Abstract
Differentially methylated regions (DMRs) are stable epigenetic features within or in proximity to imprinted genes. We used this feature to identify candidate human imprinted loci by quantitative DNA methylation analysis. We discovered a unique DMR at the 5′-end of FAM50B at 6p25.2. We determined that sense transcripts originating from the FAM50B locus are expressed from the paternal allele in all human tissues investigated except for ovary, in which expression is biallelic. Furthermore, an antisense transcript, FAM50B-AS, was identified to be monoallelically expressed from the paternal allele in a variety of tissues. Comparative phylogenetic analysis showed that FAM50B orthologs are absent in chicken and platypus, but are present and biallelically expressed in opossum and mouse. These findings indicate that FAM50B originated in Therians after divergence from Prototherians via retrotransposition of a gene on the X chromosome. Moreover, our data are consistent with acquisition of imprinting during Eutherian evolution after divergence of Glires from the Euarchonta mammals. FAM50B expression is deregulated in testicular germ cell tumors, and loss of imprinting occurs frequently in testicular seminomas, suggesting an important role for FAM50B in spermatogenesis and tumorigenesis. These results also underscore the importance of accounting for parental origin in understanding the mechanism of 6p25-related diseases.
INTRODUCTION
Genomic imprinting results in the differential expression of the two inherited alleles of a gene in a parent-of-origin specific manner. Imprinted genes play important roles in embryonic growth and development as well as in placental function. Genetic and epigenetic disruption of imprinted genes are also associated with a wide range of diseases, including cancer, behavioral disorders and congenital disorders (1,2).
Approximately 60 genes are known to be imprinted in humans (3,4). Genome-wide methods used to screen for imprinted genes have relied on the differential abundance of the maternal and paternal alleles using subtractive hybridization (5,6), detection of differential expression using marker polymorphisms (7,8) or transcriptome sequencing (9). Global screens for novel imprinted loci based on differential methylation between paternal and maternal alleles have also been performed, and include the use of methylation-sensitive representational difference analyses (10,11) or detection of differentially methylated regions (DMRs) in the gametes (12). Each of these methods has resulted in the identification of a relatively small number of novel imprinted genes. Recently, we performed a genome-wide screen using a bioinformatics approach that resulted in the prediction of 156 candidate imprinted genes in humans (13); however, the imprint status of many of these genes has not yet been experimentally confirmed.
Perhaps the most consistent and experimentally amenable feature associated with imprinted genes is the presence of DMRs located adjacent to or within these genes. Many such DMRs are established in the gametes, the only time in development when the maternal and paternal genomes are in distinct cellular compartments. Methylation is therefore established in a manner reflecting the sex of the parental individual in which the gametes reside. This methylation pattern is normally maintained in all somatic tissues throughout life, irrespective of gene expression levels (14,15). In the study described in this article, we used quantitative methylation analysis to search for novel human DMRs associated with the genes we predicted to be imprinted on chromosome 6p (13). Imprinted genes have not previously been identified at 6p, but a parent-of-origin effect in psoriasis is linked to this region of the genome (16). Loss of heterozygosity (LOH) at chromosome 6p25 is also reported in a variety of malignancies, including prostate cancer and cervical carcinoma (17–19). Trisomy or monosomy of 6p25 results in a variety of congenital malformations that include neuronal defects, psychomotor retardation, craniofacial defects and abnormalities in organogenesis (20–24). Most of these characteristics are apparent at birth, indicating important roles of the involved genes in early development.
Herein, we report the identification of a novel DMR in the 5′-region of the FAM50B locus (GenBank accession no.: NM_012135) at chromosome 6p25.2. FAM50B originated from a retrotransposon, and encodes for the protein XAP-5-like (X5L), which is highly expressed during spermatogenesis (25). We show that FAM50B transcripts are monoallelically expressed from the paternal allele in a variety of human tissues. Using phylogenetic comparisons in vertebrates, we found FAM50B to be absent in chicken and platypus, but present and biallelically expressed in the opossum and mouse. Importantly, our results also show that DMR methylation at the FAM50B locus is significantly reduced in testicular germ cell tumors (TGCTs), frequently resulting in the dysregulation of FAM50B expression, including loss of imprinting in seminomas.
MATERIALS AND METHODS
Tissues
Human conceptus tissues were obtained from the National Institutes of Health-funded Laboratory of Human Embryology at the University of Washington, Seattle. Surgically resected tumor samples and the pathological diagnoses of tumor types were obtained from the Department of Pathology, Duke University Medical Center and the National Development and Research Institutes, Inc (NDRI, New York, NY, USA). Eleven TGCTs were used in this study, including two embryonal carcinomas, one seminoma mixed with embryonal carcinoma, five seminomas and three mixed germ cell tumors. Normal adult human tissues were provided by NDRI and BioChain Institute (Hayward, CA, USA). All human tissue specimens were anonymized and used under a protocol approved by the Duke University Institutional Review Board.
Gray short-tailed opossum (Monodelphis domestica) kidney, liver, placenta, brain and embryo samples were kindly provided by Dr Kathleen Smith (Duke University, Durham, NC, USA). Mouse liver, brain, kidney and testis samples were obtained from the F1 generation of a cross between C57BL/6J and CAST/Ei mouse strains (Jackson Laboratory, Bar Harbor, ME, USA). These animal tissues were used under protocols approved by the Duke University Institutional Animal Care and Use Committee.
Genomic DNA purification and sodium bisulfite conversion
Total DNA was isolated from multiple tissues using buffer ATL, proteinase K and RNase A (Qiagen, Inc., Valencia, CA, USA) followed by phenol–chloroform extraction and ethanol precipitation. Bisulfite conversion of DNA was carried out using the Epitect Bisulfite Kit (Qiagen Inc.).
Northern blot analysis of FAM50B antisense transcript (FAM50B-AS)
Northern blot analysis of total RNA from multiple human tissues was performed using Dig Northern Starter Kit (Roche Diagnostic, Indianapolis, IN, USA) according to the manufacturer’s protocol. Two different probes to the antisense transcript were made using two primer pairs (FAM-18F/18R; FAM19-F/18R) respectively by PCR. T7 promoters in the primers FAM-18R and FAM19-R were used to generate DIG-labeled RNA complementary to FAM50B-AS by in vitro transcription. Total RNA was isolated from multiple human conceptus tissues using RNA-Stat 60 as recommended by the manufacturer (Tel-Test, Inc., Friendswood, TX, USA). Adult human total RNA was provided by BioChain Institute.
5′- and 3′-rapid amplifications of cDNA ends
RNAs from human conceptus brain and adult testis were analyzed to locate the 5′- and 3′-termini of the FAM50B-AS using the 5′/3′-rapid amplifications of cDNA ends (RACE) kit according to the manufacturer’s suggested protocol (second generation, Roche Diagnostics). For 5′-RACE, first-strand cDNA was generated using primer FAM-12F and purified using the Roche Diagnostics PCR Product Purification kit (Roche Diagnostics). The 5′-end of the purified cDNA was further modified by a tailing reaction. The tailed cDNA was amplified using two primers: oligo dT-anchor primer and FAM-13F. The DNA was re-amplified with a set of nested primers including the PCR anchor primer and FAM-14F (Supplementary Table S1).
For 3′-RACE of FAM50B-AS RNA, first-strand cDNA was generated using the oligo dT-anchor primer. The reverse-transcribed cDNA was amplified using semi-nested PCR: the first round with the PCR anchor primer and FAM-14R, the second round with the same PCR anchor primer and FAM-15R (Supplementary Table S1). For all specimens, control reactions were performed using cDNA prepared in the absence of primers to eliminate the possibility of genomic DNA and RNA contamination. These PCR products were then sequenced on an ABI 3730 DNA sequencer (Applied Biosystems, Foster City, CA, USA).
Quantitative methylation analysis
Quantitative methylation analysis of DNA in the FAM50B promoter was performed using MassARRAY EpiTYPER assays (Sequenom, San Diego, CA, USA). Primers for human FAM50B (Supplementary Table S1, FAM-1F/1R to FAM-6F/6R) were designed using Epidesigner (Sequenom; http://www.epidesigner.com) to cover the CG-rich regions with amplicons in a target range of 400–600 bp. Each reverse primer was designed to contain a T7 promoter sequence tag (5′-CAG TAA TAC GAC TCA CTA TAG GGA GAA GGC T-3′) for in vitro transcription, and each forward primer incorporated a 10-mer tag (5′-AGG AAG AGA G-3′) to balance the primer annealing temperature with that of the primer containing the T7 tag. Polymerase chain reaction amplification was performed using HotStarTaq (Qiagen, Inc.) with the following parameters: polymerase activation at 95°C for 5 min, followed by denaturation at 94°C for 20 s, annealing at 60°C for 25 s and extension at 72°C for 1 min for a total of 40 cycles, with a final incubation at 72°C for 5 min. After dephosphorylation of unincorporated dNTPs, the processed PCR products were used in in vitro transcription reactions (T-cleavage assay) according to the manufacturer’s standard protocol (Sequenom). The transcription products were conditioned to remove bilvalent cation adducts by dilution with 20 µl H2O and addition of 6 mg of Clean Resin (Sequenom). The samples were then spotted on a 384-pad Spectro-CHIP (Sequenom) using a MassARRAY Nanodispenser (Samsung, Irvine, CA, USA), followed by spectral acquisition on a MassARRAY analyzer compact MALDI-TOF MS (Sequenom). Fragments containing CpG sites were analyzed with EpiTyper software (Sequenom) to generate quantitative methylation fractions at these sites. The MassARRAY analysis was repeated three times for each sample and the results were averaged.
