Abstract
The ability of mammalian cytidine deaminases encoded by the APOBEC3 (A3) genes to restrict a broad number of endogenous retroelements and exogenous retroviruses, including murine leukemia virus and human immunodeficiency virus (HIV)-1, is now well established. The RNA editing family member apolipoprotein B (apo B)-editing catalytic subunit 1 (APOBEC1; A1) from a variety of mammalian species, a protein involved in lipid transport and which mediates C–U deamination of mRNA for apo B, has also been shown to modify a range of exogenous retroviruses, but its activity against endogenous retroelements remains unclear. Here, we show in cell culture-based retrotransposition assays that A1 family proteins from multiple mammalian species can also reduce the mobility and infectivity potential of LINE-1 (long interspersed nucleotide sequence-1, L1) and long-terminal repeats (LTRs) retrotransposons (or endogenous retroviruses), such as murine intracisternal A-particle (IAP) and MusD sequences. The anti-L1 activity of A1 was mainly mediated by a deamination-independent mechanism, and was not affected by subcellular localization of the proteins. In contrast, the inhibition of LTR-retrotransposons appeared to require the deaminase activity of A1 proteins. Thus, the AID/APOBEC family proteins including A1s employ multiple mechanisms to regulate the mobility of autonomous retrotransposons in several mammalian species.
INTRODUCTION
The ability of polynucleotide cytidine deaminases encoded by the mammalian APOBEC3 (A3) genes to restrict a variety of retroelements is now well established (1,2). A3 molecules belong to a family of proteins that also includes apolipoprotein B (apo B)-editing catalytic subunit 1 (APOBEC1; A1), activation-induced cytidine deaminase (AID), APOBEC2 (A2) and APOBEC4 (A4) (3–5). These proteins have deaminase activities that can modify cytosine bases to uracils (C–U) on DNA and/or RNA. A1, the catalytic component of a complex that deaminates apoB mRNA in gastrointestinal tissues, is the original member of this family and remains the best characterized (6,7). AID, a DNA-editing enzyme that is the second member to be identified, has been shown to play key roles in the diversification of antibody genes in activated B cells (8). The human genome encodes seven A3 proteins, from A3A to A3H, while the mouse genome contains a single A3 gene (9,10). Since only AID- and A2-like but not A3- or A1-related sequences are encoded in the genome of non-mammalian vertebrates such as birds and fishes, AID as well as A2 is thought to be the evolutionary precursor to the various AID/APOBEC family proteins (10).
Several A3 proteins have been shown to possess the capacity to reduce the mobility and infectivity potential of autonomous retrotransposon LINE-1 (long interspersed nucleotide sequence-1, L1) (11–16). In contrast, it was suggested that neither AID nor A2 possess any activity against the mobility of retroelements in cell culture assays (12,17,18), but this issue is controversial (19–21). Similar to A3, several reports including ours indicated that A1 inhibits a wide range of exogenous retroviruses such as HIV (human immunodeficiency virus)-1 (17,22–24). The inhibitory activities against HIV-1 observed with A1s from rodent and rabbit were based, at least in part, on cytidine-deamination of the viral genomic RNA, and analysis of mutational hot spots indicated that the molecular mechanisms for intra-virion editing of HIV-1 genomic RNA and apoB mRNA overlap (23,24). Thus, it is conceivable that A1 also affects the mobility of the autonomous retroelements. Here, we investigated the effects of A1 proteins on the retrotransposition of LINE-1 and long terminal repeats (LTRs) retrotransposons such as murine intracisternal A-particle (IAP) and MusD sequences. We showed that A1 proteins do possess the capacity to inhibit the transposition of L1 and endogenous retrovirus sequences. The anti-L1 activity does not require cytidine deaminase activity, and is not affected by subcellular localization of the A1 proteins. In contrast, A1 inhibits the replication of murine IAP and MusD through a DNA deamination-dependent mechanism. Together, the data suggest that the A1 proteins can also function in innate defense against endogenous retroelements, exerting their effects using multiple mechanisms in several mammalian species.
MATERIALS AND METHODS
DNA constructs
Plasmids coding for human L1 (pL1RP-EGFP and pCEP4/L1mneoI/ColE1) were kindly donated by E.T. Luning Prak and N. Gilbert, respectively (25,26). Plasmids coding for murine L1 (pCMV L1Md-Gf21neoTET), MusD (pCMV L1Mus-6DneoTNF) and IAP (pGL3-IAP92L23neoTNF) have been described (27–30). HA-tagged human A3A, A3B and A3G expression plasmids, HA-tagged A1 from mammalian species and rabbit A1 with catalytic site mutant (E63Q, E63A) expression plasmids have been described (16,23,31). Rabbit and rat A1 catalytic mutants N57A and P29T were constructed with QuickChange® XL Site-Directed Mutagenesis Kit (Stratagene) using oligonucleotide primers (rabbit A1 N57AF; 5′-CGC AGC TCG GGC AAG GCC ACC ACC AAT CAC GTG-3′, rabbit A1 N57AR; 5′-CAC GTG ATT GGT GGT GGC CTT GCC CGA GCT GCG-3′, rat A1 P29TF; 5′-GTC TTC TTT GAC ACC CGG GAA CTT-3′, rat A1 P29TR; 5′-AAG TTC CCG GGT GTC AAA GAA GAC-3′, rabbit A1 P29TF; 5′-GTC TTC TTT GAC ACC CAA GAA CTG CG-3′, rabbit A1 P29TR; 5′-CGC AGT TCT TGG GTG TCA AAG AAG AC-3′), and inserted into pCAGGS vector. To clone a full-length murine AID cDNA, total RNA from mouse germinal center B-cells was prepared using TRIzol reagent and synthesis of cDNA was performed with Superscript II RT kit using random primers. The cDNA encoding the AID gene was amplified using primer sets, 5′-GGGGGATCCATGGACAGCCTTCTGATGA-3′ and 5′-GCTCTAGATCAAAATCCCAACATACGAAA-3′. The AID cDNA fragment was inserted into the BamHI–XbaI sites of pBS-3x Flag vector and confirmed by DNA sequencing. The Flag-fused AID fragment was isolated from ‘SalI-NotI’ sites and transfer into the cloning sites in pEF-BOS-derived mammalian expression vector. The untagged AID gene was amplified with 5′-GGCTCGAGATGGACAGCCTTCTGATGAAGC-3′ and 5′-GGGGAATTCTCAAAATCCCAACATAC-3′, inserted into EcoRI site of pCXN2 vector, a derivative of pCAGGS that carries a fragment of the tk promoter and neo gene (32).
L1 retrotransposition assay
The enhanced green fluorescent protein (EGFP)-based human L1 retrotransposition assay was performed as previously described (16). Total of 3 × 105 293T cells were co-transfected with 0.5 µg of the respective APOBEC expression vector or empty vector (pCAGGS) together with 1.5 µg of EGFP-based human L1 reporter vector pL1RP-EGFP or pIRESpuro vector (CLONTECH) using Effectene® (Qiagen). The amount of plasmid DNA for transfection was normalized to 2.0 µg. Twenty-four hours post-transfection, cells were subjected to puromycin (1.0 µg/ml) selection. After 7–9 days of puromycin selection, EGFP expression resulting from retrotransposition was verified by flow cytometry. Another L1 retrotransposition assay was performed by cotransfection of HeLa cells (5 × 105 cells) with 0.4 µg of the respective APOBEC expression plasmids and 1.2 µg of neomycin-resistant (neor)-based human L1 reporter vector pCEP4/L1mneoI/ColE1 or neor-based murine L1 reporter vector pCMV L1Md-Gf21neoTET along with 0.4 µg of pIRES-EGFP (CLONTECH) using the FuGENE technology (Roche Applied Science), as previously described (16,33). After 72 h, 5 × 105 cells were equivalently re-seeded onto 100 mm dishes for G418 (0.5–0.75 mg/ml) selection and maintained. After 12–14 days of selection, resultant G418-resistant (G418R) colonies were fixed, stained with crystal violet (SIGMA), and counted. Retrotransposition frequencies were calculated as a number of G418R colonies/transfection efficiency (percent of GFP+ cells). Data are shown by the values relative to the pCAGGS control vector. The average of three experiments with standard deviation is indicated.
