics and an An60 Ti rotor, using standard approaches. The partial specific volume at this temperature was determined from the amino acid composition to be 0.7452 ml/g for the H3/H4, using the program SEDNTERP, version 1.09. Proteins were dialyzed against either high salt buffer containing 10 mM Tris–HCl, pH 7.5, 2 M NaCl, 1 mM EDTA and 1 mM TCEP or low salt buffer containing 10 mM Tris-HCl, pH 7.5, 150 mM KCl, 2 mM MgCl2 and 0.05% Brij-35 prior to analysis. The density of the high salt buffer was 1.07 767 g/ml and 1.00 504 g/ml for the low salt buffer, and the viscosity was 0.01219 P and 0.0102 P for the high salt buffer and low salt buffer, respectively, at 20°C as calculated with SEDNTERP.
Analytical sedimentation velocity and equilibrium studies were conducted with the purified H3/H4 complex dialyzed against buffer. For sedimentation velocity experiments, the sample was loaded into Epon double-sector cells and data were collected at 280 nm in a continuous scan mode at 20°C at 42 000 rpm. The sedimentation velocity data were analyzed using SEDFIT (44), floating the parameters for meniscus, bottom, time independent noise and frictional ratio. For the sedimentation equilibrium experiments, the sample was loaded into Epon charcoal-filled six-channel centerpiece. The complex was sedimented to equilibrium at three rotor speeds: 15 K, 25 K and 30 K rpm at 20°C. The sedimentation equilibrium data were analyzed using the programs SEDFIT and SEDPHAT (45).
Fluorescence assays
The X. laevis H3C110A mutant (xH3C110A) was used to mimic the yeast histone dimerization surface and to avoid any unwanted cysteine / dye reaction (46). X. laevis histone H4T71C mutant (xH4T71C) was fluorescently labeled (26,46) (Supplementary Figure S1) with the Qsy9 fluorescence quencher or fluorescein (FM) (Invitrogen) as previously described (46). The xH3C110A and Qsy9 or FM labeled xH4T71C were then prepared as tetramers yielding either H3/H4*Q or H3/H4*FM. yAsf1 was labeled with Alexa Fluor 532 (Invitrogen) yielding yAsf1*. The position of the labeled cysteine is shown in Supplementary Figure S1, while the other two cysteines in yAsf1 (Cys30 and Cys99) are not near the histone-binding interface and are not labeled efficiently in this procedure (as verified by mass spectrometry, data not shown). Measurements were performed at a yAsf1 concentration of 1 nM in buffer [10 mM Tris-HCl (pH 7.5), 150 mM KCl, 2 mM MgCl2, 1% glycerol and 0.05% Brij-35]. Double measurements were made with an integration time of 0.5 s on a Horiba Fluorolog-3 spectrometer at 20°C, using a 0.5 cm path-length cuvette. The excitation wavelength was 528 nm, with a slit width of 7 mm; emission was recorded at 547 nm with a slit width of 7 mm. H3/H4 labeled with Qsy9 was titrated to the cuvette containing yAsf1* and the decrease of yAsf1* fluorescence was monitored. Control samples with buffer or unlabeled histones were analyzed likewise to discount buffer effects on fluorescence. The buffer was scanned in the same range used in all experiments for background contributions to the readings and was corrected for in each spectrum. Varying incubation times (0–30 min) confirmed that the fluorescence signal had reached equilibrium by 5 min. The reactions were allowed to equilibrate at 20°C for at least 5 min prior to measurement, and at least three independent experiments were performed for each sample. Data were fitted with the ligand-depleted binding model [Equation (1)] in cases where the concentration of yAsf1* was within 10-fold of the Kd value.
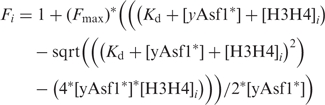 |
(1) |
with the variable i indicating the varying concentrations of H3/H4 that were titrated into the yAsf1*.
RESULTS
Asf1 increases the incorporation of histones into disomes
Asf1–H3/H4 complexes supply histones to nucleosome assembly complexes CAF-1 and HIRA in a form that is competent for nucleosome assembly (13,21,22,27). However, under a variety of conditions in vitro, H3/H4 can interact productively with DNA to form tetrasomes in the absence of histone chaperones (47,48). Furthermore, the modification of H3 by acetylation at K56 produces tetrasomes that are less stable than those formed with wild-type histones (32), and the K56Q substitution has similar effects on the dynamics and remodeling of nucleosomes as acetylation of H3 at K56 (35,37,49). Therefore, we tested the assembly properties of yAsf1 in this minimal system to learn about the intrinsic properties of Asf1 in tetrasome and disome assembly in vitro.
