Abstract
The early lytic phase of Kaposi’s sarcoma herpesvirus infection is characterized by viral replication and the global degradation (shutoff) of host mRNA. Key to both activities is the virally encoded alkaline exonuclease KSHV SOX. While the DNase activity of KSHV SOX is required for the resolution of viral genomic DNA as a precursor to encapsidation, its exact involvement in host shutoff remains to be determined. We present the first crystal structure of a KSHV SOX–DNA complex that has illuminated the catalytic mechanism underpinning both its endo and exonuclease activities. We further illustrate that KSHV SOX, similar to its Epstein–Barr virus homologue, has an intrinsic RNase activity in vitro that although an element of host shutoff, cannot solely account for the phenomenon.
INTRODUCTION
Kaposi’s sarcoma associated herpesvirus (KSHV, also known as Human Herpes Virus-8), is a member of the γ-herpesvirus family of double-stranded DNA viruses. KSHV infects both B and endothelial cells where it is the main causative agent of Kaposi’s sarcoma (KS) with epidemic or AIDS-related KS being the most common cancer associated with HIV infection (1). In addition to KS, KSHV has also been linked to a number of lymphoproliferative disorders that include primary effusion lymphoma and Multicentric Castleman’s disease (2–4). While KS can be successfully treated in the early stages of HIV infection by anti-retroviral drugs, there are no effective treatments for the advanced disease or KSHV-associated disorders.
The KSHV life cycle, similar to all herpesviruses, can be divided into a latent and lytic phase. In the majority of infected cells, the virus remains in a latent state that is characterized by a highly restricted pattern of gene expression. This contrasts with the lytic phase that is dominated by the over expression of genes required for host cell lysis and viral replication. During the initial stages of the lytic phase, a highly conserved alkaline exonuclease is expressed that is encoded by ORF 37 in KSHV. The precise function of the viral alkaline exonucleases in the context of viral genomic DNA replication is unclear; however, evidence [mostly obtained from Herpes simplex virus-1 (HSV-1)] suggests that they are involved in the processing of viral genomic DNA prior to encapsidation and facilitate the egress of viral particles from the nucleus to the cytoplasm (5–7). Consistent with this, three-way viral genomic DNA junctions have been observed during HSV infection (8,9). In the absence of alkaline exonuclease activity, these junctions accumulate in the nucleus resulting in a marked reduction in the number of mature viral particles released from infected cells (5). In support of this ‘resolvase’ role, the viral alkaline exonucleases exhibit Mg2+ or Mn2+ dependent 5′–3′ exonuclease activities in vitro (in addition to a less prevalent endonuclease activity) processing both double- and single-stranded DNA substrates (10–13). Owing to the presence of a PD-(D/E)XK motif, the viral alkaline exonucleases have been classified as members of the type II restriction endonuclease-like superfamily (14), which has been confirmed by the recent crystal structures of the native KSHV enzyme and its Epstein–Barr virus (EBV) homologue BGLF5 (15,16). Both reveal conservation of motifs key to DNase activity (7) that were first observed in the restriction endonucleases EcoRI, BamHI and HincII (17–19), together with a ‘bridge’ spanning the N- and C-terminal lobes that is also present in bacteriophage λ exonuclease (20). Bridge or arch structures are observed in a large variety of nucleases that in addition to type II restriction enzymes include members of the flap endonuclease family where they are thought to function in substrate recognition (21,22).
Onset of the lytic phase in the herpesviruses not only coincides with viral replication, but also the rapid and global degradation of host mRNA. This is thought to be one of several mechanisms utilized by these viruses to evade immune detection (23). A key protagonist of host shutoff in the γ-herpesvirus subgroup is the alkaline exonuclease that has been renamed SOX (shutoff and exonuclease) in recognition of its dual role. While host shutoff has also been observed in HSV infection and extensively studied in HSV-1, it appears to be mechanistically dissimilar to that of the γ-herpesviruses owing to the requirement for the UL41-encoded viral host shutoff protein (vhs) that actively degrades mRNA and is unrelated to the SOX homologues (24). Although the exact mechanism in the γ-herpesviruses remains to be fully elucidated, the analysis of various SOX mutants has shown that the DNA processing and shutoff activities are genetically separable and occur in different cellular compartments (25). Until recently, it was thought that the global degradation of host mRNA could not be explained by an intrinsic RNase activity of the SOX homologues, since none had been detected in vitro. Alternatively, it was proposed to originate from subversion of a host cellular pathway involving the erroneous activation or targeting of proteins participating in mRNA degradation pathways (26,27). A Mn2+-dependent RNase activity has, however, been recently characterized for BGLF5, where cleavage of both RNA and DNA are dependent on the same catalytic machinery (16). Since the available crystal structures show that the majority of non-catalytic residues implicated in host shutoff map to the N-terminal region of the molecule, and are therefore distant from the catalytic centre, it has been speculated that recognition of RNA and DNA substrates might require different structural motifs (16). To probe the molecular basis underlying the DNase activities of KSHV SOX, we have determined the crystal structure of a KSHV SOX–DNA complex involving a linear duplex. This first structure of an alkaline exonuclease complex provides important insights into the catalytic mechanism utilized by these enzymes and suggests that the bridge functions in targeting substrates for endo or exonucleolytic cleavage. We further demonstrate that similar to BGLF5, KSHV SOX also has an intrinsic RNase activity, however, site-directed mutagenesis studies reveal that it is unlikely to be uniquely responsible for host shutoff.
MATERIALS AND METHODS
Cloning and expression
The gene-encoding KSHV SOX (GenBank protein ID: AA62609) was re-engineered to optimize codon usage for expression in Escherichia coli. The codon-optimized SOX gene was then PCR amplified using the forward and reverse primers CCATGGAAGCCACCCCAACG and TCTAGACTCGAGTTACGGGGAATGCGGCAC, incorporating NcoI and XhoI restriction sites, respectively (underlined). The resulting PCR product was then sub cloned into a pCR®-Blunt II- TOPO® vector (Invitrogen) and following excision with NcoI and XhoI, the insert cloned into pETM6T1 (28) that had been previously digested using the same enzymes. This produced a 6His-NusA-6His-(TEV site)-fusion protein that after cleavage with TEV protease had the sequence GA N-terminal to the first amino acid. pETM6T1-SOX was co-transformed into BL21(DE3) StarTM (Invitrogen) with a plasmid encoding the rare tRNAs extracted from RosettaTM 2(DE3) cells (Novagen). The cells were inoculated into 100 ml of 2YT media [1.6% (w/v) bacto-tryptone (Invitrogen), 1% bacto-yeast extract (Invitrogen) and 0.5% NaCl, adjusted to pH 7.2] containing chloramphenicol (34 µg/ml) and kanamycin (25 µg/ml) and grown at 37°C overnight. A 1/100 dilution of overnight culture was then inoculated into 2YT medium and grown to an A600 of 1.0 at 37°C before induction with 1 mM isopropyl 1-thio-β-d-galactopyranoside and incubated overnight at 18°C. The cells were then harvested by centrifugation washed with buffer A (200 mM NaCl, 25 mM Tris pH 8.5) and stored at −80°C.
