Abstract
Pyrrolo[2,1-c][1,4]benzodiazepine (PBD) dimers are synthetic sequence-selective DNA minor-groove cross-linking agents that possess two electrophilic imine moieties (or their equivalent) capable of forming covalent aminal linkages with guanine C2-NH2 functionalities. The PBD dimer SJG-136, which has a C8–O–(CH2)3–O–C8′′ central linker joining the two PBD moieties, is currently undergoing phase II clinical trials and current research is focused on developing analogues of SJG-136 with different linker lengths and substitution patterns. Using a reversed-phase ion pair HPLC/MS method to evaluate interaction with oligonucleotides of varying length and sequence, we recently reported (JACS, 2009, 131, 13 756) that SJG-136 can form three different types of adducts: inter- and intrastrand cross-linked adducts, and mono-alkylated adducts. These studies have now been extended to include PBD dimers with a longer central linker (C8–O–(CH2)5–O–C8′), demonstrating that the type and distribution of adducts appear to depend on (i) the length of the C8/C8′-linker connecting the two PBD units, (ii) the positioning of the two reactive guanine bases on the same or opposite strands, and (iii) their separation (i.e. the number of base pairs, usually ATs, between them). Based on these data, a set of rules are emerging that can be used to predict the DNA–interaction behaviour of a PBD dimer of particular C8–C8′ linker length towards a given DNA sequence. These observations suggest that it may be possible to design PBD dimers to target specific DNA sequences.
INTRODUCTION
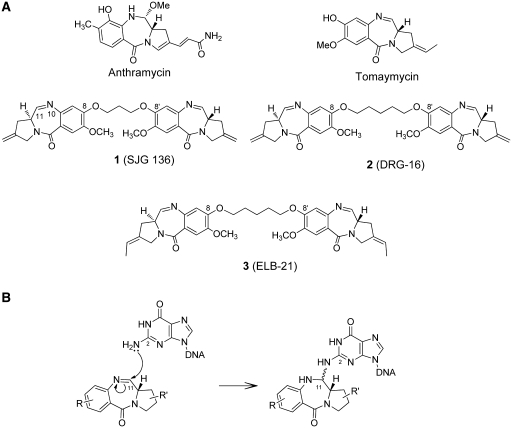
Pyrrolobenzodiazepine (PBD) dimers [e.g. 1–3, Figure 1] are synthetic sequence-selective DNA-interactive agents based on the naturally occurring anthramycin family of antitumour antibiotics (e.g. the PBD monomers anthramycin and tomaymycin, Figure 1) that target the minor groove of DNA (1–3). The PBD dimers contain two C8-linked PBD monomer units, both of which contain an electrophilic imine moiety at the N10-C11 position. These imine moieties are known to bond covalently to C2-NH2 functional groups of guanine bases in the DNA minor groove thus providing the opportunity for DNA cross-linking. Previous studies have shown that PBD dimers with odd numbers of methylenes (e.g. n = 3 or 5) in the central C8–O–(CH2)n–O–C8′ linker are the most potent interstrand cross-linking agents (4,5). One example with a C8–O–(CH2)3–O–C8′ linker, 1 (SJG-136, Figure 1), has shown clinical activity in early clinical trials (6–8), and phase II studies in leukaemia and ovarian cancer are currently underway.
Figure 1.
(A) Structures of the naturally occurring mono-alkylating PBD monomers anthramycin and tomaymycin, and the synthetic cross-linking PBD dimers, SJG-136 (1), DRG-16 (2) and ELB-21 (3). (B) Diagram showing the mechanism of covalent interaction of an electrophilic PBD unit with the nucleophilic C2-NH2 functional group of a guanine base resulting in formation of an aminal bond.
We recently developed an ion pair reversed-phase high-performance liquid chromatography/mass-spectrometry (HPLC/MS) assay to evaluate the interaction of DNA-binding agents with oligonucleotides of varying length and sequence (9–12). Studies with 1 (SJG-136) showed that the agent can form three types of adducts depending on the DNA sequence and the space between reacting guanine bases. In addition to the previously reported interstrand cross-linked adducts formed with Pu-GATC-Py binding sites (i.e. Seq-1/Seq-1; Table 1) (13,14), it was found that the agent could also form intrastrand cross-links with Pu-GATG-Py sequences (i.e. Seq-2/Seq-3; Table 1), although the interstrand adduct was predominant (see Figure 2 for graphical representation of various adduct types) (12). However, insertion of an additional base between the reactive guanines reversed this preference, with intrastrand cross-links formed at Pu-GAATG-Py sequences (Seq-6/Seq-7; Table 1) preferred over the equivalent interstrand cross-links at Pu-GAATC-Py sequences (Seq-4/Seq-5; Table 1). Crucially, competition experiments showed that, given a choice between the four sequences described above, 1 had a faster rate of reaction with the extended intrastrand cross-linking sequence Pu-GAATG-Py.
Table 1.
Sequences of oligonucleotides used in the study
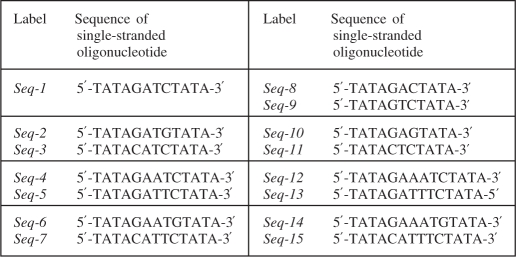 |
Complementary pairs of oligonucleotides are shown within boxes (except for Seq-1 which is self-complementary).
Figure 2.
Representations of the various types of cross-linked adducts formed between the different DNA sequences (see Table 1 for oligonucleotides used) and the PBD dimers SJG-136 (1), DRG-16 (2) and ELB-21 (3). The cross-linking base sequences are shown in red, SJG-136 in blue, and DRG-16/ELB-21 in magenta.
It is known from the literature that monomeric PBDs can stabilize duplex DNA species through the formation of covalent adducts (15). PBD dimers, which form inter- or intrastrand cross-linked adducts, can stabilize DNA to an even greater extent (13). For both PBD monomers and dimers, this stabilizing property has been shown to correlate well with their in vitro cytotoxicity (13,15). In particular, previous studies with SJG-136 (1) have shown that intrastrand adduct formation can stabilize a 12-mer duplex DNA oligonucleotide by up to 44°C, whereas interstrand adduct formation can stabilize an isomeric DNA sequence (differing only by having the guanines on opposite strands) by up to 47°C (12).
The aim of the current study was to predict the binding behaviour of PBD dimers towards a given DNA sequence with a view to eventually designing novel PBD-based agents that might produce predominantly one type of DNA adduct in cells. For the HPLC/MS studies we selected PBD dimers containing C8–O–(CH2)x–O–C8′ linkers where X = 3 (i.e. SJG-136, Figure 1) or 5 (i.e. DRG-16, 2 (5), and ELB-21, 3 (16,17); Figure 1), and DNA oligonucleotides with between one to four AT base pairs between reacting guanines (Table 1). Two molecules containing a C8–O–(CH2)5–O–C8′ linker were studied because ELB-21 has additional methyl substituents at the C2/C2′-positions, and so a comparison between these two molecules could begin to address the question of whether cross-linking preference is entirely a function of C8/C8′-linker length in the PBD family of molecules, or whether other factors such as substitution patterns play a role.
