Abstract
Upon HIV-1 infection of a target cell, the viral reverse transcriptase (RT) copies the genomic RNA to synthesize the viral DNA. The genomic RNA is within the incoming HIV-1 core where it is coated by molecules of nucleocapsid (NC) protein that chaperones the reverse transcription process. Indeed, the RT chaperoning properties of NC extend from the initiation of cDNA synthesis to completion of the viral DNA. New and effective drugs against HIV-1 continue to be required, which prompted us to search for compounds aimed at inhibiting NC protein. Here, we report that the NC chaperoning activity is extensively inhibited in vitro by small methylated oligoribonucleotides (mODN). These mODNs were delivered intracellularly using a cell-penetrating-peptide and found to impede HIV-1 replication in primary human cells at nanomolar concentrations. Extensive analysis showed that viral cDNA synthesis was severely impaired by mODNs. Partially resistant viruses with mutations in NC and RT emerged after months of passaging in cell culture. A HIV-1 molecular clone (NL4.3) bearing these mutations was found to replicate at high concentrations of mODN, albeit with a reduced fitness. Small, methylated ODNs such as mODN-11 appear to be a new type of highly potent inhibitor of HIV-1.
INTRODUCTION
Once a target cell is infected by the human immunodeficiency virus type 1 (HIV-1), the process of viral DNA synthesis by reverse transcriptase (RT) initially takes place within the virion core structure after its entry into the cytoplasm (1), which then undergoes structural modifications to become the reverse transcription complex or RTC (2). The process of reverse transcription, whereby the single-stranded genomic RNA serves as the template for the synthesis of the double-stranded viral DNA flanked by long-terminal repeats (LTRs), is a complex series of reactions that requires obligatory strand annealing and transfer. The genomic RNA is found within the HIV-1 core structure where it is extensively coated by about 1500 molecules of the nucleocapsid protein (NC) (3–6), a C-terminal product of the Gag polyprotein precursor generated by protease-mediated maturation. NC has since long been recognized as an essential co-factor of RT, chaperoning numerous key steps of the reverse transcription, namely the initiation of cDNA synthesis and the two obligatory strand transfers (1,4–9). NC is also a key determinant in genomic RNA packaging and virus assembly (10,11) and was recently found to exert a tight control on the timing of reverse transcription, by halting cDNA synthesis during virus formation (6). In addition to the currently available drugs targeting the HIV-1 enzymes RT, protease and integrase, new candidate anti-HIV-1 drugs and microbicides are in constant demand due to the emergence of drug resistant strains. This prompted us to search for compounds aimed at inhibiting the NC protein (12,13). Here, we report that the chaperoning activity of NC can be extensively inhibited in vitro by small methylated oligoribonucleotides (mODN) mimicking the LTR end sequences. These mODN were delivered intracellularly using a cell-penetrating peptide (CPP) (14–17) and found to impede HIV-1 replication in primary human T lymphocytes and macrophages at concentrations below 1 nM. Moreover, mODN were found to act as microbicides on cell-free virus. Extensive analysis and time-of-addition assays showed that the early phase of HIV-1 replication was severely impaired by mODN, notably viral cDNA synthesis. Viruses partially resistant to mODN emerged after 6 months passaging in cell culture and contained mutations in NC and RT. A HIV-1 molecular clone (NL4.3) bearing these mutations was found to replicate in the presence of 100 nM mODN, albeit at a reduced rate.
MATERIALS AND METHODS
CPP and ODNs
Peptide
The peptide Pep-2 (KETWFETWFTEWSQPKKKRKV-Cya) which is a potent carrier for the delivery of PNAs through a non-covalent approach (14,15) was used throughout the experiments.
DNA oligonucleotides
ODNs used for DNA annealing and strand transfer assays corresponded to the HIV-1 repeated R sequences, in the sense and anti-sense orientations, previously described in refs. (18,19) (see Supplementary Data), respectively.
Modified ODNs
To prepare 2′-O-methylated oligonucleotides containing aliphatic amino groups at their 3′-end, commercially available 2′-O-methylribonucleoside phosphoramidites and 3′-amino-modifier C3 CPG (Glen Research) were used. The oligonucleotides were synthesized by the phosphoramidite method on an automatic ABI 394A DNA synthesizer (Applied Biosystems) and purified by electrophoresis in a 20% polyacrylamide/7 M urea gel in TBE. The mODN sequences, methylated on each residue, were as follows:
mODN-11: 2′-O-Me-(GGUUUUUGUGU-NH2); mODN-9: 2′-O-Me-(GGUUUUUGU-NH2) (missing the 3′ GU); mODN-9U: 2′-O-Me-(UUUUGUGUU-NH2); methylated random ODN: 2′-O-Me-(UCUCACAACUC)
ODN TG (non-methylated): GGTTTTTGTGT
In vitro synthesized RNA
RNAs used for the reverse transcription assays were synthesized as described in refs. (18,19).
HIV-1 NC protein and other viral proteins
HIV-1 NC(1–55), NC(11–55) and NC(12–53) were synthesised and purified as described in ref. (19). All other retroviral NC proteins, RSV NCp12, MuLV NCp10 and FIV NCp8 were synthesized and purified as described in ref. (18–20). NCs were stored in their zinc bound form and their concentrations determined as described in ref. (20–22). HIV-1 Tat protein was synthesized as reported in ref. (18,19).
Peptide synthesis and purification
Pep-2 was synthesized by solid phase using AEDI-expansin resin with a 9050 Pep Synthesizer (PioneerTM, Applied Biosystems, Foster City, CA) (Millipore, Wartford, UK) using the Fmoc/tBoc method and purified as described (14,15). Pep-2 was N-acetylated and bears a cysteamide group at the carboxy-terminus (–NH–CH2–CH2–SH), which is required for efficient cellular uptake and delivery. Pep-2 was purified by RP-HPLC on a C18 column (Interchrom UP5 WOD/25M Uptispere 300 5 ODB, 250 × 21.2 mm). Electrospray ionization mass spectra indicated that the peptide was highly pure.
Cells
Human primary monocytes were obtained from peripheral blood mononuclear cells of healthy donors at the EFS de Lyon. Monocytes were purified and differentiated to macrophages and PBLs isolated and activated as already described (23–25). HeLaP4 cells (containing an integrated copy of β-galactosidase under the control of the viral LTR) and 293T cells were maintained in complete DMEM with 10% FCS. Chronically infected MOLT/HIV-1NL4-3 cells (26,27), and SUPT1 cells were maintained in RPMI 1640 with 10% FCS.
