Abstract
The J-binding protein 1 (JBP1) is essential for biosynthesis and maintenance of DNA base-J (β-d-glucosyl-hydroxymethyluracil). Base-J and JBP1 are confined to some pathogenic protozoa and are absent from higher eukaryotes, prokaryotes and viruses. We show that JBP1 recognizes J-containing DNA (J-DNA) through a 160-residue domain, DB-JBP1, with 10 000-fold preference over normal DNA. The crystal structure of DB-JBP1 revealed a helix-turn-helix variant fold, a ‘helical bouquet’ with a ‘ribbon’ helix encompassing the amino acids responsible for DNA binding. Mutation of a single residue (Asp525) in the ribbon helix abrogates specificity toward J-DNA. The same mutation renders JBP1 unable to rescue the targeted deletion of endogenous JBP1 genes in Leishmania and changes its distribution in the nucleus. Based on mutational analysis and hydrogen/deuterium-exchange mass-spectrometry data, a model of JBP1 bound to J-DNA was constructed and validated by small-angle X-ray scattering data. Our results open new possibilities for targeted prevention of J-DNA recognition as a therapeutic intervention for parasitic diseases.
INTRODUCTION
Base J (1), β-d-glucosyl-hydroxymethyluracil, is a hypermodified base present in all kinetoplastid flagellates studied (2), including the Trypanosoma, Leishmania and Crithidia genera and in Euglena (3), but absent from other eukaryotes, prokaryotes and viruses [reviewed in (4,5)]. J is a minor base replacing about 0.5% of T in the nuclear DNA of kinetoplastida (6,7) and is mainly present in the telomeric repeat sequence (GGGTTA)n (2,8). Small amounts of J are also found in other repetitive sequences of Trypanosoma brucei, such as the expression sites of variant surface glycoprotein (VSG) genes (9) and in sequences between transcription units (10).
Synthesis of the J nucleotide (11) and J-base containing (J-DNA) oligonucleotides by chemical methods (12,13), allowed the identification of a 93 kDa J-binding protein 1 (JBP1) in extracts of T. brucei, Leishmania species and Crithidia fasciculata (14). JBP1 binds specifically to base J in duplex DNA (15). JBP1 is essential in Leishmania (16), but not in T. brucei. The absence of JBP1 in T. brucei has no effect on growth, DNA repeat stability or gene expression, but does result in a 20-fold decrease in J level relative to wild-type cells (17). This led to the hypothesis that JBP1 catalyzes the first and rate-limiting step in J biosynthesis, the hydroxylation of T in DNA. This hypothesis was supported by the identification of a weak sequence similarity between JBP1 and Fe2+ and 2-oxoglutarate (2-OG)-dependent hydroxylases (dioxygenases) (18). Indeed, replacement of each of the four amino acids known to be essential for hydroxylase activity resulted in mutant proteins unable to complement JBP1 function in either T. brucei or Leishmania, but still able to bind to J-DNA. This showed that thymidine hydroxylase activity and J-DNA binding are independent functions of JBP1 (18,19).
There is one other protein in kinetoplastid flagellates which partially shares sequence similarity with JBP1, J-binding protein 2 (JBP2). JBP1 and JBP2 share 34% identity in their N-terminal halves (20), which contains the thymidine hydroxylase function of JBP1 (18) and of JBP2 (21,22). The C-terminal half of JBP2, but not of JBP1, contains a region similar to proteins with SWI2/SNF2-like chromatin remodeling activity. Although JBP2 contributes to J biosynthesis (10,21,22), it is not essential in T. brucei or in Leishmania and there is no evidence that JBP2 can bind to J-DNA (20).
Although JBP1 and JBP2 are unique proteins, a distant homolog of the JBP1/2 hydroxylase domain was recently identified in the mammalian protein TET1 (23,24), a fusion partner of the MLL gene in acute myeloid leukemia. TET1 and the related TET2 and TET3 proteins catalyze the conversion of 5-methylcytosine in DNA to 5-hydroxymethylcytosine, a reaction that may play an important role in the epigenetic control of gene expression (23). JBP and TET proteins have been grouped together in the TET/JBP subfamily of dioxygenases (25).
The binding of JBP1 to J-DNA has been studied by various methods. Using J-containing duplex oligonucleotides in a gel retardation assay, it was shown that JBP1 binds to J-containing oligonucleotides with an affinity between 40 and 140 nM (15). A fluorescence anisotropy polarization assay (FP) (26) yielded affinities as low as 13 nM. Binding to J-DNA is highly specific, since competition assays using gel-retardation indicated that a 500-fold excess of T-DNA could not out-compete J-specific DNA binding (27). The FP assay showed an affinity for T-DNA ∼100 times lower than that for J-DNA (1370 nM compared to 13 nM). The mode of interaction of JBP1 with J-DNA has been probed with several biochemical methods (15,27). Substitution of the hydroxymethylU in the J-base by hydroxymethylC, resulted in a 17-fold decrease in J-binding, showing that the pyrimidine base to which the glucose is attached co-determines binding affinity. At least 5 bp on both sides of J-base are required for optimal binding of JBP1, although critical contacts are restricted to two bases: major and minor groove contacts with base J and a sequence-independent major groove contact with the base immediately 5′ of base J on the same strand (position J-1) (27). Subsequent studies in which the sugar moiety of base J was systematically varied, have suggested a specific role for nucleotide J-1: its non-bridging phosphoryl oxygen hydrogen bonding to the equatorial 2- and 3-hydroxyl groups of the pyranosyl ring of the glucose of base J and locking the glucose in an ‘edge-on’ conformation perpendicular to the plane of the major DNA groove (26).
To determine exactly how JBP1 binds to J-DNA, three-dimensional molecular structures are required. As the first step toward this goal, we have identified a 160-residue autonomous folding unit (domain), DNA-Binding JBP1 domain (DB-JBP1), which binds to J-DNA with approximately the same affinity and specificity as full-length JBP1. We have determined the crystal structure of DB-JBP1, revealing a novel ‘helical bouquet’ fold and suggesting a model for the interaction with J-DNA. Hydrogen/deuterium exchange (HDX) mass spectrometry (MS) experiments probing the interaction between full length JBP-1 and J-DNA, showed that all significant changes upon binding of J-DNA were all within specific regions of the DB-JBP1 domain only. With site-directed mutagenesis and binding studies, we show that a single aspartate residue in JBP1 is essential for specific J-base recognition in vitro and for the function of JBP1 in vivo. Finally, based on all the data above, we constructed a structural model for the binding of DB-JBP1 to J-DNA, which was confirmed by small angle X-ray scattering (SAXS) measurements of the complex between JBP1 and J-DNA in solution.
MATERIALS AND METHODS
Expression and purification of recombinant proteins
The JBP1 coding sequences from the Leishmania species tarentolae, major, aethiopica, donovani, infantum and from T. brucei and C. fasciculata were amplified by PCR methods using Pfu polymerase (Stratagene) and inserted by ligation independent cloning in our in-house pET vector derivative, containing a hexa-histidine tag and an HRV 3C protease site. Clones were tested for the production of soluble protein in a heterologous Escherichia coli expression system. While JBP1 from C. fasciculata (Cf-JBP1) was obtained in large amounts, as expected from previous experience (15), the only other JBP1 made in E. coli in soluble form was the one from Leishmania tarentolae (Lt-JBP1) from a codon optimized gene.
For large-scale preparations, vectors were transformed to Bl21(DE3)T1R cells and grown in the presence of kanamycin. Protein production was induced with 0.3 mM IPTG at 15°C at an optical density of ∼0.8 for 16–18 h. Cell pellets were obtained by centrifugation at 4000 rpm and lysed with the pneumatic homogenizer EmulsiFlex-C5 in 20 mM Hepes pH 7.5, 350 mM NaCl, 1 mM TCEP, 10 mM Imidazole (Buffer A), plus a tablet of Complete EDTA-free (Roche) at 4°C. After clearing the supernatant by centrifugation, the proteins were bound in batch to Ni-chelating sepharose beads for 30 min at 4°C (GE Healthcare), washed with Buffer A and eluted in Buffer A containing 400 mM Imidazole. Fractions were analyzed by SDS–PAGE.
The JBP1 elution fractions were cleaved with 3C protease overnight at 4°C and applied to a S75 16/60 gel filtration column. Yields were between 5 and 50 mg/l of culture, depending on the construct. All proteins were >95% pure as judged by SDS–PAGE.