Bisulfite sequencing of cloned alleles
Genotyping of rs2239713 in the FAM50B promoter region was carried out to allow for allele-specific determination of methylation status. PCR was performed with primers FAM-7F and FAM-7R (Supplementary Table S1), followed by BigDye sequencing (Applied Biosystems) on an ABI 3730 DNA sequencer (Applied Biosystems). The sense DNA sequence was amplified from bisulfite converted genomic DNA from conceptus tissues using primers FAM-2F and FAM-4R. The antisense DNA sequence of heterozygous individuals was amplified from bisulfite converted genomic DNA from conceptus tissues and sperm specimens using the following primers: FAM-8F and FAM-8R. PCR products were directly ligated into the PCR® 2.1 vector using the TOPO TA Cloning Kit (Invitrogen). Clones for each specimen were directly sequenced using an ABI BigDye Terminator 3.1 Cycle Sequencing Kit (Applied Biosystems) on an ABI 3730 DNA sequencer (Applied Biosystems).
Genotyping and imprinting expression analyses in human tissues
Each individual was genotyped for polymorphism rs6597007 in FAM50B by sequencing genomic DNA. Genotyping was carried out by PCR with primers FAM-9F and FAM-9R, followed by BigDye sequencing (Applied Biosystems) on an ABI 3730 DNA sequencer (Applied Biosystems). RNA from heterozygous individuals was analyzed to determine whether expression was monoallelic or biallelic. Total RNA was isolated from multiple human tissues using RNA-Stat 60 as recommended by the manufacturer (Tel-Test, Inc., Friendswood, TX, USA). First-strand cDNA was synthesized from Dnase I-treated RNA using Superscript II (Invitrogen), as recommended by the manufacturer using gene-specific primer FAM-10R. PCR was performed for 40 cycles with primers FAM-9F and FAM-9R. For all specimens, control reactions were performed using cDNA prepared in the absence of reverse transcriptase to eliminate the possibility of genomic DNA contamination. The PCR products were gel-extracted (GenElute, Sigma Chemical Co, St Louis, MO, USA) and directly sequenced on an ABI 3730 DNA sequencer (Applied Biosystems).
Allele-specific expression of FAM50B and FAM50B-AS transcripts
Strand-specific RT–PCR was performed to distinguish between FAM50B and FAM50B-AS for allele-specific expression using SNP rs6597007. First strand cDNA was generated from Dnase I-treated RNA from human tissues using the oligo dT-anchor primer and the 3′-RACE kit as described above. For detection of FAM50B transcripts, PCR was performed using primer FAM-9F and the PCR anchor primer. For detection of FAM50B-AS, PCR was first performed using primer FAM-14R and the PCR anchor primer, followed by nested PCR using primers FAM-15F and FAM-9R.
SNP rs3196512 was used to determine allelic expression of FAM50B in TGCTs. Genotyping of genomic DNA was carried out by PCR with primers FAM-11F and FAM-11R followed by sequencing. Individuals heterozygous for polymorphism rs3196512 were further analyzed to distinguish between monoallelic and biallelic expression. First strand cDNA was generated from Dnase I-treated RNA using the oligo dT-anchor primer as described above. RT–PCR was performed using primer FAM-11F and the PCR anchor primer.
For all specimens, control reactions were performed using cDNA prepared in the absence of primers to eliminate the possibility of genomic DNA and non-specific RNA contamination. These PCR products were sequenced directly. Sequences for all primers used are provided in Supplementary Table S1.
Relative quantitative RT real-time PCR analysis of FAM50B and FAM50B-AS
Relative quantification of both FAM50B and FAM50B-AS mRNA expression levels was performed on an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems). First-strand cDNA synthesis was performed with an oligo dT primer on Dnase I-treated RNA. Real-time reactions were carried out in duplicate with QuantiFast SYBR Green PCR Master Mix (Qiagen, Inc.) in a total reaction volume of 20 µl, using thermal conditions recommended by the manufacturer. For detection of FAM50B transcripts, real-time PCR was performed using primers FAM-16F and FAM-16R. For detection of FAM50B-AS transcript levels, real-time PCR was performed using primers FAM-17F and FAM-17R that anneal to sequence within intron 1 of FAM50B. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous RNA control. Target and endogenous control amplifications were carried out in separate tubes. Results were analyzed using SDS RQ Manager 1.2 software. Each ΔCt value (ΔCt = Ct target − Ct control) was transformed to a percentage (percent target = 2−ΔCt × 100).
Analysis of opossum and mouse FAM50B orthologs
Opossum and mouse were screened for genomic DNA polymorphisms by PCR amplification using primer pairs (Supplementary Table S2, Opossum-1F/1R to -3F/3R; Mouse-1F/1R to -3F/3R) covering the entire coding region, followed by sequencing. Total RNA was isolated from multiple tissues of heterozygotes by homogenization in RNA-Stat 60 (Tel-Test). First strand cDNA was synthesized from Dnase I-treated RNA using strand-specific primers (Supplementary Table S2, Opossum-RT; Mouse- RT). cDNA fragments amplified with primers Opossum-1F/1R and Mouse-4F/4R were purified, and directly sequenced on an ABI 3730 DNA sequencer (Applied Biosystems). Quantitative DNA methylation analysis of the FAM50B promoter was performed using MassARRAY EpiTYPER assays as described above (primers shown in Supplementary Table S2, Opossum-4F/4R to -7F/7R and Mouse-5F/5R to -18F/18R).
Statistical analysis
Statistical analysis was carried out using GraphPad Prism 5 software (GraphPad Software, Inc.; La Jolla, CA, USA). Average CpG methylation and expression between TGCTs and control tissues were compared by ANOVA and t-test. Results were considered statistically significant at the 5% significance level.
RESULTS
Identification of FAM50B antisense transcripts
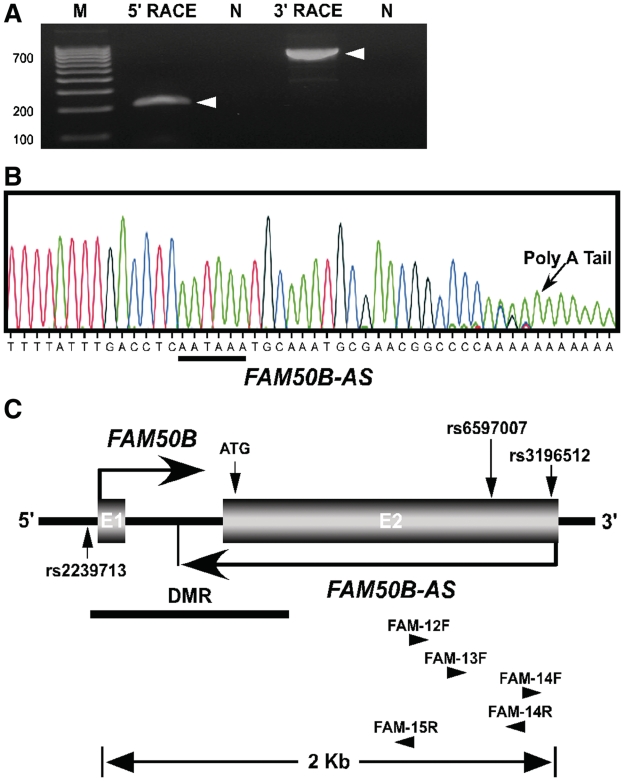
Many imprinted genes are associated with non-coding RNAs, often taking the form of antisense transcripts (26,27). We therefore tested the candidate imprinted gene FAM50B (13) for the presence of an antisense transcript using northern blot analysis and RACE. Such a transcript was detected at this locus, and hereafter referred to as FAM50B antisense (FAM50B-AS) (Figure 1A and Supplementary Figure S1). Using RNA probes specific to the putative FAM50B-AS sequence, an appropriately sized band (∼1.7 kb) was detected by northern blotting, with probes to different sequences in FAM50B-AS detecting the same band (Supplementary Figure S1). We then identified the 5′-end of FAM50B-AS at +1506 relative to the FAM50B translational start codon (Figure 1C). This is also the nucleotide position where the sense strand mRNA terminates (GenBank accession no.: NM_012135.1). The results from 3′-RACE demonstrated the terminus of FAM50B-AS occurs within intron 1 of FAM50B, at position −237 relative to the translational start codon for FAM50B. Also, when sequencing the RT–PCR product of 3′-RACE, the polyA sequence is evident (Figure 1B) in addition to a correctly positioned consensus AAUAAA motif that has been shown to be essential for cleavage and polyadenylation (27 nt upstream, within the 10–30 nt range; underlined, Figure 1B) (28). A CA dinucleotide motif which defines the polyadenylation site for most genes is also present at the polyadenylation site identified for the FAM50B-AS. These results confirm the presence of FAM50B-AS with length of 1743 bp.
Figure 1.