Assays for nucleic acid binding, annealing and strand transfer
Stem–loop destabilization by NC. We used two labeled ODNs, cTAR carrying 6-carboxyrhodamine (Rh6G) and a fluorescence quencher 4-((-4-(dimethylamino)-phenyl)-azo)-benzoic acid (DABCYL) on the 5′- and 3′-termini, respectively, to monitor stem-loop destabilization, and cTAR labeled with carboxytetramethylrhodamine (TMR) and carboxyfluorescein (Fl) at its 5′- and 3′-termini, respectively, for the annealing experiments. Inhibition of cTAR melting by NC(11–55) was monitored by adding increasing concentrations of mODN to a mixture of 25 nM Rh6G-5′-cTAR-3′-DABCYL and 275 nM NC(11–55) in 25 mM Tris–HCl, 0.2 mM MgCl2 and 30 mM NaCl, pH 7.5. Excitation and emission wavelengths were at 520 and 550 nm, respectively. Inhibition of cTAR/dTAR annealing by NC(11–55) was investigated by monitoring the annealing kinetics of 5 nM TMR-5′-cTAR-3′-Fl with 50 nM dTAR, in the presence of a 11-fold molar excess of NC(11–55) over cTAR and dTAR (28). The mODNs were added at a concentration of 440 nM to the pre-equilibrated mixtures of NC with either cTAR or dTAR for 5 min to ensure that binding equilibrium was achieved. Then, cTAR–dTAR hybridization was initiated by manual mixing of the two solutions and Fl fluorescence (excitation 480 nm, emission 520 nm) was continuously monitored (28). Extinction coefficients at 260 nm of 523 440 and 96 210 M−1 cm−1 were used for cTAR and the undecanucleotides, respectively. Emission spectra and kinetic traces were recorded at 20°C with a Fluorolog spectrofluorometer (Jobin Yvon, Horiba) equipped with a thermostated cell compartment.
The DNA and RNA binding assays, and the DNA strand exchange assays in the presence or absence of mODN-11, mODN-9, mODN-9U or ODN (TG) were run as reported in ref. (18,19).
Annealing by NCp7(1–71). Tar(+) and 32P-Tar(–) ODNs (see Supplementary Data) were incubated (0.03 pmol each) in 10 µl of buffer A at NC protein concentrations with or without methylated or non-methylated ODN as indicated in the figure legends. Reactions were performed at 37°C for 5 min except for the positive control that was incubated at 65°C. To stop the reaction and denature the protein in order to release it from the 32P-ODN, we added 5 µl of a solution containing 20% glycerol, 20 mM EDTA pH 8.0, 2% SDS, 0.25% bromophenol blue and 0.4 mg/ml calf liver tRNA. Samples were resolved by 8% native PAGE in 50 mM Tris–Borate pH 8.3, 1 mM EDTA at 4°C. Subsequently, gels were autoradiographed and the amounts of labeled single- and double-stranded DNA were assessed by PhosphorImaging.
Time-of-addition experiments
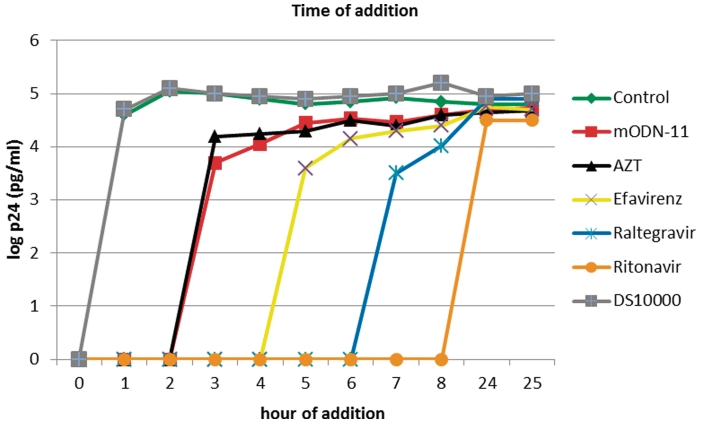
MT4 cells were infected with HIV-1 (IIIb) at a multiplicity of infection (MOI) of 0.5, and the compounds were added at different time points after infection (0, 1, 2, 3, 4, 5, 6, 7, 8, 24 and 25 h). Viral p24 antigen production was determined at 31 h post-infection (HIV-1 p24 core profile ELISA; duPont, Dreieich, Germany) (see Figure 2, ordinate values). The reference compounds (DS, Efavirenz, AZT, an Integrase Strand Transfer Inhibitor CHI/1043 or Raltegravir and ritonavir) were added at standardized concentrations corresponding to 100 times the EC50 value (half maximal effective concentration), determined at an MOI of 0.01. The mODN-11 in the form of nanoparticles upon mixing with a 10-fold excess of Pep-2, was added at 13 nM, which is 50–80× its EC50 value.
Figure 2.
Time-of-addition assays. MT-4 cells were infected with HIV-1(IIIB) at an MOI of 0.5, and the test compounds were added at different times post infection (see ‘Materials and Methods’ section). Viral p24 antigen production was determined at 31 h post infection (log p24). Compounds used were DS 10000 (15 µg/ml), Efavirenz (5 µg/ml), AZT (0.4 µM), Raltegravir (1 µM), Ritonavir (2.5 µg/ml) and mODN-11 (13 nM). The results are from a representative experiment that was repeated at least once. The antiviral drugs DS 10000 (grey) targeting entry, mODN-11 (red), the NRTI AZT (black), the NNRTI Efavirenz (yellow), the anti-IN Raltegravir (blue) and the anti-PR Ritonavir (orange) were added individually at the indicated time points. Results show that mODN-11 inhibited HIV-1 replication in MT4 cells in manner very similar to AZT (red and black, respectively) and earlier than Efavirenz (yellow).
Lentiviral vector production and purification
Production of the HIV-1, HIV-2, the simian immunodeficiency virus type 1 (SIVmac251) and the murine leukemia virus retroviral vectors was as described in refs (23–25).
HIV-1 production and treatment of virions with mODNs
Supernatant from MOLT/HIV-1 NL4-3 cells was collected, passed through a 0.45 µm PVDF filter and purified as described in ref. (27). Wild-type or mutant HIV-1 viruses were produced by DNA transfection of 293T cells with the NL4-3 plasmid DNA with mutations in the NC and RT coding sequences. Virus titers were assessed by RT-test (27) against standards of known infectivity. For direct drug treatment of virions, HIV-1 particles were mixed in 20 µl (10 µl of virus + 10 µl of 2× mODN solution in PBS) for 20 min at 37°C with the anti-NC inhibitor mODN-11 or mODN-9 complexed with Pep-2 (1:10) or with the nucleotide analog AZT (from the AIDS Reagent and Reference Program, NIH, USA). Then either RT activity of the virions was directly monitored by in vitro RT-test (27), or they were added to HeLaP4 cells in 200 µl of medium (10× dilution of mODN) for 1 h to allow virus attachment and infection. The HeLaP4 cells were covered with 1 ml more of medium (60× final dilution of mODN) and virus infectivity was assessed 48 h later by coloring the infected cells (26).