All truncation constructs were amplified by PCR using Pfu polymerase (Stratagene) and cloned for expression in the same vector, allowing cleavable His-Tag expression. Site-specific mutants were created using the QuickChange (Stratagene). Expression and purification experiments were carried out as for full-length JBP1.
Multi-angle laser light scattering
Multi-angle laser light scattering (MALLS) experiments were performed in a Superdex 200 HR 10/30 column attached to an Akta FPLC and coupled to a miniDAWN light scattering detector (Wyatt Technology) and a Dn-1000 differential refractive index detector (WGE Dr Bures). Hundred microliter of purified Lt-JBP1 at a concentration of ∼5.0 mg/ml were injected onto the column. Data analysis were carried out with the program Astra using a dn/dc value of 0.185.
Analytical size exclusion chromatography
Size exclusion chromatography experiments were performed with a Superdex 200 3.2/30 column attached to an Akta Purifier system from GE Healthcare. For Cf-JBP1 alone, we diluted 10 µl of a ∼50 µM stock of protein (0.5 nmol) in a total volume of 50 µl of buffer (20 mM Hepes/HCl pH 7.0 and 200 mM NaCl) to a concentration of ∼10 µM and injected that in the column. The elution volume (1.50 ml) indicated a size of approximately 100 kDa. Subsequently, a J-containing oligonucleotide with the sequence TAACCCJAACCCCTA was annealed with its complementary strand and concentrated to about 50 µM. A small amount of protein (1 µl of the protein stock above, ∼0.1 nmol) was mixed with a five times molar excess of the concentrated oligonucleotide (5 µl, ∼0.5 nmol) in a total buffer volume of 50 µl as above and injected onto the column under identical conditions. The molar excess of oligonucleotide was used to ensure that all of JBP1 was bound to J-DNA. The elution volume of the JBP1:J-DNA complex (1.48 ml) was very close to that of JBP1 alone (1.50 ml). However, the shift in the absorbance at 260 and 280 nm wavelength, clearly indicated that in the JBP1:J-DNA mixture, the peak corresponds to a protein:DNA complex. Using the theoretical molar extinction coefficients for JBP1 (e = 101 855) and for the oligonucleotide (e = 169 000), we calculate a stoichiometric ratio (28) of 1:1.01, that clearly indicated that Cf-JBP1 recognizes J-base containing DNA as a monomer.
Fluorescence polarization anisotropy assays
A single-stranded J-containing oligonucleotide with the sequence 5′-GGCAGCJGCAACAA-3′ was synthesized as we previously described (26), together with its T-containing equivalent GGCAGCTGCAACAA. A common complementary oligonucleotide was synthesized labeled in the 5′ with tetramethylrhodamine. Both complementary pairs of oligonucleotides were dissolved in water and heat-annealed to yield J-DNA or T-DNA and the duplex DNA was purified over a Superdex-75 10/30 Hi-Load gel filtration column. Dye concentration was determined through UV–VIS measurements and the double-stranded oligonucleotides were calibrated against the standard curve. The binding reaction was carried out with 1 nM TAMRA-labeled DNA, 20 mM HEPES pH 7.5, 140 mM NaCl (unless otherwise indicated for the salt dependence experiments), 1 g/l chicken ovalbumin, 2 mM MgCl2 and 1 mM TCEP. The maximum possible amount of all JBP1 protein variants was added and subsequent dilutions were achieved by serial 1:1-dilutions in three repeats. The reaction was incubated for 10 min at 4°C and room temperature before the measurements. Measurements were performed on an EnVision 2101 multilabel reader (Perkin Elmer) using 96-well optiplates (Perkin Elmer). The excitation filter was a Perkin Elmer X531 with a CWL of 531 nm, while the P and S emission filter were M579p with a CWL of 579 nm. All measurements were preformed at room temperature.
Reverse footprinting
Cf-JBP1 alone or mixed with J-DNA were prepared in µg amounts and digested with several proteases and in various conditions. Fragments of the Cf-JBP1:J-DNA mixture resistant to proteolysis, which were not present when JBP1 alone was digested under identical conditions, were obtained using a ratio of trypsin:Cf-JBP1 of 1:50 and an incubation of 30 min on ice.
The digested mixture was applied to a 1% agarose gel in TBE and run for 30 min at 100 V. Ethidium bromide and Coomassie blue were used to visualize the DNA and protein, respectively, in the same gel. The region of the agarose gel containing the protein bound to shifted DNA was isolated and the protein was eluted by melting the agarose gel slice under mild conditions. The eluate was size-fractionated by polyacrylamide gel electrophoresis in a 12% tris–glycine gel and transferred on a PVDF membrane in Tris–Glycine–SDS–Methanol buffer for 1 h at 100 V. The membrane was stained with Ponceau S and the band was cut out and sent for N-term sequencing; it showed the unique sequence (-)VTSSG (Cambridge Peptides, UK).
Crystallization of DB-JBP1
DB-JBP1 was crystallized using a robotic setup we have described previously (29). Clustered needle-like crystals grew at 4°C overnight in 20% w/v PEG 8000, 0.2 M MgCl2 and 0.1 M Tris pH 8.5, were crashed and used as seeds for a new screening round that gave bigger single crystals in 20% w/v PEG 3350 and 0.2 M potassium nitrate. These new crystals were used as seeds for a new screening round with selenomethionine substituted protein to obtain thick hexagonal plates with sharp edges in 15–20% w/v PEG 6000 and 0.2 M potassium nitrate. These crystals were transferred into a solution containing mother liquor supplemented with 20% glycerol, mounted in litho-loops and vitrified in liquid nitrogen for diffraction studies. Crystals from the various rounds of improvement are shown in Supplementary Figure S1.
Crystallographic structure determination of DB-JBP1
Crystallographic statistics are shown in Table 1. X-ray diffraction data were collected at a wavelength of 0.978 Å at the SLS beamline PXI, integrated with XDS (30) and scaled using SCALA (31). Positions of the selenium atoms were identified with phenix.hyss (32) that picked up seven sites. Substructure refinement and completion was performed in PHASER (33). Phase probability distributions were modified using DM; at this point the enantiomeric space group P6122 was chosen as correct. Automated model building with ARP/wARP 7.0 (34) using the SAD likelihood refinement function (35) resulted in 136 residues distributed in two chains. The model was completed manually using COOT (36), interspersed with reciprocal space refinement cycles in REFMAC5 (37), using bulk solvent scaling, non-polar hydrogens and four TLS groups. The final model was validated using WHATCHECK (38) and the MolProbity (39) server and is of excellent quality without any Ramachandran outliers, ranking in the 95th best percentile of PDB structures in that resolution. The model is deposited in the PDB with code 2XSE.
Table 1.
Crystallographic data collection and refinement statistics
| Data collection | |
| Space group | P6122 |
| Cell dimensions | |
| a, b, c (Å) | 67.34, 67.34, 186.76 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 20.0–1.90 (2.00–1.90) |
| Rmerge | 0.090 (0.663) |
| I/σI | 10.9 (2.3) |
| Completeness (%) | 99.8 (98.3) |
| Redundancy | 5.2 (5.4) |
| Refinement | |
| Resolution (Å) | 20.0–1.90 |
| No. reflections | 20 595 |
| Rwork/Rfree | 0.176/0.200 |
| No. atoms | |
| Protein | 1368 |
| Ligand/Ion | 14 |
| Water | 163 |
| B-factors | |
| Protein | 43.5 |
| Ligand/ion | 57.8 |
| Water | 49.9 |
| RMSD | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.27 |
Structure similarity searches
Structure similarity searches were carried out with two methods. SSM searches (40) against the whole PDB, after changing default search values as to the server suggestions, returned some similar structures with low significance scores. The top-hit, however (3FDQ), displayed an RMSD between Ca-atoms of matched residues of 3.25 Å over a length of 84 residues and a statistical significance Z-score of 1.3, despite a very low sequence identity of 6% in the aligned regions and a P-score (minus logarithm of the probability of achieving the same or better quality of match at a chance) of zero. All other SSM hits had Z-scores below 1.0. DALI searches, however (41), returned the same hit in the second position in the list ranked by Z-score, with a significance Z-score of 5.3 and similar alignment metrics as SSM. All other DALI hits were helical proteins but with different connectivity than our query structure. Manual inspection of the SSM and DALI structural alignments clarified that both the MogR structure (3FDQ) and DB-JBP1 had identical connectivity in the core four helices and we thus considered the two domains as structural homologs.