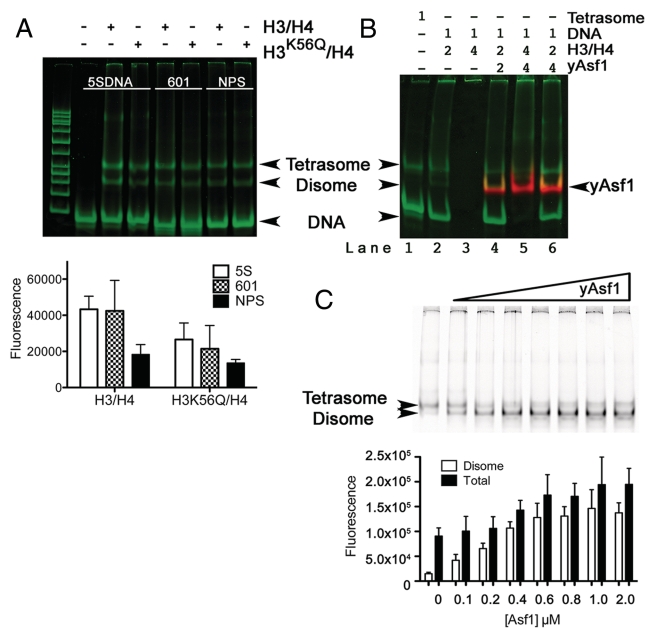
We used electrophoretic mobility shift assays (EMSA) with a minimal system comprising fluorescently labeled S. cerevisiae Asf1 (yAsf1*) (Supplementary Figure S1) in combination with labeled X. laevis H3/H4 carrying an H3 cysteine 110 to alanine substitution (H3/H4*FM) or with an additional substitution of H3 lysine 56 to glutamine (H3K56Q/H4*FM) and 80 bp DNA. In this system, yAsf1* migrates slightly faster than the tetrasome but slower than the free DNA, and the yAsf1*–H3/H4*FM complex does not enter the gel (Supplementary Figure S2). Figure 1A shows that both H3/H4*FM and H3K56Q/H4* FM associate with 80 bp DNA fragments of the naturally occurring nucleosome positioning sequence from the X. borealis 5SRNA gene (5SDNA) (50) and the selected high affinity nucleosome positioning sequence Widom 601 (601) (34,43) as well as a non-positioning DNA sequence (a region of the Asf1 gene that we named NPS). At near physiological ionic strength, one of the species migrates at the position of tetrasomes produced by classic salt dialysis methods. There is also a faster migrating species that we are calling a disome (51), based on its observation in previous studies and the measured histone:DNA ratio of one half of the tetrasome complex (Supplementary Figure S3), which would correspond to one H3/H4 dimer bound to each DNA fragment. Other than a slightly decreased amount of histone–DNA complexes observed for the non-positioning sequence relative to 601 and 5SDNA, there is not a significant difference in tetrasome or disome formation between the types of histones under these experimental conditions. This is consistent with the recently measured affinity of 601 and 5SDNA for histones (32).
Figure 1.
Asf1-assisted exchange of histone dimers onto DNA. (A) Tetrasome and disome formation in the absence of yAsf1. The 601, 5SDNA and a non-positioning sequence (NPS) 80 bp DNA fragments at 0.4 µM concentration were incubated with a 2-fold excess of H3/H4*FM or H3K56Q/H4*FM dimers prior to analysis by non-denaturing PAGE. The gel was scanned to obtain the H3/H4*FM fluorescence before the gel was stained with SYBR Green I nucleic acid stain and then rescanned, as described in Supplementary Figure S2. The DNA and histones are shown in green and the position of each species is indicated with an arrow (top panel). The products were analyzed by non-denaturing PAGE and scanned for FM fluorescence. The data from at least three independent experiments were quantitated and are presented in graphical form (bottom panel). (B) Effect of excess of yAsf1 on the formation of tetrasomes and disomes in the presence of an excess of histones. The molar ratios of each component of the mixture are indicated above the non-denaturing PAGE image, which was produced as described in (A). (C) Effect of prior addition of Asf1 on tetrasome and disome formation. H3/H4*FM histones at 0.8 µM were incubated in the absence and presence of increasing concentrations of unlabeled yAsf1 (0, 0.1, 0.2, 0.4, 0.6, 0.8, 1.0, or 2.0 µM) for 30 min at 20°C prior to addition of 0.4 µM concentration 80 bp DNA fragments of 5SDNA. After further incubation for 60 min at 20°C the products were analyzed by non-denaturing PAGE, scanned for FM fluorescence and the data from between three and six independent experiments are presented in graphical form.
As a histone chaperone, Asf1 is expected to aid in the maintenance of histones in a stable unaggregated form (22,32,48). Under conditions where histones were in excess of the DNA, such as lane 3 in Figure 1B, the histones typically precipitated the DNA. A similar excess of Asf1 improved the histone stability such that tetrasome and disome species were formed, but a majority of the DNA remained as insoluble aggregates with H3/H4, which suggested that Asf1 protects histones from aggregation. Under conditions where the histones were not in excess over the DNA, an increased concentration of Asf1 facilitated the assembly of tetrasomes and disomes (compare lanes 4 and 6 in Figure 1B). The formation of tetrasomes and disomes in the presence and absence of yAsf1 suggest that the histones prefer binding to DNA over yAsf1.
In order to examine more closely the effect of Asf1 on assembly of histones onto DNA, we examined the effect of pre-incubating H3/H4*FM histones and Asf1 before addition of DNA. The EMSA and graph of the quantitated results in Figure 1C show that pre-incubation of Asf1 with H3/H4*FM greatly increases the formation of disomes in a manner that is dependent on the dose of Asf1 up to the point where the Asf1 concentration was equal to the H3/H4 dimer concentration, whereas the formation of tetrasomes remained largely unchanged. Interestingly, the quantity of total histone–DNA complexes (tetrasomes and disomes together) is increased nearly 2-fold, and this is due to the additional disomes that are formed. The assembly of histones onto 146 bp of the Widom 601 fragments showed similar results, although it was not possible to distinguish between disomes, tetrasomes and potentially higher ratios of histone dimers associated with the DNA (Supplementary Figure S5). Therefore, Asf1 assists the assembly of histones onto DNA through the increased assembly of disomes with no significant effect on tetrasome formation.