The results show that for PBD dimers with C8–O–(CH2)3–O–C8′ and C8–O–(CH2)5–O–C8′ linkers, the type and distribution of adducts formed appear to depend on (i) the positioning of the two reactive guanine bases on the same or opposite strands, (ii) their separation (i.e. the number of base pairs, in this case ATs, between them) and (iii) the length of the C8/C8′-linker connecting the two PBD units. Although these results are presently limited to three PBD dimers, a set of rules appears to be emerging that has the potential to predict the binding behaviour of a PBD dimer of particular C8/C8′-linker length towards a given DNA sequence. For example, for the interaction of a PBD dimer containing a C8–O–(CH2)3–O–C8′ linker (i.e. SJG-136) with binding sequences containing two guanines on the same or opposite strands separated by two A/T base pairs, there is a preference for interstrand rather than intrastrand cross-link formation, whereas for shorter binding sequences (i.e. two guanines separated by a single A/T base pair), interstrand cross-linking is still preferred over intrastrand although the rate of reaction is significantly slower. For longer sequences (i.e. two guanines separated by three A/T base pairs), compared to intrastrand cross-links are preferred with a higher rate of formation compared to interstrand cross-links. Crucially, similar rules appear to apply to PBD dimers containing a C8–O–(CH2)5–O–C8′ linker (i.e. ELB-21 and DRG-16) but in relation to DNA sequences with two guanines separated by 2, 3 or 4 rather than 1, 2 or 3 A/T base pairs. Molecular modelling can be used to rationalize these observations, with the two extra methylenes in the C8–C8′ linker of DRG-16 (2) and ELB-21 (3) equating to approximately one additional A/T base pair in terms of length of the DNA helix. Although SJG-136 (1) and DRG-16 (2) differ only in their C8–C8′-linker length and so can be directly compared in a study of this type, the observation that ELB-21 (3), which has modified C2/C2′-substituents, behaves in an almost identical manner to DRG-16, suggests that the C8–C8′ linker is the key determinant of sequence preference as predicted by molecular modelling. However, further studies on PBD dimers with different C-ring substituents will be required to confirm this. It also remains to be confirmed that the DNA–interaction of PBD monomers and dimers towards short oligonucleotides as observed in studies such as this is predictive of their behaviour in cells where the DNA is compacted in the genome and associated with histone proteins. However, these results, and the simple rules that appear to explain them, suggest that it may be possible to design PBD dimers to target DNA sequences with reacting guanines positioned on the same or opposite strands separated by a predetermined number of A/T bases.
MATERIALS AND METHODS
HPLC/MS assay
Single-stranded oligonucleotides
The single-stranded (SS) oligonucleotides were purchased from Eurogentec Ltd (UK) and AtdBio Ltd (Southampton, UK) in a lyophilized form.
Double-stranded oligonucleotides
Each oligonucleotide was dissolved in 100 mM ammonium acetate (Sigma–Aldrich, UK) to form a stock solution of 2 mM which was later diluted to 1 mM by addition of annealing buffer (10 mM Tris/50 mM sodium chloride/1 mM ethylenediaminetetraacetic acid (EDTA)). Solutions of double-stranded DNAs were prepared by heating the complementary SS oligonucleotides (1 mM) in annealing buffer (pH 8.5) to 70°C for 10 min in a heating/cooling block (Grant Bio, UK). The solutions were then allowed to cool slowly to room temperature (RT) followed by storage at −20°C overnight to ensure completion of the annealing process. Working solutions of DNA duplexes of 50 µM were prepared by diluting the stored solutions with 20 mM ammonium acetate.
PBD dimers SJG-136 (1), DRG-16 (2) and ELB-21 (3)
The PBD dimers 1, 2 and 3 were supplied by Spirogen Ltd (Batch Numbers: SG2000.003, SG2057.002 and SG 2060.002, respectively) and were dissolved in 50/50 v/v methanol/water to form stock solutions of 3 mM which were stored at −20°C for no longer than 4 months. Working solutions of 200 µM were prepared by diluting the stock solutions with nuclease-free water. These were stored at −20°C for not more than 1 week, and thawed to RT for use when required.
Preparation of ligand/DNA complexes
Ligand/DNA complexes were prepared by adding a PBD dimer working solution to a duplex oligonucleotide solution in a 4:1 molar ratio at room temperature. This incubation mixture was then agitated for 5–10 s using a vortex mixer.
Ion-pair reversed-phase HPLC
Analysis was performed on a Thermo Electron HPLC system equipped with a 4.6 × 50 mm Xterra MS C18 column packed with 2.5 µM particles (Waters Ltd, UK), an UV 1000 detector, an AS3000 autosampler, an SCM1000 vacuum degasser and Chromquest software (Version 4.1). A gradient system of 150 mM triethyl ammonium bicarbonate (TEAB) as buffer A and 40% acetonitrile in water (HPLC grade, Fischer Scientific, UK) as buffer B was used. For buffer A, a 1 M pre-formulated solution of TEAB was purchased from Sigma-Aldrich (UK) and diluted to the required concentration with HPLC grade water (Fischer Scientific, UK). The gradient was ramped from 90% A at 0 min to 50% at 20 min, then 35% at 30 min and finally to 10% at 45 min. Ultraviolet (UV) absorbance was monitored at 254 nm, and fractions containing separated components were collected manually, combined when appropriate, lyophilized and then analysed using either electron impact mass spectrometry (EIMS) or matrix-assisted laser desorption/ionization time of flight (MALDI–TOF)-MS.
Lyophilization of HPLC fractions
Single or combined HPLC fractions were lyophilized using two different methods depending on the final volume. For smaller volumes (<0.5 ml), lyophilizations were carried out in a SpeedVac (Thermo Electron) using a temperature-free 4-h program. For larger volumes, the solvent was initially frozen in a glass vial using liquid nitrogen, and then freeze dried (Heto Lyolab 3000) for 2 h.
Mass spectrometric analysis
MALDI-TOF
An Applied Biosystems Voyager DE-Pro Biospectrometry Workstation MALDI–TOF mass spectrometer (Framingham, MA, USA) was used to obtain MALDI–TOF spectra of components within lyophilized fractions. Samples from fractions containing single components were prepared by diluting with matrix (37 mg THAP in 1 ml ACN, 45 mg ammonium citrate in 1 ml water—mixed 1:1) either 2:1, 1:1 or 1:5 (sample:matrix) prior to MALDI-TOF analysis. One microlitre of sample was spotted onto the MALDI target plate and allowed to dry. Samples were then analysed in positive linear mode using delayed extraction (500 ns) and an accelerating voltage of 25 000 V. Acquisition was between 4000 and 15 000 Da with 100 shots/spectrum.
Molecular modelling
In order to examine the structural feasibility of the various cross-linked DNA adducts, molecular models were constructed. To test the integrity of the structures under energetic conditions, dynamics simulations were carried out at room temperature (300 K). Minimized structures of SJG-136 (1), DRG-16 (2) and ELB-21 (3) were constructed with ChemBioOfficeTM (Cambridgesoft) and exported in PDB format. Missing residue and chain records were added, and atom names were made unique. The AMBER package (18) was used for subsequent conversion to the ‘mol2’ format applying Gasteiger charges (antechamber), with missing parameters added through the ‘parmchk’ routine. The DNA duplexes were made with AMBER, and the adducts constructed manually in the first instance using ‘Xleap’ while maintaining the S-configuration at the C11 point of covalent attachment of individual PBD units. Structures were then exported for minimization, initially restraining the DNA atoms with a high force constant to allow the bound ligand to find an optimal conformation within the minor groove without distorting the overall DNA structure. Subsequent minimization steps were applied while reducing the level of restraint on the DNA atoms to zero in a stepwise manner. The generalized Born/surface area (GB/SA) implicit solvent model was used with monovalent electrostatic ion screening simulated with the SALTCON parameter set to 0.2 (M), and with a long-range non-bonded cut-off employed.