HDX MS
Solution phase amide HDX experiments were performed with a LEAP technologies (Carroboro, NC, USA) CTC HTS twin PAL autosampler interfaced with an electrospray ionization (ESI) linear ion trap mass spectrometer (Thermo Electron, San Jose, CA, USA) (42). Of a 11 µM JBP1 stock solution (0.7 M NaCl, 15 mM KCl, pH 7.0), 4 µl were diluted with 16 µl of an equivalent composition D2O buffer and incubated for 1, 30, 60, 900 and 3600 s to allow H/D exchange to occur. Following the allotted time period, the sample was diluted to 50 µl with cold (1.5°C) 2 M urea containing 0.1% TFA. Protein was then passed through an immobilized pepsin column (2 mm × 2 cm made in-house) held at 1.5°C and the resultant peptides were captured on a C8 peptide micro trap (Michrom Bioresources, Inc., Auburn, CA, USA). Peptides were then eluted across an HPLC column (Betasil C18 50 × 2.1 mm, 5 µm) into the ESI source with a gradient of 2% CH3CN increasing to 50% CH3CN over 18 min. For the JBP1 + J-DNA experiment, 11 µM JBP1 stock solution was incubated with a J-base containing synthetic oligonucleotide prior to mixing with D2O buffer and subsequent analysis. Data represent the average of three replicates and were processed with in-house developed software (43).
Surface plasmon resonance assays
Surface plasmon resonance (SPR) spectroscopy was performed at 25°C on a Biacore T100. Oligonucleotides were prepared as described for the FP assays, but with a biotin-TEG labeled complementary strand, that was used to immobilize them on a streptavidin chip (SA, GE Healthcare). Concentrations series of all proteins were injected across the chip in a PBS buffer pH 7.4, with 0.05% (w/v) Tween-20 at a flow rate of 30 μl/min. The concentrations used were between 32 and 0.25 μM for all DB-JBP1 variants. Binding curves were recorded for each protein, using an empty flow cell as reference. All experiments were repeated at least twice in a non-sequential manner to exclude systematic errors. The Biacore T100 evaluation software was used for the initial data analysis.
Testing the function of the JBP1 D525A mutant in vivo
JBP1 is essential in L. tarentolae for making J-base (16) and this complicates the testing of mutant versions of JBP1. We have used the procedure that was described in (18) to test the hydroxylation activity of JBP1 in a rescue experiment. The principle of this experiment is schematically outlined in Supplementary Figure S2. One of the two alleles of the JBP1 gene in L. tarentolae was disrupted first with a JBP1 KO puromycin construct. The resulting JBP1+/− cells were selected with puromycin and subsequently transfected with plasmids containing either the wild-type JBP1 or JBP1 D525A mutant. Transfected cells were then selected with paromomycin, as these plasmids also contain a neomycin marker. Next, the second JBP1 allele was targeted with a JBP1 KO hygromycin construct. JBP1−/− cells were then selected with puromycin and hygromycin and paromomycin was added to ensure retention of the rescue plasmid containing either wild-type or mutant JBP1.
Numerous colonies were obtained with the cells containing the rescue plasmid with wild-type JBP1. NcoI restriction digests of the DNA of some of these colonies were hybridized with a radioactive probe, corresponding to the 3′-UTR of the JBP1 gene that is not present in the rescue gene of the plasmids. The blots were used to identify the endogenous JBP1 gene or the rescue constructs as outlined in Supplementary Figure S3. In six out of the eight clones, the band corresponding to the endogenous JBP1 gene was not present (Supplementary Figure S3), indicating a complete disruption of both JBP1 alleles and has a reasonable efficiency of the rescue plasmid. Only few clones were obtained when the JBP1 D525A rescue plasmid was used for rescue, already indicating that this mutant is inefficient in rescuing the endogenous JBP1 gene disruption. All of these clones had retained a wild-type JBP1 gene band, as shown in Supplementary Figure S3 (lanes 11–18); duplication of the endogenous wild-type JBP1 gene, is an expected reaction of Leishmania, as reported (16,18).
To verify that JBP1 protein was actually produced in sufficient amounts in the transfected cells, we quantified the amount of JBP1 by western blotting. Supplementary Figure S4 shows that some of the mutant rescue clones produced high amounts of JBP1. The localization of that protein was subsequently assessed by fluorescence microscopy. For fluorescence microscopy, wild-type L. tarentolae and cells overproducing the D525A mutant protein were fixed on the same slide with 1% formaldehyde in PBS, blocked with 1× blocking solution (Roche). Primary (α-JBP1) and secondary (Alexa 488 conjugated goat α-rabbit) antibodies were incubated in 3% BSA/PBS at 37°C. Slides were mounted in Vectashield solution (Vector Laboratories). The DNA was stained with Topro3 DNA stain (invitrogen). Images were captured with a Leica TCS SP2 AOBS confocal microscope. Details of the procedure are described in (22); the α-JBP1 antibody was described in Ref. (6).
Modeling of the DB-JBP1 complex
The complex of MogR with DNA (3FDQ) was used as the template to create a model of the binding of DB-JBP1 to J-DNA. First, the recognition helix of DB-JBP1 (517–525) was superposed to the recognition helix of MogR using COOT (36). Then, a model for J-DNA was created as a standard 20-mer B-DNA with the sequence used for the SAXS experiment. A 3 bp fragment of J-DNA from the molecular dynamics simulations that suggested the specific conformation of J-base in the major groove (26) was then grafted to the 20-mer DNA to create a J-DNA 20-mer, with J at position 12. The geometry of that structure was idealized in REFMAC5 (37) using interval restraints (44) for describing the hydrogen bonds of the J-base to the N-1-phosphoryl oxygen and between all other Watson–Crick base pairs. The 7 nt bracketing the J-base in the J-containing strand (9–15) of this idealized 20-mer were then superposed to the DNA in the MogR complex, ‘sliding’ it in various positions to find a placement that would allow the J-base to be located close to Asp525 of the recognition helix; a unique suitable solution was found and was used to create the DB-JBP1:J-DNA complex. In that complex, both C4 and C6 hydroxyls were very close to Asp525, whereas Arg517, Lys522, Lys524 were all close to DNA and available for interactions. That encouraged us to model the loop missing from the structure, residues 529–534, in a conformation where Arg532 was also in contact with DNA; a suitable stereo-chemically allowed conformation was easily identified using COOT. The final model geometry was idealized with REFMAC5 and was then compared against the SAXS experimental data and showed an excellent fit.
SAXS data collection and analysis
The same DB-JBP1 protein as used for crystallization was also used for the SAXS experiments. DB-JBP1 and double stranded J-DNA oligonucleotides with the sequence 5′-CAGAAGGCAGCJGCAACAAG-3′ were created as described above, mixed at a 1:1 molar ratio and the complex was purified by size exclusion chromatography. Two concentrations of DB-JBP1 alone (1.48 and 2.58 mg/ml) and three concentrations of the complex between DB-JBP1 and J-DNA (2.1, 2.5 and 6 mg/ml) were measured at the X13 beamline at the EMBL-Hamburg and all data were possessed by PRIMUS (45). The agreement between the experimental scattering curves and the structure of DB-JBP1, the model of DB-JBP1 bund to J-DNA and J-DNA alone were evaluated using CRYSOL (46). The ab initio model for the DB-JBP1:J-DNA complex was created by DAMMIF (47).
RESULTS
JBP1 binds to J-DNA ten thousand times stronger than to T-DNA
We have produced large amounts of JBP1 from C. fasciculata (Cf-JBP1) (15) and from L. tarentolae (Lt-JBP1) using a synthetic gene. Both Lt-JBP1 and Cf-JBP1 were purified with standard chromatographic methods as described in Materials and Methods. The high quality proteins obtained were used for all subsequent assays. Initial characterization experiments were performed with Cf-JBP1; most subsequent experiments were done with Lt-JBP1.
The molecular mass of both Cf-JBP1 and Lt-JBP1 was determined by MALLS, showing that both are monomers in solution (Supplementary Figure S5A). JBP1 also binds to DNA as a monomer, as indicated by size exclusion chromatography experiments using Cf-JBP1 (Supplementary Figure S5B).