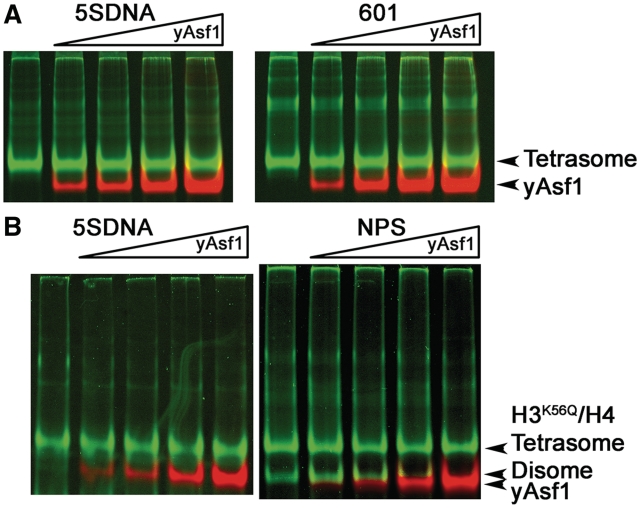
Asf1 does not remove H3/H4 from tetrasomes
Asf1 has a functional role in eviction of histones from the promoters of some activated genes (14,19,52–54), and the Asf1–H3/H4 structure suggests a mechanism by which Asf1 may access histones H3/H4 after H2A/H2B have been displaced from the nucleosome (26,28). In order to determine whether Asf1 has any intrinsic ability to bind to histones H3/H4 in the tetrasome, we conducted a series of binding studies of yAsf1 and tetrasomes that were assembled using well-established salt-gradient dialysis methods (41) (Supplementary Figure S2). Under these conditions the free histones remained in the wells. As yAsf1* was titrated over a concentration range from 0.4 to 4.0 µM up to 5-fold molar excess of tetrasomes and far above the expected Kd of yAsf1 for histones, no binding of yAsf1* to the tetrasome was observed, as might be indicated by the appearance of a supershifted tetrasome–Asf1 complex (Figure 2). Furthermore, yAsf1* was unable to cause the loss of tetramers from the DNA in this assay (Figure 2). The EMSA and graph of the quantitated results for experiments with the 5SDNA, which is representative of all of the DNA fragments tested (Supplementary Figure S4), shows that Asf1 has no effect on the formation of tetrasomes or disomes when they are pre-assembled. These results suggest that yAsf1* alone does not stably bind to the tetrasome and is incapable of dissociating histones from these tetrasomes.
Figure 2.
yAsf1 does not bind to or dissociate tetrasomes by itself. (A) Titration of yAsf1* into 0.4 µM 5SDNA or 601 80 bp tetrasomes. yAsf1* was added at 0, 0.4, 0.8, 2 and 4 µM. The images show the fluorescence of yAsf1* in red and histones H3/H4*FM and SYBR Green I stained DNA in green. (B) Titration of yAsf1 into 0.4 µM 5SDNA or non-positioning sequence (NPS) 80 bp tetrasomes formed with H3K56Q/H4*FM histones. yAsf1* was added at 0, 0.4, 0.8, 2 and 4 µM. The images were produced as in (A).
With the recent knowledge that H3 acetylated at K56 weakens nucleosomes (32), we tested whether Asf1 would be able to dissociate these tetrasomes. ‘Weak’ tetrasomes composed of H3K56Q/H4* FM tetramers and either 5S DNA or the 80 bp NPS DNA (Figure 2B) also did not show any binding of yAsf1* to the tetrasomes or any tetrasome dissociation with the addition of yAsf1*. These results show that yAsf1* alone does not stably bind nor dissociate tetrasomes, even when they are in a weakened state by use of non-positioning DNA sequences or histones that have a neutral substitution for the lysine at H3 K56.
It is formally possible that yAsf1 could associate with H3/H4 in the presence of DNA, even though it cannot dissociate the histones from the DNA, if a kinetic barrier to normal dynamic histone dissociation from DNA exists. In order to challenge the stability of the tetrasome and assess the ability of yAsf1 to capture free histones, the reactions were heated, which facilitates their repositioning by weakening histone DNA contacts (41). Supplementary Figure S6 shows that although incubation at 37°C, 46°C and 55°C disrupts the tetrasomes imperceptibly, in the presence of yAsf1* there is still no disruption and yAsf1* denatured by itself at 55°C. These results were seen for both the 5SDNA and 601 80 bp tetrasomes (Supplementary Figure S6). Therefore, the addition of thermal energy did not facilitate the dissociation of tetrasomes, demonstrating the tetrasome is a stable complex even in the presence of the histone chaperone yAsf1.
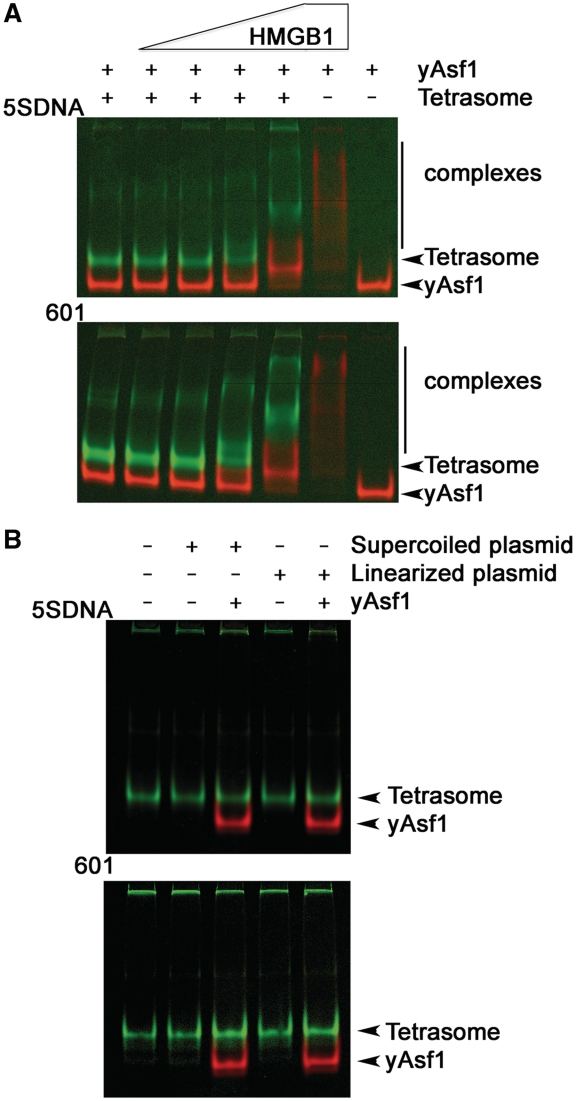
High-mobility group box protein 1 (HMGB1) is an abundant, non-histone architectural chromosomal protein with functional roles in chromatin remodeling (55–57). Although HMGB1 binds the minor groove of DNA with moderate affinity and little sequence specificity, it interacts in a structure-specific manner with DNA at its entry and exit of the nucleosome (58–60). HMGB1 destabilizes nucleosomes and allows bending of DNA that facilitates gene transcription. In order to determine whether yAsf1 could dissociate tetrasomes that have undergone distortions caused by HMGB1, the effects of Asf1 on tetrasomes were studied in the presence of HMGB1. Addition of HMGB1 to the 5SDNA and 601 tetrasomes results in HMGB1–tetrasome complexes that migrate more slowly in the gel, indicating that HMGB1 bound to the tetrasomes (Figure 3A). Even in the presence of yAsf1*, dissociation of the tetrasomes was not evident. Interestingly, yAsf1* and HMGB1 formed non-specific complexes that migrated more slowly than yAsf1* in the gel, but this is not surprising given the strength of the electrostatic interactions expected from these oppositely charged proteins. Therefore, HMGB1 bound to the tetrasomes, but did not facilitate the transfer of H3/H4 to yAsf1. At 10-fold molar excess of HMGB1 relative to tetrasomes, yAsf1* mobility in the gel was altered, but Asf1 was observed to bind to HMGB1 even in the absence of tetrasomes.