Viral infections
For single-round infections with lentiviral vectors, HeLaP4 cells, macrophages and PBLs were seeded at 105 cells/well. PBLs and macrophages were infected at the second and fifth day post-activation or differentiation, respectively. Cells were infected at a MOI of 1 (HeLaP4 cells) or 5 (PBLs, macrophages) 1 h after drug addition to the medium. Infection efficiency was examined 3–4 days later by flow cytometry as a function of eGFP expression. Infections were performed in the presence of either mODN-11 or mODN-9 complexed with Pep-2 (1:10), the nucleoside analog zidovudine (AZT) or non-nucleoside analog nevirapine (provided by the AIDS Reagent and Reference Program). For productive infection of PBLs with wild-type NL4-3 HIV-1, activated lymphocytes were incubated with either one of the inhibitor 1 h before being infected at an MOI of 0.5. After 16 h, cells were washed and the medium replaced together with the inhibitor. PBLs were cultured in the presence of IL-2. Virus production in the cell medium was detected by RT tests 12 days post-infection.
PCR and real-time PCR
HeLaP4 cells (5 x 105) were infected with HIV-1 based LV at a MOI of 1, 1 h after drug addition to the medium. For PCR, cells were lysed at 3, 6 and 24 h post-infection. Aliquots from the lysates were serially diluted (three 5-fold dilutions were made) and dilution with no signal saturation upon PCR amplification is shown. The following set of primers was used (5′–3′-nt within parentheses refer to the complete HIV-1 sequence; GenBank accession no. M38432): full-length (FL) AC37, CACTCC-CAACG-AAGA-CAAG (nt 9100–9120) and AC38, CAGCA-AGCCGA-GTCCT-GCGT (nt 699–708), as described in ref. (24). Quantitative RT–PCR of HIV-1 RNA was performed as described in the Supplementary Data.
Selection of HIV-1 resistant to mODNs and generation of mutant viruses
The selection of resistant variants was adapted from a previously described protocol (29). Multiple parallel virus passaging experiments were performed using the lymphotropic HIV-1 strain NL4-3. The selection was initiated by infecting SupT1 lymphocytes at an MOI of 0.1 in 24-well plates. Next, either none or one of the mODNs was added at an initial concentration of 0.1 nM. Cell cultures were monitored daily for the appearance of syncytia. When full-blown syncytia were observed, virus supernatants were used to initiate the next cell culture passage on naive SupT1 cells. The mODN concentration was gradually raised in the course of virus passages from 0.1 to 500 nM during a time period of 6 months. Virus resistance was assessed (by RT test) on naïve SupT1 cells infected with supernatants from the passaged viruses in the presence of increasing concentrations of the mODN. HIV-1 RNA was extracted from such cells where HIV-1 was replicating in the presence of the highest drug concentration (cells were with a clear cytopathic effect) as follows: cells were resuspended in TRIZOL reagent (Invitrogen) and total RNA/DNA was extracted following the manufacturer’s protocol. RT was performed using random hexamers and the Super Script II® kit (Invitrogen). PCR was then performed using the subsequent cDNA and primers corresponding to the genes of interest. PCR products were then subcloned in pGEMT Easy vector and unique colonies were sent for sequencing and genotypic analysis (see Supplementary Data).
The mutations found in the NC and RT domains were inserted into HIV-1 pNL4-3. To this end a fragment containing part of the gag and the pol coding sequences was cloned into a pBluescript vector in which mutations were inserted, and then cloned back into pNL4-3. Two mutagenesis procedures were used: the first, including three successive PCR has already been described (30); the second used a site-directed mutagenesis kit by StratageneR. To generate the HIV-1 double mutants RT L100I and K103N by site-directed mutagenesis the following oligonucleotides were used: sense 5′-CCACATC-CTGCAGG-GATAAA-AAAGAA-TAAATC-AGTAAC-AGTAC-TGG-3′ and antisense 5′-CCAGTA-CTGTTAC-TGATTTA-TTCTTT-TTTATCCC-TGCA-GGAT-GTGG-3′. Mutations in NC were inserted as exactly described in ref. (10). The resulting HIV-1 clones were confirmed by sequencing, viruses were produced in 293T cells by DNA transfection, and analyzed for infectivity, replication and mODN resistance in SupT1 cells.
RESULTS
Inhibition of HIV-1 NC chaperone activities by methylated oligoribonucleotides
Small 2′-O-methylated oligoribonucleotides (mODN) mimicking the LTR end sequences were found to inhibit the HIV-1 NC chaperone activities in vitro. We first examined NC ability to destabilize the secondary structure of the viral TAR stem-loop in presence of mODN (‘Materials and Methods’ section). To that end we used cTAR carrying Rh6G (6-carboxyrhodamine) and a fluorescence quencher DABCYL (4-4-(dimethylamino)-phenyl)-azo)-benzoic acid) on the 5′- and 3′-termini, respectively. We monitored the Rh6G fluorescence increase of the doubly labeled Rh6G-5′-cTAR-3′-DABCYL stem–loop by the non-aggregating NC(11–55) peptide, with or without one of the mODN (see ‘Materials and Methods’ section). Results show that mODN-11 was very efficient in inhibiting NC-directed TAR destabilization in vitro (data not shown), most probably by binding NC protein (Supplementary Figure 1).