Subsequent dynamics simulations over 2 ns were then performed under similar solvent conditions. With application of the SHAKE algorithm to C–H bond vibrations, a time step of 2 fs was used. Individual energy terms were saved every 200 steps (every 0.4 ps), and the recorded energies were used to plot graphs of the total potential energy (PE) of the constructs against the simulation time. The average PE for the whole simulation was also recorded. Conformational variation in the course of the dynamics simulations was examined by performing a rigid body root mean square (RMS) fit for the atoms of each frame of the simulation to the first frame and plotting this parameter against simulation time.
RESULTS AND DISCUSSION
HPLC/MS studies
The study utilized previously reported HPLC/MS analytical methodology (9–12) and the 15 oligonucleotides listed in Table 1. The oligonucleotide duplexes were designed to have reactive guanines at appropriate positions on the same or opposite strands to allow intrastrand or interstrand cross-links to form but with varying numbers of base pairs (i.e. 1-4 A/T base pairs) between the guanines to evaluate the effect of inter-guanine distance. Incubation of the PBDs with these annealed oligonucleotide duplexes for specific time periods followed by injection of the reaction mixture onto the HPLC column provided distinct PBD/DNA adduct peaks that could be collected, lyophilized and subjected to MALDI-TOF-MS analysis to confirm identity. Unreacted SS oligonucleotides and annealed duplexes were also analysed in the same HPLC system to provide reference peaks. As with previously reported studies, a 4:1 molar ratio of ligand:oligonucleotide was used, as it ensured completion of cross-link formation within a reasonable time frame.
Cross-linked and monoalkylated adducts were characterized based on both HPLC retention times and MALDI–TOF data as previously reported for SJG-136 (12). In this original study, interstrand, intrastrand and monoalkylated adducts were demonstrated to appear at distinct retention times with this particular HPLC system, and each was identified and characterized through a series of guanine replacement studies and MALDI–TOF MS data. The former involved replacement of key guanines in various oligonucleotide duplexes with inosine residues which lack the nucleophilic C2-NH2 functionality of normal guanines. This allowed individual adduct peaks at particular retention times to be positively identified through a process of successive ‘knock-out’.
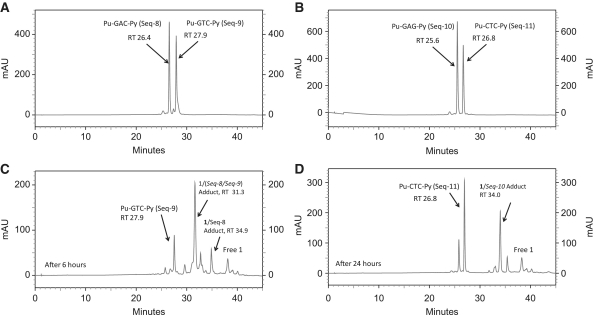
Following on from the previously reported study with 1 (SJG-136) and the Pu-GATC-Py, Pu-GATG-Py, Pu-GAATC-Py and Pu-GAATG-Py sequences (Supplementary Figures S1–S14) (12), the first experiment focused on the shortened interstrand 5′-Pu-GAC-Py (Seq-8/Seq-9 duplex; Table 1) and intrastrand 5′-Pu-GAG-Py sequences (Seq-10/Seq-11 duplex; Table 1). Surprisingly, given the difference in length between the molecule and reacting guanines, the HPLC experiments showed that 1 is capable of reacting with both sequences (Figure 3), although it reacted faster with 5′-Pu-GAC-Py indicating that it prefers interstrand cross-link formation for these shorter binding sequences. The reaction with 5′-Pu-GAC-Py was nearly complete within 6 h (Figure 3C), whereas reaction with the intrastrand 5′-Pu-GAG-Py sequence was still not complete after 24 h (Figure 3D). Interestingly no duplex adduct was observed in the HPLC assay after reaction with the Pu-GAG-Py duplex (Figure 3D), with the single stranded 1/Seq-10 adduct (RT = 34 min) being the predominant product. This relative instability of the duplex adduct under the HPLC conditions is consistent with a higher level of structural distortion compared to adducts formed from sequences with more A/T base pairs between the guanines which presumably leads to a greater rate of dissociation of the two DNA strands. A control experiment demonstrated that 1 does not react with Seq-10 (Pu-GAG-Py) alone, confirming that the observed 1/Seq-10 adduct must have resulted from dissociation of the initially formed duplex adduct. Although the rate of adduct formation with 5′-Pu-GAC-Py was much faster than for 5′-Pu-GAG-Py, it was significantly slower than for 5′-Pu-GATC-Py. The identification of all adduct peaks was confirmed using MALDI-TOF MS (Supplementary Table S1).
Figure 3.
HPLC chromatograms; (A) Annealed Pu-GAC-Py duplex (Seq-8/Seq-9); (B) Annealed Pu-GAG-Py duplex (Seq-10/Seq-11); (C) 6 hours after incubation of SJG-136 (1) with the Seq-8/Seq-9 duplex showing appearance of the 1/(Seq-8/Seq-9) interstrand cross-linked adduct at RT 31.3 min and a small amount of 1/(Seq-8) monoalkylated adduct at RT 34.9 min (resulting from partial denaturation of 1/(Seq-8/Seq-9), (D) 24 h after incubation of SJG-136 (1) with the Seq10/Seq-11 duplex showing appearance of the 1/Seq-10 monoalkylated adduct at RT 34.0 min resulting from complete denaturation of the initially formed 1/(Seq-10/Seq-11).