Protein purification and mutagenesis
Cell extracts were prepared by re-suspending the pellets in buffer A supplemented with DNase I (10 µg/ml final concentration) and an EDTA-free protease inhibitor cocktail tablet (Roche). After sonication on ice, the lysates were clarified by centrifugation (46 000g for 1 h at 4°C) and the supernatant filtered through a 0.45 µm filter prior to loading onto a 5 ml HisTrapTM FF column (GE-Healthcare). The column was washed with 20 column volumes (CVs) of buffer A containing 50 mM imidazole and the protein eluted using 5 CVs of buffer B (buffer A supplemented with 500 mM imidazole). The eluate was directly loaded onto a 5 ml HiTrap TM Q FF column (GE-Healthcare) previously equilibrated in 25 mM Tris, 50 mM NaCl, pH 8.5. The fusion protein was then eluted using a 50–1000 mM NaCl linear gradient and the tag removed by the addition of TEV protease followed by overnight dialysis in a buffer comprising 25 mM Tris pH 8.5, 200 mM NaCl, 1 mM DTT. The solution was then applied to a 5 ml HiTrapTM Q FF column (pre-equilibrated in the same buffer) and the un-tagged protein eluted using a 50–500 mM NaCl linear gradient. Fractions containing the purest protein were pooled and concentrated using a 10 kDa cut-off Vivaspin centrifugal concentrator (Vivascience). The protein was then loaded onto a gel filtration column (Superdex75 HR 10/60) that had been pre-equilibrated in a buffer consisting of 25 mM Tris pH8.5, 300 mM NaCl, 10% glycerol. Fractions were analysed on 15% SDS–PAGE gels and those containing protein of the highest purity concentrated to 4 mg/ml using a 10 kDa cut-off spin cartridge (VivaScience), aliquoted and stored at −80°C. All mutants were generated using the QuikChangeTM mutagenesis kit (Agilent Technologies) and were expressed using the same protocol described for the wild-type protein.
Oligonucleotides
The oligonucleotides used in the assays and for crystallization are shown in Table 1. 6-FAM and FL refer to those 3′ substituted with the fluorophores 6-FAM and fluorescein. P-5′ refers to oligonucleotides labelled with a 5′ phosphate group. All duplexes were initially dissolved in water and heat annealed.
Table 1.
Oligonucleotides used in assays and crystallization trials
| dsDNA | 5′-GGGGATCCTCCCAGTCGACC-3′ |
| FAM-3′-CCCCTAGGAGGATCAGCTGG-5′ | |
| dsDNA-5′P | P-5′-GGGGATCCTCCCAGTCGACC-3′-FL |
| 3′-CCCCTAGGAGGATCAGCTGG-5′ | |
| dsRNA-5′P | P-5′-UGGUUUACAUGUUCCAAUAUU-3′-FL |
| 3′-ACCAAAUGUACAAGGUUAUAA-5′ | |
| ssRNA-5′P | P-5′-UGGUUUACAUGUUCCAAUAUU-3′-FL |
| Crystallization | |
| CAdsDNA | 5′-GGGGATCCTCCCAGTCGACC-3′ |
| 3′-CCCCTAGGAGGATCAGCTGG-5′ | |
Cleavage assays
DNase and RNase assays were executed according to Buisson et al. (16) with some modifications. Exonuclease assays were performed using plasmid and linear duplex substrates. For those involving plasmids, 20 pmol of pETM6T1 vector linearized using EcoRI, were incubated with 150 pmol of KSHV SOX (or SOX mutants) for 1 h in a buffer comprising 50 mM Tris–HCl pH 9.0, 200 mM NaCl, 20 mM β-mercaptoethanol (βME), 10 mM MgCl2. For the linear substrates, 20 pmol of dsDNA or dsDNA-5′P (Table 1) were incubated with 150 pmol of KSHV SOX in the buffer used for the plasmid studies. The DNA substrates were substituted for 20 pmol of ssRNA-5′P in the RNase assay (Table 1), where reactions were performed using the incubation buffer described for BGLF5 (16). The DNA and RNA reactions were halted by the addition of 50 mM EDTA and 7.5 µl of each reaction mixture were combined with 7.5 µl of Novex TBE urea sample buffer (Invitrogen) prior to loading onto 15% TBE–urea gels (Invitrogen).
Fluorescence anisotropy
Fluorescence anisotropy assays were conducted at 25°C using a Fluoromax-3 spectrofluorimeter (Jobin Yvon Horiba), and the data fitted using GraFit (Erithacus Software). Serial dilutions of SOX were incubated for 30 min with 50 nM of dsDNA, dsDNA-5′P, dsRNA-5′P or ssRNA-5′P (Table 1) in a binding buffer comprising 25 mM Tris–HCl pH 8.5, 200 mM NaCl and 10% glycerol in a final volume of 60 µl. These were then transferred to 60 µl quartz cuvettes for anisotropy measurements that were repeated six times at each concentration of SOX. Experiments were conducted with a slit width of 5 nm with the excitation (λex) and emission (λem) wavelengths used for the various substrates as follows: dsDNA/dsDNA-5′P (λex: 492 nm, λem: 515 nm), ssRNA-5′P (λex: 493 nm, λem: 520 nm), dsRNA-5′P (λex: 494 nm, λem: 521 nm). The changes in anisotropy following titration of KSHV-SOX into the various DNA or RNA oligonucleotides (whose anisotropies varied between 0.04 and 0.1) were used to calculate the affinity constants. All data were fitted to a one site binding equation.
Crystallography
KSHV SOX at a concentration of 4 mg/ml was incubated at 4°C for 3 h with CAdsDNA (Table 1) in a protein:DNA ratio of 1:1.2 in an incubation buffer comprising 50 mM Tris–HCl pH9.0, 50 mM NaCl and 1.6 mM MgCl2. The complex was then concentrated to 8 mg/ml (KSHV SOX) in a 0.5 ml Millipore 3 kDa cut-off centrifugal concentrator and used to set up hanging drop crystallization trials in which 2 µl drops containing equal volumes of protein–DNA complex and mother liquor solution were incubated at 16°C. Single diffraction quality crystals of SOX grew overnight in 180 mM ammonium formate, 19% PEG 3350. These were then cryoprotected in mother liquor solution containing 22% ethylene glycol and flash frozen in liquid nitrogen.