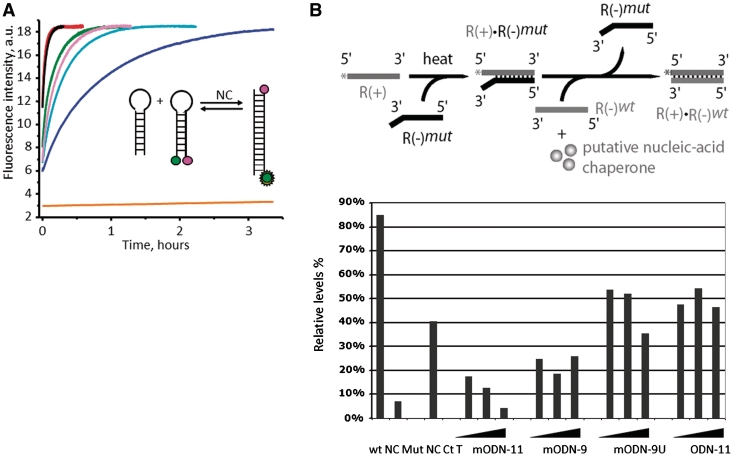
Next, NC-directed annealing of complementary TAR+ TAR– sequences was monitored in a real-time manner using cTAR labeled with TMR and Fl at its 5′- and 3′-termini, respectively (TMR-5′-cTAR-3′-Fl) and its complementary counterpart dTAR (Figure 1A; see inset for explanation). We found that mODN-11 (with a 2′-O-methyl at each residue) was a potent inhibitor of NCp7 annealing activity (Figure 1) by binding to NC (see below and Supplementary Figure S1). In fact, fluorescence increases, due to the increase of the interchromophore distance subsequent to the formation of the cTAR/dTAR extended duplex (ED), is accelerated ∼104 times by NC (Figure 1A, inset). However, addition of mODN-11 at a mODN to NC ratio of 0.75:1 drastically slowed down cTAR/dTAR annealing by NC (Figure 1A and B). The non-methylated DNA counterpart (GT) of mODN-11 appeared far less efficient, indicating that methylation of the sugars is important for inhibition (Figure 1A). Last, mODN-9U, missing the 5′ GGU of mODN-11, and mODN-9 missing the 3′GU of mODN-11 and the random methylated sequence had little inhibitory effect on NC-directed annealing of cTAR/dTAR, indicating some sequence specificity in the mODN inhibition observed. These results are in agreement with the fact that the NC Zinc Fingers (ZF) have a preference for GU in vitro (31–34) and indicate that mODN-11 inhibits the NC chaperoning activity (Figure 1), most probably by binding to the protein. In fact, binding of mODN-11 to NC was demonstrated by titration curves (Supplementary Data and Supplementary Figure S1), where the binding of mODN-11 to NC(11–55) was monitored through the fluorescence quenching of the Trp37 residue (32,34). Fitting of the titration curves yielded a binding constant of 1.4 ± 0.1 × 10e7 M−1.
Figure 1.
In vitro mODN-mediated inhibition of NC nucleic acids chaperoning activity. (A) Inhibitory activity of mODN and related oligonucleotides on NC(11–55)-directed cTAR–dTAR hybridization. The annealing kinetics were performed with 5 nM TMR-5′-cTAR-3′-Fl, 50 nM dTAR and 605 nM NC(11–55) in the absence (red) or in the presence of 440 nM of the random methylated sequence (black), mODN-9U (green), mODN-9 (pink), ODN GT (light blue) and mODN-11 (dark blue) (see ‘Materials and Methods’ section). The orange curve represents the annealing of TMR-5′-cTAR-3′-Fl with dTAR in the absence of NC(11–55). Formation of the extended cTAR/dTAR duplex was monitored in real time by Fl fluorescence (excitation at 480 nm and emission at 520 nm). Inset: principle of the FRET-based hybridization assay: in green the donor dye (fluorescein), and in magenta, the acceptor dye (TMR). Note that the random methylated ODN had no effect while mODN-11 had the strongest inhibitory activity. (B) The inhibitory activity of the mODNs and a related non-methylated ODN on NCp7(1–71)-directed HIV-1 DNA strand transfer was investigated in vitro using 32P-labeled R+, mutated R– and wt R– (see Scheme and Supplementary Data), in conditions previously reported (see ‘Materials and Methods’ section). The assays contained 10 nM R+:R– mut and 10 nM wt R–, 100 nM NCp7(1–71) and increasing concentrations of the mODN from 100 to 300 nM. After 5 min at 37°C, samples were processed and analyzed by native PAGE as reported before (18,19). Upper panel: schematic illustration of the DNA strand transfer reaction, namely formation of the ds DNA R+:R– mut bearing mutations, and subsequently addition of the wt R– (green) and NCp7 resulting in the formation of the complete R+:R– DNA. Lower panel: quantitative data of the DNA strand transfer assays with extensive strand transfer (wt NC), inactive NC (Mut NC) and partial strand transfer by heating for 1 h at 65°C (Ct T). The mODN-11 caused extensive inhibition while mODN-9 was partially inhibitory while the non-methylated ODN (TG) was inactive. Note that mODN complexed with Pep-2 were inactive in such in vitro annealing assays (data not shown). Sequences of the ODNs were as follows: mODN-11, 2′-O-Me-(GGUUUUUGUGU-NH2); mODN-9, 2′-O-Me-(GGUUUUUGU-NH2), thus missing the 3′ GU; mODN-9U, 2′-O-Me-(UUUUGUGUU-NH2); methylated random ODN: 2′-O-Me-(UCUCACAACUC); ODN TG (non-methylated): GGTTTTTGTGT.
The influence of mODN on NC chaperoning activities was also investigated using the full-length aggregating NCp7 protein and in vitro canonical assays namely TAR:cTAR hybridization, and viral DNA strand exchange (9,18,19,35,36). Results in Supplementary Figure S2 show that mODN-11 inhibited NCp7 annealing activity, while mODN-9 (missing the 3′ GU of mODN-11) was moderately active and the mODN-9U (missing the 5′GGU of mODN-11) and the non-methylated ODN were poorly active. The DNA strand exchange assay based on R sequences (Figure 1B, top and Supplementary Data) confirmed that mODN-11 was capable of inhibiting NC chaperoning activity, while mODN-9U and ODN-11 were poorly active (Figure 1). HIV-1 NC is also known to control the specificity of reverse transcription by preventing false initiation in vitro (5–7,37). In the presence of mODN-11, false initiation of reverse transcription was restored, indicating an inhibition of NCp7 activity with little or no influence on the RT enzyme (data not shown). The same chaperoning assays were conducted with other nucleic acid chaperone proteins such as HIV-1 Tat, RSV NCp12, MoMuLV NCp10 and FIV NCp8 (19–22,37). None of these nucleic acid chaperone proteins were inhibited by mODN-11 and mODN-9 (data not shown).
Inhibition of HIV-1 replication by methylated ODNs
The seemingly HIV-1-specific nature of mODN, notably the presence of GU sequences and 2′-O-methyl, prompted us to examine their influence on HIV-1 replication in human cells. To that end, mODNs were formulated with a CPP at an ODN to CPP ratio of 1:10 as described in refs. (14,15), which generates nanoparticles ensuring direct delivery of the active compound into the cytoplasm of the target cells. First, we determined the IC50 (half maximal inhibitory concentration) and CC50 (half maximal cytotoxicity concentration) of the formulated mODNs on HIV-1 replication in MT4 cells, which showed that mODN-11 strongly inhibited HIV-1 replication with an IC50 of ∼0.3 nM, while mODN-9 was 100 times less efficient. The CC50 of mODN-11 and mODN-9 was between 7.7 and 13.4 µM and thus a 1000-fold higher than the IC50 for mODN-11 formulated with Pep-2 (see ‘Materials and Methods’ section). Under these conditions the IC50 and CC50 of AZT were 4.5 nM and 50 µM, respectively (data not shown). We then investigated the mode of action of mODN-11 by means of time of addition assays on MT4 cells infected by HIV-1 as described in ref. (38). Results revealed that mODN-11 inhibited the early steps in a manner very similar to the nucleoside analog AZT (Figure 2).