Bacterial mutator assay
To generate the bacterial-expression vectors encoding the various APOBECs, APOBEC fragments in the relevant pCAGGS-based plasmid, including the C-terminal HA tag (23), were PCR-amplified by using oligonucleotides (sense orientation, human A3G; 5′-CTC GAG ATG AAG CCT CAC TTC AGA AAC ACA GTG G-3′, human A1; 5′-CTC GAG ATG ACT TCT GAG AAA GGT CCT TCA ACC G-3′, ferret A1; 5′-CTC GAG ATG GCT TCT GAC AAA GGT CCT TCA GC-3′ rabbit A1; 5′-CTC GAG ATG GCT TCC GAG AAA GGT CCT TCA AA-3′, rat A1; 5′-CTC GAG ATG AGT TCC GAG ACA GGC CCT G-3′, XhoI site underlined) and another primer (antisense orientation, 5′-CTG CAG TCA AGC GTA ATC TGG AAC ATC GTA TGG GTA-3′, PstI site underlined), digested with XhoI and PstI restriction enzymes, and inserted into XhoI and PstI sites present in the bacterial expression plasmid pTrcHisA (Invitrogen). All constructs were verified by DNA sequencing. The ung (uracil DNA glycosylase)-deficient Escherichia coli strain BW310 [(34,35); donated by E. coli Genetic Resource Center, Yale University] was transformed with the pTrcHisA parental plasmid and vectors encoding the various APOBEC cDNAs. Transformed bacteria were then selected overnight on LB plates containing ampicillin. Twenty colonies were pooled into 2 ml of LB medium plus ampicillin plus 1 mM IPTG and cultures grown overnight at 37°C. One hundred microliters of the saturated culture was then plated on LB plates containing 100 µg/ml of rifampicin, and the total number of rifampicin-resistant (RifR) colonies per plate was counted 24 h later. For viable cells, appropriate dilution was plated onto LB plate containing ampicillin and mutation frequencies were caluculated as RifR colonies per viable cell. To verify protein expression, 100 µl of the saturated IPTG-induced culture was lysed and analyzed by western blot analysis as described above.
Immunofluorence microscopy and confocal analysis
A total of 2×104 HeLa cells were seeded onto 8-well Lab-Tek Chamber Slide (Nalge Nunc International) and immunofluorescence studies were performed at 24 h after transient transfection using FuGENE. Cells were subsequently fixed with 4% formaldehyde in PBS(−) for 30 min, permeabilized with 0.1% Triton-X100 in PBS for 2 min at room temperature, and subsequently washed 3 times in PBS. Then, the cells were treated with 0.1 M glycine/PBS for quenching and 0.3% BSA/PBS for blocking. For APOBECs staining, coverslips were incubated in a humid chamber at 37°C for 1 h with anti-HA antibody (HA.11, Covance; 1:1000 dilution) in 0.3% BSA/PBS. A fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Sigma; 1:300) in 0.3% BSA/PBS was then added and incubation continued for an additional hour. Subsequently, 4,6-diamidino-2-phenylindole staining (Invitrogen; 1 µg/ml) was performed for 5 min. The coverslips were mounted with Fluorescent Mounting Medium (Dako). Fluorescence pattern were visualized with a Zeiss LSM 700 laser-scanning confocal microscopy. The images were captured using IPLab and processed using Adobe PhotoShop 4.0 software.
IAP and MusD retrotransposition assay
IAP and MusD retrotransposition assay was performed by cotransfection of HeLa cells with 0.2–0.4 µg of the respective APOBEC expression plasmids or empty vector (pCAGGS) together with 0.6 µg of neor-based murine IAP-reporter vector pGL3-IAP92L23neoTNF or 1.2 µg of neor-based murine MusD-reporter vector pCMV Mus-6DneoTNF along with 0.4 µg of pIRES-EGFP using FuGENE, as previously described (33). After 72 h, 1 × 105–5 × 105 cells were re-seeded onto 100 mm dishes for G418 (0.75–1.0 mg/ml) selection and maintained. Twelve to fourteen days after selection, resultant G418R colonies were fixed, stained with crystal violet, and counted. Retrotransposition frequencies were calculated as described above.
Quantitation of de novo L1 copy number
Copy number of retrotransposed L1 elements was estimated by real-time PCR targeting the EGFP gene as described previously (16). Briefly, 6–7 days after transfection as described above, total cellular DNA was extracted from 293T cells by using the QIAamp DNA Blood Mini Kit (Qiagen) which is able to recover both chromosomal and episomal DNA. Real-time PCR reactions were then performed using TaqMan Gene Expression Master Mix by 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). L1 DNA levels are presented as copies per 106 cells.
Fast protein liquid chromatography analysis
The 293T cells were co-transfected with 1.5 µg of pIRESpuro or pL1RP-EGFP and 0.5 µg of respective pCAGGS-APOBEC-expression vectors. At 24 h post-transfection, the transfected cells were harvested and then lysed by Fast protein liquid chromatography (FPLC) buffer [50 mM HEPES pH 7.2, 125 mM NaCl, 10% glycerol, 0.1% NP-40 and protease inhibitor cocktail (Sigma-Aldrich)]. Twenty micrograms of the total protein was separated by sequentially adding 300 µl of the FPLC buffer into 10 fractions on Sepharose 4B (Sigma-Aldrich) packed serological column (Fisher). The each eluted fraction was analyzed by western blot. To test RNase sensitivity, the cell lysates were treated with RNase A (50 µg/ml, QIAGEN) at 37°C for 1 h, before fractionation.
RNA immunoprecipitation assay and real-time quantitative RT–PCR
To examine the physical association of A1 protein with L1 and cellular RNAs, RNA immunoprecipitation assay was carried out as previously described (36), with minor modification. Briefly, 293T cells were co-transfected with 1.5 µg of pL1RP-EGFP and 0.5 µg of expression plasmids for APOBEC family proteins, and 24 h later, cells were harvested and then lysed by FPLC buffer. The cell lysates were cross-linked and then immunoprecipitated with control rabbit IgG (Cell Singnaling) or anti-HA rabbit IgG (Cell Singnaling) conjugated to Dynabeads® M-280 Sheep anti-Rabbit IgG (Invitrogen). The precipitated RNAs with APOBEC proteins were extracted from coprecipitated samples and analyzed by real-time quantitative RT–PCR (qRT–PCR) with primers specific for L1-EGFP, 7SL RNA, GAPDH and eIF4G2 (37). Isolated samples were treated with TURBOTM DNase (Ambion) and then reverse transcribed using High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems). qRT–PCR was performed using Applied Biosystems 7500 (Applied Biosystems) and MESA Blue qPCR MasterMix (EUROGENTEC). The target sequences were amplified using the following primer set. L1-EGFP, forward; 5′-TCC AGG AGC GCA CCA TCT T-3′ and reverse; 5′-ATG CCC TTC AGC TCG ATG C-3′. 7SL RNA, forward; 5′-ATC GGG TGT CCG CAC TAA G-3′ and reverse; 5′-CAC CCC TCC TTA GGC AAC CT-3′. GAPDH, forward; 5′-GCA AAT TCC ATG GCA CCG T-3′and reverse; 5′-TCG CCC CAC TTG ATT TTG G-3′. eIF4G2, forward; 5′-ACA AAT GCC AGG TAG CGG AA-3′ and reverse; 5′-TTG CCT CCC ATC TCT CCA AA-3′.
Statistical analysis
The statistical significance between two groups was determined by performing the Student’s test. A value of P < 0.05 was considered to be of statistical significance.
RESULTS
A1s from multiple mammalian species possess anti-L1 activity
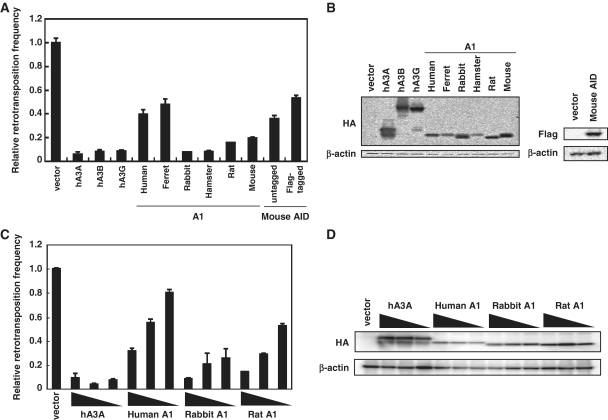
To examine the anti-L1 activities of A1s, we co-transfected 293T cells with expression plasmids encoding various A1s carrying C-terminal HA-tag and the EGFP-based L1 retrotransposon indicator construct pL1RP-EGFP (25), with hA3A, hA3B and hA3G (which are also C-terminal HA epitope-tagged) serving as controls. The pL1RP-EGFP plasmid contains a full-length human L1 genome with the EGFP gene inserted in the anti-sense orientation, as well as a ‘puromycin’-resistance gene which allows for selection. The EGFP gene is under the control of a CMV promoter and is disrupted by a γ-globin intron in the sense orientation. EGFP expression, therefore, requires transcription and splicing of the L1 transcript, followed by reverse transcription and retrotransposition into the host cell genome. After 7–9 days of puromycin selection, EGFP expression resulting from retrotransposition was verified by flow cytometry. The appearance of many GFP+ events in cultures transfected with the pL1RP-EGFP plasmid alone (7.21% with a MFI of 421.3; vector) as compared with the few events in those without (0.3% with a MFI of 49.9; Nega) demonstrates the dependence of EGFP expression on the successful occurrence of L1 retrotransposition (Supplementary Figure S1). As expected, pronounced decrease in the L1 retrotransposition was observed when hA3A and hA3B were expressed (4.6-fold and 5.1-fold, respectively). Consistent with previous observations (11–16), hA3G was also found to cause pronounced decrease in L1 mobility (5.1-fold). The anti-L1 activity of A1 proteins from different mammalian species was more variable and was dose-dependent (Figure 1C). At 0.5 µg DNA input, the reduction in L1 mobility with A1s from human and ferret was modest (2.4-fold and 1.8-fold, respectively), while those from rodents such as hamster, rat and mouse reduced L1 retrotransposition by ∼3-fold. A1 from rabbit was the most potent, with close to a 4-fold decrease in L1 retrotransposition seen with this cytidine deaminase. As AID from multiple species including primary vertebrates such as chicken and fish has also been shown to possess anti-L1 activity (21), we tested mouse AID activity against the L1 reporter construct pL1RP-EGFP in 293T cells. Although mouse AID reduced L1 retrotransposition, its activity is lower than those observed with A1s from other mammalian species (Figure 1 and Supplementary Figure S1).