Data were collected on beamline ID-23-1 at the ESRF on a single flash frozen crystal to 2.5 Å that were subsequently processed and scaled using MOSFLM and SCALA from the CCP4 suite (29). The structure was solved by Molecular Replacement using MOLREP (30), manually re-built in COOT (31) and refined using PHENIX (32) where cycles of TLS refinement were also performed. The TLS groups were defined according to the suggestions of the TLS motion determination server (33,34). The final model, comprised of a single protein monomer, 38 nt, 2 Mg2+ ions and 127 water molecules had an Rcryst of 20.89% and Rfree of 26.52%. All stereochemical parameters were well within the expected ranges for a structure at this resolution (Table 2). The co-ordinates and structure factors have been deposited in the protein data bank under the accession code 3POV.
Table 2.
Data collection and refinement statistics
| Space group | P212121 |
| Unit cell [a, b, c (Å)] | 54.52, 67.66, 174.56 |
| Resolution (Å) | 63.1–2.5 (2.6–2.5) |
| Total no. of reflections | 89560 |
| No. of unique reflections | 22693 |
| Redundancy | 3.9 (4.0) |
| Completeness (%) | 98.9 (99.4) |
| <I>/<σ(I)> | 8.7 (2.6) |
| Rmergea | 9.3 (53.3) |
| Refinement | |
| No. of protein atoms | 3467 |
| No. of DNA atoms | 775 |
| No. of Mg2+ ions | 2 |
| No. of solvent atoms | 127 |
| Rworkb/Rcrystc (%) | 20.89/26.52 |
| Maximum likelihood co-ordinate error (Å) | 0.38 |
| Overall B-factors (Å2)d | 64/55/69.0/108/51.4/79.0 |
| Deviations from ideal stereochemistry | |
| RMSD bonds (Å) | 0.002 |
| RMSD angles (°) | 0.653 |
| RMSD B-factors (Å2)e | 8.0 |
| Wilson B-factor (Å2) | 48.6 |
| Ramachandran plot analysisf | |
| Most favoured (%) | 95.85 |
| Additionally allowed (%) | 4.84 |
| Disallowed (%) | 0.69 |
Values in parentheses are for the highest resolution shell (2.6–2.50 Å).
aRmerge=Σ(|Ii – <I>|)/Σ(<I>), where the sum is calculated over all observations of a measured reflection (Ii), and <I> is the mean intensity of all the measured observations (Ii).
bRcryst=Σ (|Fobs – Fcalc|)/Σ (Fobs), Fobs are the observed structure factor amplitudes, and Fcalc those calculated from the model.
cRfree is equivalent to Rcryst but where 5% of the measured reflections have been excluded from refinement and set aside for cross-validation purposes.
dAverage B-values for all atoms/protein/Mg2+/DNA/solvent/formate ion/.
eRMSD in B-factor between covalently bonded main chain atoms and side chain atoms.
fRamachandran plot analysis was from molprobity (45).
RESULTS
Screening of duplexes for crystallization
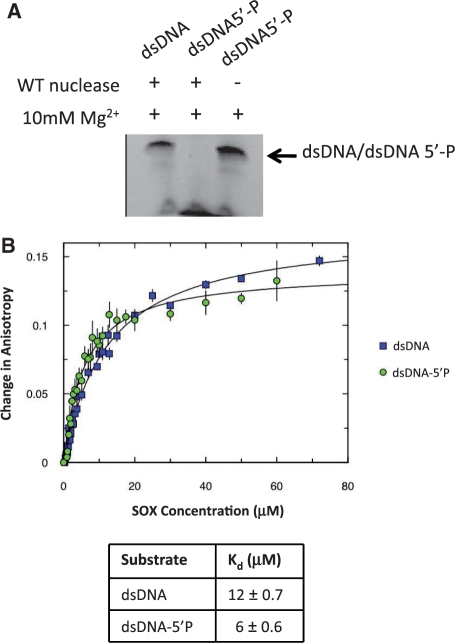
Initial studies using linear duplexes containing a centrally located CA mismatch revealed that the presence of the mismatch had no effect on either the binding affinity or cleavage activity when compared with those substituted with a G:C base pair in the analogous position (data not shown). In contrast, those labelled with a 5′ phosphate were highly susceptible to degradation while those that were lacking the modification showed little evidence of cleavage (Figure 1A). The 5′ phosphate substitution, however, had only a modest impact on overall binding affinity based on the results of fluorescence polarization anisotropy assays that indicated similar modes of binding for the unmodified or modified duplexes dsDNA and dsDNA-5′P, respectively (Table 1 and Figure 1B). The duplex CAdsDNA containing the CA mismatch but lacking the 5′ phosphate was therefore used for crystallization trials due to its apparent lack of susceptibility to cleavage.
Figure 1.
(A) Exonuclease assays involving the DNA substrates dsDNA and dsDNA-5′P illustrate that cleavage is only observed on duplexes in which one strand has been substituted with a 5′ phosphate group. (B) Fluorescence anisotropy assays indicate that both duplexes bind to SOX with similar affinities (12 µM for dsDNA and 6 µM for dsDNA-5′P) suggestive of largely equivalent modes of binding.
Overall structure
The structure of the KSHV SOX–DNA complex was determined by molecular replacement using the protein co-ordinates of the native KSHV–SOX enzyme [PDB entry 3FHD (see ‘Materials and Methods’ section)]. Initial inspection revealed density for the DNA in the vicinity of the proposed DNA binding region or ‘canyon’ that is located in the interface between the N- and C-terminal lobes of the molecule (Figure 2A). Despite containing a centrally located, wobble base paired CA mismatch (A29:C12), there is no discernable distortion of the duplex that adopts a standard B conformation (Supplementary Figure S1A). Out of the 40 nt, all could be assigned although G1 and C40 were only partially ordered. Although nucleotides 2–13 in strand 1 and 31–39 in strand 2 are stabilized by a variety of protein–DNA interactions (described in the ‘Protein–DNA interactions’ section), those remaining do not interact with the canyon of KSHV–SOX and form few interactions with symmetry related protein molecules in the crystal lattice (Supplementary Figure S1B). As a consequence, these nucleotides are considerably less well ordered consistent with the overall higher B-values and breaks in the phosphodiester backbone observed. Aside from these regions, the electron density is of good quality overall (see Supplementary Figure S2A).
Figure 2.
(A) Cartoon of KSHV SOX and corresponding 2mFo-DFc omit map density for the DNA (magenta, contoured at 1σ) bound at the interface or ‘canyon' between the N- and C-terminal lobes (green and yellow, respectively) of the SOX molecule. The density for nucleotides at the free end of the duplex is more fragmented due to a lack of stabilizing protein–DNA interactions. (B) Sequence alignments of the conserved DNase motifs and bridge regions in the γ-herpesvirus SOX homologues from Human herpesvirus 8 (KSHV), Bovine herpesvirus 2 (BHV2), Mouse herpesvirus 68 (MHV68), Epstein–Barr virus (BGLF5), Porcine lymphotropic herpesvirus 2 (PLHV) and UL12 from the α-herpes virus HSV-1. The alignment was performed using CLUSTALW (http://www.ebi.ac.uk/Tools/clustalw2/index.html). (C) The locations of motifs II (pink), III (grey), IV (orange) and VI (blue) within the KSHV–SOX complex that out of the seven conserved sequences mediate the observed protein–DNA interactions or are involved in catalysis. The position of the partially ordered bridge (red) is also highlighted. The second SOX nuclear localization sequence (NLS2, green), that has a role in DNA binding is also shown.