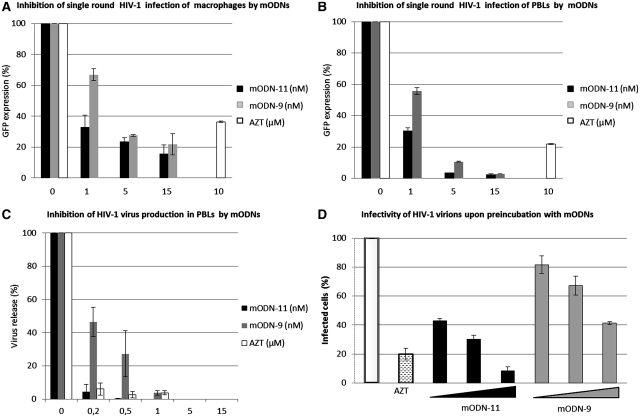
This prompted us to use a HIV-1-based lentivector encoding eGFP to infect human HeLa P4 cells with or without AZT as a control nucleoside RT inhibitor (NRTI). Inhibition of HeLa P4 cell infection at 1 nM ODN was ∼90% for mODN-11, 25% for mODN-9 and 10% for mODN-9U (Supplementary Figure S3). The mODN alone, ODN (TG) or the CPP alone did not have any effect on lentiviral infection (data not shown). Thus we focused our analysis on the impact of mODN-11 and mODN-9 on HIV-1 replication in primary human cells. Primary PBL and macrophages were infected with the HIV-1-based lentivector, and 60 and 30% inhibition of infection was achieved at 1 nM mODN-11 and mODN-9, respectively, while there was ∼60% inhibition by 10 µM AZT (Figure 3A). In the case of PBLs, mODN-11 was again more active than mODN-9 with 70 and 50% inhibition, respectively, of lentivector infection at 1 nM while inhibition was 80% at 10 µM AZT (Figure 3B). Next, we examined the impact of mODN on the complete HIV-1 replication in PBLs, since these represent the primary target cells for HIV-1. The mODNs resulted in extensive inhibition of virus replication, with 90 and 50% inhibition for mODN-11 and mODN-9, respectively, at 0.2 nM, while AZT concentrations of 0.2–0.5 µM were required to reach a similar inhibition (Figure 3C).
Figure 3.
Inhibition of HIV-1 replication by mODNs. (A and B) Cell infection by HIV-1 lentivector. Monocyte-derived macrophages (A), and activated peripheral blood lymphocytes (PBL) (B) were incubated with mODN-11 and mODN-9 at 1–15 nM, or with 10 µM AZT (as a control), 1 h prior to infection with an HIV-1-based lentiviral vector, pseudotyped with the VSVg envelope and expressing the GFP fluroescent protein (see ‘Materials and Methods’ section). Note that mODN had to be complexed with Pep2 since they were totally inactive in their free form (not shown). After 72 h post-infection, transduction efficiencies were assessed by flow cytometry as a function of GFP expression. GFP expression in the absence of drugs was considered as 100%. Note that mODN-11 was more inhibitory (95% inhibition at 5 nM) than mODN-9 on PBL (B), while both inhibited macrophage infection to similar levels (80% at 5 nM) (A) (C) HIV-1 replication in PBLs. Cells were PHA/IL-2 activated for 48 h and then infected with HIV-1 NL4-3 at an MOI of 0.5 1 h after addition of mODN-11, mODN-9 or AZT (see ‘Materials and Methods’ section). After 14 h, the cells were extensively washed, medium was replaced, drugs re-administered and cells incubated for 12 days to allow de novo virus production. The latter was analyzed by monitoring RT-activity of cell-free virions. Viral particles released in the absence of drug were considered as 100%. In the present experimental conditions, mODN-11 and AZT at 0.5 nM and 0.5 µM, respectively, were found to extensively inhibit virus production. (D) Incubation of HIV-1 virions with mODN. Virus pellets of HIV-1 NL4-3 were incubated with mODN (1, 5 and 15 nM) and AZT (10 µM) for 20 min at 37°C, and used to infect indicator HeLaP4 cells expressing β-galactosidase upon HIV-1 infection. The number of positively infected blue cells were counted and referred to as 100% for the positive cells infected with untreated virus. RT activity was directly assessed on the virions incubated with mODN or AZT for 20 min at 37°C using the RT-test (cf. ‘Materials and Methods’ section).
To investigate the impact of mODN on the infectivity of cell-free virus, HIV-1 particles were mixed for 20 min with mODN-11 or mODN-9 complexed with Pep-2, or with AZT in 20 µl final volume which was then added (and subsequently the mODN or AZT concentration was highly diluted) in the medium on HeLaP4 indicator cells. Virus infectivity was assessed 48 h later by counting the blue (infected) cells upon staining (see ‘Materials and Methods’ section). Results showed that 15 nM mODN-11 almost completely blocked virus infection while mODN-9 partially inhibited HIV-1 infection (60%) (Figure 3D). This was likely due to an inhibition of cDNA synthesis supported by the fact that exogenous RT activity from mODN-11 treated virions was significantly decreased in vitro (Supplementary Figure S4). The effect of mODNs on purified virions may be related to their capacity to enter viral cores and tightly bind the RT complex as suggested for other anti-RT inhibitors (39) and which is consistent with the in vitro data presented above.
The virus specificity of the formulated mODN-11 and mODN-9 was examined on recombinant HIV-2, SIVmac and MoMuLV infectivity in cell cultures. We found that infection of these retroviruses was not influenced by the mODNs while it was inhibited by AZT (Supplementary Figure S5).
Inhibition of cDNA synthesis by mODN
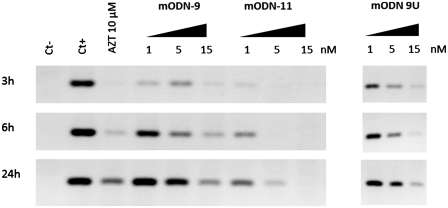
Since mODN-mediated inhibition of HIV-1 replication resembled that caused by AZT (Figure 2), we monitored cDNA synthesis at 3, 6 and 24 h post-infection in the presence of increasing concentrations of either mODN-11, mODN-9, mODN-9U or the NRTI AZT at 10 µM. Results show that mODN-11 caused an 80–90% inhibition of cDNA synthesis at 1–5 nM at 6–24 h, while mODN-9 and mODN-9U were much less inhibitory. AZT at 10 µM caused an 80–90% reduction (Figure 4; Supplementary Figure S6A). At the same time, the levels of genomic RNA in newly infected cells were not affected by mODN or AZT (Supplementary Figure S6B). Taken together these results indicate that mODNs target the reverse transcription of genomic RNA at the early stage of virus multiplication. This is in agreement with the time of addition assay where inhibition occurred if AZT was added 2 h post-infection.