Genomic map of human FAM50B locus. (A) FAM50B-AS RACE-PCR showing a 211 bp product of 5′-RACE and an 823 bp product of 3′-RACE. M, 100 bp DNA ladder. N, control reactions were performed using cDNA prepared in the absence of primers to eliminate the possibility of genomic DNA and RNA contamination. (B) Sequencing of 3′-RACE-PCR demonstrates the polyA sequence and AAUAAA motif (underlined) of FAM50B-AS. (C) The FAM50B locus spans 2 kb of genomic sequence at chromosome 6p25 and consists of two exons (E1 and E2). Horizontal arrows indicate the sense (FAM50B) and antisense (FAM50B-AS) transcripts of the gene. FAM50B-AS originates at the 3′-end of FAM50B and terminates within FAM50B intron 1. The black horizontal line indicates the position of the DMR. Vertical arrows designate positions of SNPs rs2239713 (C/T), rs6597007 (G/C), rs3196512 (G/C) and the translational start codon ATG. The position and direction of primers used for RACE is indicated by horizontal arrowheads. Primer sequences are provided in Supplementary Table S1.
Identification of a FAM50B-associated DMR
Many imprinted genes are associated with CpG-rich regions that exhibit differential methylation of the maternal and paternal alleles. For detection of DMRs, regions are identified for which multiple CpG sites show a methylated to unmethylated ratio of ∼1:1, a pattern consistent with DNA methylated on only one chromosome. To identify potential DMRs, we used the Sequenom MassARRAY Epityper (Sequenom) for quantitative DNA methylation analysis on bisulfite treated DNA. Validation tests on previously identified DMRs of known imprinted genes (PEG3, NAP1L5 and MEG3), showed methylation of 30–60% for these regions (unpublished data).
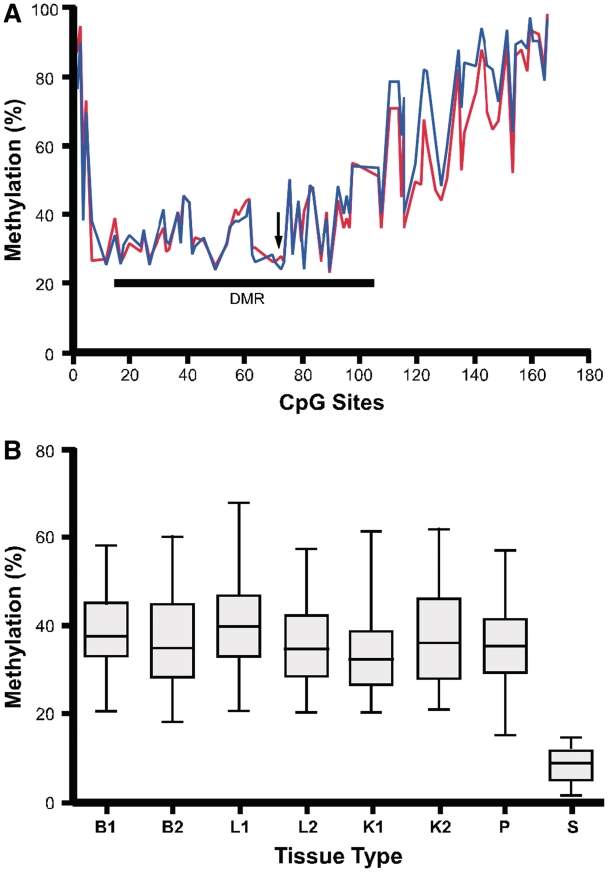
We then searched genomic regions 10-kb upstream and downstream of the candidate imprinted gene FAM50B for GC rich regions (G + C >40%). We identified a CpG island spanning the FAM50B promoter and most of the gene (−1353 to +1066 relative to the translational start codon; Chr6: 3848692-3851100, Version February 2009: GRCh37/hg19; Figure 1B). We quantified methylation at the 166 CpG sites within this region in human conceptus liver (n = 3) and brain (n = 3). The core region, from the 12th CpG to 110th CpG (−884 bp to +415 bp relative to the translational start codon), was 30% to 60% methylated in these tissues, whereas the CpG sites in the 5′- and 3′-boundary regions were more highly methylated (Figure 2A). These findings indicate the existence of a candidate DMR in the core region.
Figure 2.
CpG methylation status at the FAM50B locus. (A) The percent methylation of CpG sites 1–166 was quantified by Sequenom MassARRAY using DNA from conceptus liver (blue line, n = 3) and brain (red line, n = 3). CpG site numbers correspond to the positions within the GC rich region (−1353 to +1066 relative to the FAM50B translational start codon). Arrow indicates the position of the translational start codon. The location of a DMR is shown by a horizontal line. (B) Average methylation of CpG sites (12 to 110) does not vary significantly for conceptus tissues from the three germ layers and from different gestational ages, while sperm shows low levels of methylation at this locus. B, brain; L, liver; K, kidney; P, placenta; S, sperm; B1, L1 and K1—gestational age of 81 days; B2, L2 and K2—gestational age of 110 days.
The average methylation in brain, liver and kidney conceptus tissues (gestational ages 81 and 110 days) did not significantly vary for the region from the 12th to 110th CpG (P = 0.12, n = 2, ANOVA) (Figure 2B). We also determined the degree of methylation for this region in ejaculated spermatozoa (n = 2) and found that it was strikingly hypomethylated in comparison to the somatic tissues analyzed (Figure 2B). These findings indicate that the methylation observed in this region is either derived from the oocyte, or is established post-fertilization.
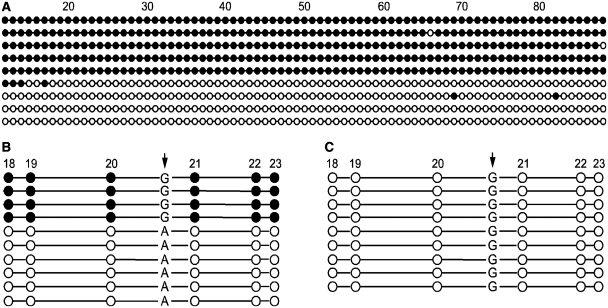
To determine if the DNA methylation present in this region is allele-specific, we performed bisulfite sequencing of individual cloned alleles from human conceptus liver (n = 2). These sequences showed that CpG sites in this region for any cloned allele were either completely methylated or unmethylated (Figure 3A). These findings are consistent with this region comprising a DMR of ∼740 bp that is likely involved in regulating imprinting at the FAM50B locus. We also used the single nucleotide polymorphism rs2239713, which is located between CpG sites 20 and 21, to distinguish between the parental alleles. On the sense strand, SNP rs2239713 is a C/T polymorphism that is indistinguishable following bisulfite conversion, so we instead designed primers to analyze the complementary DNA strand, where the polymorphism is present as a G or A. Analysis of bisulfite-modified DNA from three individual conceptus liver tissues of individuals heterozygous for this SNP who have homozygous G/G mothers showed that methylation is allele specific, with the maternally derived G allele fully methylated and the paternally derived A allele fully unmethylated (Figure 3B). Bisulfite sequencing of cloned alleles of mature sperm DNA (n = 2) showed a complete lack of methylation in this region (Figure 3C), consistent with the previously described quantitative analysis. These results confirm the presence of a DMR with maternal DNA methylation at the FAM50B locus.
Figure 3.
Allele-specific methylation at the FAM50B locus. (A) Clones obtained from bisulfite-treated conceptus liver DNA are either predominantly methylated or unmethylated from the 18th CpG site to the 66th CpG site. (B) Clones obtained from bisulfite-treated conceptus liver DNA, representing one of three individuals heterozygous for SNP rs2239713 (arrow), while their matched maternal deciduae are homozygous G/G for the SNP sites. The allele-specific distribution of methylation with respect to the G/A polymorphism demonstrates that the maternally-derived allele is methylated while the paternally derived allele is unmethylated. (C) Clones obtained from bisulfite-treated mature sperm DNA are completely unmethylated. CpG sites are represented by circles. Unfilled circle, unmethylated CpG site; filled circle, methylated CpG site.