Protein–DNA interactions
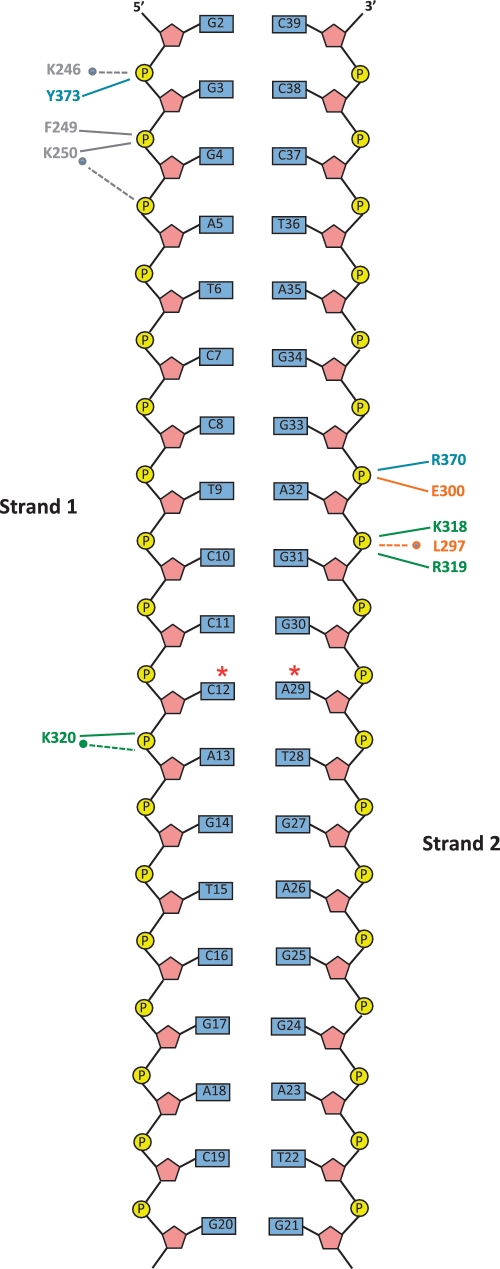
Seven conserved motifs are essential for DNase activity in the alkaline exonucleases (Figure 2B) (14). Of these, residues within motifs III, IV and VI, which together form the canyon in the SOX molecule, interact directly with the duplex (Figure 2C) along with those in a loop between motifs IV and V (residues 315–320). This region maps to the second SOX nuclear localization sequence (NLS2) identified in BGLF5 and is highly conserved among the γ-herpesvirus homologues (25,35). When compared with the native structure, the DNA binding motifs undergo modest remodelling on association with DNA (the RMSD calculated for all equivalent Cα atoms is 1.5 Å); the only exception being residues 315–318 of NLS2 that are absent in the native structure, but fully ordered in the complex (Supplementary Figure S2B).
Consistent with the lack of sequence preference exhibited by the SOX homologues, the majority of protein–DNA contacts involve the phosphodiester backbone and mostly occur in the minor groove. There are only two areas of specific contact that cluster at the 5′-end of strand 1 (close to the active site) and a more central region of the duplex involving strand 2 predominantly (Figure 3), although a plethora of van der Waals interactions involving the three DNA binding motifs and NLS2 are observed. Motif III features heavily in the recognition of the G3, G4 and A5 phosphate groups towards the 5′-end of strand 1. In this region, the Nζ group of K246, located within the strictly conserved PD-(D/E)XK sequence, interacts with O1P of G3 via a water molecule while the main chain NH groups of K250 and F249 (both highly conserved) donate hydrogen bonds to O1P of G4 (Figures 3 and 4A). The carbonyl oxygen of K250 also contributes a water mediated hydrogen bond to O1P of A5. Y373, located in motif VI, donates an additional hydrogen bond to O1P of G3 via its hydroxyl group. In contrast, recognition of the central duplex region is largely dependent on residues K318, R319 and K320 in NLS2, L297 and E300 in domain IV and R370 in motif VI (Figures 3 and 4B). Hydrogen bonds are donated to O1P and O2P of G33 by the side chain NH1 and main chain NH groups of R370 and E300, respectively. The Nζ moiety of K318 donates a hydrogen bond to O2P of A32 that is further stabilized by a water mediated hydrogen bond involving the carbonyl oxygen of L297. A32 is additionally recognized by a hydrogen bond donated by the peptide NH group of R319 to O1P. The side chain moiety of R319 is observed to protrude into the minor groove where it is able to mediate a range of van der Waals interactions with C12, C31 and A32 (Figure 4B). Within this region, K320 additionally contributes direct and water mediated hydrogen bonds via its main chain NH and carbonyl oxygen moieties, respectively, to O1P of A13. No protein–DNA interactions are observed for nucleotides G14–G20 of strand 1 and G21–G30 of strand 2.
Figure 3.
Schematic diagram of protein–DNA hydrogen bonds stabilizing the complex. Those depicted by dashed lines and asterisks are mediated by water molecules and those with unbroken lines, main or side chain groups. C12 and A29 (marked with red asterisks) correspond to the CA mismatch.
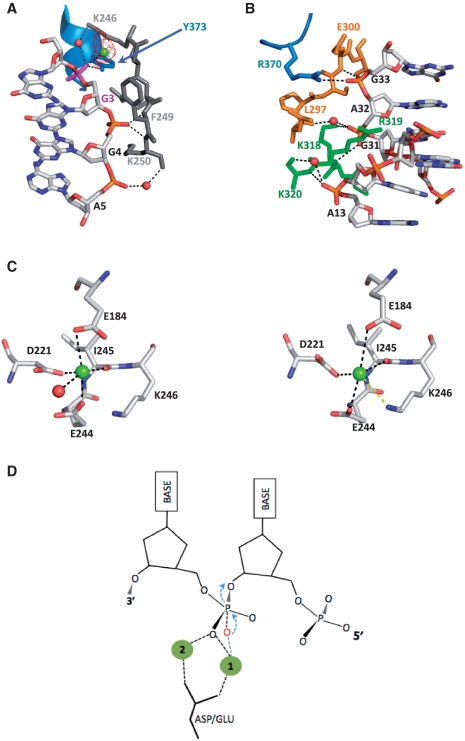
Figure 4.