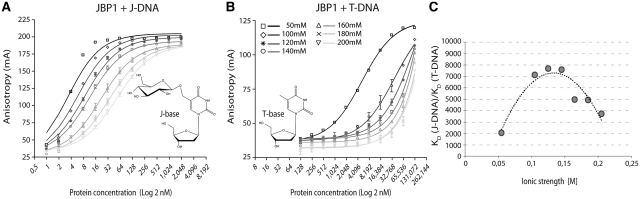
Protein–DNA binding is typically dominated by hydrogen bonding and electrostatic interactions, which are salt-sensitive. We therefore tested the dependence of J-DNA binding affinity on salt concentration. For this assay, we used a FP setup, similar to the one described in (26). The binding of JBP1 to a DNA sequence derived from the VSG gene, containing a J or a T nucleotide, exhibits a strong dependence on salt concentration (Figure 1A–C). The maximal discrimination between J-DNA and T-DNA takes place around physiological salt conditions and is about ten thousand times, with a KD for J-DNA of 11.1 nM and for T-DNA of 83 µM (Figure 1C and Supplementary Table S1).
Figure 1.
Specificity of JBP1 for J-DNA over T-DNA is salt dependent. (A and B) Binding of varying concentrations of Lt-JBP1 to J-DNA (A) and T-DNA (B) measured by the anisotropy of the signal of 1 nM labeled DNA, for seven different (buffered) concentrations of NaCl; symbols indicate the data points and vertical bars, the measures standard deviation; lines represent the fitted function that determination of the allowed KD; for very tight KD the fit is poorer since it is likely to be far from the assumed equilibrium. (C) The ratio of the calculated KD for T-DNA over J-DNA (indicating the specificity toward J-DNA) is shown as the function of the ionic strength of the buffer; the dotted line represents a polynomial fit to the data without assuming a physical model.
A small domain of JBP1, DB-JBP1, is sufficient for specific J-DNA recognition
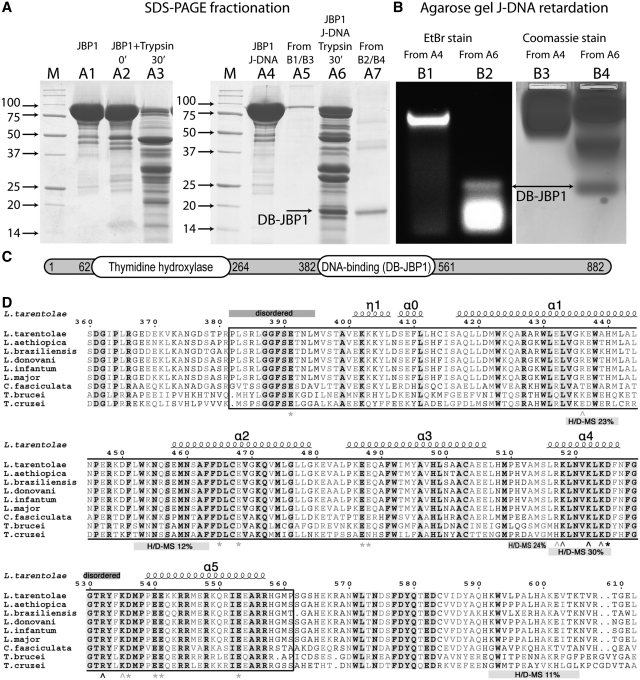
To determine whether a specific domain of JBP1 is responsible for J-DNA recognition, we performed a ‘reverse footprint’ experiment. Cf-JBP1 was mixed with J-DNA in microgram amounts and treated with limiting amounts of protease in an attempt to identify a protein domain that becomes resistance to proteolysis upon J-DNA binding. We found conditions in which J-DNA-bound JBP1 produced fragments resistant to proteolysis, which were not present when JBP1 alone was digested under identical conditions (Figure 2A). The digested mixture was used in a non-radioactive agarose gel retardation assay with J-containing oligonucleotide. We identified a clear band of shifted J-DNA (Figure 2B). This band migrated faster than the control J-DNA bound by full-length JBP1 and overlapped with a protein band (Figure 2B), suggesting that a small proteolysis-resistant fragment was responsible for the band shift. This fragment was identified by N-terminal protein sequencing to start at residue 382, while a combination of molecular weight information, secondary structure predictions and alignments allowed us to also define the most likely C-terminus at residue 561 (Figure 2C and D). This domain was termed DNA Binding JBP1 domain (DB-JBP1).
Figure 2.
Identification of the DB-JBP1 domain. (A) SDS–PAGE separation before and after limited proteolysis of the Cf-JBP1 protein alone and bound to J-DNA; the JBP1 and DB-JBP1 bands recovered after elution from the gel after the retardation assay are also shown and DB-JBP1-1is indicated by arrows. (B) Agarose gel DNA mobility retardation assay for JBP1 mixed with J-DNA before and after limited proteolysis, stained with ethidium bromide and with Coomassie blue; DB-JBP1 is indicated by arrows. (C) A schematic drawing of the JBP1 sequence with the N-terminal thymidine hydroxylase domain and the DB-JBP1 domain indicated. (D) A multiple sequence alignment of JBP1 from Leishmania, Trypanosoma and Crithidia; the DB-JBP1 domain is boxed; fully conserved residues are in black and bold on a gray background; conserved residues are in gray and bold; the secondary structure elements and residue numbers correspond to the L. tarantolae protein; disordered regions and peptides identified to be in contact with DNA in the HDX-MS experiment are annotated with labeled bars; wedge indicate Lys/Arg mutations and asterisks indicate D/E mutations (gray for no effect, black for affecting DNA binding).
The DB-JBP1 domain was produced in E. coli and yielded large amounts of soluble protein. We compared the binding of the Leishmania DB-JBP1 and full-length Lt-JBP1 to J-DNA using the fluorescent polarization assay (Supplementary Figure S6 and Table S1). DB-JBP1 binds to J-DNA with an affinity about three times less than full-length JBP1 (32.9 nM compared to 11.1 nM) and to T-DNA with a marginally lower affinity (129 µM compared to 83 µM; Supplementary Table S1). The discrimination between J-DNA and T-DNA at physiological salt concentrations is about the same for both JBP1 and DB-JBP1 (Supplementary Table S1).
The crystal structure of the DB-JBP1 domain
The DB-JBP1 structure was determined using Se-Met substituted DB-JBP1 in a single-wavelength anomalous dispersion (SAD) experiment and refined to yield a model with all residues in favorable regions of the Ramachandran plot and an Rfree of 20.3% (Table 1). The final model does not contain the first 13 residues (382–394) and six residues in the loop between residues 528 and 535, which could not be modeled in the electron density and are presumably disordered.