Figure 3.
Effect of HMGB1 or supercoiled DNA on tetrasome dissociation by Asf1. (A) Effect of HMGB1 on stability of tetrasomes in the presence of yAsf1. HMGB1 was added to give a final concentration of 0, 0.4, 0.8, 2 and 4 µM with 0.4 µM 5SDNA and 601 tetrasomes. The gels were scanned and processed as in Supplementary Figure S1, with fluorescence of H3/H4*FM and SYBR Green I stained DNA shown in green, and yAsf1* in red. (B) DNA competition assay. Supercoiled or linear 5SDNA or 601 plasmid DNA was added at 0.02 µM concentration to tetrasomes in the absence or presence of yAsf1. The images were produced as in (A).
It is also possible that there may be transient dissociation of the histones from DNA in the normal dynamic equilibrium that yAsf1 might assist, but which might not be evident in the previous experiments. To determine whether yAsf1 assists in transient tetrasome disassembly, the same Asf1-tetrasome reactions were carried out in the presence of an excess of supercoiled or linear plasmid DNA (48). Figure 3B shows that histones did not transfer from the short 80 bp DNA to the longer linear or supercoiled DNA plasmid in the absence or the presence of excess yAsf1*. Taken together, these experiments clearly demonstrated that Asf1 by itself is unable to remove histones from DNA, even under circumstances where H2A/H2B are not present, the histones are modified by a glutamine substitution at K56 of H3, and the DNA is short enough to offer the best opportunity for histone removal.
Oligomeric status of H3/H4 histones
The results from these assembly and disassembly studies, as well as the recent measurements of histone affinities for DNA of Luger, et al. (32), suggested that the histone DNA complex may have a substantially higher affinity than yAsf1 does for histones. As the affinity of yAsf1 for histones has not been determined, we sought to measure this binding affinity under physiological conditions. Like others (61–63), we found that the H3/H4 complexes are stable at high salt concentrations (>1 M NaCl), but at low salt concentrations H3/H4 were relatively unstable due to aggregation and they were unsuitable for thermodynamics studies. However, as it has been possible to stabilize histones at low ionic strength and low concentrations <1 µM using non-ionic detergents or osmolytes (62,64), we identified conditions to stabilize the histones at physiological ionic strength.
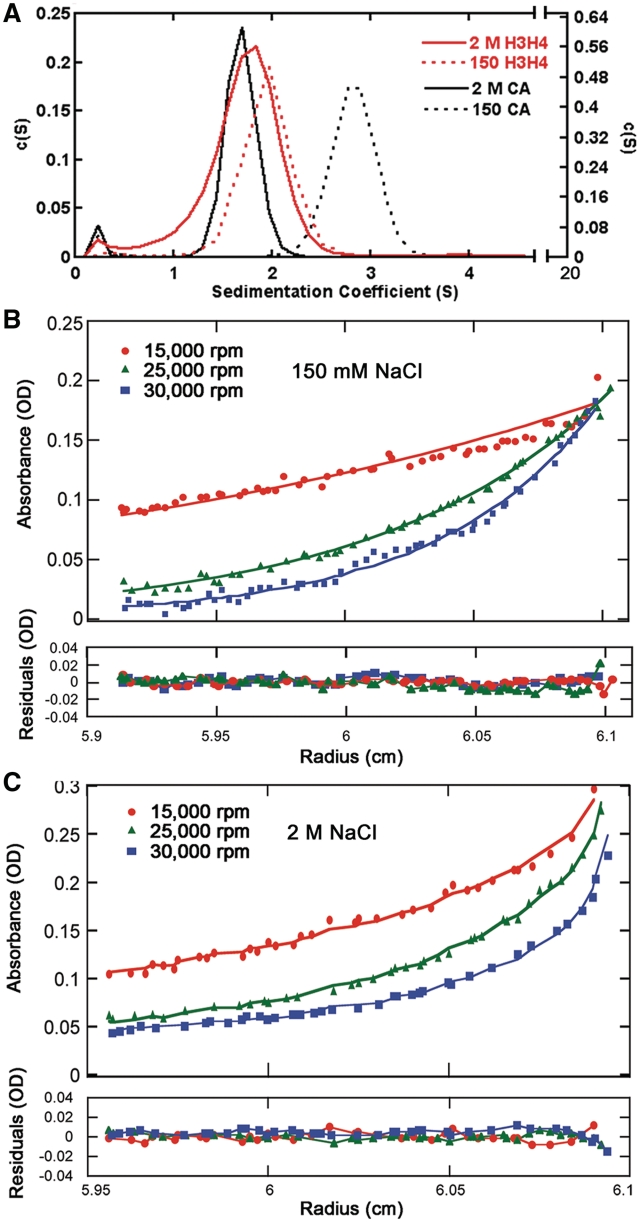
The self-associative behavior of the H3/H4 histone complex was then analyzed in low-ionic and high-ionic strength conditions by AUC. Given the highly charged character of histones H3/H4, we hypothesized that H3/H4 tetramers would be favored at high ionic concentrations, and at physiological ionic concentrations histones H3/H4 might exist predominantly as dimers. AUC using sedimentation velocity (AUC-SV) (Figure 4A) showed that H3/H4 exist mostly as single species and sediment at 1.70 and 1.93 Svedberg units in both the high-salt and low-salt buffers, respectively (Table 1). Estimating molecular weights from the AUC-SV data can be misleading, because the buffer conditions were vastly different, the histones themselves are asymmetric and unstructured proteins (nearly one-third of the N- and C-terminal tails are natively unfolded), and the shape of the H3/H4 dimer and H3/H4 tetramer are not the same. Therefore, carbonic anhydrase (CA), which is a 29 kDa globular protein and protein standard for hydrodynamic studies, was used as a control. The relative S20,w values and the sedimentation coefficients (Table 1) of the histones compared to CA suggested that H3/H4 in low-salt buffer are smaller than CA, and the S20,w values for H3/H4 in high-salt buffer is larger than CA, which supported the hypothesis that H3/H4 is a tetramer in the presence of 2 M NaCl and a dimer in the presence of 150 mM NaCl.