DNA recognition. (A) Stabilization of the duplex in the region closest to the catalytic centre exclusively involves strand 1 and is dominated by residues in motif III. The protein–DNA contacts are a combination of water, main and side chain mediated hydrogen bonds involving residues K246, F249, K250 and Y373 (colour coded according to their motif locations as in Figure 2C) and the phosphate groups of G3, G4 and A5 together with a range of van der Waals interactions. mFo-DFc omit map density (red, contoured at 3σ although present at 5σ) attributable to a second metal ion (green) was also identified that is absent in the native KSHV structure. (B) The protein–DNA interactions observed for the second region of contact involves a more mixed distribution of residues over the conserved motifs compared with the first. Here residues R370 (motif VI), K318 and K320 (NLS), E297 and E300 (motif IV) mediate water, main and side chain hydrogen bonds with G31, A32 and G33 of strand 2 and A13 of strand 1. The side chain moiety of R319 protrudes into the minor groove where it makes a range of van der Waals contacts with C12, G31 and A32. (C) Mg1 and surrounding co-ordination sphere in the wild-type (left) and DNA bound (right) KSHV SOX structures. The loss of tight ligands contributed by E184 and a water molecule (disordered in our complex) results in a weaker association as a result of a shift of Mg1. In this position, Mg1 is within hydrogen bonding distance of the Nζ group of K246. (D) Schematic diagram of the classical two metal nucleophilic attack mechanism. The oxygen atom of the activated water is shown in red.
Active site configuration and cleavage geometry
Essential to the endo/exonuclease activities of the viral SOX homologues are residues within the conserved PD-(D/E)XK sequence where the D and consecutive D/E residues are located within motifs II and III, respectively, and correspond to D221 and E244 in KSHV SOX (25). In the native structure, these together with E184, which is also strictly conserved in the SOX homologues, contribute ligands to a bound Mg2+ ion (Mg1) via their carboxylate groups (15). Mg1, though slightly shifted and less well ordered, is also present in our complex and, as a result of this re-positioning, falls within hydrogen bonding distance of the Nζ group of K246 (Figure 4C). Since the predominant activity of SOX is that of a 5′–3′ exonuclease, it has been suggested that the scissile bond, located at the 3′-end of the 5′ nt, should be within hydrogen bonding distance of Mg1. This is inconsistent with the position of G2 in our complex, the site of exonucleolytic cleavage, whose partially ordered phosphate group is angled away from the catalytic region and >7 Å from Mg1.
While cleavage at G2 based on our structure would be highly unlikely, G3 is more favourably juxtaposed with respect to the catalytic region. In our complex, the phosphate group of G3 forms water mediated hydrogen bonds with K246 of the PD-(D/E) XK motif as previously described and also contacts a second Mg2+ ion (Mg2, present at a level of 5σ in mFo-DFc omit maps, Figure 4A) absent in the native KSHV structure. Mg2 is located a distance of ∼5 Å from Mg1 and is further liganded to Oε1 of E184, a water molecule and the peptide NH group of C247. The presence of two metal ions in the catalytic region is suggestive of a classical two metal SN2 mechanism [reviewed in (36,37)]; however, the phosphate group of G3 is not in the canonical position for endonucleolytic processing. In most examples of two metal mechanisms, the metal ions are co-ordinated between the non-bridging oxygens of the scissile phosphate (Figure 4D) and are further liganded by a highly conserved aspartate. In this configuration, one metal ion acts to deprotonate a water molecule for generation of a hydroxyanion attacking nucleophile, while the other stabilizes the resultant pentavalent transition state intermediate. Even though Mg1 and Mg2 are both liganded to E184 that could substitute for an aspartate, only Mg2 is in direct contact with the phosphate group of G3 (Figure 5A). Mg1, located ∼7 Å from G3, would therefore be unable to generate a hydroxyanion for nucleophilic attack of the G3 P-O3 bond.
Figure 5.
(A) Stereofigure showing the active site geometry in relation to G3, the nucleotide whose phosphate group is in contact with K246 of the PD-(D/E)XK and the second metal ion, Mg2. The relative juxtaposition of G3 to Mg1 and a possible catalytic water molecule are inconsistent with a configuration that would promote cleavage at this site based on the classical two metal mechanisms. (B) Rotation of G1 out of the duplex stack such that a modelled 5′ phosphate group would be able to hydrogen bond to S146 and S219 (in the rotamer conformations observed in the native KSHV structure) that form a conserved ‘serine cluster’ (brown), is sufficient to align the scissile phosphate G2 with Mg1 and Mg2 for cleavage in a near classical two metal mechanism configuration. In this conformation, the two metal ions straddle the stereospecific oxygen O1P and the water molecule bound to K246 (A, light blue sphere) is well placed for inline attack.
Although Mg1 is proposed to act as a hydroxyanion generator, structural and mutagenesis studies have suggested that highly conserved amino acid residues such as lysine, tyrosine or serine might fulfil this role [reviewed in (37)]. Our complex suggests that in principle, K246 could activate the water molecule bound to O1P of G3 and has been shown to have a key catalytic role in support of this (38). However, the so generated nucleophile would be poorly positioned for inline attack. Alternatively, the hydroxyl group of Y373 that is also hydrogen bonded to O1P of G3 could be deprotonated to form an oxyanion that could in turn generate a phosphotyrosyl covalent intermediate. This can be rejected on the basis of site directed mutagenesis studies that show Y373 is a non-essential residue (data not shown). Although there is still the possibility that KSHV–SOX might utilize a novel variant of a two metal mechanism in which G3 could be well placed for catalysis, the fact that the duplex used in our crystallization studies remains intact and that it fails to undergo processing in our in vitro assay strongly suggests that G3 is intractable to cleavage, similar to G2. This is strengthened by the observation that endonucleolytic processing appears to be restricted to plasmid substrates (7,39) and has not been reported for linear duplexes. The inability of linear duplexes lacking a 5′ phosphate to be efficiently targeted for exonucleolytic processing or undergo endonucleolytic cleavage can therefore be explained by the association of these oligonucleotides with KSHV-SOX resulting in potential scissile phosphate groups being inappropriately positioned for cleavage as observed in our complex.
5′ phosphate recognition: the key to exo and endonucleolytic cleavage
Although the relative juxtapositions of the scissile phosphate, metal ions and attacking nucleophile are important factors for cleavage, substrate stabilization is also essential. For the exonucleolytic reaction, this would additionally involve recognition of the 5′ phosphate. A putative 5′ phosphate binding site, encompassing residues S144, S145 and S146 of motif I together with S219 of motif II, has been proposed for the SOX homologues based on the findings that these residues are associated with a sulphate ion and anion in the native KSHV and BGLF5 structures, respectively (15,16) (we also note a formate ion bound in this region of our complex). The importance of this ‘serine cluster’ is further underpinned by mutagenesis studies where substitution of S219 in both the HSV-1 and baculovirus SOX homologues results in significant attenuation of exo and endonuclease activities (7,39). In order for a modelled 5′ phosphate group on G1 in our complex to contact the cluster for exonucleolytic cleavage, the nucleotide would have to be rotated towards motif 1 from its position stacked against G2 (implied by the partially ordered density) to enable the formation of hydrogen bonds with S146 and S219. These in turn, would only be required to adopt the side chain rotamers observed in the native KSHV structure (Figure 5B). Surprisingly, manipulating G1 into this position results in optimal placement of G2 as the scissile phosphate. In this configuration, Mg1 and Mg2 have the classical geometry in which both are liganded to the stereospecific, non-bridging oxygen O1P. In addition, the water molecule hydrogen bonded to both K246 and O1P (though out of the range for contacts with either Mg1 or Mg2), is now near optimally positioned as a nucleophile given an inline attack angle of 147°. These results suggest that for exonucleolytic processing, stabilization of the 5′ phosphate group is key to the correct positioning of the scissile bond for cleavage.