Figure 1.
Inhibition of L1 retrotransposition by A1 proteins. (A) 293T cells were co-transfected with 0.5 µg of the expression plasmids for APOBEC family proteins and 1.5 µg of L1 retrotransposition indicator construct pL1RP-EGFP. After 24 h, cells were subjected to puromycin (1.0 µg/ml) selection. GFP expression within the transfected 293T cells was analyzed on flow cytometry, after 7–9 days of puromycin selection. Relative retrotransposition frequency in the absence of APOBEC proteins (vector) was set as 1.0. The histogram bars represent the mean of three independent cultures, and the standard deviation is shown. (B) Western blot analysis was performed by using extracts from 293T cells transfected by the expression plasmids for APOBEC family proteins and detected by using antibodies specific for the epitopes present in the test proteins. (C) The transposition frequency of L1 in 293T cells co-transfected with 1.5 µg of pL1RP-EGFP along with the expression plasmids for APOBEC family proteins as described in (A), except for that the amounts of APOBEC-expression plasmids were varied (0.5, 0.25 and 0.125 µg). The histogram bars represent the mean of three independent cultures, and the standard deviation is shown. (D) Western blot analysis of the protein expression levels from the experiment in (C).
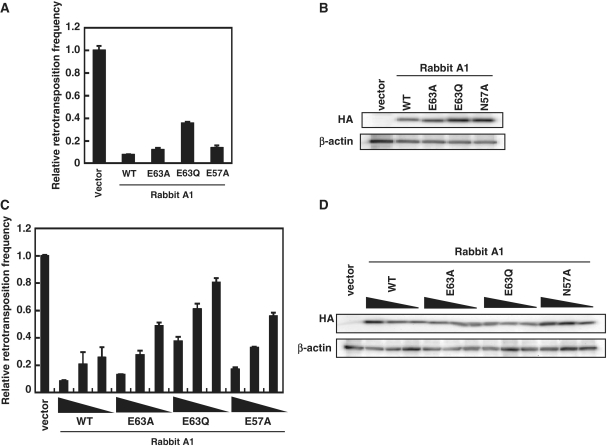
To assess whether the editing activity of A1 is necessary for inhibition of L1 retrotransposition, we tested the rabbit A1 catalytic active site mutant in which the critical glutamic acid at Position 63 (Figure 2) was changed to glutamine (E63Q; 23). Furthermore, because a murine AID catalytic mutant in which the critical asparagine at Position 51 was changed to alanine (N51A) was reported to result in the complete loss of the DNA deamination activity (38), we introduced similar mutation at N57 (Figure 2) of rabbit A1, a site homologous to N51 of AID, and examined the effect on L1 retrotransposition. Results showed that the E63A, E63Q, E57A mutants and WT proteins were expressed at comparable levels, but the anti-L1 activity of rabbit A1 was moderately, not significantly altered by these mutations (Figure 3 and Supplementary Figure S1). Sequence analysis of HIV-1 genomic RNA in the presence of rabbit A1 WT or N57A mutant revealed that the editing activity of E57A mutant was almost disappeared (Supplementary Figure S2 and Table S1). Therefore, we concluded that the inhibitory activities of A1s against the L1 retrotransposition are not largely dependent on cytidine-deaminating activity.
Figure 2.
Comparison of the predicted amino acid sequence of mammalian A1 proteins. Amino acids sequence alignment of A1 from primates (human; GenBank accession number NM001644), mustelas (ferret; AB425821), lagomorphs (rabbit; U10695) and rodents (hamstar; AF176577, rat; NM012907 and mouse; NM031159). The numbers are predicted amino acid residue positions. The putative bipartite nuclear targeting signals are shown in dark gray boxes. The active site core motif in the cytidine deaminase domain is indicated in gray boxes. The amino acid residue Pro(P)-29, which is shown to be critical for the nuclear localization (42), is denoted by dot. The amino acid residue Asn(N)-57, a site homologous to Asn(N)-51 of murine AID, which is shown to be critical for the DNA deamination activity (38), is denoted by reverse triangle. The Glu(E)-63 residue of active site serving an essential role in catalysis as a proton shuttle, is indicated by asterisks.
Figure 3.
Inhibition of L1 retrotransposition by rabbit A1 with catalytic site mutations. (A) 293T cells were co-transfected with 0.5 µg of the expression plasmids for rabbit A1 catalytic site mutants and 1.5 µg of pL1RP-EGFP. GFP expression within the transfected 293T cells were analyzed on flow cytometry as described. Relative retrotransposition frequency in the absence of APOBEC proteins (vector) was set as 1.0. The histogram bars represent the mean of three independent cultures, and the standard deviation is shown. (B) Western blot analysis was performed by using extracts from 293T cells transfected by the expression plasmids for rabbit A1 catalytic site mutants and detected by using antibodies specific for the epitopes present in the test proteins. (C) The transposition frequency of L1 in 293T cells co-transfected with 1.5 µg of pL1RP-EGFP along with decreasing amounts (0.5, 0.25 and 0.125 µg) of the expression plasmids for rabbit A1 catalytic site mutants as described in (A). The histogram bars represent the mean of three independent cultures, and the standard deviation is shown. (D) Western blot analysis of the protein expression levels from the experiment in (C).
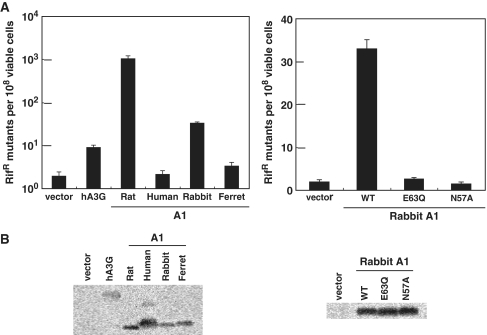
A1s from rat and rabbit exhibit DNA mutator activities in bacteria
Expression of A1 protein from rat in E. coli has previously been shown to greatly enhance the frequency of the E. coli RNA polymerase B gene (rpoB) mutations (39,40). Mutations in rpoB are then detected by screening for the frequency of Rifr colonies. This demonstrated that the apoB mRNA editing enzyme A1 from certain mammalian species can act as a powerful DNA mutator which edits DNA cytidine in bacteria. This previously described DNA mutation assay in bacteria was used to test whether A1s from small animal species have the ability to edit dC residues to dU on single-stranded DNA templates. Consistent with previous observations (35,39), expression of rat A1 and hA3G greatly (1000-fold) and modestly (8.9-fold) enhance, respectively, the frequency of rpoB mutations. Similarly, expression of A1 from rabbit was found to increase the number of Rifr colonies by 33-fold, while A1 from human and ferret had a negligible effect (Figure 4). Furthermore, at comparable expression levels, the rabbit A1 catalytic site mutants E63Q and N57A failed to elicit activity in E. coli (Figure 4). These data strongly suggest that DNA mutator activity of A1s from rat and rabbit in bacteria is cytidine-deaminase dependent.
Figure 4.
Mutations of E. coli genomic DNA by various A1 proteins. (A) The abilities of APOBEC family proteins to enhance mutagenesis levels in bacteria were analyzed. Plasmid encoding the indicated proteins was introduced into bacteria, and their expression was induced. The level of mutagenesis was then assessed by plating the bacteria on medium containing rifampicin and counting the number of RifR colonies. The median mutation frequencies of twelve independent cultures with the standard deviation are indicated as the number of RifR colonies per viable cells. (B) Western blotting of APOBEC family proteins expression in the bacterial strain analyzed in (A) was performed using an HA-tag-specific antibody.