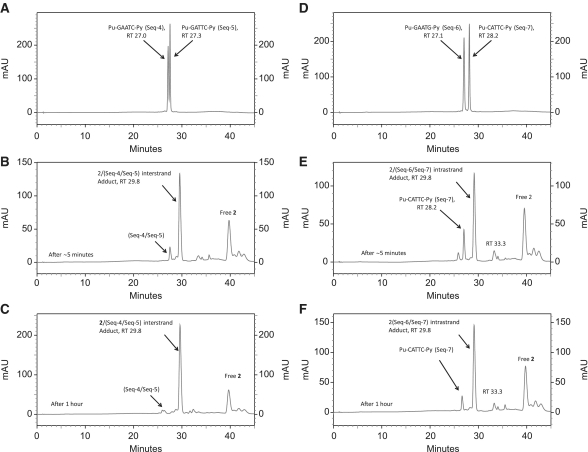
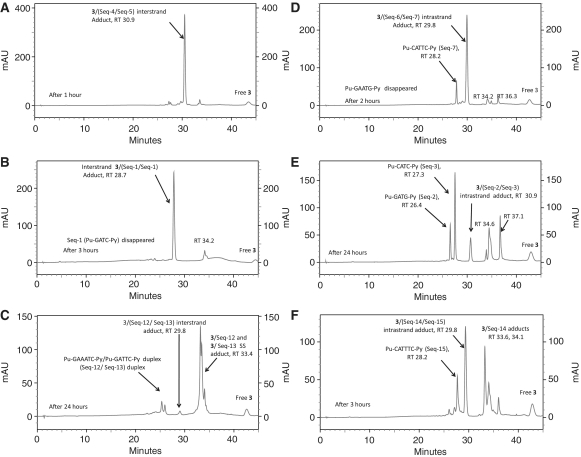
Next, DRG-16 (2, C8–O–(CH2)5–O–C8′) was studied with the 5′-Pu-GAATC-Py duplex (Seq-4/Seq-5, Table 1) containing an additional A/T base pair between reacting guanines compared to the 5′-Pu-GATC-Py SJG-136 interstrand cross-linking sequence (Figure 4). For DRG-16, this was previously suggested to be the ideal separation between reactive guanines for an interstrand cross-link based on molecular modelling studies (19). Analysis of the annealed duplex formed from Seq-4 and Seq-5 resulted in two peaks at RT 27.0 and 27.3 min identified by MALDI–TOF MS as denatured SS Seq-4 and Seq-5, respectively (Figure 4A). Mixing the annealed duplex with 2 followed by immediate analysis showed the partial (∼85%) formation of an adduct peak at RT 29.8 min as the major product (Figure 4B). A time course study showed that interstrand adduct formation was almost complete (∼95%) within 1 h (Figure 4C). Intrastrand cross-linking with the same separation between reacting guanines was then investigated using the Seq-6/Seq-7 duplex in which the cytosine-9 of Seq-4 (5′-TATAGAATCTATA-3′) had been mutated to a guanine (5′-TATAGAATGTATA-3′) (Table 1). Analysis of the annealed duplex alone using identical HPLC conditions resulted in two peaks at RT 27.1 and 28.2 min, confirmed by MALDI–TOF MS to be the SS Seq-6 and Seq-7, respectively (Figure 4D). Addition of 2 to the annealed Seq-6/Seq-7 duplex followed by immediate HPLC analysis showed the rapid emergence of a peak at RT 29.8 min, the identity of which was confirmed by MALDI–TOF MS as the intrastrand 1:1 2/(Seq-6/Seq7) duplex adduct (Figure 4E). A time course study showed the emergence of a minor peak at RT 33.3 min (see below), although the main 29.8 min peak remained prominent with time and accounted for more than 90% of the products at the end of the experiment. The rate of intrastrand adduct formation with Seq-6/Seq7 was surprisingly rapid and complete within 1 h, with the peak corresponding to Seq-6 completely disappearing (Figure 4F). In the 0 h (i.e. ∼5 min) chromatogram (Figure 4E), the ratio of peaks relating to the unreacted SS DNA species (approximately equal in Figure 4D before the addition of 2) had changed in favour of Seq-7 (28.2 min), suggesting that 2 had reacted preferentially with Seq-6. However, the smaller peak at 33.3 min had a m/z of 4609 consistent with the 1:1 2/Seq-6 monoalkylated adduct, indicating that a proportion of the 1:1 2/(Seq-6/Seq7) adduct, once formed, had dissociated on the HPLC column to the 1:1 2/Seq-6 monoalkylated adduct and Seq-7, thus explaining the predominance of the latter.
Figure 4.
HPLC chromatograms; (A) Annealed Pu-GAATC-Py duplex (Seq-4/Seq-5); (B) Immediately (i.e. ∼5 min) after mixing DRG-16 (2) with the Seq-4/Seq-5 duplex showing appearance of the 2/(Seq-4/Seq-5) interstrand adduct at RT 29.8 min; (C) 1 h after mixing 2 with the Seq-4/Seq-5 duplex followed by incubation at room temperature showing completion of reaction; (D) Annealed Pu-GAATG-Py duplex (Seq-6/Seq-7); (E) Immediately (∼5 min) after mixing DRG-16 (2) with the Seq-6/Seq-7 duplex showing appearance of the intrastrand 2/(Seq-6/Seq-7) duplex adduct at RT 29.8 min and a small amount of monoalkylated adduct at RT 33.3 min; (F) 1 h after mixing 2 with the Seq-6/Seq-7 duplex showing the reaction nearing completion.
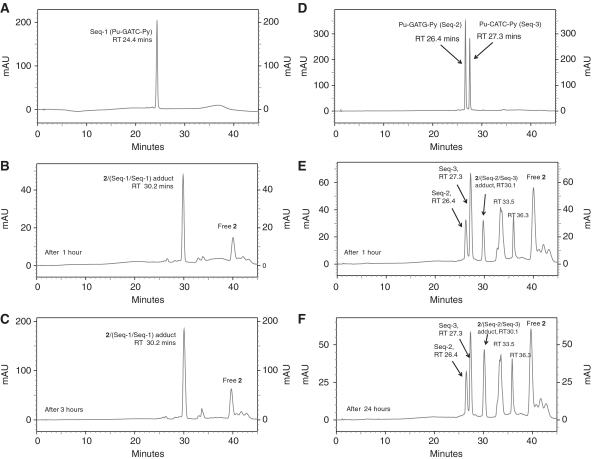
Similar HPLC experiments were conducted on the interaction of 2 with the self-complementary Pu-GATC-Py duplex (Seq1/Seq1), originally reported as the preferred binding site for 1 (13), and the corresponding intrastrand Pu-GATG-Py duplex (Seq-2/Seq-3) (Figure 5). It was observed that 2 reacted faster with the interstrand Pu-GATC-Py sequence (Figure 5A–C) compared to the Pu-GATG-Py intrastrand sequence (Figure 5D–F). Reaction with Seq-2/Seq-3 was complete within 3 h (Figure 5C), whereas reaction with Seq-2/Seq-3 was incomplete even after 24 h (Figure 5F). In the case of reaction with Pu-GATG-Py, two additional peaks were observed at RT 33.5 and 36.3 min. The first was identified from MS and retention time as the cross-linked SS 2/Seq-2 arising from denaturation of the duplex intrastrand cross-linked adduct 2/(Seq-2/Seq-3), and also explaining the excess of Seq-3 at 27.3 min (Figure 5E). The second additional peak at 36.3 min was tentatively identified by similar means based on previously reported inosine experiments (12) as monoalkylated 2/Seq-2. It is noteworthy that the rates of reactions for the interstrand cross-linking duplexes Pu-GATC-Py and Pu-GAATC-Py were comparable but the reaction rate was significantly slower for the intrastrand cross-linking Pu-GATG-Py duplex compared to the longer Pu-GAATG-Py. This result clearly shows that 2 prefers interstrand cross-link formation with both the Pu-GATC-Py and Pu-GAATC-Py sequences compared to the equivalent intrastrand cross-linking sequences. The identification of all peaks was confirmed from MALDI–TOF–MS data (see Supplementary Data, Supplementary Table S2).
Figure 5.
HPLC chromatograms; (A) 5′-Pu-GATC-Py-3′ (Seq-1/Seq-1) duplex denatured under HPLC conditions, RT 24.4 min. (B) 1 h after mixing DRG-16 (2) with the Seq-1/Seq-1 duplex showing the appearance of an interstrand 2/(Seq-1/Seq-1) Pu-GATC-Py duplex adduct at RT 30.2 min. (C) 3 h after mixing 2 with Seq-1/Seq-1 showing completion of reaction. (D) Pu-GATG-Py (Seq-2/Seq-3) duplex denatured under HPLC conditions. (E) 1 h after mixing 2 with the Seq-2/Seq-3 duplex showing the appearance of intrastrand 2/(Seq-2/Seq-3) Pu-GATG-Py duplex adduct at RT 30.1 min. Two additional peaks were observed at RT 33.5 and 36.3 min. (F) 24 h after mixing 2 with Seq-2/Seq-3 showing that reaction was still not complete.