Imprinting and expression analysis of the FAM50B locus in human tissues
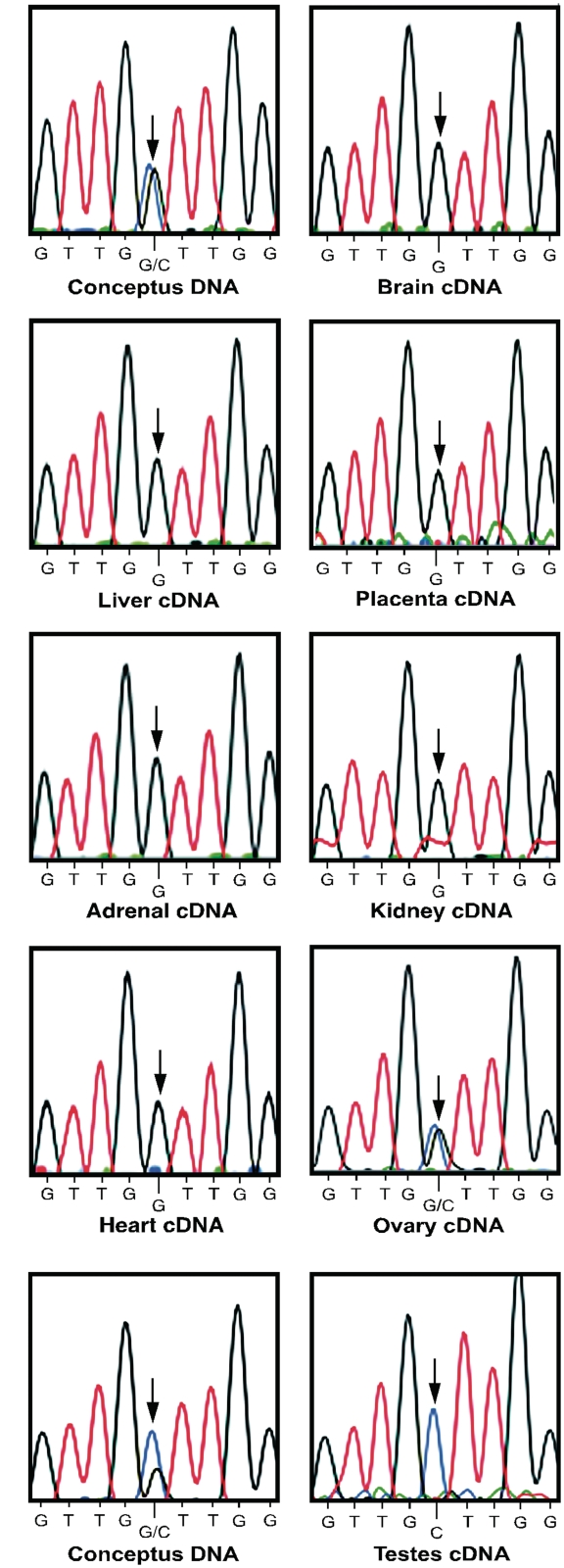
To determine if transcripts associated with the FAM50B locus are monoallelically expressed during early development, RT–PCR was performed followed by nucleotide sequencing of the cDNA in a panel of conceptus tissues from multiple individuals with gestational ages ranging from 81 to 113 days. We analyzed a 206 bp region containing a single nucleotide polymorphism (rs6597007) located in exon 2. For human conceptuses showing heterozygosity at the polymorphic site, monoallelic expression of FAM50B was observed in nearly all fetal tissues analyzed including brain (n = 5), liver (n = 5), placenta (n = 6), adrenal gland (n = 2), kidney (n = 1), heart (n = 1) and testis (n = 2), while expression was biallelic in the ovary (n = 1) (Figure 4). Furthermore, RT–PCR followed by nucleotide sequencing of cDNA from adult testis (n = 1) and liver (n = 1) from individuals heterozygous for SNP rs6597007 demonstrated that FAM50B monoallelic expression was maintained in adulthood (Supplementary Figure S2A).
Figure 4.
Imprinting analysis of human FAM50B. Genomic DNA from human conceptuses (A and B) is heterozygous for polymorphism rs6597007 (G/C). Sequencing of cDNA demonstrates monoallelic expression of FAM50B transcripts in multiple conceptus tissues, including brain, liver, placenta, adrenal gland, kidney, heart and testis. In contrast, FAM50B expression is biallelic in fetal ovary. The position of SNP rs6597007 used to determine allelic expression is indicated by the arrows.
Matched maternal decidual tissues of informative conceptuses were genotyped to determine the parental origin of the expressed FAM50B allele in humans. DNA from the maternal decidua for a conceptus polymorphic at site rs6597007 (G/C) was homozygous C/C for this SNP (Supplementary Figure S2B). Transcripts in the conceptus were derived only from the allele containing G at this SNP position, indicating paternal expression. Identical results for parental allele-specific expression were obtained for three additional matched sets of decidua and conceptus tissues, demonstrating that only the paternally inherited allele was expressed.
To determine if both the FAM50B and FAM50B-AS transcripts are monoallelically expressed, we performed strand-specific RT-PCR by amplifying the 3′-end of FAM50B and FAM50B-AS containing SNP rs6597007 using 3′-RACE-derived cDNA (Supplementary Figure S3A). We demonstrated that the FAM50B sense and antisense transcripts were both monoallelically expressed from the same allele in conceptus brain (n = 1), adult liver (n = 1) and adult testis (n = 1) (Figure 4 and Supplementary Figures S2 and S3B).
The level of expression of FAM50B and FAM50B-AS in human tissues was determined by relative quantitative RT–PCR using cDNA derived from a panel of conceptus and adult tissues (n = 2). FAM50B exhibits the highest expression in pre- and post-natal testis tissue, while FAM50B-AS expression is highest in conceptus testis and brain (Supplementary Figure S4A and S4B).
Analysis of genes neighboring FAM50B in human tissues
Imprinted genes are often present as gene clusters in the genome. We therefore determined if genes neighboring FAM50B also exhibit imprinted expression. In the human genome, there are six genes in a 200 kb region centered around FAM50B. The three pseudogenes, RPS25P7, TDGF4 and LOC728344 are not transcribed, and there are no annotated SNPs useable for allele-specific expression analysis of LOC100289494 and LOC100289591. The remaining gene, C6orf145, was analyzed for allele-specific expression, and we found that both parental alleles were expressed at approximately equivalent levels from all conceptus tissues analyzed, including brain (n = 3), heart (n = 1), kidney (n = 2), muscle (n = 2), lung (n = 1) and placenta (n = 5) (Supplementary Figure S5, SNP rs226959) These findings cannot, however, exclude the possibility that C6orf145 is imprinted in a highly tissue- and/or developmental stage-specific manner.
Evolutionary analysis of FAM50B
FAM50B, also known as XAP-5 like gene (X5L), encodes a 325 amino acid (AA) protein and arose from the X-linked XAP-5 gene, most likely by retrotransposition (25). XAP-5 is a conserved protein found in a wide range of eukaryotes ranging from the roundworm (Caenorhabditis elegans) to the human. Amino acid alignment of XAP-5 and X5L demonstrates that they are 77% identical in the human.
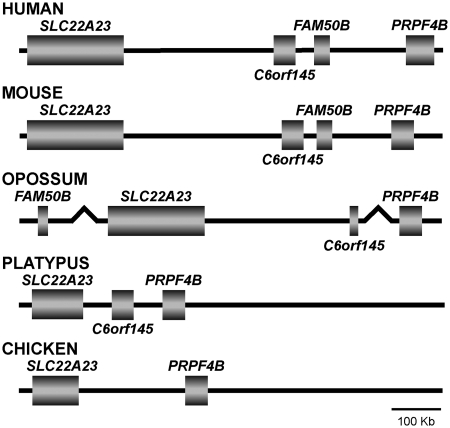
To determine the timing of the retrotransposition event responsible for creating the FAM50B locus during vertebrate evolution, we compared the region encompassing FAM50B in human and mouse (Eutherian), opossum (Metatherian), platypus (Prototherian) and chicken (Aves) using the UCSC and Ensembl genome browsers (http://genome.ucsc.edu and http://www.ensembl.org/, respectively). In the mouse, FAM50B is near C6orf145 and between SLC22A23 and PRPF4B (Figure 5). It encodes a 334 AA protein with 83% sequence similarity to the human ortholog. BLAST analysis of the platypus and opossum genomes (http://www.ncbi.nlm.nih.gov/Genomes) showed that the opossum, but not the platypus, contains a gene orthologous to FAM50B. Unlike mouse and human, the opossum FAM50B ortholog is not located between SLC22A23 and PRPF4B, but rather resides 78 Mb from the 3′-end of SLC22A23. The opossum FAM50B gene also has an intronless open reading frame, and encodes a 338 AA protein (Gene ID: LOC100027108; GenBank accession no.: XM_001370796) with 76% similarity to the human ortholog. Interestingly, both FAM50B and C6orf145 are absent in the chicken genome. These findings are consistent with FAM50B having undergone retrotransposition from the X chromosome source gene after Therians split from Prototherians.
Figure 5.
Phylogenetic comparison of the genomic structure of FAM50B and flanking genes. FAM50B is flanked by C6orf145 and PRPF4B in human (chromosome 6) and mouse (chromosome 13), but is 78 Mb upstream from the SLC22A23 ortholog in the opossum (chromosome 3). FAM50B orthologs are not present in the platypus (UltraContig Ultra 352) and chicken genomes (chromosome 2).
As in the human and mouse, FAM50B is located between C6orf145 and PRPF4B (SuperContig Scaffold54 from Ensembl genome browser) in the African elephant (Loxodonta africana), a member of the most ancestral placental super order, Afrotheria. Thus, FAM50B appears to have integrated into the genomic region between C6orf145 and PRPF4B after the divergence of marsupials from ancestral Eutherian mammals.