Figure 4.
Analytical ultracentrifugation of H3/H4 at different ionic strengths. (A) Sedimentation velocity AUC analysis of H3/H4. The normalized c(s) distribution curves calculated from the raw data using SEDFIT (44) are shown for H3/H4 at 10 µM (dimer in red) and carbonic anhydrase (CA in black) in buffers containing 2 M NaCl (solid line) and 150 mM NaCl (dotted line), respectively. (B and C) Sedimentation equilibrium concentration profiles of H3/H4 were measured in buffers with 150 mM (B) and 2 M (C) NaCl concentrations, respectively. The absorption profile was recorded at 280 nm at three rotor speeds each: 15 000 rpm (red circle), 25 000 rpm (green triangle), and 30 000 rpm (blue square). The continuous line is the non-linear least-squares global fit to a single exponential function, with single species having a molecular masses shown in Table 2, as analyzed with SEDPHAT (45).
Table 1.
Analytical ultracentrifugation sedimentation velocity analysis of H3/H4
| [NaCl] M | Protein | c(s)a S | s20,wb |
|---|---|---|---|
| 0.15 | H3/H4 | 1.96 | 2.03 |
| 2.0 | H3/H4 | 1.70 | 2.70 |
| 0.15 | CA | 2.81 | 2.89 |
| 2.0 | CA | 1.72 | 2.68 |
aSedimentation coefficient calculated using the continuous distribution c(s) Lamm equation.
bSedimentation coefficient has been corrected to 20.0°C and the density of water, which enables a comparison of results from experiments performed in different buffers and temperatures.
In order to determine the molecular weights and the oligomeric status of H3/H4 under in the different buffer conditions, we used sedimentation equilibrium AUC (AUC-SE). The analysis performed using SEDPHAT (45) (Figures 4B and C) gave molecular weights of 28 771 Da for H3/H4 in low-salt buffer and 49 214 Da for H3/H4 in high-salt buffer (Table 2). These values are within 10% of the theoretical molecular weights of 26 509 Da and 53 218 Da for H3/H4 dimers and tetramers, respectively. The sedimentation equilibrium analysis of CA under the same conditions in the high- and low-salt buffer also gave molecular weights that were within 10% of the theoretical value. These results clearly demonstrated that H3/H4 used here at 10 µM in high-salt buffer formed tetramers but in the low-salt buffer formed dimers.
Table 2.
Analytical ultracentrifugation sedimentation equilibrium analysis of H3/H4
| [NaCl] M | [H3/H4] µM | vbar | Density (g/cm3) | Calculated mol. wt (Da) | Theoretical mol. wt (Da) |
|---|---|---|---|---|---|
| 0.15 | 10.2 | 0.7452 | 1.00 504 | 28 771 | 26 509 |
| 2.0 | 12.8 | 0.7452 | 1.07 767 | 49 214 | 53 018 |
The affinity of yAsf1 for histones
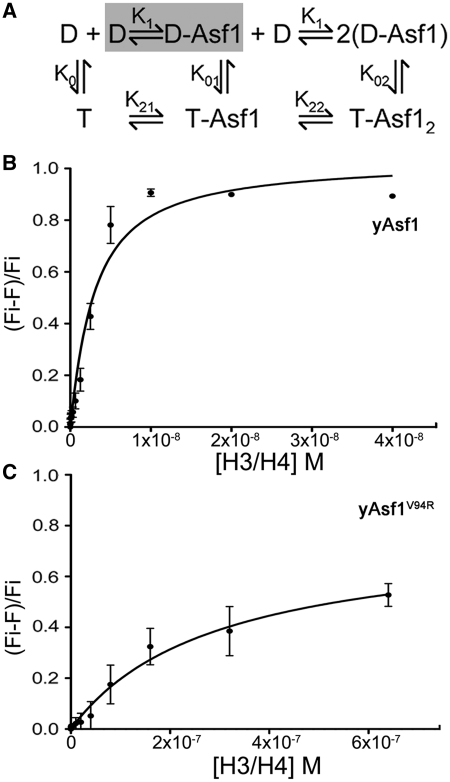
The finding that the histones were dimers under the quantitative binding conditions used in this study greatly simplified the analysis required to obtain affinity measurements of yAsf1–histone complexes. Since the H3/H4 complexes were predominantly, if not all, in the dimer form, the determination of the association constants for the Asf1–histone interaction did not take into account the complete histone dimer-tetramer equilibrium (Figure 5A). We used fluorescence quenching of labeled yAsf1* with quencher-labeled H3/H4*Q to measure the histone-binding affinity of yAsf1. The measured Kd for yAsf1 was 2.5 ± 0.7 nM (Figure 5B). Buffer did not quench yAsf1* fluorescence, indicating that the quenching signal comes from the association of yAsf1 with the histones. To account for potential non-specific association of the proteins, we also tested the yAsf1 mutant V94R (yAsf1*V94R), which is defective in binding to histones due the disruptive substitution at the Asf1–H3 interface (65). At a much higher concentration of histones yAsf1*V94R exhibited a lower degree of quenching (Figure 5C), giving rise to a measured Kd of 290 ± 53 nM.