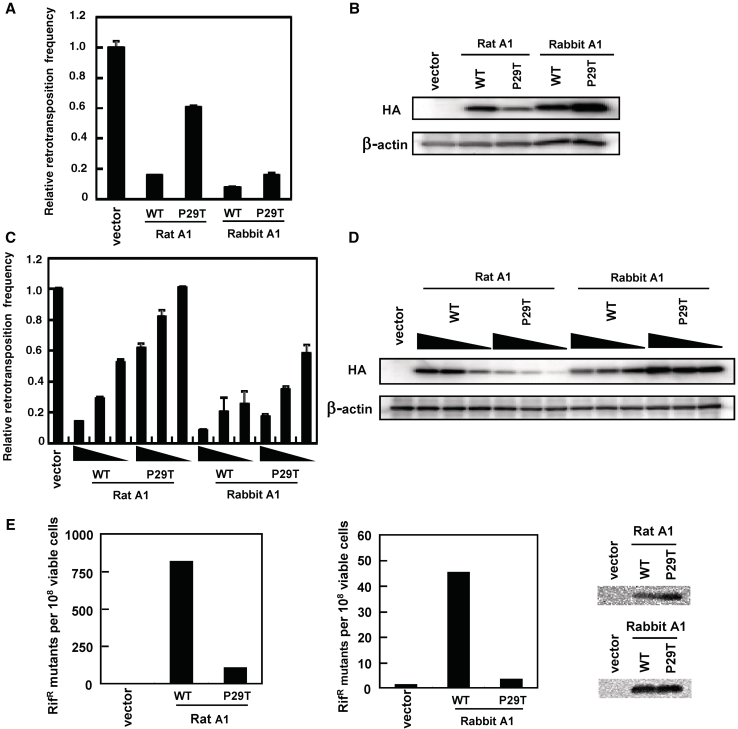
Subcellular localization mutants of A1s inhibit retrotransposition of L1
Previous studies demonstrated both nuclear and cytoplasmic localization of A1s in transfected cells (41,42). The nuclear localization signal (NLS) of A1 interacts with importin α, while the C-terminal leucine-rich nuclear export signal (NES) associates with exportin 1 during transport. Mutation of proline 29 to threonine (P29T; Figure 2) results in the abolishment of nuclear targeting (42). To examine the effects of subcellular localization on the anti-L1 activities of A1s, expression plasmids encoding P29T mutant forms of rat and rabbit A1 proteins were generated and co-transfected into 293T cells with the pL1RP-EGFP. Results showed that while the ability of the rat A1 P29T mutant to restrict the mobility of L1 was severely impaired, the P29T mutant forms of rabbit A1 were equally active against L1 (Figure 5A). However, western blot analysis of lysates from 293T transfected cells revealed a large discrepancy between the steady-state levels of the two proteins, with much lower levels of rat A1 P29T protein expression (Figure 5B, upper panel). When increasing amounts of rat A1 P29T expression plasmid were transfected into 293T cells together with a fixed amount of the pL1RP-EGFP, such that the level of expression is similar to that of wild-type (0.5 µg P29T plasmid), the rat A1 mutant P29T protein displayed ∼2-fold inhibition (Figure 5C and D). Furthermore, when 2.0 µg plasmid was used, the rat A1 mutant P29T inhibited L1 to the same extent as the wild-type (Supplementary Figure S1B). Thus, the inability of rat A1 P29T to restrict L1 retrotransposition appears to reflect low protein expression levels in transfected cells, and that the significantly different expression levels of the rabbit and rat A1 mutant P29T may explain the observed disparity in their anti-L1 activity.
Figure 5.
Effect of rat and rabbit A1 with subcellular localization mutations on L1 retrotransposition. (A and C) 293T cells were co-transfected with 1.5 µg of pL1RP-EGFP together with 0.5 µg of the expression plasmids for HA-tagged rat and rabbit A1 (WT; wild-type) or their subcellular localization mutants (P29T). The decreasing amounts (0.5, 0.25 and 0.125 µg) of APOBEC expression plasmids were used in (C). EGFP-based L1 retrotransposition assay was performed as described. Relative retrotransposition frequency in the absence of APOBEC proteins (vector) was set as 1.0. The histogram bars represent the mean of three independent cultures, and the standard deviation is shown. (B and D) Western blot showing expression of the HA-tagged proteins from a representative experiment from (A and C). (E) Escherichia coli-based RifR mutation assay was carried out using rat and rabbit A1 or their subcellular localization mutants. The median mutation frequencies of twelve independent cultures with the standard deviation are indicated as the number of RifR colonies per viable cells. Western blotting of HA-tagged protein expression in the bacterial strain analyzed was performed using an HA-tag-specific antibody.
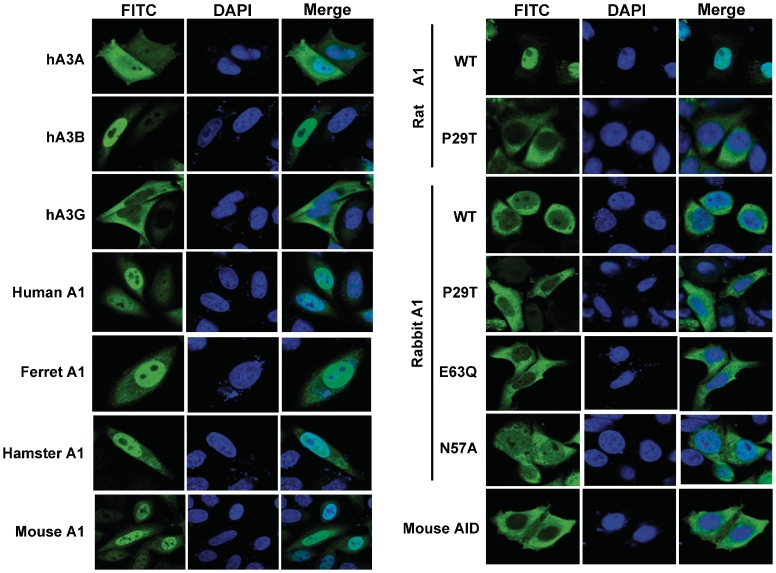
We next confirmed the intracellular localization of the wild-type and P29T mutant rat and rabbit A1 proteins transiently expressed in HeLa cells, with hA3A, hA3B, hA3G and human A1 serving as controls. Consistent with previous reports, hA3A and human A1 exhibited both nuclear and cytoplasmic localizations (12,41,42), while hA3B was found exclusively in the nucleus (11) and hA3G localized predominantly in the cytoplasm (Figure 6). Nucleus and cytoplasm localizations were also observed for ferret A1, mouse A1, hamster A1 and catalytic mutants of rabbit A1 (E63Q and N57A), whereas the murine AID protein was found exclusively in the cytoplasm (Figure 6).
Figure 6.
Subcellular localization of the APOBEC family proteins. HA-tagged APOBEC proteins were expressed in HeLa cells, fixed and immunofluorescence staining was performed using an HA-tag-specific antibody and a FITC-conjugated goat anti-mouse IgG (in green). Cell nuclei were visualized by 4,6-diamidino-2-phenylindole (DAPI) staining (in blue), and the samples staining by confocal microscopy.
For both wild-type and P29T mutant A1 from rabbit, localization is largely cytoplasmic, but wild-type A1 from rat exhibited profound nuclear accumulation that was abolished with the P29T mutation (42). This latter observation for the P29T mutant A1 from rat suggests that the anti-L1 activity of A1s is largely not affected by subcellular localization of the proteins.
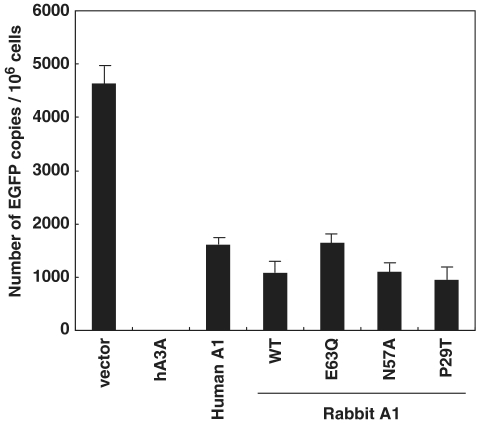
A1 suppress de novo L1 DNA synthesis
Our findings thus far suggested that A1s from multiple mammalian species have the potential to attenuate L1 retrotransposition. However, how A1 acts against the retrotransposition of L1 remains unclear, and it is not known whether the anti-L1 function involves additional cellular co-factor(s). We, therefore, measured the presumed copy number of retrotransposed L1 elements by performing real-time PCR using total DNA which was isolated from the cells used in Figures 1, 3 and 5 in order to determine if A1 is indeed effective on retrotransposition inhibition. As noted above, the retroposition cassette contains an intron in the EGFP gene that can only be removed during a retrotransposition event. A real-time PCR targeting the intronless EGFP, therefore, should distinguish the EGFP-based L1 retrotransposon indicator construct and any endogenous L1 DNA that might have been newly synthesized in the cells from a de novo pL1RP-EGFP-mediated retrotransposition event (16). As shown in Figure 7, EGFP could be amplified in cells transfected with pL1RP-EGFP and the empty vector control. In contrast, when pL1RP-EGFP was transfected into the cells together with hA3A expression plasmid, only background levels of EGFP were detected. Analysis using human A1, rabbit A1 wild-type, catalytic mutants (E63Q and N57A) and subcellular localization mutant (P29T) showed that results obtained with this assay were largely consistent with those observed in flow cytometry analysis (Figures 1, 3 and 5). Thus, results obtained by detecting not only integrants (Figure 7), but also reverse transcripts are similar to those in which levels of EGFP expression represent L1 retrotransposition (Figures 1, 3 and 5). We, therefore, conclude that A1 as well as hA3A are indeed able to inhibit the accumulation of nascent L1 DNA, suggesting interference with L1 reverse transcription and/or integration or intracellular movement of L1 ribonucleoprotein (RNP). Furthermore, the suppressive activity against de novo L1 DNA synthesis was mainly in a deamination-independent manner, and was not affected by subcellular localization of the proteins.