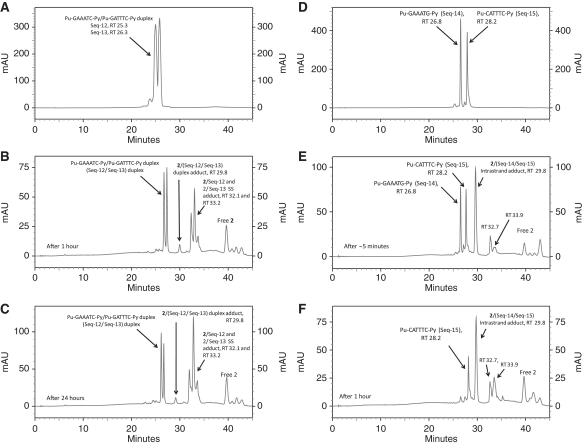
To investigate the effect of inserting an additional base between reactive guanines, two duplex oligonucleotides, Seq-12/Seq-13 and Seq-14/Seq-15, were designed to contain the extended interstrand (Pu-GAAATC-Py) and intrastrand (Pu-GAAATG-Py) cross-linking sites, respectively. After incubation of 2 with the Seq-12/Seq-13 duplex, subsequent HPLC analysis showed the emergence of a minor peak at RT 29.8 mins and major peaks at RT 32.1 and 33.2 min (Figure 6B). MALDI–TOF–MS data confirmed the minor 29.8 min peak to be the interstrand 2/(Seq-12/Seq-13) adduct peak, whereas the RT 32.1 and 33.2 min peaks were identified as closely eluting 1:1 2/Seq-12 and 1:1 2/Seq-13 monoalkylated adducts, respectively (see Table 2 for MS data). A time course study showed that adduct formation was still not complete after 24 h, with the 29.8 min peak (i.e. the interstrand cross-linked adduct) remaining as the minor peak throughout the experiment (Figure 6C). These results indicate that 2 has a poor reactivity towards this extended interstrand cross-linking site and forms a high proportion of monoalkylated adducts instead, presumably due to the two PBD imine moieties failing to stretch effectively between the two nucleophilic guanine sites. However, interaction of 2 with the extended intrastrand cross-linking sequence Pu-GAAATG-Py (Seq-14/Seq-15) resulted in the rapid (i.e., ∼5 mins) emergence of a major new peak at RT 29.8 min along with two minor peaks at RT 32.7 and 33.9 min (Figure 6E). As anticipated, the RT 29.8 min peak was identified from retention time and MALDI–TOF–MS data as the intrastrand 1:1 2/(Seq-14/Seq-15) duplex adduct. The minor RT 32.7 and RT 33.9 min peaks gave MS data corresponding to single-stranded 1:1 2/Seq-14 adducts. Based on retention times from previous studies with inosine-containing oligonucleotides (12), the 32.7 min peak was assigned as the 1:1 intrastrand cross-linked 2/Seq-14 adduct, and the 33.9 min peak as the 1:1 2/Seq-14 monoalkylated adduct. However, the intrastrand cross-linked 1:1 2/(Seq-14/Seq-15) duplex adduct was clearly the most abundant species. Crucially, reaction with the intrastrand duplex Pu-GAAATG-Py was complete within 1 h (Figure 6F), whereas reaction with the interstrand Pu-GAAATC-Py duplex had proceeded to only a small extent after 24 h (Figure 6C). It is noteworthy that the rate of reaction for the interstrand cross-linking duplex Pu-GAAATC-Py was significantly slower than for the shorter Pu-GATC-Py duplex but the rate of intrastrand cross-linking with the Pu-GAAATG-Py duplex duplex was faster compared to Pu-GATG-Py. Together, these data clearly demonstrate the preference of 2 to form intrastrand cross-links with an extended DNA sequence (i.e. four AT base pairs between reacting guanines), thus mirroring the behaviour of the shorter SJG-136 (1) molecule towards sequences containing three AT base pairs between reacting guanines. This result is consistent with the molecular modelling studies (see below) which suggest that the length of the extra two methylenes of the C8–O–(CH2)5–O–C8′ central linker of 2 compared to the C8–O–(CH2)3–O–C8′ linker of 1 equate to approximately one base pair spacing within the DNA minor groove.
Figure 6.
HPLC chromatograms; (A) Seq-12/Seq-13 Pu-GAAATC-Py duplex denatured under HPLC conditions. (B) 1 h after mixing DRG-16 (2) with the Seq-12/Seq-13 duplex showing appearance of a minor interstrand Pu-GAAATC-Py duplex adduct at RT 29.8 min, along with single-stranded 2/(Seq-12) and 2/(Seq-13) adducts at RT 32.1 and 33.2 min, respectively. (C) Negligible change after 24 h except for a small increase in 2/(Seq-12) and 2/(Seq-13) adducts at RT 32.1 and 33.2 min in relation to the unreacted Seq-12 and Seq-13 peaks at 25.3 and 26.3 min, respectively. (D) Seq-14/Seq-15 duplex denatured under HPLC conditions. (E) Immediately (i.e. ∼5 min) after mixing 2 with the Seq-14/Seq-15 duplex showing appearance of a major intrastrand 2/(Seq-14/Seq-15) Pu-GAAATG-Py duplex adduct peak at RT 29.8 min. Additional minor peaks are visible at RT 32.7 and 33.9 min, identified as the single-stranded 1:1 intrastrand cross-linked 2/Seq-14 adduct, and the 1:1 2/Seq-14 monoalkylated adduct, respectively. (F) Reaction is complete after 1 h with disappearance of the Seq-14 oligonucleotide at RT 26.8 min.
Table 2.
Theoretical and observed masses of the double-stranded oligonucleotides and their 1:1 adducts with SJG-136 (1), DRG-16 (2) and ELB-21 (3)
| DNA sequence of duplex oligonucleotide | Theoretical mass of DNA duplex alone | Observed (1:1) DNA/SJG-136 adduct mass (DNA mass + 556.61) | Observed (1:1) DNA/DRG-16 adduct mass (DNA mass + 584.66) | Observed (1:1) DNA/ELB-21 adduct mass (DNA mass + 612.72) |
|---|---|---|---|---|
| Pu-GATC-Py (Seq-1/Seq-1) | 7287.4 | 7841.3 | 7871.2 | 7902.45 |
| Pu-GATG-Py (Seq-2/Seq-3) | 7286.74 | 7842.06 | 7873.2 | 7903.0 |
| Pu-GAATC-Py (Seq-4/Seq-5) | 7904.13 | 8458.3 | 8485.6 | 8517.8 |
| Pu-GAATG-Py (Seq-6/Seq-7) | 7904.13 | 8459.1 | 8488.7 | 8516.0 |
| Pu-GAC-Py (Seq-13/Seq-14) | 6669.17 | 7225.78 | N/A | N/A |
| Pu-GAG-Py (Seq-10/Seq-11) | 6669.17 | 7225.61 | N/A | N/A |
| Pu-GAAATC-Py (Seq-12/Seq-13) | 8522.05 | N/A | 9108.2 | 9136.3 |
| Pu-GAAATG-Py (Seq-14/Seq-15) | 8522.05 | N/A | 9107.3 | 9136.4 |
N/A - Not available, as the sequence is either too short or too long to form cross-links with respective agents.
Finally, in order to assess whether a minor modification (i.e. addition of methyl groups) to the C2/C2′-substituents of DRG-16 (2) might influence its cross-linking properties, a similar set of HPLC experiments was conducted with the PBD dimer ELB-21 (3) in which it behaved in an identical manner to 2. Figures 7A–F show the HPLC profiles for reaction of 3 with the key six duplex sequences at the point at which reaction had gone to completion, except in the case of Figure 7C (Pu-GAAATC-Py) and E (Pu-GATG-Py), the least preferred sequences, in which reaction was incomplete after 24 h. The identification of each peak was confirmed using MALDI–TOF–MS data (Supplementary Table S3, see Supplementary Data).