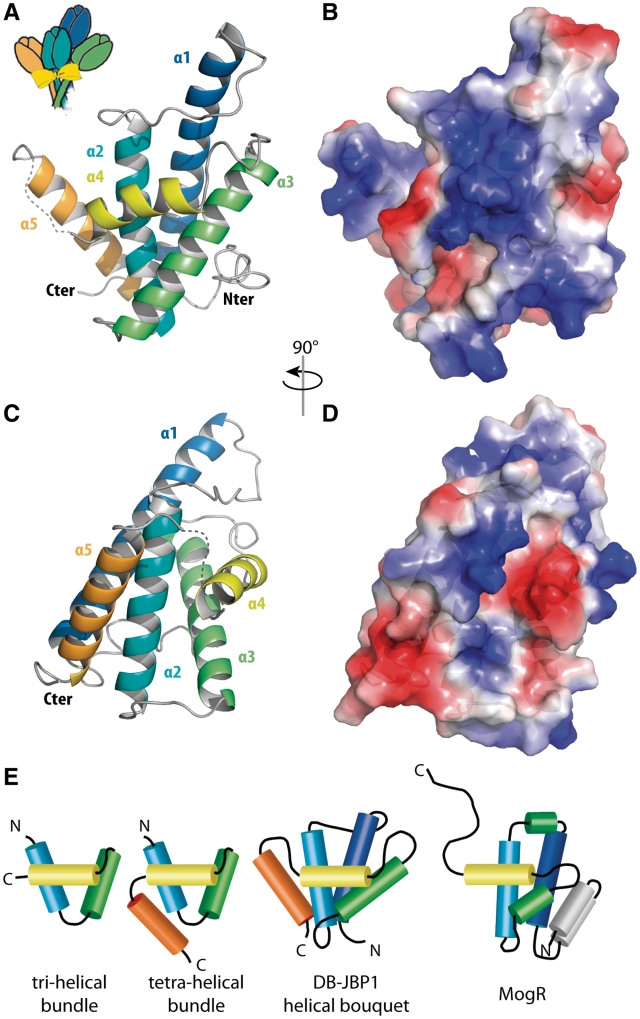
The structure of DB-JBP1 is a helical bundle in an arrangement we call a ‘helical bouquet’ (Figure 3A and B). The fold is made by five helices, of which the four longest, α1, α2, α3 and α5 run antiparallel in the same approximate orientation (the ‘flowers’ of the bouquet), with their axes at angles of about 15°–45°. Helix α4 is perpendicular to this arrangement, creating a ‘ribbon’ running across the front. The helical bouquet appears to be a divergent variant of the aberrant three-helical bundle helix-turn-helix (HTH) domains, closest to the prokaryotic tetra-helical bundle (Figure 3E) (48). The ribbon helix in the DB-JBP1 helical bouquet corresponds to the recognition helix of the core three-helical bundle (α2–4) preceded N-terminally by a long 28-residue helix (α1) and followed C-terminally by a 19-residue helix (α5). Notably, the supporting α3 helix and the recognition α4 (ribbon) helix are not connected by the sharp turn characteristic of HTH motifs, but by an unusually extensive loop of nine residues. The conserved ‘small-hydrophobic-small’ signature motif in the turn between helix-2 and helix-3 of the core HTH structure (48) is conserved in the long connecting loop of the JBP1 family and is directly preceding the recognition helix (Ala-Met-Ser in Leishmania and Crithidia and Ser-Met-Gly or Ala-Val-Gly in the Trypanosoma proteins). The ‘charged-hydrophobic-small’ motif in the supporting helix of HTH domains is absent in JBP1.
Figure 3.
The structure of DB-JBP1. (A) Cartoon representation of DB-JBP1, in the orientation reminiscent of a flower bouquet; each helix is in a different color and labeled; the recognition helix is in yellow. (B) The same cartoon after a 90° rotation along the vertical axis. (C and D) Surface representations (in the same orientation as A and B respectively) colored according to the electrostatic surface potential (−7 to 7 kT). (E) Schematic drawing of typical HTH domains, DB-JBP1 and MogR, with helices drawn as cylinders; structurally equivalent helices are drawn in the same colors used for the structure of DB-JBP1.
Searches for structural homologs of DB-JBP1 in the PDB, using either SSM (40) or DALI (41), returned a significant, albeit weak, structural similarity with the HTH-domain structure of the MogR transcriptional repressor of the bacterial pathogen Listeria monocytogenes (49) (Figure 3E and Supplementary Figure S7). The MogR structure has been determined in complex with AT-rich DNA, with the recognition helix mediating major groove contacts as expected. The long C-terminal wing that mediates minor groove interaction with the AT-rich DNA in MogR, is in DB-JBP1 the 6-residue disordered loop leading to the α5 helix that is absent in MogR. By analogy to MogR, this disordered loop likely gets ordered when DB-JBP1 gets in contact with DNA and could mediate minor groove interactions.
The electrostatic surface potential of DB-JBP1 shows a large positive patch in the surface where the recognition helix lies, also suggesting that this is the primary area for DNA binding (Figure 3C and D). Given, however, the low sequence conservation (6% identity) and the large differences between superposed Cα atoms (3.2 Å) between DB-JBP1 and MogR, its closest HTH domain structural homolog, it was impossible to reliably predict in further detail the mode of JBP1 binding to DNA and explain the preference for J-DNA. We therefore used other methods to analyze the contacts of J-DNA with the JBP1 protein.
HDX MS demonstrates the DB-JBP1 regions involved in DNA-binding
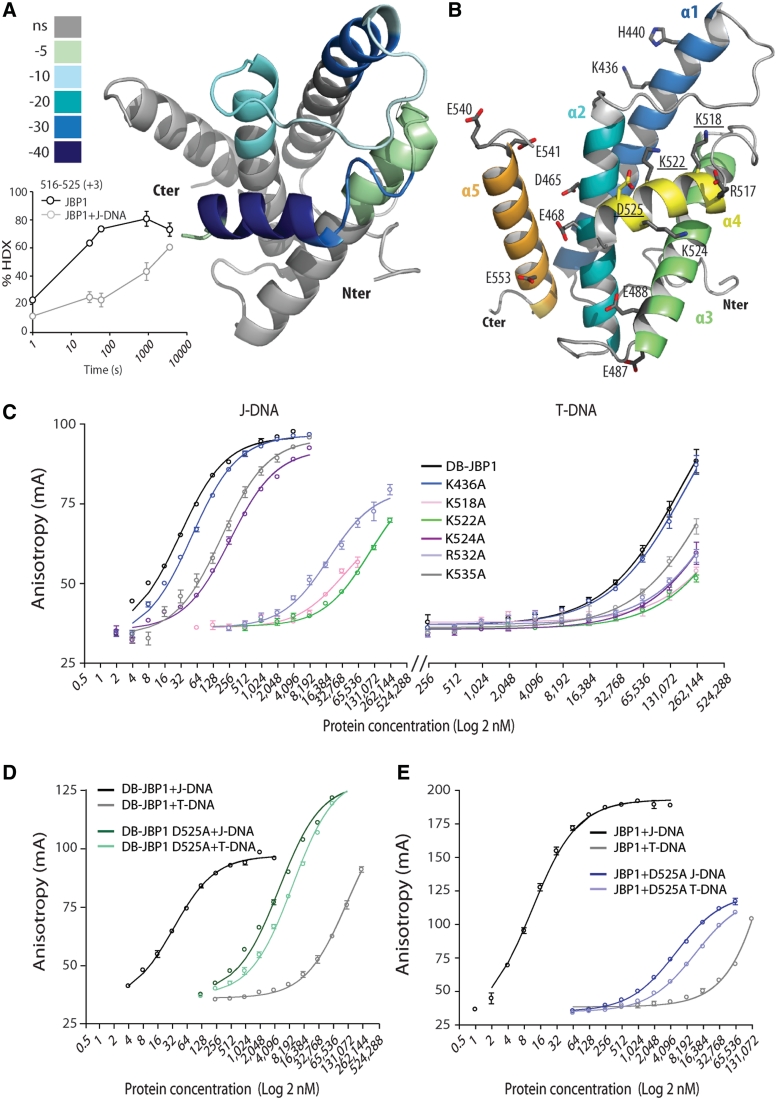
We used HDX-MS to determine which regions of full-length JBP1 interact with DNA. The HDX-MS experiment measures the rate of exchange of protein backbone amide hydrogen with deuterium. The rate of this exchange is influenced primarily by hydrogen bonding and therefore operates as a sensor for the local chemical environment of the amide hydrogen. The deuterium exchange is plotted as a function of time for the protein alone and is then compared to the values obtained for protein bound to DNA. The peptides that show reduced exchange rates upon complex formation with DNA are, therefore, involved in DNA binding, either directly or through allosteric perturbation of the protein structure/dynamics. We show that only a few peptides exhibit reduced H/D exchange rates upon binding of JBP1 to J-DNA, all spanning the DB-JBP1 domain (Figures 2C, 4A and B and Supplementary File S1 for detailed results). The most prominent changes occurred in the peptides 516–525 (helix α4, 30% reduction in exchange rate, Figure 4A). The other peptides that showed a significant change upon J-DNA binding, 434–441 (helix α1, 23%), 510–515 (the loop between helices α3 and α4, 124%) and 452–463 (helix α2 and the loop before, 12%) (Supplementary Figure S8), are also within the DB-JBP1 domain. The peptide spanning residues 593–605 that showed a modest change (11%, Supplementary Figure S8) is just C-terminal to DB-JBP1. Eight different peptides that were identified in the HDX-MS experiment corresponding to the N-terminal half of the supporting helix α3 (∼484–494), which often mediates some contacts with DNA, did not show any significant changes (−2% to 2%). A few more changes, all within the DB-JBP1 domain, had values below 10%, while no change above 5% was observed elsewhere in JBP1 (Supplementary Figure S9).
Figure 4.