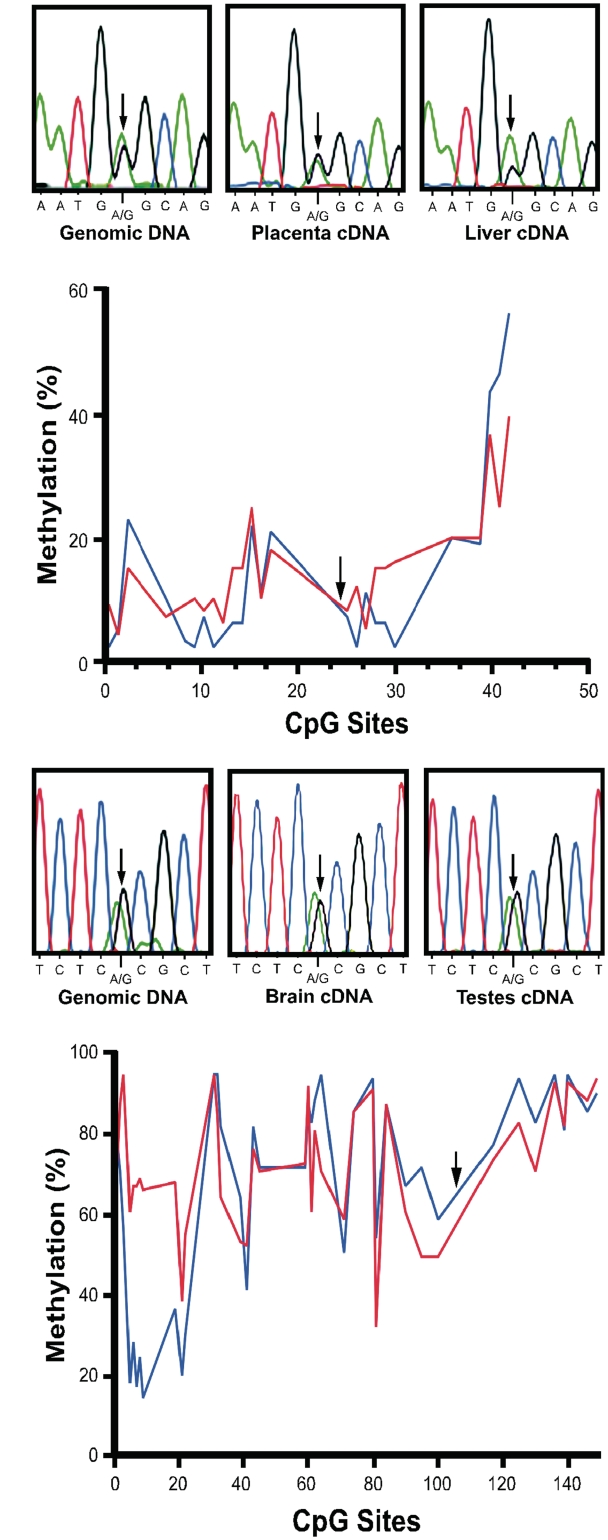
We determined that transcripts from the opossum FAM50B ortholog have high levels of expression in kidney, liver, embryo and placenta, but expression was not detected in the brain (Supplementary Figure S6A). Transcripts of the mouse FAM50B ortholog were expressed in liver, kidney, brain and testes (Supplementary Figure S6B). Nevertheless, expression of FAM50B in the opossum (Figure 6A) and mouse (Figure 6C) was biallelic in all tissues investigated. We did not test for the presence of an antisense transcript in the opossum and mouse. Moreover, CpG methylation of the genomic region orthologous to the DMR in the human is comparatively hypomethylated in the opossum (Figure 6B), and hypermethylated in the mouse (Figure 6D).
Figure 6.
Imprinting analysis of FAM50B in the opossum and the mouse. (A) cDNA sequencing demonstrates biallelic expression of FAM50B in opossum placenta and liver. Arrows indicate the position of the SNP used to determine the allelic expression patterns. (B) Methylation at CpG dinucleotides is <20% for most of the region that encompasses the opossum FAM50B translational start codon, from −340 to 972 bp. Blue line, DNA from liver; red line, DNA from kidney. Arrow indicates the position of the translational start codon. (C) FAM50B cDNA sequencing demonstrates biallelic expression of the FAM50B in mouse brain and testis. Arrows indicate the position of the SNP used to determine the allelic expression patterns. (D) Methylation is >60% at most CpG sites in proximity to the translational start codon of murine FAM50B (from −8800 to 1120 bp relative to translational start codon). Arrow indicates the position of the translational start codon. Blue line, DNA from kidney; red line, DNA from liver.
Loss of the methylation imprint in human TGCTs
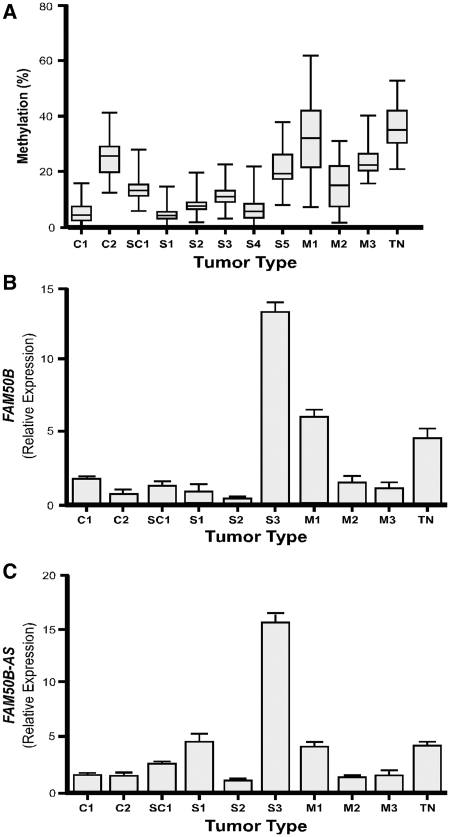
DMR methylation and allele-specific expression of FAM50B in TGCTs was determined to assess the role of this gene in cancer formation (Figure 7A). Methylation of the FAM50B DMR was significantly reduced (P < 0.001, unpaired t-test) in 10 of 11 TGCTs specimens we investigated, including two embryonic carcinomas (C1, 5.1 ± 3.9%; C2, 25.8 ± 7.0%) one seminoma mixed with embryonic carcinoma (SC1, 14.5 ± 5.3%), five seminomas (S1, 4.5 ± 2.8%; S2, 8.1 ± 3.8%; S3, 11.7 ± 4.3; S4, 6.2 ± 4.7%; S5, 21.0 ± 7.0%) and mixed germ cell tumors (M2, 15.7 ± 8.4%; M3, 24.2 ± 6.5%) relative to that in normal testis (TN, 36.6 ± 7.7%, n = 3). DMR methylation was not significantly altered in a mixed germ cell tumor (M1, 32.4 ± 14.7%; P = 0.07).
Figure 7.
Methylation status and gene expression of FAM50B locus in TGCTs. (A) Percent methylation of the FAM50B DMR in each tumor and in normal testis controls (n = 3). (B) Relative expression levels, normalized to GAPDH, of FAM50B transcripts in TGCTs and in normal testis controls (n = 3). (C) Relative expression levels, normalized to GAPDH, of the FAM50B-AS transcripts in TGCTs and in normal testis controls (n = 3). TGCTs include: embryonic carcinoma (C1, C2), seminoma mixed with embryonic carcinoma (SC1), pure seminoma (S1, S2, S3 and S4, S5), and mixed germ cell tumor (M1, M2 and M3). Normal testis tissue (TN, n = 3) was used for comparison.
Quantitative PCR demonstrated that the decreased level of methylation at the FAM50B DMR in TGCTs was frequently associated with a reduced level of FAM50B and FAM50B-AS expression (Figure 7B and C). Only seminoma S3 demonstrated a significant increase in expression of both FAM50B (P < 0.001) and FAM50B-AS (P = 0.002) relative to that in normal testis. These findings indicate that DMR methylation status alone is insufficient for regulation of FAM50B and FAM50B-AS transcript levels in some TGCTs.
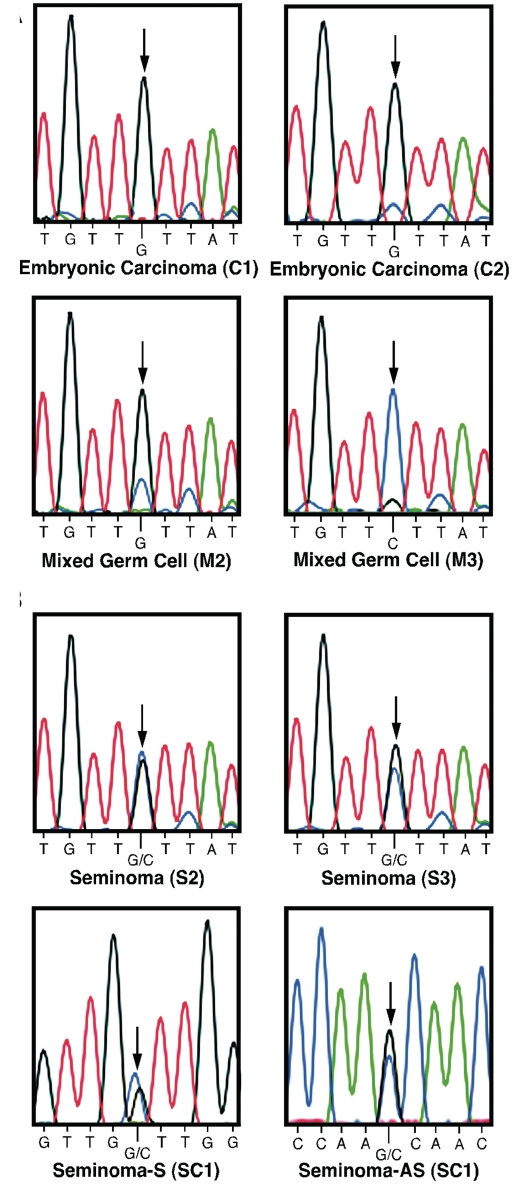
To determine if FAM50B imprinting is maintained in TGCTs, we analyzed tumors heterozygous for SNP rs3196512 (C/G), which included embryonic carcinomas C1 and C2, seminomas S2 and S3, and mixed germ cell tumors M2 and M3, to distinguish the allelic expression pattern for the FAM50B transcript. We also analyzed the seminoma mixed with embryonic carcinoma (SC1), which is heterozygous for SNP rs6597007 (C/G), to determine the allelic expression pattern for both the sense (S) and antisense (AS) transcripts (Figure 8). In the embryonic carcinomas (C1 and C2) and mixed germ cell tumors (M2 and M3) investigated, monoallelic expression of FAM50B was maintained (Figure 8A); however, in the seminomas (S2 and S3) both alleles of FAM50B were expressed (Figure 8B). Furthermore, both alleles of FAM50B and FAM50B-AS were expressed in the seminoma mixed with embryonic carcinoma (SC1) (Figure 8B). These results indicate that loss of imprinting at the FAM50B locus is a common event in seminomas.