Figure 4.
Influence of mODNs on reverse transcription. To evaluate the effect of the mODNs on HIV-1 replication, proviral DNA synthesis was examined at different times post-infection by PCR. HeLaP4 cells were grown in the presence of AZT, mODN-11, mODN-9 or mODN-9U for 1 h and then infected with HIV-1 LV. At 3, 6 and 24 h post-infection, the cells were lysed and PCR was performed to evaluate the complete cDNA (after the second-strand transfer, see ‘Materials and Methods’ section). The negative control (ct–) represents cells, which were not infected and untreated, whereas the positive control (ct+) represents infected but untreated cells. Note that mODN-11 was clearly more inhibitory than mODN-9 and mODN-9U (see lane 5 nM).
HIV-1 resistance to mODN
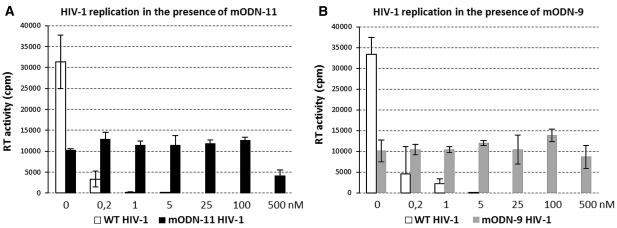
In an attempt to identify the targets of mODN directed inhibition of virus replication, selection experiments were carried out using the HIV-1 strain NL4-3 and SupT1 lymphocytes in the absence of mODN or in the presence of mODN-11 or mODN-9 at an initial concentration of 0.1 nM. Parallel cell cultures, with or without mODN, were monitored daily for the appearance of syncytia. When full-blown syncytia were observed, virus supernatants were used to initiate the next cell culture passage on naive SupT1 cells. The mODN concentration was gradually raised in the course of the passages from 0.1 to 500 nM during a time period of 6 months. The virus resistance was assessed by RT activity on naïve SupT1 cells infected with supernatants from the passages in the presence or absence of mODN. Viruses passaged at increasing concentrations of mODN-11 (Figure 5A) or mODN-9 (Figure 5B) were replicating in concentrations as high as 100 and 500 nM albeit at reduced rate, while virus passaged without mODN was almost completely inhibited at 0.2 and/or 1 nM. (Figure 5). Genotypic analysis of the viral genes was then performed in search for resistance conferring mutations (see ‘Materials and Methods’ section).
Figure 5.
Generation of mODN resistant HIV-1 upon multiple virus passages in cell culture. (A and B) HIV-1 wt was grown in SupT1 cells in the absence or presence of rising concentrations of mODN-11 or mODN-9 for 6 months when resistance to high concentrations of mODN occurred as observed by syncytia formation under the microscope. To validate these observations, supernatants from the resistant and passaged wild-type viruses were collected and used for de novo infection of naïve SupT1 cells in the presence of the inhibitors. To that end, SupT1 cells were incubated for 1 h with or without mODN and then infected with the same MOI of resistant or wt virus overnight. Cells were extensively washed, mODN re-added and de novo virus production was assessed after 4 days by RT testing. Panel shows RT activity as cpm (counts per minute) of radioactive dTTP 32P which is proportional to virus production in the cell supernatant. Note that both passaged viruses were only partially resistant to mODN, with a fitness reduced by ∼60–65% (panels A and B).
Nucleotide characterization of the HIV-1 strains resistant to mODNs
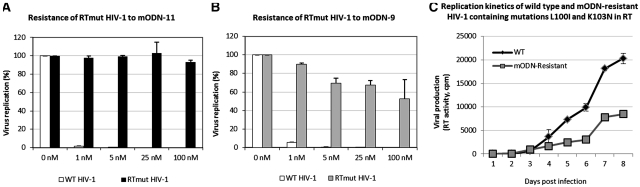
The genotypic analysis revealed that two mutations were repeatedly found in the RT sequence. These were L100I and K103N, corresponding to U>A and A>U, respectively, known to confer a high level of virus resistance to non-nucleoside RT inhibitors (NNRTI) such as Efavirenz, Delavirdine and Nevirapine (39–43). Two G>A mutations (namely, R32K and M46I) were sometimes found in NC, while notably none were found in the integrase (IN) and envelope (Env) coding sequences. The two RT mutations were inserted into HIV-1 NL4.3 by site-directed mutagenesis (see ‘Materials and Methods’ section) and the resulting HIV-1 mutant was found to replicate in SupT1 cells in the presence of mODN at concentrations as high as 100 nM (Figure 6A and B), but viral fitness was reduced since replication levels were ∼30% of the wild-type NL4.3 (Figure 6C). The NC mutations were also inserted into HIV-1 NL4-3 but did not confer resistance to mODNs (data not shown). Their insertion into the HIV-1 NL4-3 clone containing already the two RT mutations did not influence virus resistance to mODNs or fitness (data not shown).
Figure 6.
Replication of a virus clone resistant to mODN-11 and mODN-9. The molecular clone HIV-1 NL4-3 (WT HIV-1) or HIV-1 NL4-3 with the resistance conferring mutations in RT (RTmutHIV-1) was used to infect SupT1 cells in the presence of increasing concentrations of either mODN-11 (A) or mODN-9 (B). Fitness of both viruses was also assessed (C): the time course represents the viral replication in SupT1 cells up to 8 days post-infection at the same MOI (referred to the RT activity). Virus replication was monitored by measuring RT activity in the supernatants (see ‘Materials and Methods’ section and legend to Figure 5).