Figure 7.
Real-time PCR targeting spliced EGFP genes. Total cellular DNA was extracted at 6–7 days post-transfection from the cells used in Figures 1 and 3, subjected to real-time PCR analysis using a probe designed for the detection of spliced EGFP. Histgram bars represent the mean of three independent experiments, and the standard deviation is shown.
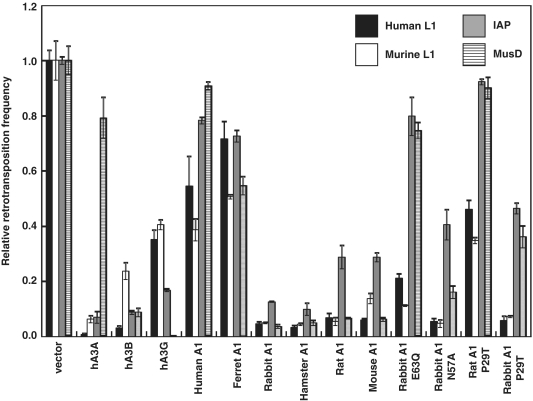
A1s from multiple mammalian species can also inhibit the retrotransposition of murine L1
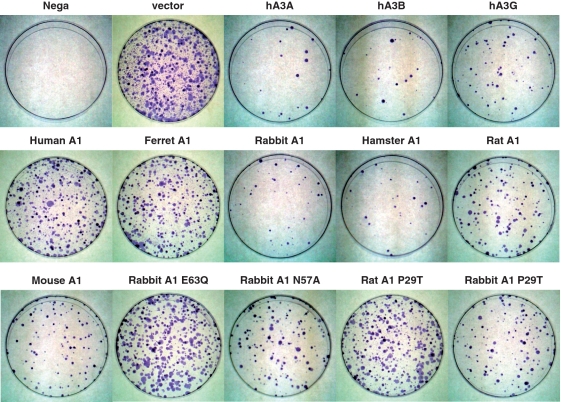
To confirm results of the EGFP-based L1 retrotransposition assay, we next performed a different type of retrotransposition assay, using L1 vector which carries neor gene instead of the EGFP gene. L1 retrotransposition events could be detected upon G418 selection of the target HeLa cells, with no G418R clones detected with the control plasmid (data not shown). As shown in Figure 8 and Supplementary Figure S3A, results obtained with the neor-based retrotransposition assay using human L1 pCEP4/L1mneoI/ColE1 plasmid are consistent with those seen using EGFP-based assay. Mouse L1s are generally homologous to human L1s but differ significantly in their 5′UTR and the 5′ half of ORF1. Thus, the effect of A1 proteins on the retrotransposons of mouse L1 was examined using murine L1 pCMV L1Md-Gf21neoTET plasmid. As expected, hA3G caused modest decreases in retrotransposition of both human and murine L1s, and more pronounced impairment in retrotransposition of L1s were observed when hA3A and hA3B were co-transfected (Figure 8 and Supplementary Figure S3B). A modest reduction in the mobility of human L1 was also found with murine AID (Figure 1 and Supplementary Figure S1A). These findings are in agreement with recent analysis by MacDuff et al. (21), showing that AID from a panel of vertebrates including chicken and zebrafish is able to inhibit the replication of L1 and MusD through a DNA deamination-independent mechanism. A1s from rabbit and rodents were found to markedly impair retrotransposition of both human and murine L1 (Figure 8 and Supplementary Figure S3A and B). Analysis using rabbit A1 catalytic mutants (E63Q and N57A) showed that the cytidine deaminase activity is not required for the anti-L1 activity. Furthermore, we revealed that the inhibition of L1 retrotransposition was not affected by subcellular localization of A1 proteins. These multiple lines of investigations indicate that, similar to hA3A, hA3B and hA3G, A1s from multiple mammalian species can function on both human and murine L1, presumably through a deamination-independent mechanism.
Figure 8.
Effects of APOBEC family proteins on a range of autonomous retroelements using neor-based retrotransposition assay. HeLa cells were co-transfected with 1.2 µg of L1 retrotransposition indicator construct human L1 pCEP4/L1mneoI/ColE1, murine L1 pCMV L1Md-Gf21neoTET, MusD retrotransposition indicator construct murine MusD pCMV Mus-6DneoTNF or 0.6 µg of IAP, murine IAP retrotransposition indicator construct pGL3-IAP92L23neoTNF, together with 0.4 or 0.2 µg of respective APOBEC expression plasmids and 0.4 µg of pIRES-EGFP vector. After 72 h, cells were trypsinized, re-seeded onto 100 mm dishes, and subjected to G418 (0.5, 0.75 or 1.0 mg/ml) selection. After 12–14 days of selection, resultant G418R colonies were fixed, stained with crystal violet and counted to determine the level of autonomous retrotransposition. Retrotransposition frequency was calculated as the number of G418R clones/transfection efficiencies (percent of GFP+ cells). Data are presented by the values relative to samples containing retroelement alone (defined by quantifying G418R clones). The average of three experiments with standard deviation is indicated.
A1s from multiple mammalian species can inhibit the retrotransposition of murine IAP and MusD
A3s have been demonstrated to restrict the intracellular transposition of a series of retroelements, including the autonomous retrotransposons that bear LTR such as murine IAP and MusD sequences, and this restriction appears to be accompanied by hypermutation in the retroelement genome that are attributed to the cytidine deaminase activity of the A3 proteins (33,43). To examine the ability of A1s from multiple mammalian species to inhibit the murine IAP and MusD in human cells, we co-transfected HeLa cells with expression plasmids encoding various AID/APOBEC family proteins and the previously described IAP and MusD retrotransposition indicator constructs IAP pGL3-IAP92L23neoTNF, murine MusD pCMV Mus-6DneoTNF. This constructs contain full-length IAP and MusD genome with the neor gene inserted in the antisense orientation, which is spliced out of the RNA intermediate, resulting in a functional gene after reverse transcription and integration (43).
As demonstrated in Figures 8 and 9, hA3A, hA3B and hA3G expression indeed reduced IAP retrotransposition significantly, as previously described (43). Interestingly, MusD was found to be more sensitive to hA3G, but relatively resistant to hA3A than IAP. The A1s from rabbit and rodents were found to be highly active against the retrotransposition of IAP and MusD, reducing their mobility >100-fold in some experiments. In contrast to A1s from rabbit and rodent, A1s from human and ferret exhibited no significant activity against IAP and MusD retrotransposition. Furthermore, in contrast to the activity against the L1 retrotransposition, the ability of the catalytic mutants (E63Q, N57A) of rabbit A1 to restrict the IAP and MusD retrotransposition were significantly, but not completely impaired, suggesting the existence of a deaminase-independent restriction mechanism by A1. To verify the findings with the rabbit A1 mutant, hamster, rat and mouse A1 catalytic mutants E63A and E63Q were constructed and their inhibitory activity against retrotransposition of human L1 pCEP4/L1mneoI/ColE1, or murine MusD pCMV Mus-6DneoTNF was examined.
Figure 9.
Inhibition of murine IAP LTR retrotransposons by A1 proteins. HeLa cells were co-transfected with the expression plasmids for APOBEC family proteins and the neomycin-resistant (neor)-based murine IAP reporter vector pGL3-IAP92L23neoTNF along with pIRES-EGFP. Cells were subjected in G418 and resistant colonies counted 12–14 days after transfection. This experiment is representative of the data compiled in Figure 8.
Results showed that the ability of these catalytic mutants to restrict the MusD retrotransposition was significantly impaired, while the activity against the L1 retrotransposition was relatively conserved (Supplementary Figures S3D, E and 4 A). Hyperediting of MusD genome by rat and rabbit A1s was examined by differential DNA denaturation PCR (3D-PCR) (24) to further demonstrate a deaminase-dependent restriction mechanism. Sequencing of cloned 3D-PCR products obtained with rat and rabbit A1 showed that guanosine bases were extensively edited, yielding G–A hypermutations in plus-strand DNA (Supplementary Figure S5). On the other hand, sequence analysis of de novo L1 insertions reveals no obvious editing (Supplementary Figure S6). Interestingly, the ability of the rabbit A1 P29T mutant to restrict the mobility of IAP and MusD were severely impaired, although this rabbit A1 P29T mutant was as active as wild-type against both human and murine L1. The inhibitory activities against murine IAP and MusD observed in A1s from rabbit were based on, at least in part, cytidine-deaminating activity on IAP and MusD proviral DNA, since rabbit A1 P29T mutant exhibit a negligible DNA mutator activities in bacteria (Figure 5E). On the other hand, the inability of rat A1 P29T to restrict murine IAP and MusD retrotransposition appears to reflect low protein expression levels in transfected cells (Figure 5 and Supplementary Figure S4B), as discussed above.