Figure 5.
Affinity of Asf1 and H3/H4 determined by fluorescence quenching. (A) Schematic diagram of model equilibria for yAsf1 association with H3/H4 complexes. D and T indicate H3/H4 dimers and tetramers, respectively. (B) H3/H4*Qsy9 binding to yAsf1* was observed by fluorescence quenching. H3/H4*Qsy9 was titrated into 1.0 nM yAsf1*. The data were fitted with a ligand-depleted binding model (Equation 1; GraphPad Prism) because the concentration of yAsf1* was within 10-fold of the Kd value. (C) H3/H4*Qsy9 binding to yAsf1*V94R was observed by fluorescence quenching. H3/H4*Qsy9 was titrated into 1.0 nM yAsf1*V94R. The curve was fitted (GraphPad Prism) with a Langmuir single-site binding isotherm.
DISCUSSION
We determined that under conditions of physiological ionic strength and at moderate concentration (≤10 µM) H3/H4 exist primarily as dimers. This is in contrast to our expectation from the literature that the histones would either form exclusively H3/H4 tetramers or exist as a mixture of dimers and tetramers due to the expected association constant for the dimer–tetramer equilibrium. Previous studies have found that the dimer–tetramer equilibrium association constant for chicken erythrocyte H3/H4 is in the 10 to 20 µM range depending on the buffer conditions (66). Recently, H3/H4 tetramers were found to be the predominant species under conditions used for pulsed EPR studies, which employed nearly identical Xenopus histones with a Cys110Ala substitution in H3, but these studies were conducted at a 10-fold higher concentration of histones than used here (67). Others have reported that H3/H4 exists in the tetramer form, but the size-exclusion chromatography experiments were conducted at a salt concentration of 0.75 M, where the equilibrium between dimer and tetramer would have been shifted toward the tetramer form (28,66).
Although in vitro at moderate H3/H4 concentrations and physiological ionic strength, the histones exist primarily as dimers, the form of free histones in the cell will be dependent on the relative affinities of the many histone chaperones for the dimer versus tetramer form of H3/H4 in addition to concentration of free histones. Our finding here that the affinity of yAsf1 for H3/H4 is in the low nanomolar range together with the previously measured association constant for the H3/H4 dimer–tetramer equilibrium, which is in low micromolar range (66), indicates that H3/H4 probably rarely if ever exist in the cell as a free H3/H4 tetramer, and that the tetramer form would exist only when H3/H4 are bound to DNA or to particular histone chaperones that promote tetramer formation. Thus, these thermodynamic results are consistent with findings in an earlier study, which showed that free histones in yeast appear to exist primarily in the dimer form in vivo (23).
The finding that yAsf1 binds histones tightly, but is not sufficient by itself to dissociate the tetrasome, is consistent with a role as a protector of histone when they are either in excess over DNA or are not in the vicinity of DNA. Our observation that yAsf1 can guide the assembly of H3/H4 with DNA into histone–DNA complexes by preventing non-specific aggregation of the histones with DNA is analogous to recent findings with Nap1 (32). Interestingly, we also found that Asf1 was even able to assist in the direct deposition of H3/H4 dimers onto DNA. Although the action of Asf1 directly in assembly of disomes was not necessarily anticipated, the relatively high affinity of the complex suggests that Asf1 will be a dominant H3/H4 binding protein in the cell, and this would be expected to bias the species of histones that are available for direct deposition, if they are not assembled through normal chromatin assembly pathways involving HIRA or CAF-1.
Asf1 binds H3/H4 with higher affinity than Nap1, but in contrast to Nap1, Asf1 is quite specific for H3/H4, and not other histones. As such Asf1 serves as a scaffold that directs H3/H4 toward different cellular fates through the formation of varied multi-protein complexes and acquisition of various histone post-translational modifications. Asf1–H3/H4 complexes can carry a variety post-translational histone modifications in the H4 tail K5K12ac2, the H3 tail at H3 K9me1, H3 K14ac and H3 K18ac, as well as acetylation of H3 lysine 56 in the core of H3 (17,68–72). Evidence suggests that acetylation of lysine 56 in general depends on the association of the histones with Asf1 (36,73,74). Whether Asf1 is also necessary for the introduction of other modifications or not, these modifications direct the association of the Asf1–H3/H4 complexes and/or the histones themselves to downstream chromatin assembly machineries (21,75–77).
Although yAsf1 does not directly interact with the tetrasome with measurable affinity, nor does yAsf1 promote histone release from DNA, our finding that histones H3/H4 bind yAsf1 only slightly more weakly than they bind to DNA places Asf1 in a pivotal position for direct participation in the nucleosome disassembly processes. The difference in affinity between the Asf1–H3/H4 interaction and the ≤1 nM affinity of H3/H4 for DNA is also subject to modulation by post-translational modifications (32). While all of the relevant histone and Asf1 modifications are still not known, the effect of histone post-translational modifications on the histone–DNA equilibrium in the case of acetylation of H3 K56 shown here and recently by the Luger lab (32) clearly illustrate that single modifications have real, albeit incremental effects on tetrasome stability. It is intriguing to speculate that Asf1 modifications, ATP-dependent chromatin remodeling complexes or additional histone modifications may tip the balance toward less stable histone–DNA association and enable Asf1 to compete for histones and facilitate tetrasome disassembly.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (R01 GM079154 to M.E.A.C.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We are grateful to Karolin Luger for providing the histone gene plasmids, Karl Maluf for assistance with the AUC analyses, Sarah Roemer for providing the purified HMGB1, Cassandra Vogel for assistance in the early stages of this study and Jessica Tyler for helpful discussions.