Figure 8.
Allelic expression of FAM50B in TGCTs. (A) cDNA sequencing of TGCTs demonstrates maintenance of monoallelic expression of the FAM50B transcript in embryonic carcinoma C1 and C2 and mixed germ cell tumor M2 and M3. (B) cDNA sequencing of TGCTs demonstrates biallelic expression of the FAM50B transcripts in the seminomas S2 and S3 and biallelic expression of both FAM50B and FAM50B-AS in seminoma mixed with embryonic carcinoma SC1. The SNP rs3196512 (G/C) was used to determine allelic expression of FAM50B in sample C1, C2, M2, M3, S2 and S3. The SNP rs6597007 (G/C) was used to determine allelic expression of FAM50B and FAM50B-AS in sample SC1. The position of the SNP used to determine allelic expression is indicated by the arrows.
To determine if the DNA methylation in the DMRs is involved in regulating imprinting of the FAM50B locus, we performed bisulfite sequencing of individual cloned alleles of S2 (Supplementary Figure S7A) and S3 (Supplementary Figure S7B). Our results showed that allele-specific methylation was partially lost in S2 and S3. These results were associated with the loss of imprinted expression of FAM50B transcripts in seminomas, indicating that this DMR is required for maintaining the imprinted status of this gene.
DISCUSSION
In our search for imprinting regulatory regions on chromosome 6p, we identified a novel DMR at the FAM50B locus, a gene we previously predicted to be imprinted (13). The unique parent-of-origin-dependent expression of imprinted genes is in large part regulated by the methylation status of associated DMRs. There are two distinct types of DMRs that regulate the expression of genomically imprinted genes: germline DMRs that are established during gametogenesis and somatic DMRs that are formed post-fertilization, sometimes in a tissue-specific manner (29). Using the Sequenom MassARRAY system, we performed methylation analysis at the FAM50B locus in human tissues derived from the three germ layers. These methylation analyses, combined with those of mature human spermatozoa, identified a novel DMR at the promoter region of FAM50B. The FAM50B DMR showed uniform differential methylation in embryonic tissues of different germ layer origin. It was maternally methylated in conceptus liver and unmethylated in sperm. Thus, this DMR is either a maternal methylation imprint mark established in oocytes or it is acquired post-fertilization. Our results therefore support the use of the Sequenom MassARRAY as an effective tool for experimentally identifying novel DMRs of imprinted genes.
We identified both sense and antisense transcripts of FAM50B, and confirmed the imprint status of both, with each transcript displaying paternal expression in normal human tissues. Thus, we demonstrated for the first time that FAM50B is an imprinted gene in humans. The function of the gene products from both FAM50B and its X-linked resource gene XAP-5 are currently unknown. However, phylogeny indicates related genes with XAP5 domains in both Brassica and Arabidopsis, indicating that the origin of FAM50-related genes occurred early in the development of eukaryotes. Hints as to function may come from Arabidopsis XCT (XAP-5 circadian timekeeper), which regulates the circadian clock in response to light, acting as a global regulator of growth (30).
XAP-5 contains runs of CG-rich trinucleotide repeats (CCG) in the 5′-untranslated region, and therefore is considered a candidate disease gene (31); however, this triplet repeat is not present in FAM50B. We found high expression of FAM50B in embryonic tissues, indicating an important role in prenatal development. We also found that FAM50B is highly expressed in testis, consistent with the prior observation that X-derived retrogenes are more frequently expressed in spermatocytes than autosomally-derived retrogenes (32).
A considerable proportion of imprinted genes produce both sense and antisense transcripts, which was our impetus for searching for a FAM50B antisense transcript. Non-coding antisense transcripts are broadly classified into large non-coding RNAs and small regulatory RNAs, acting in cis or in trans, respectively, to repress the flanking protein-coding imprinted genes (26,27). FAM50B-AS, together with IGF2AS (IGF2 antisense) (33), APEG3 (PEG3 antisense) (34), SLC22A18AS (SLC22A18 antisense) (35) and PEG1-AS (PEG1 antisense) (36), may be more suitably classified into a new RNA group due to their specific features. First, all are paternally expressed from the same allele with their sense transcripts, which differs from the reciprocal imprinting pattern of large and small non-coding RNAs (for example, paternally expressed antisense Air and maternally expressed sense IGF2R; and maternally expressed miRNA MiR-431 and paternally expressed sense RTL1). Second, the length of the antisense transcripts is 1–2.5 kb, much shorter than the large non-coding RNAs (usually 10–100 kb), but much longer than the small non-coding RNAs (<300 bases). The biological functions of these antisense transcripts, whether involved in regulating the imprinting of their sense transcripts or in maintaining normal tissue function, remain elusive. Interestingly, imprinted FAM50B-AS exhibits the highest level of expression in fetal brain, indicating that FAM50B-AS is potentially involved in preserving normal brain function.
Multiple transcribed and functional retrotransposed genes that originated from X-linked housekeeping genes have been uncovered in the genomes of mammals (37,38). The percentage of expressed retrotransposons that originate from X chromosome genes is higher than those originating from autosomal chromosomes. Five of these X-derived retrotransposed genes in mice (i.e. MCTS2, NAP1L5, U2AF1-RS1, Inpp5f_v2 and Peg12) and three orthologues in humans (i.e. MCTS2, known as PSIMCT-1, INPP5F_V2 and NAP1L5) are known to be imprinted, and all are paternally expressed (39,40). In contrast, KLF14 retrotransposed from an autosomal chromosome, and is maternally expressed (41). Of particular interest is RB1, which is not a retrotransposed gene; however, the preferential maternal expression of the RB1 transcript and the paternal expression of an alternative transcript of this gene is linked to a retrotransposed pseudogene derived from an autosomal chromosome (42). In the case of RB1, the pseudogene is not transcribed, but rather acts as a weak promoter for an alternate RB1 transcript, regulated by a DMR in the pseudogene (43). Clearly, retrotransposed elements play an important role in parental allele silencing of imprinted genes. It has been hypothesized that the retrotransposition of X-derived genes is a mechanism to insure continued activity for essential genes that are otherwise silenced by transient X-chromosome inactivation during spermatogenesis (32,44). FAM50B is another example of a maternally silenced imprinted gene derived from an X-linked gene by retrotransposition. This adds further support to the hypothesis that imprinting functions in part as a dosage compensation mechanism (45). However, unlike other known retrotransposed imprinted genes, FAM50B is not located within an intron of a host gene.
Comparison of X-inactivation and retrotransposed genes between marsupials and eutherians may be highly informative for understanding the evolution of imprinted gene silencing, especially with regard to the idea that retrotransposition of X-linked genes compensates for X-inactivation during spermatogenesis. The recent discovery of X-chromosome inactivation during spermatogenesis in opossum has led to a model for the origins of X-chromosome silencing for dosage compensation in females. In female marsupials, it is always the paternal X chromosome that is silenced. Persistence of X-inactivation that was originally established during spermatogenesis might explain this paternal-specific X chromosome silencing in females. If this is the ancestral method of mammalian X-inactivation, then the reactivation and subsequent random inactivation seen in eutherians is a modification of the simpler marsupial mechanism, possibly providing resiliency by not forcing dependency on one X chromosome (46).
It may be possible to elucidate the mechanism by which retrotransposed imprinted genes acquired parent-of origin expression by investigating their evolution in mammals. Only a small number of imprinted genes in eutherians are also monoallelically expressed in marsupials (47), and none of the known eutherian imprinted genes that originated from retrotransposition events are imprinted in marsupials. The imprinting of MCTS2, INPP5F_V2 and NAP1L5 is conserved in the human and mouse, indicating retrotransposition occurred before radiation of the human and rodent lineages (42). U2af1-rs1 and Peg12 are postulated to have been created by retrotransposition during mouse evolution after the divergence of mice and humans since their orthologs are not present in the human (48). Phylogenetic comparison of the orthologs of the FAM50B locus indicates that this gene was integrated into the opossum genome in a position different from that in human, mouse and elephant. Although it is expressed in a variety of opossum tissues, we showed that FAM50B is not imprinted in the opossum. Furthermore, FAM50B in the mouse does not contain a DMR in the promoter region, and it is also not imprinted. Hence imprinted expression of human FAM50B appears to have originated from a species-specific retrotransposition event that occurred subsequent to the radiation of the human and rodent lineages. These findings demonstrate that the repertoires of imprinted genes vary between species, which is consistent with previous reports indicating that imprinted genes did not arise at a single point during evolution (49).
To obtain insight into the importance of FAM50B in normal spermatogenesis, we analyzed its methylation profile and expression pattern in seminomatous and non-seminomatous TGCTs. TGCTs have a pluripotent nature and display histology reflecting that of primordial germ cells (PGCs) to embryonal and somatic cell differentiation. Seminomas recapitulate some aspects of spermatogenesis, whereas non-seminomas demonstrate embryonal and extraembryonal differentiation patterns (50). Besides histological distinctions, they have epigenetic differences. Seminomas have widespread hypomethylation, both in CpG islands and globally, unlike non-seminomas, which have higher methylation overall (51). This hypomethylation is a significant similarity between seminomas and PGCs, which also have very low methylation levels. All these similarities distinguish seminomas from other types of TGCTs, and suggest that seminomas originate from a more primitive, less differentiated cell type that may be more prone to tumorigenesis.