Mapping the DB-JBP1 regions that bind DNA. (A) The peptides that show hydrogen–deuterium exchange rate differences upon J-DNA binding are colored according to the magnitude of the differences and mapped in the DB-JBP1 structure; the insert depicts the change in hydrogen–deuterium exchange rate of the JBP1 peptides 516–525 (which corresponds to the recognition helix α4) in the unbound and J-DNA bound state. (B) Location of the DB-JBP1 point mutants used to identify the residues involved in J-DNA binding. (C) Binding of varying concentrations of the various Arg/Lys mutants of DB-JBP1 to J-DNA (left) and T-DNA (right); colors are used purely to indicate the different curves more clearly. (D) Binding of DB-JBP1 and (E) JBP1 and the corresponding D525A mutants to T-DNA and to J-DNA.
This experiment suggests that DB-JBP1 is the only JBP1 domain involved in J-DNA binding and that it is sufficient for specific J-DNA recognition. The biggest change in hydrogen–deuterium exchange rates upon J-DNA binding overlaps perfectly with helix α4, confirming that residues in this helix are the primary site for DNA interaction.
DB-JBP1 recognizes DNA through lysine and arginine residues in helix α4
To find the residues that interact with the DNA backbone we created seven point mutants in DB-JBP1 (Figure 4B), based on structural data and the HDX-MS experiments: R517A, K518A, K522A, K524A targeting each positively charged residues in helix α4; R532A and K535A targeting the intrinsically disordered loop downstream helix α4 which had an appreciable 8% decrease in the HDX-MS data and K436A lying at the end of helix α1 that also showed some changes in the HDX-MS data. All mutant proteins were expressed and purified and were used in the FP assay (Figure 4C). The R532A, K518, K522A and to a lesser extent K524A proteins, showed decreased affinity for both J-DNA and T-DNA, suggesting that these residues, all but one lying in the α4 ribbon helix, are involved in DNA binding, but do not specifically recognize the J-base. K436A and K535A showed no appreciable change in affinity.
A single aspartate residue is responsible for J-binding specificity
The glucose of the J-base lies in the major groove of the DNA and should be recognized specifically by DB-JBP1 residues. We hypothesized that aspartate or glutamate residues could recognize the hydroxyl groups of the glucose moiety and these should be fully conserved in the JBP1 family. Ten such residues were located and we created eight point mutants (since there are two pairs of glutamates): E391A, D465A, E468A, E487/488A, D525A, D536A, E540/541A, E553A. Five of these mutants produced soluble protein in E. coli and were assayed in a SPR experiment in comparison with wt DB-JBP1. The D525A mutant protein exhibited no observable binding to J-DNA under the SPR experimental conditions, while all other mutants showed no significant differences compared to DB-JBP1 (Supplementary Figure S10 and Table S1).
The D525A DB-JBP1 mutant was characterized in more detail in the FP assay. This mutant protein has strikingly lost nearly all discrimination for J-DNA, since its affinity for J-DNA is only 5.1 µM, similar to its affinity for T-DNA, 9.7 µM (Figure 4D and Supplementary Table S1). The affinity of the D5252A mutant for T-DNA (9.7 µM) has increased about 10 times compared to wild-type DB-JBP1 (∼83 µM); a possible explanation for this observation is the removal of the repulsion between the negatively charged aspartate carboxylates and the DNA backbone phosphates. Consistently, the full-length JBP1 D525A mutant has the same low affinity for J-DNA (5.1 µM) and its affinity for T-DNA is 11.7 µM (Figure 4E and Supplementary Table S1). These results strongly indicate that a single residue, Asp525, is necessary and sufficient for conferring specificity toward J-DNA.
Aspartate 525 is important for in vivo function of JBP1
We tested whether the JBP1 D525A mutant can functionally replace wild-type JBP1 in L. tarentolae, using an approach we previously described (18) and outlined in Supplementary Figure S2. Leishmania tarentolae cells in which one of the two JBP1 alleles had been inactivated by insertion of a drug resistance marker were transfected with plasmids expressing either wild-type or mutant JBP1. Then, the second chromosomal JBP1 allele was inactivated with a targeting construct containing a second drug resistance marker. Whereas both alleles of chromosomal JBP1 could be easily disrupted in the presence of the rescue plasmid containing wild-type JBP1, JBP1 D525A completely failed to rescue. Clones were obtained containing both drug resistance markers, but in these clones an extra copy of the wild-type gene had appeared (Supplementary Figure S3). Generation of additional wild-type gene copies is the standard response of Leishmania, if attempts are made to disrupt both alleles of an essential gene (16,50). The negative result with the mutant JBP1 is not due to defective synthesis or routing of the mutant protein, which is made in high amounts (Supplementary Figure S4). The protein is reaching the nucleus, as our anti-JBP1 antibody detects more JBP1 in the nucleus of cells containing the rescue plasmid with mutant JBP1 than with wild-type JBP1 (Supplementary Figure S4). We conclude that mutant JBP1 is unable to provide sufficient JBP1 function for L. tarentolae to survive.
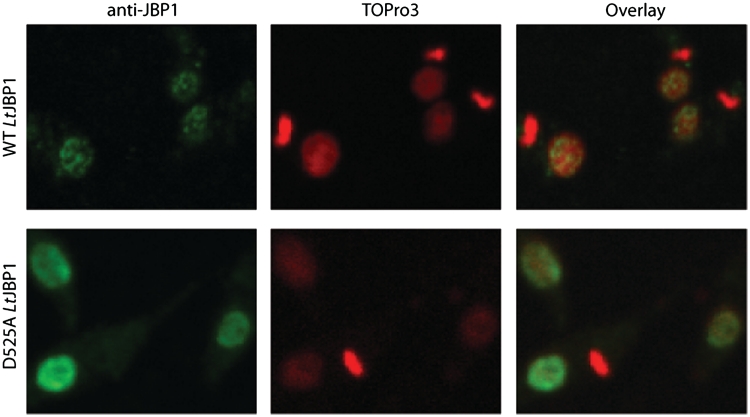
Interestingly, the distribution of wild-type and D525A mutant JBP1 in the nucleus is different. Wild-type JBP1 is located in discrete spots (Figure 5), presumably the J-rich telomeres of L. tarentolae, which are known to be present in about 16 clusters in Leishmania (51). In contrast, JBP1-D525A exhibits a more diffuse nuclear staining pattern (Figure 5) indicating that it has lost its ability to specifically bind J-DNA also in vivo.
Figure 5.
The D525A JBP1 mutant protein fails to localize in J-rich nuclear regions. Fluorescence microscopy images of L. tarentolae cells fixed on microscope slides and stained with anti-JBP1 antibody and TOPro3 (DNA). JBP1 in the wild-type cells (upper panel) localizes in discrete spots in the nucleus, but the cells producing also the D525A mutant JBP1 (lower panel) show a more diffuse pattern. The bright staining DNA ovals in the cells are the kinetoplastid DNA networks; the nuclei stain less intense. The ‘doughnut-like’ staining with anti-JBP1 antibody in the lower panel is probably due to weaker staining of the large nucleolus, creating an apparent hole in the nucleus.