DISCUSSION
Taken together, the data presented herein show for the first time that short 2′-O-methylated oligoribonucleotides rich in GU such as mODN-11 can extensively inhibit the HIV-1 NC chaperoning activities in vitro (Figure 1 and Supplementary Figure S1). These mODN show a certain degree of specificity as indicated by the extensive inhibition of TAR DNA annealing by NCp7 caused by mODN-11 as compared to mODN-9 and mODN-9U (Figure 1A and B). The mechanism of mODN-11 action is mediated by NC binding (Supplementary Figure S1), notably that of the hydrophobic plateau formed by the zinc fingers (ZFs) to the single-stranded mODN rich in GUs (33,34). Thus, mODN-11 would represent a potent anti-HIV-1 drug targeting the early steps of HIV-1 replication in PBL and macrophages (Figures 2 and 3A and B). In fact, the time of additon assays show that mODN-11 inhibits HIV-1 replication in a manner similar to the NRTI AZT (Figure 2) by impairing viral DNA synthesis (Figure 4; Supplementary Figure S6) suggesting that it targets the RTC. Moreover, addition of mODN-11 together with AZT at sub-nanomolar concentrations results in a complete inhibition of HIV-1 infection, indicating that mODN-11 targets an element of the RTC different from AZT, possibly NC as expected from the in vitro results (Figure 1; Supplementary Figure S7). In addition, mODN-11 can target cell-free virions by inhibiting their replication (Figure 3D), and thus would represent a potential microbicide. Upon virus passaging in the presence of increasing concentrations of mODN-11 or mODN-9, virus became resistant to the mODN (Figure 5). Genetic analysis of the emerging mODN-resistant viruses allowed to characterize mutations in the RT and NC coding sequences but not in other Gag and Pol sequences (see ‘Results’ section). Inserting the two most frequent mutations in RT, in the HIV-1 NL4.3 molecular clone revealed that these mutations conferred phenotypic resistance to mODN-11 and mODN-9 at 100 nM (Figure 6).
The above in vitro and ex vivo findings on such modified mODNs further support the view that the RT enzyme cooperates with NC during viral DNA synthesis in newly infected cells. This is in agreement with prior in vitro results where NC was found to be an indispensable partner of the RT enzyme (4,44,45). Furthermore, ex vivo evidences clearly show that HIV-1 containing ZF mutations disrupting the NC hydrophobic plateau failed to complete viral DNA synthesis (4,7,44–46). Therefore, it is tempting to speculate that the NC binding site on RT encompasses the NNRTI binding pocket, which lies close to the polymerase primer grip and the polymerase active site driving cDNA synthesis (47,48), a question that is presently receiving further attention.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European TRIOH Consortium within the FP6 (Europe); Agence Nationale de Recherches sur le SIDA (ANRS France); Sidaction (France); FINOVI (Lyon, France); the Russian Foundation for Basic Research (projects 08-04-01252 and 08-04-01293); Fondation pour la Recherche Medicale, France (ACE20051206242 to S.A.). Funding for open access charge: INSERM, ANRS, FINOVI.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Thanks are due to Roland Ivanyi-Nagy, Andrea Cimarelli and Mike Rau for their comments and suggestions, to Barbara Van Remoortel (KU Leuven, Belgium) for performing the time-of-addition assays and to the AIDS Reagent and Reference Program (National Institutes of Health, USA).
REFERENCES
- 1.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Sowder RC, Barsov E, Hood BL, Fisher RJ, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iordanskiy SN, Bukrinsky MI. Analysis of viral and cellular proteins in HIV-1 reverse transcription complexes by co-immunoprecipitation. Methods Mol. Biol. 2009;485:121–134. doi: 10.1007/978-1-59745-170-3_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darlix J, Garrido JL, Morellet N, Mély Y, de Rocquigny H. Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv. Pharmacol. 2007;55:299–346. doi: 10.1016/S1054-3589(07)55009-X. [DOI] [PubMed] [Google Scholar]
- 4.Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 5.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 6.Mougel M, Houzet L, Darlix J. When is it time for reverse transcription to start and go? Retrovirology. 2009;6:24. doi: 10.1186/1742-4690-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas JA, Gorelick RJ. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134:39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 9.Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7:754–774. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigorov B, Décimo D, Smagulova F, Péchoux C, Mougel M, Muriaux D, Darlix J. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology. 2007;4:54. doi: 10.1186/1742-4690-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Souza V, Summers MF. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 12.Goldschmidt V, Miller Jenkins LM, de Rocquigny H, Darlix J, Mély Y. The nucleocapsid protein of HIV-1 as a promising therapeutic target for antiviral drugs. HIV Therapy. 2010;4:179–198. [Google Scholar]
- 13.de Rocquigny H, Shvadchak V, Avilov S, Dong CZ, Dietrich U, Darlix J, Mély Y. Targeting the viral nucleocapsid protein in anti-HIV-1 therapy. Mini Rev. Med. Chem. 2008;8:24–35. doi: 10.2174/138955708783331603. [DOI] [PubMed] [Google Scholar]
- 14.Morris MC, Chaloin L, Choob M, Archdeacon J, Heitz F, Divita G. Combination of a new generation of PNAs with a peptide-based carrier enables efficient targeting of cell cycle progression. Gene Ther. 2004;11:757–764. doi: 10.1038/sj.gt.3302235. [DOI] [PubMed] [Google Scholar]
- 15.Morris MC, Gros E, Aldrian-Herrada G, Choob M, Archdeacon J, Heitz F, Divita G. A non-covalent peptide-based carrier for in vivo delivery of DNA mimics. Nucleic Acids Res. 2007;35:e49. doi: 10.1093/nar/gkm053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crombez L, Morris MC, Deshayes S, Heitz F, Divita G. Peptide-based nanoparticle for ex vivo and in vivo drug delivery. Curr. Pharm. Des. 2008;14:3656–3665. doi: 10.2174/138161208786898842. [DOI] [PubMed] [Google Scholar]
- 17.Morris MC, Deshayes S, Heitz F, Divita G. Cell-penetrating peptides: from molecular mechanisms to therapeutics. Biol. Cell. 2008;100:201–217. doi: 10.1042/BC20070116. [DOI] [PubMed] [Google Scholar]
- 18.Kuciak M, Gabus C, Ivanyi-Nagy R, Semrad K, Storchak R, Chaloin O, Muller S, Mély Y, Darlix J. The HIV-1 transcriptional activator Tat has potent nucleic acid chaperoning activities in vitro. Nucleic Acids Res. 2008;36:3389–3400. doi: 10.1093/nar/gkn177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanyi-Nagy R, Lavergne J, Gabus C, Ficheux D, Darlix J. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 2008;36:712–725. doi: 10.1093/nar/gkm1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali MB, Chaminade F, Kanevsky I, Ennifar E, Josset L, Ficheux D, Darlix J, Fossé P. Structural requirements for nucleocapsid protein-mediated dimerization of avian leukosis virus RNA. J. Mol. Biol. 2007;372:1082–1096. doi: 10.1016/j.jmb.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 21.De Rocquigny H, Gabus C, Vincent A, Fournié-Zaluski MC, Roques B, Darlix JL. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl Acad. Sci. USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moscardini M, Pistello M, Bendinelli M, Ficheux D, Miller JT, Gabus C, Le Grice SFJ, Surewicz WK, Darlix J. Functional interactions of nucleocapsid protein of feline immunodeficiency virus and cellular prion protein with the viral RNA. J. Mol. Biol. 2002;318:149–159. doi: 10.1016/S0022-2836(02)00092-X. [DOI] [PubMed] [Google Scholar]