While mutagenesis studies focused on S219 have shown that the serine cluster has a more prominent role in exonucleolytic cleavage, as evidenced by the higher level of attenuation compared with that observed for endonucleolytic processing (7), it is nonetheless key to both reactions. Based on our model for exonucleolytic cleavage, the association of a phosphate group with this site in the context of a continuous duplex could only occur if the helix were locally denatured. In keeping with this, the recent studies of Mikhailov et al. demonstrate that endonucleolytic processing is only observed on single-stranded minicircle or supercoiled plasmid substrates that, in an attempt to relieve torsional stress, develop local regions of single-stranded DNA that are the likely targets for cleavage (7,39). Though difficult to speculate on how supercoiled/mini-circle plasmids or indeed the natural substrate would be accommodated, comparison of our complex with the BGLF5 SOX homologue strongly implicates the bridge as being essential to this process. Bridge architectures, based on studies involving the flap endonucleases, were originally proposed to have a pivotal role in exonucleolytic cleavage by inducing strand separation for targeting of the scissile strand. It has further been suggested that they function as DNA ‘clamps’ stabilizing specific types of DNA architecture (22,40,41). In both our complex and native KSHV SOX, the bridge (that spans amino acid residues 154–180, Figure 2B) is only partially ordered. The analogous region, however, is completely intact in the BGLF5 SOX mutant D203S that shares ∼42% sequence identity with the KSHV enzyme.
Superposition of the BGLF5 D203S co-ordinates with those of our complex reveals that while both structures have a high degree of similarity overall (see Supplementary Figure S2C), the duplex could not be accommodated in the canyon of the BGLF5 mutant without substantial re-modelling of the 5′-end of strand 1. This would be due to severe steric clashes between bridge residues G156 to V159 in BGLF5 (equivalent to A174 and L177 in KSHV SOX, absent in both our complex and the native enzyme) and all base pairs 5′ to G4 (Figure 6A). These unfavourable contacts arise from an ∼5.5 Å lateral movement of residues A160 and F161 (equivalent to A178 and F179 in KSHV SOX) towards the C-terminal lobe of the SOX molecule, away from the active site. The nucleotides 5′ to G4 could only be favourably accommodated in the context of the BGLF5 bridge conformer if they were no longer stacked in a regular duplex but instead situated in largely single-stranded regions of DNA. The conformation of the bridge observed in the D203S BGLF5 mutant, therefore, seems more consistent with endonucleolytic cleavage in which major local distortion of the DNA appears to be required for recognition in contrast to the exonuclease reaction where only relatively minor changes focused on the 5′-nt are required. This suggests that conformational re-modelling of the bridge may be the trigger for switching SOX between endo and exonucleolytic cleavage modes.
Figure 6.
(A) Superposition of the KSHV DNA complex with the D203S BGLF5 mutant reveals a number of severe steric clashes involving residues G156-V159 (BGLF5) at the C-terminus of the bridge and all base pairs 5′ to G4. This is as result of repositioning of residues A160 and F161 that move ∼5.5 Å towards the C-terminal lobe of the SOX molecule. Only a denatured duplex could interact with the serine cluster (highlighted in green) and be favourably accommodated by the bridge in this alternative conformation. (B) RNase assay of wild-type KSHV SOX in the presence of 10 mM MgCl2 or MnCl2. (C) Fluorescence anisotropy binding assays of SOX involving single- (ssRNA-5′P) and double-stranded (dsRNA-5′P) RNA oligonucleotides. The results obtained for dsDNA-5′P are included for comparison.
RNase activity and host shutoff
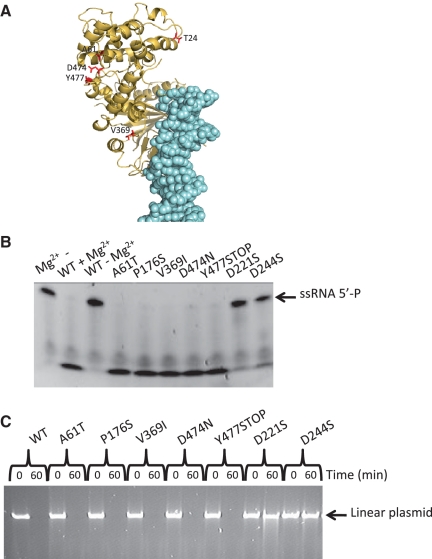
Although it has been proposed that host shutoff results from subversion of host mRNA degradation pathways, recent data have shown that in EBV, BGLF5 has an intrinsic, Mn2+ dependent RNase activity. It has been demonstrated that this activity involves the catalytic residues utilized for DNA processing (16). To establish whether KSHV SOX also shares this intrinsic activity, we performed a similar RNase assay using a uracil rich ssRNA 20-mer (Figure 6B). Our results show unequivocally that KSHV SOX possesses an RNase activity that cannot be attributed to contamination since experiments performed with the D221S and E244S mutants lacking DNase activity but competent in DNA binding (data not shown) reveal its complete abolition. These results are similar to those reported for BGLF5 confirming that both DNA and RNA processing utilize the same catalytic machinery and is consistent with the in vivo studies of Glaunsinger et al. (25). Unlike BGLF5, however, the RNase activity of KSHV SOX does not appear to be Mn2+ dependent since similar activities are observed in assays performed in the presence of Mg2+ alone at the same concentration. Fluorescence Anisotropy studies (Figure 6C) further reveal that SOX is able to associate with single- and double-stranded RNA with similar affinities (Kd’s of 87 and 95 µM, respectively) that is suggestive of engagement with only one strand. Interestingly, the affinity for RNA is considerably weaker than for DNA (∼15- to 16-fold weaker when ssRNA-5′P and dsRNA-5′P are compared with dsDNA-5′P, Figure 6C).
In addition to those involved in catalysis, residues within the N-terminal lobe of SOX (Figure 7A) have also been shown to have an essential role in host shutoff (25). To ascertain whether these non-catalytic shutoff residues contribute to the RNase activity of KSHV SOX, the mutants T24I, A61T, P176S, V369I, D474N and Y477STOP, shown to significantly attenuate or abolish global degradation of host mRNA, were generated (with the exception of T24I that was insoluble) and screened using our RNase and DNase assays (Figure 7B and C). All mutants exhibited wild-type RNase and DNase activities. Our results, therefore, suggest that host shutoff in KSHV cannot be entirely explained by the intrinsic RNase activity of KSHV SOX which would in turn support the view that SOX mediates host shutoff by interacting with as yet unidentified cellular factors.