A structural model for DB-JBP1 binding to J-DNA and validation by SAXS
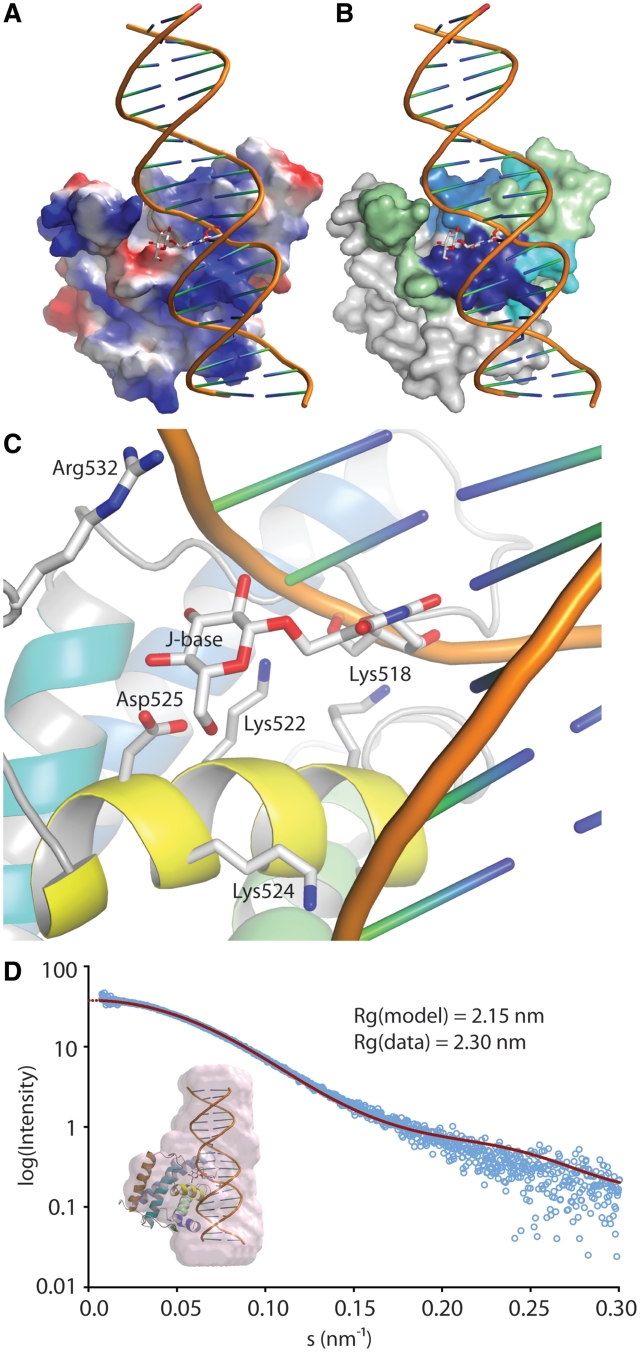
In the absence of a crystal structure of DB-JBP1 in complex with J-DNA, we created a model based on the structure of MogR with DNA (49), using interactive graphics (see Experimental Methods). The modeled J-DNA runs along the positive patch in JBP1 (Figure 6A) and along the patch identified by the HDX-MS experiments (Figure 6B). In addition, Asp525 is in contact with the J-base, while the Arg/Lys residues important for general DNA recognition are in contact with the J-DNA backbone (Figure 6C).
Figure 6.
A model of JBP1 binding to J-DNA. (A) JBP1 is shown as a surface colored by electrostatic potential; J-DNA is shown as a cartoon with the J-base as stock model; while the DNA backbone runs along the positive (blue) patch in the JBP1 surface, the J-base faces a negative (red) ‘island’. (B) Same as in A but the JBP1 surface is colored by the propensity of peptides to interact with J-DNA according to the HDX-MS experiments, like in Figure 4A. (C) A close up of the interaction of JBP1 with the J-base; the residues important for interaction according to mutagenesis studies are shown as stick models and are labeled. (D) Validation of the model against SAXS data; the graph shows the intensity calculated from the model (red line) and the experimental data (open cyan circles) against the scattering amplitude; in the inlet picture the pink surface represent an ab initio beads model calculated from the SAXS data and superposed onto the JBP1:J-DNA complex model.
To validate this model, we measured SAXS curves for DB-JBP1 alone and in complex with J-DNA in solution, to determine their low-resolution shape. The radius of gyration calculated from the SAXS data for the DB-JBP1 in complex with J-DNA (2.3 nm) fits well with that calculated from our model (2.15 nm). In addition, the calculated scattering curves from the structure of DB-JBP1 and from our model of the complex between DB-JBP1 and J-DNA, both fit very well only to their corresponding experimental SAXS data (Figure 6D and Supplementary Figure S11). Furthermore, a low resolution ab initio beads model created based on the complex SAXS data is also in good agreement with our model (Figure 6D).
DISCUSSION
Under physiological conditions, JBP1 shows a remarkable 10 000-fold binding preference for J-DNA over T-DNA with the same sequence. This is in line with the results of Sabatini et al. (15) who showed that a 500-fold excess of T-DNA was unable to out-compete binding of J-DNA to JBP1. In salt concentrations lower than physiological, this specificity is compromised, explaining previous reports that the preference for J-DNA is only 100-fold (26).
We show that the DB-JBP1 domain, which has affinity and specificity close to that of full-length JBP1, is the only region that mediates binding of JBP1 to J-DNA. The two functions of JBP1 are therefore separated in two different parts of the protein: the thymidine hydroxylase resides in the N-terminal half and the J-DNA binding in the C-terminal half. Cross et al. (14) raised the possibility that a HTH motif located in the N-terminal half of JBP1 (residues ∼250–260), would be involved in DNA binding, as this motif had some similarity with the Myb homeodomain. This hypothetical binding site is not corroborated by our experiments.
The DB-JBP1 fold is a divergent variant of the HTH domains family, a motif that can be traced back in 6–9 copies, with divergent functionality in the last universal common ancestor (LUCA), and it is a common module in many proteins and multiprotein assemblies (48). The HTH domain in DB-JBP1 is characterized by an unusually long 9-residue ‘turn’ and the long helices that bracket the core three-helical bundle. This helical bouquet is not readily classifiable in the HTH families described. This raises puzzling questions as to its ancestry and incorporation into the JBP1 multidomain scaffold.
The recognition helix of the HTH motif, the ribbon of the helical bouquet, bears a single aspartate residue (Asp-525) that appears to be the sole determinant of J-DNA versus T-DNA recognition. It has been suggested (26) that the C2 and C3 hydroxy groups of the J-base glucose form hydrogen bonds with the non-bridging pro-R phosphoryl oxygen of the J-1 nucleotide phosphate group. In this model, the C4 and C6 hydroxy groups of the glucose moiety are free to engage in additional contacts. We speculate that Asp-525 is recognizing one or both of the C4 or C6 hydroxy groups.
Our model does not point to any direct binding contacts between JBP1 and the pyrimidine heterocycle of base J. However, it has been shown that replacement of the hydroxymethyluracil of base J with hydroxymethylcytosine reduces binding to JBP1 ∼15 times (27). This result argues that JBP1 directly recognizes the base heterocycle, putting our model at apparent odds with the experimental data. However, chemical intuition suggests that a potential intramolecular hydrogen bond can be formed between the C4 amino group of the cytosine heterocycle and the C6 hydroxy group of the pendant β-d-glucosyl, an interaction that cannot be established in the native base J. This hydrogen bond would result in a change in the overall geometry of the ‘edge on’ conformation of the β-d-glucose group in the major groove, a critical determinant of JBP1 binding (26). Therefore, in the specific case of hydroxymethylcytosine, the reduction in JBP1 binding can be accounted for without invoking any recognition of the heterocycle by JBP1. The binding of JBP1 to in vitro glucosylated hydroxymethylcytosine–DNA is of special interest, as it has recently been used in a new and efficient method, to isolate DNA containing hydroxymethylcytosine (52). This should help to locate this new mammalian epigenetic marker base (24,53) more precisely in DNA.
The D525A mutant of JBP1, in contrast to wild-type JBP1, fails to rescue the targeted JBP1 gene deletion in L. tarentolae. Previous work with T.brucei has shown that introduction of wild-type JBP1 into trypanosomes without any J in their DNA results in J synthesis (10). JBP1 is, therefore, able to start from scratch, i.e. hydroxylate some T-residues in T-DNA, to which it binds poorly. Once a few initial J molecules have been made, hydroxylation can apparently accelerate sufficiently to introduce substantial amounts of J into the DNA (10). In contrast, with the D525A mutant of JBP1 the sluggish introduction of some J is evidently not followed by a rapid increase in J, because mutant JBP1 is unable to bind strongly and specifically to J and hydroxylate more T's in an efficient fashion. Highly specific J-binding is therefore an essential prerequisite for JBP1 function. From previous experiments (54), we have deduced that even 10–15% of wild-type JBP1 function is sufficient for normal multiplication of L. tarentolae: it follows that D525A JBP1 provides less than 10% of wild-type function and that DNA binding alone cannot restore the complete JBP1 function without the specific recognition and strong binding of JBP1 to pre-existing J.
Having established that Asp525 is a residue crucial for JBP1 function both in vitro and in vivo, we constructed a model for JBP1 binding to J-DNA, where the ribbon helix is positioned along the major groove of J-DNA. The low-resolution shape of our model is consistent with solution X-ray scattering experiments. Although the atomic interactions in this model are in agreement with all our mutagenesis data, the details remain to be structurally validated.