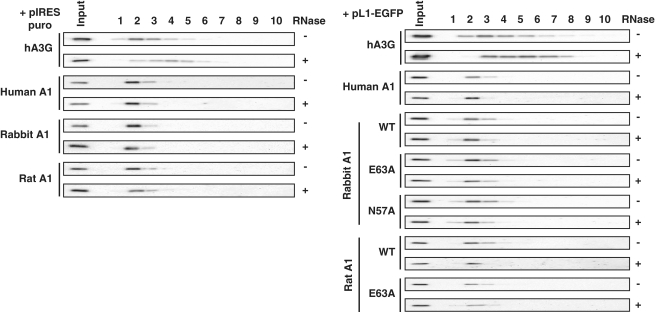
Formation of intracellular high-molecular-mass A1 complexes
It was previously reported that hA3G can interact with cellular mRNAs (44–47) to form high-molecular-mass (HMM) RNP complexes (46,48) and to associate with stress granules, staufen granules, or P bodies. However, it had been reported that the inhibitory activity of human A3 proteins against L1 retrotransposition does not correlate with intracellular HMM formation or P-body association (18). To explore A1 proteins are associated with HMM complexes, the expression plasmids encoding various A1s carrying C-terminal HA-tag, with hA3G (which are also C-terminal HA epitope-tagged) serving as controls, were co-transfected into 293T cells in the absence or presence of the L1 retrotransposition indicator construct pL1RP-EGFP. After 24 h, the transfected cells were lysed and subjected to FPLC analysis. To test RNase sensitivity, the cell lysates were treated with RNase A, before fractionation. Each eluted fraction was subjected to western blotting with HA antibody. Consistent with previous observations (46,48), hA3G assembled into HMM complexes that converts to a low-molecular-mass (LMM) form by RNase treatment (Figure 10), A1 proteins were found to exist in a HMM form both in the absence or presence of pL1RP-EGFP (Figure 10). Notably, and in sharp contrast to hA3G, A1 distribution was not affected by RNase treatment (Figure 10), suggesting that this single-domain cytidine deaminase may be able to interact differently and/or more strongly with host RNA(s).
Figure 10.
Formation of HMM A1 complexes. 293T cells were co-transfected with the expression plasmids for APOBEC family proteins and the L1 retrotransposition indicator construct pL1RP-EGFP or pIRES-EGFP. After 24 h, the transfected cells were lysed and subjected to FPLC analysis. To test RNase sensitivity, the cell lysates were treated with RNase A at 37°C for 1 h, before fractionation. The each eluted fraction was subjected to western blot with HA antibody.
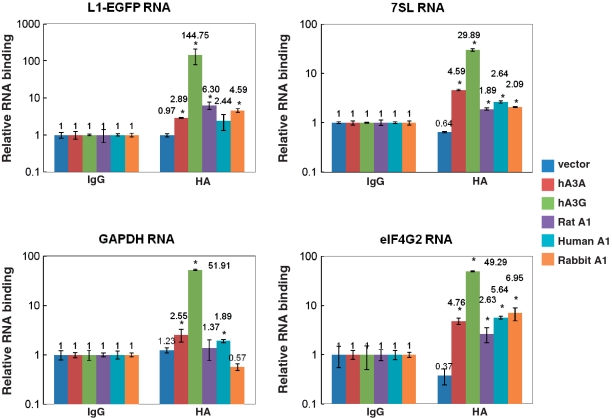
Selective interaction of A1 proteins with L1 and cellular RNAs
To examine the physical association of A1 protein with L1 and cellular RNAs, RNA immunoprecipitation assay was carried out as previously described (36), with minor modification. 293T cells were co-transfected with the expression plasmids for APOBEC family proteins carrying C-terminal HA-tag and the L1 retrotransposition indicator construct pL1RP-EGFP, cross-linked and then immunoprecipitated with anti-HA antibody conjugated to Dynabeads. Cell lysates from transfected 293T cell lysates were also immunoprecipitated side by side with control IgG as the negative control to determine non-specific binding of various RNAs in the assay system. The precipitated RNAs with APOBEC proteins were extracted from co-precipitated samples and analyzed by qRT–PCR with primers specific for L1-EGFP, 7SL RNA, GAPDH and eIF4G2(NAT1; novel APOBEC-1 target no.1). The latter has been identified as a novel translational repressor mRNA that is extensively edited at multiple sites in rabbit A1 transgenic mice (37). Copy numbers of these co-precipitated RNAs with control IgG were then set as 1, and the graphs indicate RNA copy numbers relative to the control. An efficient interaction of hA3G with L1-EGFP RNA was detected in this assay system (Figure 11). Furthermore, the efficient interactions between hA3G and Polymerase III (Pol III)-derived 7SL RNA, Polymerase II (Pol II)-transcribed GAPDH, and elF4G2 RNA were also detected. A relatively efficient interaction was also detected between A1s from multiple mammalian species with L1-EGFP RNA and elF4G2. However, the copy number of L1-EGFP RNA co-precipitated with rat and rabbit A1s were 23- and 32-fold lower than those seen with hA3G (Figure 11). It is possible that this weaker interaction can be explained by the fact that A1s are single-domain cytidine deaminases, while hA3G have double deamination domains. Although we show that rat and rabbit A1s were able to interact with L1 RNA, further studies will be required to verify the exact step(s) of L1 replication affected by these APOBEC family proteins.
Figure 11.
Selective interaction of APOBEC family proteins with L1 and cellular RNAs. 293T cells were co-transfected with the expression plasmids for APOBEC family proteins and the L1 retrotransposition indicator construct pL1RP-EGFP, cross-linked and then immunoprecipitated with control IgG or anti-HA antibody conjugated to Dynabeads. The precipitated RNAs with APOBEC proteins were extracted from co-precipitated samples and analyzed by qRT-PCR with primers specific for L1-EGFP, 7SL RNA, GAPDH and eIF4G2. The graphs indicate RNA binding relative to IgG control. The histogram bars represent the mean of three independent experiments, and the standard deviation is shown. *P < 0.05.
DISCUSSION
A large portion of the mammalian genome is composed of endogenous retrotransposons, with L1 elements contributing to over 35% of the mass of the human and mouse genomes (49,50). Around 100 copies of L1 elements in humans and 3000 copies in mice appear to be active (29,51,52). These elements modified mammalian genomes not only by creating insertions, but also by indirect replication of short interspersed elements (SINEs) and processed pseudogenes. L1 retrotransposition results in human diseases such as Haemophiliae VIII/IX and Duchene muscular dystrophy, and in the generation of novel polymorphisms (53,54). Obviously, cellular machineries have evolved to support a balance between new L1 insertions that cause deleterious gene disruptions and those that confer beneficial genetic diversity.
Members of the A3 cytidine deaminases are almost certainly one class of the cellular machineries that play roles as innate restriction factors against endogenous retroelements (1,2). Here, we demonstrate that mammalian A1s from multiple species are also capable of inhibiting the replication of the autonomous non-LTR and LTR retrotransposons in cell culture-based retrotransposition assays.
It has been indicated that A3 gene(s) arose from an AID-like ancestral gene through a series of gene duplication and diversification (5,10,55,56). The A3 genes are specific to placental mammals and non-placental mammals such as opossum and platypus genomic sequences appear to lack A3 genes (55). However, marsupial opossum has been reported to express functional A1 gene (57). Thus, the studies of the restriction activities of A1 proteins against retroelements should be required to fully understand the complex evolutionary history of APOBEC genes in intrinsic immunity.
Human APOBEC3A (hA3A) and APOBEC3B (hA3B) were found to be the most potent inhibitors of the L1 retrotransposon, whereas only hA3B inhibits HIV-1 replication. The mechanism of anti-retroviral and anti-retrotransposon potency differ and the latter appears to be independent of the enzymatic activity (14). Whether these A3 proteins bind to L1 encoded open reading frames ORFs and/or specific sequences in L1 RNA remains open. A ∼6 kb L1 element contains an internal promoter in the 5′ UTR (58), followed by ORF1 and ORF2 (59–62). ORF1 encodes RNA-binding protein that has nucleic acid chaperone activity (63–65) and is required for cytoplasmic RNP particle formation and for downstream steps in L1 retrotransposition (66). ORF2 encodes protein with endonuclease and reverse transcriptase activities, both ORF1 and ORF2 are critical for retrotransposition by a ‘copy and paste’ mechanism (59,60). L1 DNA synthesis in the nucleus is based on ‘target-primed reverse transcription (TPRT)’ in which ORF2p nicks target chromosomal DNA, using the resultant 3′-OH to prime the reverse transcription of L1 RNA as a template (67,68). At an early phase before TPRT, L1 encoding polyadenylated RNA forms a RNP complex as a retrotransposition intermediate by associating with ORF1p and ORF2p in the cytoplasm (61,62,69,70). It was recently reported that A3s do not require direct interactions with ORF1p for inhibiting the L1 retrotransposition (71). On the other hand, the interactions of ORF2p with A3 proteins have not been addressed, thus far, since ORF2p was shown to be difficult to detect within the cells (72). Thus, the exact step of L1 retrotransposition affected by A3 molecules is still unknown.