Figure 7.
(A) Cartoon showing the locations of the non-catalytic host shutoff residues (red) within the complex (the DNA is shown in light blue). (B) RNase assays of all mutants (excluding T24I) known to reduce/abolish host shutoff. (C) Plasmid exonuclease assays of catalytic and non-catalytic shutoff mutants.
DISCUSSION
We report the first crystal structure of a viral alkaline exonuclease in complex with a DNA duplex. Our complex has confirmed the involvement of the D-(D/E)XK motif and residues in conserved motifs III, IV, VI (previously implicated in DNase activity) and NLS2 in DNA binding. Although the duplex used in our studies did not undergo processing, the structure has provided key insights into the cleavage geometry utilized by the SOX nucleases. To this end, we have identified two Mg2+ ions bound within the catalytic region and shown that alignment of the putative 5′ phosphate binding site (serine cluster) with the 5′ phosphate of the substrate results in a near classical two metal ion cleavage configuration (36,37,42). A potential nucleophilic water molecule has also been identified based on this model that has near perfect geometry for inline attack. Interestingly, the association between this water molecule and K246 together with its overly long distance from either of the two metal ions is highly suggestive of K246 acting as the general base. Although this residue is strictly conserved among the SOX homologues and the equivalent lysine in the endonuclease MutH binds to a nucleophilic water molecule in a similar configuration (43), a major drawback in this assignment lies in its positive charge at pHs <10.5. This would prevent abstraction of a proton and subsequent generation of an attacking nucleophile at the optimal pH of 8.5–9.0. The juxtaposition of K246 to Mg1 in our structure, however, would substantially lower its pKa rendering it capable of generating a hydroxyanion. Alternatively, it is possible that the two metal ions approach each other more closely than the 5 Å observed in our complex since 4 Å has been cited as the optimum separation distance for maximal transition state stabilization (37). Following such movement, Mg1 might be well placed to assist in nucleophile generation.
In addition to addressing the cleavage geometry, our structure has also shown that endonucleolytic processing could only occur on highly distorted duplexes containing regions of single-stranded DNA in keeping with the reported biochemical studies. They also suggest that these structures are likely to be stabilized by the bridge and that conformational re-modelling of this region may be the trigger for their recognition. Although bridge or arch architectures were originally thought to function as strand separators forming an aperture through which a single DNA strand could be threaded for high processivity, there is no evidence of this in our complex or that it would be required for catalysis. It appears that exonucleolytically processed substrates are ostensibly bound as duplexes [as suggested by Dahlroth et al. (15)], and that the only re-arrangements required are those necessary for recognition of the 5′ phosphate group. This is in good agreement with the similar Kd’s observed for the duplex used in our structural studies and the same duplex substituted with a 5′ phosphate group. It is also difficult to envisage how a threading mechanism would address the endonuclease reaction satisfactorily with stabilization of highly distorted DNA structures by the bridge appearing a more likely scenario. This is in keeping with the bridge acting as a DNA ‘clamp’ that has also been proposed (22). In order to fully characterize the function of the bridge and further analyse the significance of the conformational states observed, additional biochemical and structural studies will have to be performed using more physiological branched substrates although there is still uncertainty regarding their exact nature.
While the DNase activity of the γ-herpesvirus SOX homologues is important in the maturation of virus particles, it has also been shown to compromise the integrity of the host genome in EBV (44). This combined with the rapid and global degradation of host mRNA will ultimately culminate in cell death. Although the exact mechanism of host shutoff remains to be determined, it was initially thought not to involve an intrinsic RNase activity. Recently, it was reported that the EBV SOX homologue BGLF5 possesses an in vitro RNase activity (16). In the current study, we were able to characterize a similar but non-identical metal ion-dependent RNase activity that interestingly was not attenuated by the non-catalytic mutations associated with shutoff. While RNase activity is clearly an important element of shutoff, our results indicate that SOX is not solely responsible for the phenomenon and that additional factors, most likely involving protein–protein interactions (as initially suggested), are required for degradation of host mRNA. This is further supported by the relatively low affinity of SOX for RNA obtained from our fluorescence polarization studies that is suggestive of a requirement for an interacting partner to enhance binding and possibly increase processivity. Given the burden of disease caused by herpesviruses in general and KSHV specifically, understanding the molecular basis underlying their intrinsic DNase and RNase activities, together with the identification and characterization of potential interacting partners, will provide alternative strategies for the design of novel therapeutics. Our KSHV–SOX–DNA complex and preliminary characterization of the KSHV SOX RNase activity are therefore important steps towards this goal.
ACCESSION NUMBER
3POV.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding open access charge: Medical Research Council project grant (grant number G0701236 to T.B.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Nora Cronin for her assistance with data collection and acknowledge the ISMB Biophysics facility (Birkbeck/UCL) in which the Fluorescence Anistropy studies were conducted.