The FAM50B DMR showed hypomethylation in seminomatous and the majority of non-seminomatous TGCTs; however, complete loss of imprinted expression of FAM50B and FAM50B-AS was only observed in seminomas. Although hypomethylation of the reciprocally imprinted genes, H19 and IGF2, is also observed in TGCTs, null expression, monoallelic expression and biallelic expression of these genes are observed in both seminomatous and non-seminomatous TGCTs (52,53). Thus, the methylation status of the DMR for H19 and IGF2 is not always correlated with their allelic expression. Similarly, imprinted gene expression at the FAM50B locus is not always associated with the methylation status of its DMR in non-seminomatous TGCTs. This indicates that factors other than methylation may contribute to transcriptional regulation of imprinting of FAM50B in non-seminomatous TGCTs. In support of this idea, a recent study suggests that histone methylation may act as the principal regulator of gene expression in non-seminomas (54). Determination of histone modifications at the FAM50B locus may help clarify the mechanism regulating imprinted expression of FAM50B in non-seminomatous TGCTs. Analysis of copy number variations in TGCTs has also been extensively studied (55). A number of chromosomal aberrations have been observed, but none of them are associated with the 6p25 locus containing FAM50B. Additionally, the similarities of seminomas to undifferentiated PGCs suggest that seminomas have reduced epigenetic regulation besides the observed hypomethylation, which would be consistent with the loss of FAM50B imprinting in only this tumor type. The high frequency of loss of imprinting of FAM50B and FAM50B-AS in the genesis of seminomas also indicates that imprinted expression of this gene needs to be tightly regulated during normal spermatogenesis.
Both segmental duplications and deletions at 6p25 have been associated with diseases and developmental disorders (56–59). Genetic loss or gain sometimes detrimentally alters expression of biallelically expressed genes. In contrast, when imprinted genes are present at the location of copy number variation, gene dosage is usually altered dramatically, ranging from complete loss of expression to overexpression. Our finding that the FAM50B locus is imprinted stresses the importance of determining parent-of-origin inheritance patterns, in order to better understand the pathogenesis of diseases linked to chromosome location 6p25.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institute of Health (R01 ES015165, R01 DK085173 and R01 ES016772), the Ester B. O’Keeffe Foundation Award and Fred and Alice Stanback. Funding for open access charge: The Ester B. O'Keeffe Foundation Award.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We sincerely thank Autumn Bernal, Radhika Das and Daniel Hampton for providing analysis tools and materials. We thank Kathleen Smith for providing us with opossum tissue samples.
REFERENCES
- 1.Falls JG, Pulford DJ, Wylie AA, Jirtle RL. Genomic imprinting: implications for human disease. Am. J. Pathol. 1999;154:635–647. doi: 10.1016/S0002-9440(10)65309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ubeda F, Wilkins JF. Imprinted genes and human disease: an evolutionary perspective. Adv. Exp. Med. Biol. 2008;626:101–115. [PubMed] [Google Scholar]
- 3.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends. Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Maeda N, Hayashizaki Y. Genome-wide survey of imprinted genes. Cytogenet. Genome Res. 2006;113:144–152. doi: 10.1159/000090826. [DOI] [PubMed] [Google Scholar]
- 5.Miyoshi N, Kuroiwa Y, Kohda T, Shitara H, Yonekawa H, Kawabe T, Hasegawa H, Barton SC, Surani MA, Kaneko-Ishino T, et al. Identification of the Meg1/Grb10 imprinted gene on mouse proximal chromosome 11, a candidate for the Silver-Russell syndrome gene. Proc. Natl Acad. Sci. USA. 1998;95:1102–1107. doi: 10.1073/pnas.95.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaneko-Ishino T, Kuroiwa Y, Miyoshi N, Kohda T, Suzuki R, Yokoyama M, Viville S, Barton SC, Ishino F, Surani MA. Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat. Genet. 1995;11:52–59. doi: 10.1038/ng0995-52. [DOI] [PubMed] [Google Scholar]
- 7.Mizuno Y, Sotomaru Y, Katsuzawa Y, Kono T, Meguro M, Oshimura M, Kawai J, Tomaru Y, Kiyosawa H, Nikaido I, et al. Asb4, Ata3, and Dcn are novel imprinted genes identified by high-throughput screening using RIKEN cDNA microarray. Biochem. Biophys. Res. Commun. 2002;290:1499–1505. doi: 10.1006/bbrc.2002.6370. [DOI] [PubMed] [Google Scholar]
- 8.Pollard KS, Serre D, Wang X, Tao H, Grundberg E, Hudson TJ, Clark AG, Frazer K. A genome-wide approach to identifying novel-imprinted genes. Hum. Genet. 2008;122:625–634. doi: 10.1007/s00439-007-0440-1. [DOI] [PubMed] [Google Scholar]
- 9.Babak T, Deveale B, Armour C, Raymond C, Cleary MA, van der KD, Johnson JM, Lim LP. Global survey of genomic imprinting by transcriptome sequencing. Curr. Biol. 2008;18:1735–1741. doi: 10.1016/j.cub.2008.09.044. [DOI] [PubMed] [Google Scholar]
- 10.Smith RJ, Kelsey G. Identification of imprinted loci by methylation: use of methylation-sensitive representational difference analysis (Me-RDA) Methods Mol. Biol. 2001;181:113–132. doi: 10.1385/1-59259-211-2:113. [DOI] [PubMed] [Google Scholar]
- 11.Kelsey G, Bodle D, Miller HJ, Beechey CV, Coombes C, Peters J, Williamson CM. Identification of imprinted loci by methylation-sensitive representational difference analysis: application to mouse distal chromosome 2. Genomics. 1999;62:129–138. doi: 10.1006/geno.1999.6022. [DOI] [PubMed] [Google Scholar]
- 12.Strichman-Almashanu LZ, Lee RS, Onyango PO, Perlman E, Flam F, Frieman MB, Feinberg AP. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res. 2002;12:543–554. doi: 10.1101/gr.224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–1730. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mann JR. Imprinting in the germ line. Stem Cells. 2001;19:287–294. doi: 10.1634/stemcells.19-4-287. [DOI] [PubMed] [Google Scholar]
- 15.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 16.Burden AD, Javed S, Bailey M, Hodgins M, Connor M, Tillman D. Genetics of psoriasis: paternal inheritance and a locus on chromosome 6p. J. Invest. Dermatol. 1998;110:958–960. doi: 10.1046/j.1523-1747.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- 17.Chatterjee A, Pulido HA, Koul S, Beleno N, Perilla A, Posso H, Manusukhani M, Murty VV. Mapping the sites of putative tumor suppressor genes at 6p25 and 6p21.3 in cervical carcinoma: occurrence of allelic deletions in precancerous lesions. Cancer Res. 2001;61:2119–2123. [PubMed] [Google Scholar]
- 18.Dumur CI, Dechsukhum C, Ware JL, Cofield SS, Best AM, Wilkinson DS, Garrett CT, Ferreira-Gonzalez A. Genome-wide detection of LOH in prostate cancer using human SNP microarray technology. Genomics. 2003;81:260–269. doi: 10.1016/s0888-7543(03)00020-x. [DOI] [PubMed] [Google Scholar]
- 19.Mazurenko NN, Bliev AI, Bidzhieva BA, Peskov DI, Snigur NV, Savinova EB, Kiselev FL. Loss of heterozygosity at chromosome 6 as a marker of early genetic alterations in cervical intraepithelial neoplasias and microinvasive carcinomas. Mol. Biol. (Mosk) 2006;40:436–447. [PubMed] [Google Scholar]
- 20.Smith BS, Pettersen JC. An anatomical study of a duplication 6p based on two sibs. Am. J. Med. Genet. 1985;20:649–663. doi: 10.1002/ajmg.1320200411. [DOI] [PubMed] [Google Scholar]
- 21.Davies AF, Mirza G, Sekhon G, Turnpenny P, Leroy F, Speleman F, Law C, van Regemorter N, Vamos E, Flinter F, et al. Delineation of two distinct 6p deletion syndromes. Hum. Genet. 1999;104:64–72. doi: 10.1007/s004390050911. [DOI] [PubMed] [Google Scholar]
- 22.Ng D, Mowrey P, Ragoussis J, Mirza G, Coll E, Di Fazio MP, Turner C, Levin SW. Molecularly defined interstitial tandem duplication 6p case with mild manifestations. Am. J. Med. Genet. 2001;103:320–325. [PubMed] [Google Scholar]
- 23.Mirza G, Williams RR, Mohammed S, Clark R, Newbury-Ecob R, Baldinger S, Flinter F, Ragoussis J. Refined genotype-phenotype correlations in cases of chromosome 6p deletion syndromes. Eur. J. Hum. Genet. 2004;12:718–728. doi: 10.1038/sj.ejhg.5201194. [DOI] [PubMed] [Google Scholar]
- 24.Stohler R, Kucharski E, Farrow E, Torres-Martinez W, Delk P, Thurston VC, Vance GH. A case of de novo partial tetrasomy of distal 6p and review of the literature. Am. J. Med. Genet. A. 2007;143A:1978–1983. doi: 10.1002/ajmg.a.31678. [DOI] [PubMed] [Google Scholar]