Figure 7.
HPLC chromatograms for ELB-21 (3); (A) 1 h after mixing 3 with the Seq-4/Seq-5 duplex showing complete formation of the interstrand 3/(Seq-4/Seq-5) Pu-GAATC-Py adduct at RT 30.9 min; (B) 3 h after mixing 3 with the Seq-1/Seq-1 duplex showing complete formation of interstrand 3/(Seq-1/Seq-1) Pu-GATC-Py duplex adduct at RT 28.7 min, along with a minor peak at RT 34.2; (C) 24 h after mixing 3 with the Seq-12/Seq-13 duplex showing reaction nearing completion with a minor peak for the interstrand 3/(Seq-12/Seq-13) Pu-GAAATC-Py duplex adduct at RT 29.8 min, along with close major peaks for 3/(Seq-12) and 3/(Seq-13) single-stranded adducts at approximately RT 33.4 min. (D) 2 h after mixing 3 with the Seq-6/Seq-7 duplex showing completion of reaction and formation of the intrastrand 3/(Seq-6/Seq-7) Pu-GAATG-Py adduct at RT 29.8 min, along with minor peaks at RT 34.2 and 36.3 min; (E) 24 h after mixing 3 with the Seq-2/Seq-3 duplex showing intrastrand 3/(Seq-2/Seq-3) Pu-GATG-Py duplex adduct at RT 30.9 min, along with additional peaks at RT 34.6 and 37.1 min (reaction still not complete), (F) 3 h after mixing 3 with the Seq-14/Seq-15 duplex showing completion of reaction and formation of major peaks for the intrastrand Pu-GAAATG-Py duplex adduct at 29.8 min and minor single-stranded adducts at 33.6 and 34.1 min.
Molecular modelling studies
In an attempt to visualize and rationalize interaction of the three PBD dimers with the different DNA sequences, molecular models were constructed to investigate the fit of 1, 2 and 3 within the various cross-linking sites while attempting to estimate the proximity of the reacting guanines to the two electrophilic N10-C11 imine moieties. According to these models, both 1 and 2 appear to be easily accommodated within their ideal sequences as determined by previous modelling studies (5,13) (i.e. two and three AT base pairs between reacting guanines, respectively), and also in those which are 1 bp shorter. As previously reported for SJG-136 and the Pu-GATC-Py/Pu-GATG-Pu sequence (13), model building shows that for B-form DNA, the C2-NH2 of a guanine occupies an almost identical mid-groove position whichever strand it is located on, albeit with the -NH2 in an opposite orientation. Thus, for DRG-16 (2), after covalently linking one end of it to guanine-5 of either the 5′-TATAGAATCTATA-3′ or 5′-TATAGAATGTATA-3′ duplexes, the C2-NH2 of the other reacting guanine (whether on the same or opposite strand) occupies an almost identical mid-groove position, supporting the feasibility of forming both inter- and intrastrand cross-links.
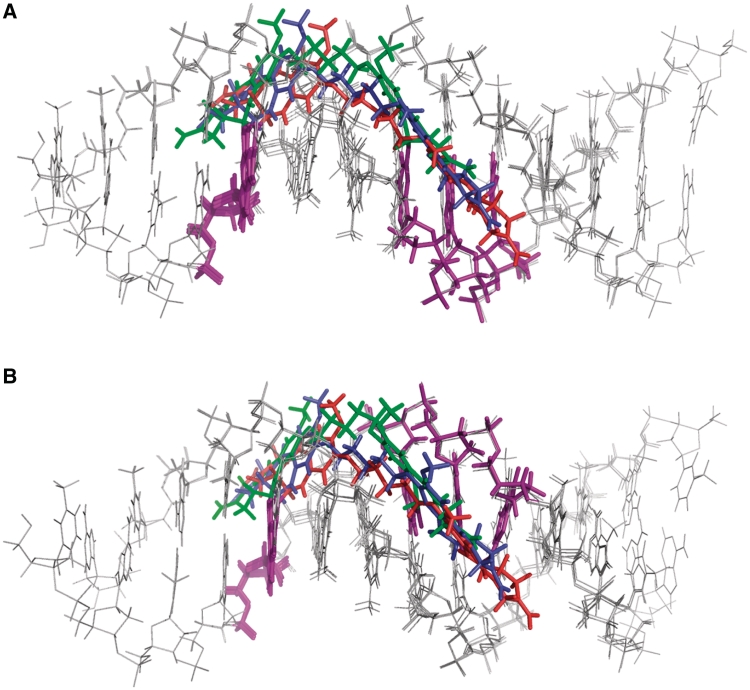
Molecular dynamics simulations were carried out for both SJG-136 (1) and DRG-16 (2) with their extended interstrand and intrastrand sequences (i.e. three and four AT base pairs between reacting guanines, respectively), and some distortion of the DNA became evident, necessary to maintain the cross-link over the given span of bases. The greater distortion observed in the extended interstrand models over the course of the molecular dynamics simulations is evident from the graphs of RMS deviation of atom coordinates of 1 and 2 when covalently linked through the guanines (see Supplementary Figures S24 and S25 of Supplementary Data). The initial minimized models used for the simulations showed how, in the case of the extended sequences, the C8/C8′ linker is forced to adopt a lower position in the minor groove. Figure 8 shows superimposed models for DRG-16 (2) covalently bound to the three interstrand (Figure 8A) and three intrastrand cross-linking sequences (Figure 8B) of different length. For both 1 (see Supplementary Figure S22 and Supplementary Data) and 2, the position of the C8/C8′-linker in the groove corresponds to whether there are 1, 2 or 3 (for SJG-136) or 2, 3 or 4 (for DRG-16) base pairs between reacting guanines, respectively, with the lowest position adopted for the longest sequence. Thus, in these models, the position of the linker with respect to the minor-groove floor directly reflects the differences in distance between the two electrophilic imine moieties of the PBD dimer and the two nucleophilic C2-NH2 groups of the reacting guanines. Analysis of the van der Waals contacts between the linkers and DNA atoms for the most extended sequences showed that the contacts were close but not overlapping in this region.
Figure 8.
Overlay of energy minimised molecular models of the different types of covalent cross-linked adducts formed by DRG-16 (2) with the duplex oligonucleotides of different lengths and cross-linking potential (i.e. inter- versus intrastrand): (A) DRG-16 interstrand cross-linking with Pu-GATC-Py (green ligand), Pu-GAATC-Py (blue ligand) and Pu-GAAATC-Py (red ligand) duplex oligonucleotides; (B) DRG-16 intrastrand cross-linking with Pu-GATG-Py (green ligand), Pu-GAATG-Py (blue ligand) and Pu-GAAATG-Py (red ligand) duplex oligonucleotides. Note: In both figures the covalently bound guanine bases are highlighted in purple.
Interestingly, for both molecules, more distortion was observed for the interstrand compared to the intrastrand adducts, consistent with the experimentally observed preference for the intrastrand forms. Furthermore, the degree of distortion of the interstrand adduct was considerably higher in the case of 2, consistent with the mono-adducts formed in the reaction between 2 and its extended interstrand sequence. The formation of mono-adducts was not observed during the reaction of 1 with its corresponding extended sequence.