REFERENCES
- 1.Schulz TF. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8): epidemiology and pathogenesis. J. Antimicrob. Chemother. 2000;45(Suppl T3):15–27. doi: 10.1093/jac/45.suppl_4.15. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis L, Mesri EA, Nador RG, Said JW, Asch AS, Knowles DM, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. [PubMed] [Google Scholar]
- 3.Boshoff C, Weiss RA. Kaposi's sarcoma-associated herpesvirus. Adv. Cancer Res. 1998;75:57–86. doi: 10.1016/s0065-230x(08)60739-3. [DOI] [PubMed] [Google Scholar]
- 4.Cesarman E, Knowles DM. The role of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in lymphoproliferative diseases. Semin. Cancer Biol. 1999;9:165–174. doi: 10.1006/scbi.1998.0118. [DOI] [PubMed] [Google Scholar]
- 5.Martinez R, Sarisky RT, Weber PC, Weller SK. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein JN, Weller SK. In vitro processing of herpes simplex virus type 1 DNA replication intermediates by the viral alkaline nuclease, UL12. J. Virol. 1998;72:8772–8781. doi: 10.1128/jvi.72.11.8772-8781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldstein JN, Weller SK. The exonuclease activity of HSV-1 UL12 is required for in vivo function. Virology. 1998;244:442–457. doi: 10.1006/viro.1998.9129. [DOI] [PubMed] [Google Scholar]
- 8.Severini A, Morgan AR, Tovell DR, Tyrrell DL. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology. 1994;200:428–435. doi: 10.1006/viro.1994.1206. [DOI] [PubMed] [Google Scholar]
- 9.Severini A, Scraba DG, Tyrrell DL. Branched structures in the intracellular DNA of herpes simplex virus type 1. J. Virol. 1996;70:3169–3175. doi: 10.1128/jvi.70.5.3169-3175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bronstein JC, Weber PC. Purification and characterization of herpes simplex virus type 1 alkaline exonuclease expressed in Escherichia coli. J. Virol. 1996;70:2008–2013. doi: 10.1128/jvi.70.3.2008-2013.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Draper KG, Devi-Rao G, Costa RH, Blair ED, Thompson RL, Wagner EK. Characterization of the genes encoding herpes simplex virus type 1 and type 2 alkaline exonucleases and overlapping proteins. J. Virol. 1986;57:1023–1036. doi: 10.1128/jvi.57.3.1023-1036.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann PJ, Cheng YC. The deoxyribonuclease induced after infection of KB cells by herpes simplex virus type 1 or type 2. I. Purification and characterization of the enzyme. J. Biol. Chem. 1978;253:3557–3562. [PubMed] [Google Scholar]
- 13.Hoffmann PJ, Cheng YC. DNase induced after infection of KB cells by herpes simplex virus type 1 or type 2. II. Characterization of an associated endonuclease activity. J. Virol. 1979;32:449–457. doi: 10.1128/jvi.32.2.449-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bujnicki JM, Rychlewski L. The herpesvirus alkaline exonuclease belongs to the restriction endonuclease PD-(D/E)XK superfamily: insight from molecular modeling and phylogenetic analysis. Virus Genes. 2001;22:219–230. doi: 10.1023/a:1008131810233. [DOI] [PubMed] [Google Scholar]
- 15.Dahlroth SL, Gurmu D, Haas J, Erlandsen H, Nordlund P. Crystal structure of the shutoff and exonuclease protein from the oncogenic Kaposi's sarcoma-associated herpesvirus. FEBS J. 2009;276:6636–6645. doi: 10.1111/j.1742-4658.2009.07374.x. [DOI] [PubMed] [Google Scholar]
- 16.Buisson M, Geoui T, Flot D, Tarbouriech N, Ressing ME, Wiertz EJ, Burmeister WP. A bridge crosses the active-site canyon of the Epstein-Barr virus nuclease with DNase and RNase activities. J. Mol. Biol. 2009;391:717–728. doi: 10.1016/j.jmb.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 17.Kim YC, Grable JC, Love R, Greene PJ, Rosenberg JM. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990;249:1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- 18.Newman M, Strzelecka T, Dorner LF, Schildkraut I, Aggarwal AK. Structure of restriction endonuclease BamHI and its relationship to EcoRI. Nature. 1994;368:660–664. doi: 10.1038/368660a0. [DOI] [PubMed] [Google Scholar]
- 19.Horton NC, Dorner LF, Perona JJ. Sequence selectivity and degeneracy of a restriction endonuclease mediated by DNA intercalation. Nat. Struct. Biol. 2002;9:42–47. doi: 10.1038/nsb741. [DOI] [PubMed] [Google Scholar]
- 20.Kovall R, Matthews BW. Toroidal structure of lambda-exonuclease. Science. 1997;277:1824–1827. doi: 10.1126/science.277.5333.1824. [DOI] [PubMed] [Google Scholar]
- 21.Nishino T, Ishino Y, Morikawa K. Structure-specific DNA nucleases: structural basis for 3D-scissors. Curr. Opin. Struct. Biol. 2006;16:60–67. doi: 10.1016/j.sbi.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 22.Tomlinson CG, Atack JM, Chapados B, Tainer JA, Grasby JA. Substrate recognition and catalysis by flap endonucleases and related enzymes. Biochem. Soc. Trans. 2010;38:433–437. doi: 10.1042/BST0380433. [DOI] [PubMed] [Google Scholar]
- 23.Zuo J, Thomas W, van Leeuwen D, Middeldorp JM, Wiertz EJ, Ressing ME, Rowe M. The DNase of gammaherpesviruses impairs recognition by virus-specific CD8+ T cells through an additional host shutoff function. J. Virol. 2008;82:2385–2393. doi: 10.1128/JVI.01946-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zelus BD, Stewart RS, Ross J. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J. Virol. 1996;70:2411–2419. doi: 10.1128/jvi.70.4.2411-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glaunsinger B, Chavez L, Ganem D. The exonuclease and host shutoff functions of the SOX protein of Kaposi's sarcoma-associated herpesvirus are genetically separable. J. Virol. 2005;79:7396–7401. doi: 10.1128/JVI.79.12.7396-7401.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee YJ, Glaunsinger BA. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol. 2009;7:e1000107. doi: 10.1371/journal.pbio.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kushner SR. mRNA decay in prokaryotes and eukaryotes: different approaches to a similar problem. IUBMB Life. 2004;56:585–594. doi: 10.1080/15216540400022441. [DOI] [PubMed] [Google Scholar]
- 28.Bagneris C, Ageichik AV, Cronin N, Wallace B, Collins M, Boshoff C, Waksman G, Barrett T. Crystal structure of a vFlip-IKKgamma complex: insights into viral activation of the IKK signalosome. Mol. Cell. 2008;30:620–631. doi: 10.1016/j.molcel.2008.04.029. [DOI] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta. Crystallogr. D Biol. Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, Grosse-Kunstleve RW, Hung LW, Loerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 33.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J. Appl.Crystallogr. 2006;39:109–111. [Google Scholar]
- 34.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62(Pt 4):439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 35.Liu MT, Hsu TY, Chen JY, Yang CS. Epstein-Barr virus DNase contains two nuclear localization signals, which are different in sensitivity to the hydrophobic regions. Virology. 1998;247:62–73. doi: 10.1006/viro.1998.9228. [DOI] [PubMed] [Google Scholar]
- 36.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 37.Yang W. Nucleases: diversity of structure, function and mechanism. Q Rev. Biophys. 2011;44(Pt 1):1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu MT, Hu HP, Hsu TY, Chen JY. Site-directed mutagenesis in a conserved motif of Epstein-Barr virus DNase that is homologous to the catalytic centre of type II restriction endonucleases. J Gen. Virol. 2003;84(Pt 3):677–686. doi: 10.1099/vir.0.18739-0. [DOI] [PubMed] [Google Scholar]
- 39.Mikhailov VS, Okano K, Rohrmann GF. Specificity of the endonuclease activity of the baculovirus alkaline nuclease for single-stranded DNA. J. Biol. Chem. 2004;279:14734–14745. doi: 10.1074/jbc.M311658200. [DOI] [PubMed] [Google Scholar]
- 40.Hosfield DJ, Mol CD, Shen B, Tainer JA. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- 41.Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 42.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 43.Lee JY, Chang J, Joseph N, Ghirlando R, Rao DN, Yang W. MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage. Mol. Cell. 2005;20:155–166. doi: 10.1016/j.molcel.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 44.Wu CC, Liu MT, Chang YT, Fang CY, Chou SP, Liao HW, Kuo KL, Hsu SL, Chen YR, Wang PW, et al. Epstein-Barr virus DNase (BGLF5) induces genomic instability in human epithelial cells. Nucleic Acids Res. 2010;38:1932–1949. doi: 10.1093/nar/gkp1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.