It is possible that A1 proteins interact with L1 encoded ORFs, specific sequences in L1 RNA and/or host proteins that facilitate retrotransposition important for initial steps in TPRT. A1 proteins might be able to interact with the cytoplasmic RNPs and interfere with subsequent RNP transport and/or nuclear import. Alternatively, A1 may function at other steps downstream in the L1 retrotransposition pathway; A1 in the nucleus might be able to access to L1 RNA and block the generating nick or subsequent priming of reverse transcription of the first strand. Furthermore, subsequent step(s) of TPRT such as the second (+) strand DNA synthesis or target site duplication formation might be blocked. We show in this study that the anti-L1 activity of A1 was not largely affected by subcellular localization of the proteins. A1 protein without nuclear localizing signal might be able to enter the nucleus with the RNP complex in which ORF2p harbors a putative NLS (72,73). The exact step(s) of L1 replication affected by these APOBEC cytidine deaminases remains to be verified, in order to clarify the molecular mechanism through which L1 retrotranspositions are inhibited, mainly by the deamination-independent manner.
In this study, the anti-L1 activity of A1 from multiple species was found to be variable, despite similar expression levels. These inter-species variations may be attributed to multiple factors such as interaction with cofactors and protein stability.
Despite the impact of L1 insertion on mammalian genome evolution, much of the process of L1 retrotransposition, especially in vivo remains unexplored. Previously, L1 mRNA expression has been documented infrequently in differentiated tissues and predominantly in germ cells (74,75), consistently L1 retrotransposition in vivo was thought to occur mainly in germ cells (76,77). The L1 transcription has been shown to be activated by the binding of the transcriptional factors such as YY1, SOX-11 and RUNX3 to the 5′ UTR (78–80). This L1 promoter-driven transcription appears to be repressed by the binding of the methyl-CpG-binding protein 2 (MeCP2) and the CpG methylation, followed by the heterochromatination, in differentiated somatic tissues (81). However, more recent studies suggested the de novo L1 retrotransposition may usually occur early in embryogenesis, not in germ cells, and be essential for early embryo preimplantation developement. L1 RNA assembled into its RNP complex appears to be stable and play a role in creating genome diversity by being carried over through fertilization and integrate during embryogenesis (82,83). This scenario indicated that the germ cells should have evolved several post-transcriptional defense mechanisms that strictly prevent L1 integration into the genome. These might include the post-transcriptional silencing via RNA interference (RNAi) (84,85) and cytidine deamination via the A3 family-mediated machinery (13,33,86) as well. Combined with these formally documented defense mechanisms, our data in this study indicate that the A1-mediated machinery may contribute to control L1 retrotransposition in germ cells.
Although the expressions of A1 in human have been formally documented only in gastrointestinal tissue thus far, there is a much larger tissue distribution for the mouse, rat and rabbit A1s, including tissues such as liver and spleen which were assumed to have little apoB mRNA (87–91). Accordingly, there are two promoters for distinct transcripts for the mouse, whereas there is a single promoter in humans (89,92,93). We confirmed that A1 mRNA is expressed in ovary and testis in mouse and rabbit (Supplementary Figure S7), placing A1 in a compartment where L1 retrotransposition may have the greatest impact in vivo (25,94,95).
The replication cycle of the endogenous retroviruses IAP and MusD is different from that of non-LTR retrotransposon L1s, in which reverse transcription occurs within the nucleus based on TPRT. We found that the retrotransposition of these endogenous retroviruses were also sensitive to A1s from multiple mammalian species. Similar to L1 expression profiles as discussed above, it has been documented that the genome of endogenous retrovirus is expressed predominantly in germ cells and generally methylated in most differentiated somatic tissues (96,97). It was suggested that the de novo IAP and MusD retrotransposition may usually occur in both germ cells and early in embryogenesis (77,98). Collectively, our functional data in this study, together with evidence for A1 expression in these tissues, combined to suggest that A1 plays role in the intrinsic immune mechanisms for preventing the spread of foreign and endogenous nucleic acids in addition to its integral roles in apoB mRNA editing. Obviously, uncontrolled expression or transposition of these autonomous retrotransposons is deleterious for the host by causing deleterious gene disruptions. In contrast, appropriate levels of retrotransposition in germ-line cells or early in embryogenesis might contribute to beneficial genetic diversity and host genome evolution.
Overall the spectrum of biological function of the AID/APOBEC family (comprising AID, A1, A2, A3 subgroups, A4 and more) of DNA/RNA cytidine deaminases in vertebrates is expanding. The several members of AID/APOBEC family inhibit the mobility of endogenous retroelements as well as the retroviral infectivity through both editing and non-editing mechanisms. These antiviral activities of A1s from several mammalian species provide insights into the evolution and diversification of AID/APOBEC family in mammalian species. Elucidation of the multitude of activities and viruses targeted by these AID/APOBEC family proteins will contribute to the understanding of how these DNA/RNA cytidine deaminases protect host genomes from invading nucleic acids.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Higo Bank; The Cooperative Research Project on Clinical and Epidemiological Studies of Emerging and Re-emerging Infectious Diseases; Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists and Grant-in-Aid for Research Activity Start-up from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) (to T.I.); Research and Education Program for Development of Therapy of Emerging and Reemerging Infectious Diseases Including AIDS; Global Centers of Excellence (COE) program Global Education Research Center Aiming at the Control of AIDS. Funding for open access charge: Higo Bank.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank C. Cheng-Mayer for critical reviewing the article and S. Harada for supports. We also thank K. Fukuda for helpful assistance; T. Izumi, K. Monde and Y. Ishizaka for discussions; E.T. Luning Prak, N. Gilbert and the National Institutes of Health AIDS Research and Reference Reagent Program. Division of AIDS, NIAID, National Institutes of Health for regents used in this research.
REFERENCES
- 1.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 2.Malim MH, Emerman M. HIV-1 accessory proteins-ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 4.Goff SP. Death by deamination: a novel host restriction system for HIV-1. Cell. 2003;114:281–283. doi: 10.1016/s0092-8674(03)00602-0. [DOI] [PubMed] [Google Scholar]
- 5.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 6.Chester A, Scott J, Anant S, Navaratnam N. RNA editing: cytidine to uridine conversion in apolipoprotein B mRNA. Biochim. Biophys. Acta. 2000;1494:1–13. doi: 10.1016/s0167-4781(00)00219-0. [DOI] [PubMed] [Google Scholar]
- 7.Keegan LP, Gallo A, O`Connell MA. The many roles of an RNA editor. Nat. Rev. Genet. 2001;2:869–878. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- 8.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 9.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 10.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger M. Evolution of the AID/APOBEC family of polynucleotide (deoxy) cytidine deaminases. Mol. Biol. Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 11.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O’Shea KS, Moran JV, Cullen BR. Cellular inhibitor of long interspersed element 1 and Alu retrotransposition. Proc. Natl Acad. Sci. USA. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 13.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Löwer J, Cichutek K, Flory E, Schumann GG, Münk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 14.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 15.Hulme A, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T, Sata T, Tokunaga K. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;35:2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 18.Niewiadomska AM, Tian C, Tan L, Wang T, Sarkis PTN, Yu XF. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J. Virol. 2007;81:9577–9583. doi: 10.1128/JVI.02800-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gourzi P, Leonova T, Papavasiliou FN. A role for activation-induced cytidine deaminase in the host response against a transforming retrovirus. Immunity. 2006;24:779–786. doi: 10.1016/j.immuni.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 20.MacDuff DA, Harris RS. Directed DNA deamination by AID/APOBEC3 in immunity. Curr. Biol. 2006;16:R186–189. doi: 10.1016/j.cub.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 21.MacDuff DA, Demorest ZL, Harris RS. AID can restrict L1 retrotransposition suggesting a dual role in innate and adaptive immunity. Nucleic Acids Res. 2009;37:1854–1867. doi: 10.1093/nar/gkp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishop KN, Holmes RK, Sheehy AM, Malim MH. APOBEC-mediated editing of viral RNA. Science. 2004;305:645. doi: 10.1126/science.1100658. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda T, Ohsugi T, Kimura T, Matsushita S, Maeda Y, Harada S, Koito A. The anti-retroviral potency of APOBEC1 deaminase from small animal species. Nucleic Acids Res. 2008;36:6859–6871. doi: 10.1093/nar/gkn802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petit V, Guétard D, Renard M, Keriel A, Sitbon M, Wain-Hobson S, Vartanian JP. Murine APOBEC1 is a powerful mutator of retroviral and cellular RNA in vitro and in vivo. J. Mol. Biol. 2009;385:65–78. doi: 10.1016/j.jmb.2008.10.043. [DOI] [PubMed] [Google Scholar]
- 25.Prak ET, Dodson AW, Farkash EA, Kazazian HH., Jr Tracking an embryonic L1 retrotransposition event. Proc. Natl Acad. Sci. USA. 2003;100:1832–1837. doi: 10.1073/pnas.0337627100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 27.Dewannieux M, Dupressoir A, Harper F, Pierron G, Heidmann T. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat. Genet. 2004;36:534–539. doi: 10.1038/ng1353. [DOI] [PubMed] [Google Scholar]