REFERENCES
- 1.Kornberg RD. Structure of chromatin. Annu. Rev. Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 2.Jorcano JL, Ruiz-Carrillo A. H3.H4 tetramer directs DNA and core histone octamer assembly in the nucleosome core particle. Biochemistry. 1979;18:768–774. doi: 10.1021/bi00572a005. [DOI] [PubMed] [Google Scholar]
- 3.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 4.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 5.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akey CW, Luger K. Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol. 2003;13:6–14. doi: 10.1016/s0959-440x(03)00002-2. [DOI] [PubMed] [Google Scholar]
- 7.Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr. Opin. Struct. Biol. 2008;18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eitoku M, Sato L, Senda T, Horikoshi M. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell. Mol. Life Sci. 2008;65:414–444. doi: 10.1007/s00018-007-7305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das C, Tyler JK, Churchill MEA. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem. Sci. 2010;35:476–489. doi: 10.1016/j.tibs.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 11.Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutton A, Bucaria J, Osley MA, Sternglanz R. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics. 2001;158:587–596. doi: 10.1093/genetics/158.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 14.Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 15.Adkins MW, Tyler JK. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J. Biol. Chem. 2004;279:52069–52074. doi: 10.1074/jbc.M406113200. [DOI] [PubMed] [Google Scholar]
- 16.Ramey CJ, Howar S, Adkins M, Linger J, Spicer J, Tyler JK. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol. Cell. Biol. 2004;24:10313–10327. doi: 10.1128/MCB.24.23.10313-10327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl Acad. Sci. USA. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanematsu F, Takami Y, Barman HK, Fukagawa T, Ono T, Shibahara K, Nakayama T. Asf1 is required for viability and chromatin assembly during DNA replication in vertebrate cells. J. Biol. Chem. 2006;281:13817–13827. doi: 10.1074/jbc.M511590200. [DOI] [PubMed] [Google Scholar]
- 19.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Nakatani Y, Ray-Gallet D, Quivy JP, Tagami H, Almouzni G. Two distinct nucleosome assembly pathways: dependent or independent of DNA synthesis promoted by histone H3.1 and H3.3 complexes. Cold Spring Harb. Symp. Quant. Biol. 2004;69:273–280. doi: 10.1101/sqb.2004.69.273. [DOI] [PubMed] [Google Scholar]
- 21.Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 22.Munakata T, Adachi N, Yokoyama N, Kuzuhara T, Horikoshi M. A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes Cells. 2000;5:221–233. doi: 10.1046/j.1365-2443.2000.00319.x. [DOI] [PubMed] [Google Scholar]
- 23.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 24.Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell. 2005;17:301–311. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 25.Tamburini BA, Carson JJ, Adkins MW, Tyler JK. Functional conservation and specialization among eukaryotic anti-silencing function 1 histone chaperones. Eukaryot. Cell. 2005;4:1583–1590. doi: 10.1128/EC.4.9.1583-1590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.English CM, Maluf NK, Tripet B, Churchill ME, Tyler JK. ASF1 binds to a heterodimer of histones H3 and H4: a two-step mechanism for the assembly of the H3-H4 heterotetramer on DNA. Biochemistry. 2005;44:13673–13682. doi: 10.1021/bi051333h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 29.Korber P, Barbaric S, Luckenbach T, Schmid A, Schermer UJ, Blaschke D, Horz W. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J. Biol. Chem. 2006;281:5539–5545. doi: 10.1074/jbc.M513340200. [DOI] [PubMed] [Google Scholar]
- 30.Kim HJ, Seol JH, Han JW, Youn HD, Cho EJ. Histone chaperones regulate histone exchange during transcription. EMBO J. 2007;26:4467–4474. doi: 10.1038/sj.emboj.7601870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams SK, Tyler JK. Transcriptional regulation by chromatin disassembly and reassembly. Curr. Opin. Genet. Dev. 2007;17:88–93. doi: 10.1016/j.gde.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell. 2010;37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrews AJ, Downing G, Brown K, Park YJ, Luger K. A thermodynamic model for Nap1-histone interactions. J. Biol. Chem. 2008;283:32412–32418. doi: 10.1074/jbc.M805918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thastrom A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 35.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams SK, Truong D, Tyler JK. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl Acad. Sci. USA. 2008;105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lavelle C, Prunell A. Chromatin polymorphism and the nucleosome superfamily: a genealogy. Cell Cycle. 2007;6:2113–2119. doi: 10.4161/cc.6.17.4631. [DOI] [PubMed] [Google Scholar]
- 39.Zlatanova J, Bishop TC, Victor JM, Jackson V, van Holde K. The nucleosome family: dynamic and growing. Structure. 2009;17:160–171. doi: 10.1016/j.str.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 40.Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Method. Mol. Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 41.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 42.Roemer SC, Adelman J, Churchill ME, Edwards DP. Mechanism of high-mobility group protein B enhancement of progesterone receptor sequence-specific DNA binding. Nucleic Acids Res. 2008;36:3655–3666. doi: 10.1093/nar/gkn249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 44.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 46.Park YJ, Dyer PN, Tremethick DJ, Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J. Biol. Chem. 2004;279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 47.Alilat M, Sivolob A, Revet B, Prunell A. Nucleosome dynamics. Protein and DNA contributions in the chiral transition of the tetrasome, the histone (H3-H4)2 tetramer-DNA particle. J. Mol. Biol. 1999;291:815–841. doi: 10.1006/jmbi.1999.2988. [DOI] [PubMed] [Google Scholar]