To further support the preference of 1 and 2 to form inter- or intrastrand cross-linked adducts depending on the spatial separation of reactive guanines, molecular dynamics simulations were carried out for each molecule cross-linked to three pairs of inter- and intrastrand sequences (Table 3), and an average potential energy (PE) was determined for each adduct. Except for the adducts of 2 with the ideal base separation (i.e. Seq-4/Seq-5 and Seq-6/Seq-7), in each case the average PE values supported the experimentally observed results, with the interstrand adducts having lower energies for the ideal and shorter length sequences, while for the extended sequences the intrastrand adducts had a significantly lower average PE value. Although the PE values for adducts of 2 with Pu-GAATG-Py and Pu-GAATC-Py were the reverse of that anticipated, they are only 3.34 kcal/mol apart, and the experimentally observed rates of formation of these intrastrand and interstrand adducts are more similar (i.e. ∼95% versus 85%, respectively, after 1 h) compared to the reaction of SJG-136 (1) with its equivalent Pu-GATG-Py and Pu-GATC-Py sequences (i.e. ∼50% reaction after 12 h and 5 min, respectively) (12), consistent with the greater difference of 8.07 kcal/mol.
Table 3.
Average potential energies for a 2000 ps molecular dynamics simulation of the different types of covalent cross-linked adducts formed by SJG-136 (1) and DRG-16 (2) with duplex oligonucleotides containing one to four AT-base pairs between reacting guanines
| DNA sequence of duplex oligonucleotide | Type of cross-link possible | Average potential energy (kcal/mol) of SJG-136 cross-linked adducts | Average potential energy (kcal/mol) of DRG-16 cross-linked adducts | Type of cross-link preferred by SJG-136 observed in HPLC studies | Type of cross-link preferred by DRG-16 observed in HPLC studies |
|---|---|---|---|---|---|
| Pu-GAC-Py (Seq-13/Seq-14) | Interstrand | −3361.86 | Sequence too short to form cross-links | Interstrand | Sequence too short to form cross-links |
| Pu-GAG-Py (Seq-10/Seq-11) | Intrastrand | −3361.57 | |||
| Pu-GATC-Py (Seq-1/Seq-1) | Interstrand | −3678.54 | −3680.54 | Interstrand | Interstrand |
| Pu-GATG-Py (Seq-2/Seq-3) | Intrastrand | −3670.47 | −3671.13 | ||
| Pu-GAATC-Py (Seq-4/Seq-5) | Interstrand | −3955.65 | −3972.74 | Intrastrand | Interstrand |
| Pu-GAATG-Py (Seq-6/Seq-7) | Intrastrand | −3980.09 | −3976.08 | ||
| Pu-GAAATC-Py (Seq-12/Seq-13) | Interstrand | Sequence too long to form cross-links | −4253.13 | Sequence too long to form cross-links | Intrastrand |
| Pu-GAAATG-Py (Seq-14/Seq-15) | Intrastrand | −4264.31 |
CONCLUSIONS
The data presented here demonstrate that preference for the type of adduct formed (i.e. interstrand versus intrastrand) between PBD dimers and DNA can be rationalized and predicted based on whether the guanines are on the same or opposite strands of a DNA duplex, the distance between the two reacting guanines, and the length of the linker between the C8/C8′-positions of the two PBD units of the dimer.
For SJG-136 (1) which has a C8–O–(CH2)3–O–C8′ linker, there is a preference for interstrand cross-linking with Pu-GAC-Py or Pu-GATC-Py sequences rather than cross-linking with the isomeric intrastrand sequences, although reaction is much slower with the shorter sequence. For the longer Pu-GAATC-Py or Pu-GAATC-Py sequences, intrastrand cross-linking is preferred, and is more rapid than the interstrand cross-linking that occurs with the shorter sequences. Also, with these longer sequences, monoalkylated adducts begin to appear as minor products, which may represent intermediate species en-route to the final cross-linked adducts. Finally, with the even longer Pu-GAAATG-Py and Pu-GAAATG-Py sequences, 1 cannot reach between the two guanines when the bases are either on the same or opposite strands, and so monoalkylated adducts are observed almost exclusively.
For DRG-16 (2) and ELB-21 (3) which contain the longer C8–O–(CH2)5–O–C8′ linker, there is a preference for interstrand cross-linking with Pu-GATC-Py or Pu-GAATC-Py sequences rather than cross-linking with the isomeric intrastrand sequences, although reaction is slower with the shorter sequence. However, for both 2 and 3, reaction with the intrastrand Pu-GAATG-Py sequence was found to be very rapid compared to the equivalent reaction of 1 with the intrastrand sequence Pu-GATG-Py, perhaps reflecting the greater flexibility of the longer central C8/C8′ linker. For the longer Pu-GAAATC-Py and Pu-GAAATG-Py sequences, intrastrand cross-linking is preferred and is more rapid than the interstrand cross-linking that occurs with shorter sequences. For these longer sequences, monoalkylated adducts begin to appear as minor products, which may represent intermediate species en route to the final cross-linked adducts. Finally, for the even longer Pu-GAAAATC-Py and Pu-and GAAAATG-Py sequences, these PBD dimers cannot reach between the two guanines when on the same or opposite strands, and so only mono-adducts are observed.
Although only three PBD dimers were investigated in this study, given that 2 and 3 behaved in a similar fashion, it is likely that preference for intrastrand or interstrand cross-linking with particular DNA sequences may be more dependent on the C8/C8′ linker length than structural differences in the C-rings.
The various types of adducts formed by PBD dimers may be relevant to the biological activity of this class of antitumour agents. At present, the interstrand cross-links produced by SJG-136 (1) at Pu-GATC-Py sequences are thought to be responsible for its mechanism of action (20,21), and these are measured in in vitro cell-based experiments and in peripheral blood lymphocytes (PBLs) during clinical trials using the COMET (22) and gamma yH2AX-foci (23) assays. However, further studies are now required to determine whether intrastrand or monoalkylation adducts also contribute to the biological activity.
Finally, these observations, and the simple rules that appear to explain them, suggest that it may be possible to design novel PBD dimers as chemical probes or therapeutic agents to produce predominantly one type of DNA adduct in cells. Further studies are underway to try to understand the effect of more significant changes to the C8/C8′-linker (such as the insertion of heteroatoms) on the pattern of adducts produced.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spirogen Ltd is thanked for providing funding for the oligonucleotides. The Commonwealth Scholarship Commission is acknowledged for a Scholarship to K.M.R (BDCA-005.01). Funding for open access charge: The Commonwealth Scholarship Commission.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Spirogen Ltd is thanked for supplying samples of SJG-136 (1), DRG-16 (2) and ELB-21 (3), as is Dr Kersti Karu for her help with the MALDI MS experiments, and Dr Emma Sharp is acknowledged for her assistance with preparing the manuscript.