- 28.Ribet D, Dewannieux M, Heidmann T. An active murine transposon family pair: retrotransposition of ‘master’ MusD copies and ETn trans-mobilization. Genome Res. 2004;14:2261–2267. doi: 10.1101/gr.2924904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodier JL, Ostertag EM, Du K, Kazazian HH., Jr A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esnault C, Casella JF, Heidmann T. A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells. Nucleic Acids Res. 2002;30:E49. doi: 10.1093/nar/30.11.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohsugi T, Koito A. Human T cell leukemia virus type I is resistance to the antiviral effects of APOBEC3. J. Virol. Methods. 2007;139:93–96. doi: 10.1016/j.jviromet.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 32.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 33.Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 34.Duncan BK, Weiss B. Specific mutator effects of ung (Uracil-DNA glycosylase) nutations in Escherichia coli. J. Bacteriol. 1982;151:750–755. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haché G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- 36.Maeda K, Singh SK, Eda K, Kitabatake M, Pham P, Goodman MF, Sakaguchi N. GANP-mediated recruitment of activation-induced cytidine deaminase to cell nuclei and to immunoglobulin variable region DNA. J. Biol. Chem. 2010;285:23945–23953. doi: 10.1074/jbc.M110.131441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamanaka S, Poksay KS, Arnold KS, Innerarity TL. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev. 1997;11:321–333. doi: 10.1101/gad.11.3.321. [DOI] [PubMed] [Google Scholar]
- 38.Shivarov V, Shinkura R, Honjo T. Dissociation of in vitro DNA deamination activity and physiological functions of AID mutants. Proc. Natl Acad. Sci. USA. 2008;105:15866–15871. doi: 10.1073/pnas.0806641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 40.Petersen-Mahrt SK, Neuberger MS. In vitro deamination of cytosine to uracil in single-stranded DNA by apolipoprotein B editing complex catalytic subunit 1 (APOBEC1) J. Biol. Chem. 2003;278:19583–19586. doi: 10.1074/jbc.C300114200. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y, Yang Y, Smith HC. Multiple protein domains determine the cell type-specific nuclear distribution of the catalytic subunit required for apolipoprotein B mRNA editing. Proc. Natl Acad. Sci. USA. 1997;94:13075–13080. doi: 10.1073/pnas.94.24.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chester A, Somasekaram A, Tzimina M, Jarmuz A, Gisbourne J, O`Keefe R, Scott J, Navaratnam N. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. EMBO J. 2003;22:3971–3982. doi: 10.1093/emboj/cdg369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esnault C, Millet J, Schwartz O, Heidmann T. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res. 2006;34:1522–1531. doi: 10.1093/nar/gkl054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, Malim MH. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P-bodies and stress granules. J. Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, Heidmann T, Green WC. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl Acad. Sci. USA. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 2006;281:29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- 47.Wichroski MJ, Robb GB, Rana TM. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2:E41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo K, Liu B, Xiao Z, Yu Y, Yu X, Gorelick R, Yu XF. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 2004;78:11841–11852. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 50.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 51.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeBerardinis RJ, Goodier JL, Ostertag EM, Kazazian HH., Jr Rapid amplification of a retrotransposon subfamily is evolving the mouse genome. Nat. Genet. 1998;20:288–290. doi: 10.1038/3104. [DOI] [PubMed] [Google Scholar]
- 53.Hulme AE, Kulpa DA, Garcia Perez JL, Moran JV. Totowa, NJ: Humana Press; 2006. The impact of LINE-1 retrotransposition on the Human genome. [Google Scholar]
- 54.Babushok DV, Kazazian HH., Jr Progress in understanding the biology of the human mutagen LINE-1. Hum. Mutat. 2007;28:527–539. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- 55.LaRue RS, Jónsson SR, Silverstein KAT, Lajoie M, Bertrand D, El-Mabrouk N, Hötzel I, Andrésdóttir V, Smith TPL, Harris RS, et al. The artiodactyls APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol. Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv. Immunol. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- 57.Fujino T, Navaratnam N, Jarmuz A, Von Haeseler A, Scott J. C→U editing of apolipoprotein B mRNA in marsupials: identification and characterization of APOBEC-1 from the American opossum Monodelphus domestica. Nucleic Acids Res. 1999;27:2662–2671. doi: 10.1093/nar/27.13.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 60.Moran JV, Holmes SE, Naas TP, DeBeradinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 61.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 62.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Jr, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kolosha VO, Martin SL. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc. Natl Acad. Sci. USA. 1997;94:10155–10160. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kolosha VO, Martin SL. High-affinity, non-sequence-specific RNA binding by the open reading frame (ORF1) protein from long interspersed nuclear element 1 (LINE-1) J. Biol. Chem. 2003;278:8112–8117. doi: 10.1074/jbc.M210487200. [DOI] [PubMed] [Google Scholar]
- 66.Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum. Mol. Genet. 2005;14:3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 67.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 68.Cost GL, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 70.Hohjoh H, Singer MF. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lovsin N, Peterlin BM. APOBEC3 proteins inhibit LINE-1 retrotransposition in the absence of ORF1p binding. Ann. NY Acad. Sci. 2009;1178:268–275. doi: 10.1111/j.1749-6632.2009.05006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goodier JL, Ostertag EM, Engleka KA, Seleme MC, Kazazian HH., Jr A potential role for the nucleolus in L1 retrotransposition. Hum. Mol. Genet. 2004;13:1041–1048. doi: 10.1093/hmg/ddh118. [DOI] [PubMed] [Google Scholar]
- 73.Kubo S, Seleme MC, Soifer HS, Perez JL, Moran JV, Kazazian HH, Jr, Kasahara N. L1 retrotransposition in non-dividing and primary human somatic cells. Proc. Natl Acad. Sci. USA. 2006;103:8036–8041. doi: 10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Branciforte D, Martin SL. Developemental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol. Cell. Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc. Natl Acad. Sci. USA. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposition. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 77.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 78.Tchénio T, Casella JF, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 2000;28:411–415. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang N, Zhang L, Zhang Y, Kazazian HH., Jr An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 2003;31:4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004;32:3846–3855. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu F, Zingler N, Schumann G, Strätling WH. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res. 2001;29:4493–4501. doi: 10.1093/nar/29.21.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beraldi R, Pittoggi C, Sciamanna I, Mattei E, Spadafora C. Expression of LINE-1 retrotransposons is essential for murine preimplantation development. Mol. Reprod. Dev. 2006;73:279–287. doi: 10.1002/mrd.20423. [DOI] [PubMed] [Google Scholar]
- 83.Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, Kazazian HH., Jr L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-defricient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 86.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Greeve J, Altkemper I, Dieterich JH, Greten H, Windler E. Apolipoprotein B mRNA editing in 12 different mammalian species: hepatic expression is reflected in low concentrations of apoB-containing plasma lipoproteins. J. Lipid Res. 1993;34:1367–1383. [PubMed] [Google Scholar]
- 88.Lau PP, Zhu HJ, Baldini A, Charnsangavej C, Chan L. Dimeric structure of a human apolipoprotein B mRNA editing protein and cloning and chromosomal localization of its gene. Proc. Natl Acad. Sci. USA. 1994;91:8522–8526. doi: 10.1073/pnas.91.18.8522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nakamura M, Oka K, Krushkal J, Kobayashi K, Yamamoto M, Li WH, Chan L. Alternative mRNA splicing and differential promoter utilization determine tissue-specific expression of the apolipoprotein B mRNA-editing protein (Apobec1) gene in mice. Structure and evolution of Apobec1 and related nucleoside/nucleotide deaminases. J. Biol. Chem. 1995;270:13042–13056. doi: 10.1074/jbc.270.22.13042. [DOI] [PubMed] [Google Scholar]
- 90.Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 91.Yamanaka S, Poksay KS, Balestra ME, Zeng GQ, Innerarity TL. Cloning and mutagenesis of the rabbit apoB mRNA editing protein. A zinc motif is essential for catalytic activity, and non-catalytic auxiliary factor(s) of the editing complex are widely distributed. J. Biol. Chem. 1994;269:21725–21734. [PubMed] [Google Scholar]
- 92.Fujino T, Navaratnam N, Scott J. Human apolipoprotein B RNA editing deaminase gene (APOBEC1) Genomics. 1998;47:266–275. doi: 10.1006/geno.1997.5110. [DOI] [PubMed] [Google Scholar]
- 93.Greeve J, Axelos D, Welker S, Schipper M, Greten H. Distinct promoters induce APOBEC-1 expression in rat liver and intestine. Arterioscler. Thromb. Vasc. Biol. 1998;18:1079–1092. doi: 10.1161/01.atv.18.7.1079. [DOI] [PubMed] [Google Scholar]
- 94.Ostertag EM, DeBerardinis RJ, Goodier JL, Zhang Y, Yang N, Gerton GL, Kazazian HH., Jr A mouse model of human L1 retrotransposition. Nat. Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 95.Brouha B, Meischl C, Ostertag E, de Boer M, Zhang Y, Neijens H, Roos D, Kazazian HH., Jr Evidence consistent with human L1 retrotransposition in maternal meiosis. Am. J. Hum. Genet. 2002;71:327–336. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl Acad. Sci. USA. 2004;101:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 98.Dupressoir A, Heidmann T. Germ line-specific expression of intracisternal A-particle retrotransposons in transgenic mice. Mol. Cell. Biol. 1996;16:4495–4503. doi: 10.1128/mcb.16.8.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.