- 23.Goujon C, Rivière L, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix J, Cimarelli A. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology. 2007;4:2. doi: 10.1186/1742-4690-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarrosson-Wuilleme L, Goujon C, Bernaud J, Rigal D, Darlix J, Cimarelli A. Transduction of nondividing human macrophages with gammaretrovirus-derived vectors. J. Virol. 2006;80:1152–1159. doi: 10.1128/JVI.80.3.1152-1159.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arfi V, Lienard J, Nguyen X, Berger G, Rigal D, Darlix J, Cimarelli A. Characterization of the behavior of functional viral genomes during the early steps of human immunodeficiency virus type 1 infection. J. Virol. 2009;83:7524–7535. doi: 10.1128/JVI.00429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grigorov B, Arcanger F, Roingeard P, Darlix J, Muriaux D. Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J. Mol. Biol. 2006;359:848–862. doi: 10.1016/j.jmb.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 27.Grigorov B, Attuil-Audenis V, Perugi F, Nedelec M, Watson S, Pique C, Darlix J, Conjeaud H, Muriaux D. A role for CD81 on the late steps of HIV-1 replication in a chronically infected T cell line. Retrovirology. 2009;6:28. doi: 10.1186/1742-4690-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Godet J, de Rocquigny H, Raja C, Glasser N, Ficheux D, Darlix J, Mély Y. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J. Mol. Biol. 2006;356:1180–1192. doi: 10.1016/j.jmb.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 29.Nijhuis M, van Maarseveen NM, Lastere S, Schipper P, Coakley E, Glass B, Rovenska M, de Jong D, Chappey C, Goedegebuure IW, et al. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med. 2007;4:e36. doi: 10.1371/journal.pmed.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mikaelian I, Sergeant A. A general and fast method to generate multiple site directed mutations. Nucleic Acids Res. 1992;20:376. doi: 10.1093/nar/20.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berglund JA, Charpentier B, Rosbash M. A high affinity binding site for the HIV-1 nucleocapsid protein. Nucleic Acids Res. 1997;25:1042–1049. doi: 10.1093/nar/25.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avilov SV, Godet J, Piémont E, Mély Y. Site-specific characterization of HIV-1 nucleocapsid protein binding to oligonucleotides with two binding sites. Biochemistry. 2009;48:2422–2430. doi: 10.1021/bi8022366. [DOI] [PubMed] [Google Scholar]
- 33.Fisher RJ, Rein A, Fivash M, Urbaneja MA, Casas-Finet JR, Medaglia M, Henderson LE. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J. Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vuilleumier C, Bombarda E, Morellet N, Gérard D, Roques BP, Mély Y. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry. 1999;38:16816–16825. doi: 10.1021/bi991145p. [DOI] [PubMed] [Google Scholar]
- 35.Roda RH, Balakrishnan M, Hanson MN, Wohrl BM, Le Grice SFJ, Roques BP, Gorelick RJ, Bambara RA. Role of the reverse transcriptase, nucleocapsid protein, and template structure in the two-step transfer mechanism in retroviral recombination. J. Biol. Chem. 2003;278:31536–31546. doi: 10.1074/jbc.M304608200. [DOI] [PubMed] [Google Scholar]
- 36.Vo M, Barany G, Rouzina I, Musier-Forsyth K. HIV-1 nucleocapsid protein switches the pathway of transactivation response element RNA/DNA annealing from loop-loop “kissing” to “zipper”. J. Mol. Biol. 2009;386:789–801. doi: 10.1016/j.jmb.2008.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lapadat-Tapolsky M, Gabus C, Rau M, Darlix JL. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J. Mol. Biol. 1997;268:250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- 38.Hombrouck A, Van Remoortel B, Michiels M, Noppe W, Christ F, Eneroth A, Sahlberg BL, Benkestock K, Vrang L, Johansson NG, et al. Preclinical evaluation of 1H-benzylindole derivatives as novel human immunodeficiency virus integrase strand transfer inhibitors. Antimicrob. Agents Chemother. 2008;52:2861–2869. doi: 10.1128/AAC.00210-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ventura M, Tarrago-Litvak L, Dollé V, Nguyen CH, Legraverend M, Fleury HJ, Litvak S. Effect of nucleoside analogs and non-nucleoside inhibitors of HIV-1 reverse transcriptase on cell-free virions. Arch. Virol. 1999;144:513–523. doi: 10.1007/s007050050522. [DOI] [PubMed] [Google Scholar]
- 40.Joly V, Descamps D, Peytavin G, Touati F, Mentre F, Duval X, Delarue S, Yeni P, Brun-Vezinet F. Evolution of human immunodeficiency virus type 1 (HIV-1) resistance mutations in nonnucleoside reverse transcriptase inhibitors (NNRTIs) in HIV-1-infected patients switched to antiretroviral therapy without NNRTIs. Antimicrob. Agents Chemother. 2004;48:172–175. doi: 10.1128/AAC.48.1.172-175.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bacheler LT, Anton ED, Kudish P, Baker D, Bunville J, Krakowski K, Bolling L, Aujay M, Wang XV, Ellis D, et al. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 2000;44:2475–2484. doi: 10.1128/aac.44.9.2475-2484.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das K, Bauman JD, Clark AD, Frenkel YV, Lewi PJ, Shatkin AJ, Hughes SH, Arnold E. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc. Natl Acad. Sci. USA. 2008;105:1466–1471. doi: 10.1073/pnas.0711209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Picado J, Martínez MA. HIV-1 reverse transcriptase inhibitor resistance mutations and fitness: a view from the clinic and ex vivo. Virus Res. 2008;134:104–123. doi: 10.1016/j.virusres.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 44.Buckman JS, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 2003;77:1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grohmann D, Godet J, Mély Y, Darlix J, Restle T. HIV-1 nucleocapsid traps reverse transcriptase on nucleic acid substrates. Biochemistry. 2008;47:12230–12240. doi: 10.1021/bi801386r. [DOI] [PubMed] [Google Scholar]
- 46.Lener D, Tanchou V, Roques BP, Le Grice SF, Darlix JL. Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J. Biol. Chem. 1998;273:33781–33786. doi: 10.1074/jbc.273.50.33781. [DOI] [PubMed] [Google Scholar]
- 47.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Götte M, Rausch JW, Marchand B, Sarafianos S, Le Grice SFJ. Reverse transcriptase in motion: conformational dynamics of enzyme-substrate interactions. Biochim. Biophys. Acta. 2010;1804:1202–1212. doi: 10.1016/j.bbapap.2009.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.