REFERENCES
- 1.Cipolla L, Araujo AC, Airoldi C, Bini D. Pyrrolo[2,1-c][1,4]benzodiazepines as a scaffold for the design and synthesis of anti-tumour drugs. Anticancer Agents Med. Chem. 2009;9:1–31. doi: 10.2174/187152009787047743. [DOI] [PubMed] [Google Scholar]
- 2.Kamal A, Rao MV, Laxman N, Ramesh G, Reddy GSK. Recent developments in the design, synthesis and structure–activity relationship studies of Pyrrolo[2,1-c][1,4]benzodiazepines as DNA-interactive antitumour antibiotics. Curr. Med. Chem. Anticancer Agents. 2002;2:215–254. doi: 10.2174/1568011023354119. [DOI] [PubMed] [Google Scholar]
- 3.Thurston DE. In: Molecular Aspects of Anticancer Drug-DNA Interactions. Neidle S, Waring MJ, editors. Vol. 1. London, UK: The Macmillan Press Ltd.; 1993. pp. 54–88. [Google Scholar]
- 4.Bose DS, Thompson AS, Smellie M, Berardini MD, Hartley JA, Jenkins TC, Neidle S, Thurston DE. Effect of linker length on DNA-binding affinity, cross-linking efficiency and cytotoxicity of C8-linked pyrrolobenzodiazepine dimers. Chem. Commun. 1992;20:1518–1520. [Google Scholar]
- 5.Gregson SJ, Howard PW, Gullick DR, Hamaguchi A, Corcoran KE, Brooks NA, Hartley JA, Jenkins TC, Patel S, Guille MJ, et al. Linker length modulates DNA cross-linking reactivity and cytotoxic potency of C8/C8' ether-linked C2-exo-unsaturated pyrrolo[2,1-c][1,4]benzodiazepine (PBD) dimers. J. Med. Chem. 2004;47:1161–1174. doi: 10.1021/jm030897l. [DOI] [PubMed] [Google Scholar]
- 6.Hochhauser D, Meyer T, Spanswick VJ, Wu J, Clingen PH, Loadman P, Cobb M, Gumbrell L, Begent RH, Hartley JA, et al. Phase I study of sequence-selective minor groove DNA binding agent SJG-136 in patients with advanced solid tumors. Clin. Cancer Res. 2009;15:2140–2147. doi: 10.1158/1078-0432.CCR-08-1315. [DOI] [PubMed] [Google Scholar]
- 7.Janjigian YY, Lee W, Kris MG, Miller VA, Krug LM, Azzoli CG, Senturk E, Calcutt MW, Rizvi NA. A phase I trial of SJG-136 (NSC#694501) in advanced solid tumors. Cancer Chemother. Pharmacol. 2010;65:833–838. doi: 10.1007/s00280-009-1088-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puzanov I, Lee W, Chen A, Calcutt MD, Hachey DL, Vermeulen WL, Spanswick VJ, Hartley JA, Rothenberg ML. Phase I, pharmacokinetic and pharmacodynamic study of SJG-136, a novel DNA sequence selective minor groove cross-linking agent, in advanced solid tumors. Clin. Cancer Res. 2011 doi: 10.1158/1078-0432.CCR-10-2056. [Epub ahead of print 23 February 2011, DOI: 10.1158/1078-0432.CCR-10-2056] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narayanaswamy M, Griffiths WJ, Howard PW, Thurston DE. An assay combining high-performance liquid chromatography and mass spectrometry to measure DNA interstrand cross-linking efficiency in oligonucleotides of varying sequences. Anal. Biochem. 2008;374:173–181. doi: 10.1016/j.ab.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 10.Rahman KM, Mussa V, Narayanaswamy M, James CH, Howard PW, Thurston DE. Observation of a dynamic equilibrium between DNA hairpin and duplex forms of covalent adducts of a minor groove binding agent. Chem. Commun. 2009;2:227–229. doi: 10.1039/b816163a. [DOI] [PubMed] [Google Scholar]
- 11.Rahman KM, Vassoler H, James CH, Thurston DE. DNA sequence preference and adduct orientation of pyrrolo[2,1-c][1,4]benzodiazepine antitumor agents. ACS Med. Chem. Lett. 2010;1:427–432. doi: 10.1021/ml100131b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahman KM, Thompson AS, James CH, Narayanaswamy M, Thurston DE. The pyrrolobenzodiazepine dimer SJG-136 forms sequence-dependent intrastrand DNA cross-links and monoalkylated adducts in addition to interstrand cross-links. J. Am. Chem. Soc. 2009;131:13756–13766. doi: 10.1021/ja902986x. [DOI] [PubMed] [Google Scholar]
- 13.Gregson SJ, Howard PW, Hartley JA, Brooks NA, Adams LJ, Jenkins TC, Kelland LR, Thurston DE. Design, synthesis, and evaluation of a novel pyrrolobenzodiazepine DNA-interactive agent with highly efficient cross-linking ability and potent cytotoxicity. J. Med. Chem. 2001;44:737–748. doi: 10.1021/jm001064n. [DOI] [PubMed] [Google Scholar]
- 14.Gregson SJ, Howard PW, Jenkins TC, Kelland LR, Thurston DE. Synthesis of a novel C2/C2′-exo unsaturated pyrrolobenzodiazepine cross-linking agent with remarkable DNA binding affinity and cytotoxicity. Chem. Commun. 1999;9:797–798. [Google Scholar]
- 15.Wells G, Martin CRH, Howard PW, Sands ZA, Laughton CA, Tiberghien A, Woo CK, Masterson LA, Stephenson MJ, Hartley JA, et al. Design, synthesis, and biophysical and biological evaluation of a series of pyrrolobenzodiazepine - poly(N-methylpyrrole) conjugates. J. Med. Chem. 2006;49:5442–5461. doi: 10.1021/jm051199z. [DOI] [PubMed] [Google Scholar]
- 16.Hadjivassileva T, Thurston DE, Taylor PW. Pyrrolobenzodiazepine dimers: novel sequence-selective, DNA-interactive, cross-linking agents with activity against Gram-positive bacteria. J. Antimicrob. Chemother. 2005;56:513–518. doi: 10.1093/jac/dki256. [DOI] [PubMed] [Google Scholar]
- 17.Hadjivassileva T, Stapleton PD, Thurston DE, Taylor PW. Interactions of pyrrolobenzodiazepine dimers and duplex DNA from methicillin-resistant Staphylococcus aureus. Int. J. Antimicrob. Agents. 2007;29:672–678. doi: 10.1016/j.ijantimicag.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, et al. AMBER 9. Amber 9. University of California, San Francisco. 2006 [Google Scholar]
- 19.Smellie M, Bose DS, Thompson AS, Jenkins TC, Hartley JA, Thurston DE. Sequence-selective recognition of duplex DNA through covalent interstrand cross-linking: kinetic and molecular modeling studies with pyrrolobenzodiazepine dimers. Biochemistry. 2003;42:8232–8239. doi: 10.1021/bi034313t. [DOI] [PubMed] [Google Scholar]
- 20.Alley MC, Hollingshead MG, Pacula-Cox CM, Waud WR, Hartley JA, Howard PW, Gregson SJ, Thurston DE, Sausville EA. SJG-136 (NSC 694501), A novel rationally designed DNA minor groove interstrand cross-linking agent with potent and broad spectrum antitumour activity: part 2: efficacy evaluations. Cancer Res. 2004;64:6700–6706. doi: 10.1158/0008-5472.CAN-03-2942. [DOI] [PubMed] [Google Scholar]
- 21.Hartley JA, Spanswick VJ, Brooks N, Clingen PH, McHugh PJ, Hochhauser D, Pedley RB, Kelland LR, Alley MC, Schultz R, et al. SJG-136 (NSC 694501), a novel rationally designed DNA minor groove interstrand cross-linking agent with potent and broad spectrum antitumour activity: part 1: cellular pharmacology, in vitro and initial in vivo antitumor activity. Cancer Res. 2004;64:6693–6699. doi: 10.1158/0008-5472.CAN-03-2941. [DOI] [PubMed] [Google Scholar]
- 22.Hartley JM, Spanswick VJ, Gander M, Giacomini G, Whelan J, Souhami RL, Hartley JA. Detection of DNA interstrand crosslinking in clinical samples using the single cell COMET assay. Ann. Oncol. 1998;9:645. [Google Scholar]
- 23.Redon CE, Nakamura AJ, Zhang YW, Ji JP, Bonner WM, Kinders RJ, Parchment RE, Doroshow JH, Pommier Y. Histone yH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers. Clin. Cancer Res. 2010;16:4532–4542. doi: 10.1158/1078-0432.CCR-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.