- 48.Jackson V. Preferential binding of histones H3 and H4 to highly positively coiled DNA. Biochemistry. 1995;34:10607–10619. doi: 10.1021/bi00033a036. [DOI] [PubMed] [Google Scholar]
- 49.Somers J, Owen-Hughes T. Mutations to the histone H3 alpha N region selectively alter the outcome of ATP-dependent nucleosome-remodelling reactions. Nucleic Acids Res. 2009;37:2504–2513. doi: 10.1093/nar/gkp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Panetta G, Buttinelli M, Flaus A, Richmond TJ, Rhodes D. Differential nucleosome positioning on Xenopus oocyte and somatic 5S RNA genes determines both TFIIIA and H1 binding: a mechanism for selective H1 repression. J. Mol. Biol. 1998;282:683–697. doi: 10.1006/jmbi.1998.2087. [DOI] [PubMed] [Google Scholar]
- 51.Peterson S, Danowit R, Wunsch A, Jackson V. NAP1 catalyzes the formation of either positive or negative supercoils on DNA on basis of the dimer-tetramer equilibrium of histones H3/H4. Biochemistry. 2007;46:8634–8646. doi: 10.1021/bi6025215. [DOI] [PubMed] [Google Scholar]
- 52.Korber P, Horz W. In vitro assembly of the characteristic chromatin organization at the yeast PHO5 promoter by a replication-independent extract system. J. Biol. Chem. 2004;279:35113–35120. doi: 10.1074/jbc.M405446200. [DOI] [PubMed] [Google Scholar]
- 53.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 54.Takahata S, Yu Y, Stillman DJ. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol. Cell. 2009;34:405–415. doi: 10.1016/j.molcel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bianchi ME, Beltrame M. Flexing DNA: HMG-box proteins and their partners. Am. J. Hum. Genet. 1998;63:1573–1577. doi: 10.1086/302170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas JO, Travers AA. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 57.Travers A. Recognition of distorted DNA structures by HMG domains. Curr. Opin. Struct. Biol. 2000;10:102–109. doi: 10.1016/s0959-440x(99)00056-1. [DOI] [PubMed] [Google Scholar]
- 58.Schroter H, Bode J. The binding sites for large and small high-mobility-group (HMG) proteins. Studies on HMG-nucleosome interactions in vitro. Eur. J. Biochem. 1982;127:429–436. doi: 10.1111/j.1432-1033.1982.tb06890.x. [DOI] [PubMed] [Google Scholar]
- 59.Nightingale K, Dimitrov S, Reeves R, Wolffe AP. Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J. 1996;15:548–561. [PMC free article] [PubMed] [Google Scholar]
- 60.An W, van Holde K, Zlatanova J. The non-histone chromatin protein HMG1 protects linker DNA on the side opposite to that protected by linker histones. J. Biol. Chem. 1998;273:26289–26291. doi: 10.1074/jbc.273.41.26289. [DOI] [PubMed] [Google Scholar]
- 61.Karantza V, Freire E, Moudrianakis EN. Thermodynamic studies of the core histones: pH and ionic strength effects on the stability of the (H3-H4)/(H3-H4)2 system. Biochemistry. 1996;35:2037–2046. doi: 10.1021/bi9518858. [DOI] [PubMed] [Google Scholar]
- 62.Banks DD, Gloss LM. Folding mechanism of the (H3-H4)2 histone tetramer of the core nucleosome. Protein Sci. 2004;13:1304–1316. doi: 10.1110/ps.03535504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banks DD, Gloss LM. Equilibrium folding of the core histones: the H3-H4 tetramer is less stable than the H2A-H2B dimer. Biochemistry. 2003;42:6827–6839. doi: 10.1021/bi026957r. [DOI] [PubMed] [Google Scholar]
- 64.Mazurkiewicz J, Kepert JF, Rippe K. On the mechanism of nucleosome assembly by histone chaperone NAP1. J. Biol. Chem. 2006;281:16462–16472. doi: 10.1074/jbc.M511619200. [DOI] [PubMed] [Google Scholar]
- 65.Mousson F, Lautrette A, Thuret JY, Agez M, Courbeyrette R, Amigues B, Becker E, Neumann JM, Guerois R, Mann C, et al. Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proc. Natl Acad. Sci. USA. 2005;102:5975–5980. doi: 10.1073/pnas.0500149102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baxevanis AD, Godfrey JE, Moudrianakis EN. Associative behavior of the histone (H3-H4)2 tetramer: dependence on ionic environment. Biochemistry. 1991;30:8817–8823. doi: 10.1021/bi00100a013. [DOI] [PubMed] [Google Scholar]
- 67.Bowman A, Ward R, El-Mkami H, Owen-Hughes T, Norman DG. Probing the (H3-H4)2 histone tetramer structure using pulsed EPR spectroscopy combined with site-directed spin labelling. Nucleic Acids Res. 2010;38:695–707. doi: 10.1093/nar/gkp1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barman HK, Takami Y, Nishijima H, Shibahara K, Sanematsu F, Nakayama T. Histone acetyltransferase-1 regulates integrity of cytosolic histone H3-H4 containing complex. Biochem. Biophys. Res. Commun. 2008;373:624–630. doi: 10.1016/j.bbrc.2008.06.100. [DOI] [PubMed] [Google Scholar]
- 69.Benson LJ, Gu Y, Yakovleva T, Tong K, Barrows C, Strack CL, Cook RG, Mizzen CA, Annunziato AT. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J. Biol. Chem. 2006;281:9287–9296. doi: 10.1074/jbc.M512956200. [DOI] [PubMed] [Google Scholar]
- 70.Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J. Biol. Chem. 2007;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- 71.Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol. Cell. Biol. 2008;28:4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jasencakova Z, Scharf AN, Ask K, Corpet A, Imhof A, Almouzni G, Groth A. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell. 2010;37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 73.Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J. Biol. Chem. 2006;281:37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- 74.Adkins MW, Carson JJ, English CM, Ramey CJ, Tyler JK. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J. Biol. Chem. 2007;282:1334–1340. doi: 10.1074/jbc.M608025200. [DOI] [PubMed] [Google Scholar]
- 75.Mello JA, Sillje HH, Roche DM, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, Yates JR, III, Kaufman PD. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.