- 25.Sedlacek Z, Munstermann E, Dhorne-Pollet S, Otto C, Bock D, Schutz G, Poustka A. Human and mouse XAP-5 and XAP-5-like (X5L) genes: identification of an ancient functional retroposon differentially expressed in testis. Genomics. 1999;61:125–132. doi: 10.1006/geno.1999.5931. [DOI] [PubMed] [Google Scholar]
- 26.Mohammad F, Mondal T, Kanduri C. Epigenetics of imprinted long noncoding RNAs. Epigenetics. 2009;4:277–286. [PubMed] [Google Scholar]
- 27.Peters J, Robson JE. Imprinted noncoding RNAs. Mamm. Genome. 2008;19:493–502. doi: 10.1007/s00335-008-9139-4. [DOI] [PubMed] [Google Scholar]
- 28.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi H, Suda C, Abe T, Kohara Y, Ikemura T, Sasaki H. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet. Genome Res. 2006;113:130–137. doi: 10.1159/000090824. [DOI] [PubMed] [Google Scholar]
- 30.Martin-Tryon EL, Harmer SL. XAP5 CIRCADIAN TIMEKEEPER coordinates light signals for proper timing of photomorphogenesis and the circadian clock in Arabidopsis. Plant Cell. 2008;20:1244–1259. doi: 10.1105/tpc.107.056655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazzarella R, Pengue G, Yoon J, Jones J, Schlessinger D. Differential expression of XAP5, a candidate disease gene. Genomics. 1997;45:216–219. doi: 10.1006/geno.1997.4912. [DOI] [PubMed] [Google Scholar]
- 32.Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jegou B, Kaessmann H. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 2008;6:e80. doi: 10.1371/journal.pbio.0060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okutsu T, Kuroiwa Y, Kagitani F, Kai M, Aisaka K, Tsutsumi O, Kaneko Y, Yokomori K, Surani MA, Kohda T, et al. Expression and imprinting status of human PEG8/IGF2AS, a paternally expressed antisense transcript from the IGF2 locus, in Wilms' tumors. J. Biochem. 2000;127:475–483. doi: 10.1093/oxfordjournals.jbchem.a022630. [DOI] [PubMed] [Google Scholar]
- 34.Choo JH, Kim JD, Kim J. Imprinting of an evolutionarily conserved antisense transcript gene APeg3. Gene. 2008;409:28–33. doi: 10.1016/j.gene.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallagher E, Mc GA, Chung WY, Mc CO, Harrison M, Kerin M, Dervan PA, Mc CA. Gain of imprinting of SLC22A18 sense and antisense transcripts in human breast cancer. Genomics. 2006;88:12–17. doi: 10.1016/j.ygeno.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Li T, Vu TH, Lee KO, Yang Y, Nguyen CV, Bui HQ, Zeng ZL, Nguyen BT, Hu JF, Murphy SK, et al. An imprinted PEG1/MEST antisense expressed predominantly in human testis and in mature spermatozoa. J. Biol. Chem. 2002;277:13518–13527. doi: 10.1074/jbc.M200458200. [DOI] [PubMed] [Google Scholar]
- 37.Kriegs JO, Churakov G, Kiefmann M, Jordan U, Brosius J, Schmitz J. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moller-Krull M, Delsuc F, Churakov G, Marker C, Superina M, Brosius J, Douzery EJ, Schmitz J. Retroposed elements and their flanking regions resolve the evolutionary history of xenarthran mammals (armadillos, anteaters, and sloths) Mol. Biol. Evol. 2007;24:2573–2582. doi: 10.1093/molbev/msm201. [DOI] [PubMed] [Google Scholar]
- 39.Cowley M, Oakey RJ. Retrotransposition and genomic imprinting. Brief. Funct. Genomics. 2010;9:340–346. doi: 10.1093/bfgp/elq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wood AJ, Roberts RG, Monk D, Moore GE, Schulz R, Oakey RJ. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 2007;3:e20. doi: 10.1371/journal.pgen.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanber D, Berulava T, Ammerpohl O, Mitter D, Richter J, Siebert R, Horsthemke B, Lohmann D, Buiting K. The human retinoblastoma gene is imprinted. PLoS Genet. 2009;5:e1000790. doi: 10.1371/journal.pgen.1000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker-Katiraee L, Carson AR, Yamada T, Arnaud P, Feil R, Abu-Amero SN, Moore GE, Kaneda M, Perry GH, Stone AC, et al. Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution. PLoS Genet. 2007;3:e65. doi: 10.1371/journal.pgen.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buiting K, Kanber D, Horsthemke B, Lohmann D. Imprinting of RB1 (the new kid on the block) Brief. Funct. Genomics. 2010;9:347–353. doi: 10.1093/bfgp/elq014. [DOI] [PubMed] [Google Scholar]
- 44.Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc. Natl Acad. Sci. USA. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walter J, Paulsen M. The potential role of gene duplications in the evolution of imprinting mechanisms. Hum. Mol. Genet. 2003;12(Spec. No. 2):R215–R220. doi: 10.1093/hmg/ddg296. [DOI] [PubMed] [Google Scholar]
- 46.Hornecker JL, Samollow PB, Robinson ES, Vandeberg JL, McCarrey JR. Meiotic sex chromosome inactivation in the marsupial Monodelphis domestica. Genesis. 2007;45:696–708. doi: 10.1002/dvg.20345. [DOI] [PubMed] [Google Scholar]
- 47.Renfree MB, Hore TA, Shaw G, Graves JA, Pask AJ. Evolution of genomic imprinting: insights from marsupials and monotremes. Annu. Rev. Genomics Hum. Genet. 2009;10:241–262. doi: 10.1146/annurev-genom-082908-150026. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Joh K, Masuko S, Yatsuki H, Soejima H, Nabetani A, Beechey CV, Okinami S, Mukai T. The mouse Murr1 gene is imprinted in the adult brain, presumably due to transcriptional interference by the antisense-oriented U2af1-rs1 gene. Mol. Cell. Biol. 2004;24:270–279. doi: 10.1128/MCB.24.1.270-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans HK, Weidman JR, Cowley DO, Jirtle RL. Comparative phylogenetic analysis of blcap/nnat reveals eutherian-specific imprinted gene. Mol. Biol. Evol. 2005;22:1740–1748. doi: 10.1093/molbev/msi165. [DOI] [PubMed] [Google Scholar]
- 50.Oosterhuis JW, Looijenga LH. Testicular germ-cell tumours in a broader perspective. Nat. Rev. Cancer. 2005;5:210–222. doi: 10.1038/nrc1568. [DOI] [PubMed] [Google Scholar]
- 51.Smiraglia DJ, Szymanska J, Kraggerud SM, Lothe RA, Peltomaki P, Plass C. Distinct epigenetic phenotypes in seminomatous and nonseminomatous testicular germ cell tumors. Oncogene. 2002;21:3909–3916. doi: 10.1038/sj.onc.1205488. [DOI] [PubMed] [Google Scholar]
- 52.Furukawa S, Haruta M, Arai Y, Honda S, Ohshima J, Sugawara W, Kageyama Y, Higashi Y, Nishida K, Tsunematsu Y, et al. Yolk sac tumor but not seminoma or teratoma is associated with abnormal epigenetic reprogramming pathway and shows frequent hypermethylation of various tumor suppressor genes. Cancer Sci. 2009;100:698–708. doi: 10.1111/j.1349-7006.2009.01102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawakami T, Zhang C, Okada Y, Okamoto K. Erasure of methylation imprint at the promoter and CTCF-binding site upstream of H19 in human testicular germ cell tumors of adolescents indicate their fetal germ cell origin. Oncogene. 2006;25:3225–3236. doi: 10.1038/sj.onc.1209362. [DOI] [PubMed] [Google Scholar]
- 54.Lambrot R, Kimmins S. Histone methylation is a critical regulator of the abnormal expression of POU5F1 and RASSF1A in testis cancer cell lines. Int. J. Androl. 2010 doi: 10.1111/j.1365-2605.2010.01063.x. doi:10.111/j.1365-2605.2010.01063.x [Epub ahead of print; 19 May 2010] [DOI] [PubMed] [Google Scholar]
- 55.Von Eyben FE. Chromosomes, genes, and development of testicular germ cell tumors. Cancer Genet. Cytogenet. 2004;151:93–138. doi: 10.1016/j.cancergencyto.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 56.Lehmann OJ, Ebenezer ND, Ekong R, Ocaka L, Mungall AJ, Fraser S, McGill JI, Hitchings RA, Khaw PT, Sowden JC, et al. Ocular developmental abnormalities and glaucoma associated with interstitial 6p25 duplications and deletions. Invest. Ophthalmol. Vis. Sci. 2002;43:1843–1849. [PubMed] [Google Scholar]
- 57.Chanda B, Asai-Coakwell M, Ye M, Mungall AJ, Barrow M, Dobyns WB, Behesti H, Sowden JC, Carter NP, Walter MA, et al. A novel mechanistic spectrum underlies glaucoma-associated chromosome 6p25 copy number variation. Hum. Mol. Genet. 2008;17:3446–3458. doi: 10.1093/hmg/ddn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caluseriu O, Mirza G, Ragoussis J, Chow EW, MacCrimmon D, Bassett AS. Schizophrenia in an adult with 6p25 deletion syndrome. Am. J. Med. Genet. A. 2006;140:1208–1213. doi: 10.1002/ajmg.a.31222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gould DB, Jaafar MS, Addison MK, Munier F, Ritch R, MacDonald IM, Walter MA. Phenotypic and molecular assessment of seven patients with 6p25 deletion syndrome: relevance to ocular dysgenesis and hearing impairment. BMC Med. Genet. 2004;5:17. doi: 10.1186/1471-2350-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.