JBP1 was assumed to have a ‘glucose-binding pocket’ that could be targeted by potential small molecule inhibitors. Unfortunately, our model renders this possibility less likely, since the glucose is recognized by the aspartate residue protruding from the ‘ribbon’ helix, much in the same manner as other helical major groove interactions in DNA recognition. Such interactions have been notoriously difficult to target by small-molecule inhibitors. However, the specific charge pattern, with the acidic aspartate surrounded by basic residues, could provide a possible target site and a positive surprise. In addition, small molecules or short synthetic peptide inhibitors based on the DB-JBP1 recognition helix, targeting J-DNA rather than JBP1, or chemicals targeting the J-DNA conformation as previously proposed (26), might present a better suited therapeutic strategy.
ACCESSION NUMBER
PDB 2XSE.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The State of Florida (financial support to M.J.C. and P.R.G.); Skaggs Insitute for Chemical Biology (postdoctoral training grant to R.K.G.). Funding for open access charge: Institutional.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Crystallography experiments were performed on the PX beamline at the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland; we thank P. Celie for data collection assistance. SAXS experiments were performed on the X33 beamline at the EMBL/DESY (Hamburg) synchrotron radiation facilities; we thank F. Groothuizen and T. Sixma for data collection and D. Svergun for local support. E.C. and A.P. thank V. De Marco for help in identifying the exact limits of the DB-JBP1 domain. We are grateful to A. Fish and the NKI Protein Facility for assistance with the SPR experiments and to R. Hibbert for assistance with the FP experiments. T.H. performed all FP and SPR experiments, purified proteins for all assays, crystallized DB-JBP1, collected diffraction data and prepared figures for this manuscript; E.C. established initial JBP1 purification protocols and identified and purified the DB-JBP1 domain; M.J.C. performed and analyzed the HDX MS experiments; S.J. performed the in vivo experiments; B.tR. assisted in characterizing the DB-JBP1 domain; R.K.G. synthesized the J-oligo phosphoramidite precursors; R.P.J. refined the JBP1 structure; D.L. helped establish the FP assays and optimize purification protocols; P.R.G. supervised the HDX MS experiments; P.W. supervised J-oligo synthesis; A.P., H.vL. and P.B. initiated the project and supervised experiments at the NKI; A.P. wrote the manuscript (with help from P.B.), determined and built the DB-JBP1 structure, created the JBP1:J-DNA model, analyzed SAXS data, prepared figures and co-ordinated experiments and Dr Phillip Ordoukhanian, Director of the Scripps Center for Protein and Nucleic Acid Research for J-oligo synthesis and purification.
REFERENCES
- 1.Gommers-Ampt JH, Van Leeuwen F, De Beer ALJ, Vliegenthart JFG, Dizdaroglu M, Kowalak JA, Crain PF, Borst P. Beta-D-glucosyl-hydroxymethyluracil: A novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993;75:1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- 2.Van Leeuwen F, Taylor MC, Mondragon A, Moreau H, Gibson W, Kieft R, Borst P. Beta-d-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proc. Natl Acad. Sci. USA. 1998;95:2366–2371. doi: 10.1073/pnas.95.5.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dooijes D, Chaves I, Kieft R, Dirks-Mulder A, Martin W, Borst P. Base J originally found in kinetoplastida is also a minor constituent of nuclear DNA of Euglena gracilis. Nucleic Acids Res. 2000;28:3017–3021. doi: 10.1093/nar/28.16.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst P, Sabatini R. Base J: discovery, biosynthesis, and possible functions. Annu. Rev. Microbiol. 2008;62:235–251. doi: 10.1146/annurev.micro.62.081307.162750. [DOI] [PubMed] [Google Scholar]
- 5.Gommers-Ampt JH, Borst P. Hypermodified bases in DNA. FASEB J. 1995;9:1034–1042. doi: 10.1096/fasebj.9.11.7649402. [DOI] [PubMed] [Google Scholar]
- 6.Toaldo CB, Kieft R, Dirks-Mulder A, Sabatini R, van Luenen HG, Borst P. A minor fraction of base J in kinetoplastid nuclear DNA is bound by the J-binding protein 1. Mol. Biochem. Parasitol. 2005;143:111–115. doi: 10.1016/j.molbiopara.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Leeuwen F, Dirks-Mulder A, Dirks RW, Borst P, Gibson W. The modified DNA base beta-d-glucosyl-hydroxymethyluracil is not found in the tsetse fly stages of Trypanosoma brucei. Mol. Biochem. Parasitol. 1998;94:127–130. doi: 10.1016/s0166-6851(98)00060-7. [DOI] [PubMed] [Google Scholar]
- 8.van Leeuwen F, Wijsman ER, Kuyl-Yeheskiely E, van der Marel GA, van Boom JH, Borst P. The telomeric GGGTTA repeats of Trypanosoma brucei contain the hypermodified base J in both strands. Nucleic Acids Res. 1996;24:2476–2482. doi: 10.1093/nar/24.13.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Leeuwen F, Wijsman ER, Kieft R, van der Marel GA, van Boom JH, Borst P. Localization of the modified base J in telomeric VSG gene expression sites of Trypanosoma brucei. Genes Dev. 1997;11:3232–3241. doi: 10.1101/gad.11.23.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cliffe LJ, Siegel TN, Marshall M, Cross GA, Sabatini R. Two thymidine hydroxylases differentially regulate the formation of glucosylated DNA at regions flanking polymerase II polycistronic transcription units throughout the genome of Trypanosoma brucei. Nucleic Acids Res. 2010;38:3923–3935. doi: 10.1093/nar/gkq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wijsman ER, van den Berg O, Kuyl-Yeheskiely E, van der Marel GA, Van Boom JH. Synthesis of 5-(Beta d-glucopyranosyloxymethyl)-2′-deoxyuridine and derivatives thereoff. A modified d-nuceloside from the DNA of Trypanosoma brucei. Recl. Trav. Chim. Pays-Bas. 1994;113:337–338. [Google Scholar]
- 12.de Kort M, Ebrahimi E, Wijsman ER, van der Marel GJ, van Boom JH. Synthesis of oligodeoxynucleotides containing 5-(&bgr;-d-Glucopyranosyloxymethyl)-2′-deoxyuridine, a modified nucleoside in the DNA of Trypanosoma brucei. Eur. J. Org. Chem. 1999;9:2337–2344. [Google Scholar]
- 13.Turner JJ, Meeuwenoord NJ, Rood A, Borst P, van der Marel GA, Van Boom JH. Reinvestigation into the synthesis of oligonucleotides containing 5-(ß-d-glucopyranosyloxymethyl)-2′deoxyuridine. Eur. J. Org. Chem. 2003;19:3832–3839. [Google Scholar]
- 14.Cross M, Kieft R, Sabatini R, Wilm M, de Kort M, van der Marel GA, van Boom JH, van Leeuwen F, Borst P. The modified base J is the target for a novel DNA-binding protein in kinetoplastid protozoans. EMBO J. 1999;18:6573–6581. doi: 10.1093/emboj/18.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabatini R, Meeuwenoord N, van Boom JH, Borst P. Recognition of base J in duplex DNA by J-binding protein. J. Biol. Chem. 2002;277:958–966. doi: 10.1074/jbc.M109000200. [DOI] [PubMed] [Google Scholar]
- 16.Genest PA, ter Riet B, Dumas C, Papadopoulou B, van Luenen HGAM, Borst P. Formation of linear inverted repeat amplicons following targeting of an essential gene in Leishmania. Nucleic Acids Res. 2005;33:1699–1709. doi: 10.1093/nar/gki304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cross M, Kieft R, Sabatini R, Dirks-Mulder A, Chaves I, Borst P. J-binding protein increases the level and retention of the unusual base J in trypanosome DNA. Mol. Microbiol. 2002;46:37–47. doi: 10.1046/j.1365-2958.2002.03144.x. [DOI] [PubMed] [Google Scholar]
- 18.Yu Z, Genest PA, ter Riet B, Sweeney K, DiPaolo C, Kieft R, Christodoulou E, Perrakis A, Simmons JM, Hausinger RP, et al. The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase. Nucleic Acids Res. 2007;35:2107–2115. doi: 10.1093/nar/gkm049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cliffe LJ, Kieft R, Southern T, Birkeland SR, Marshall M, Sweeney K, Sabatini R. JBP1 and JBP2 are two distinct thymidine hydroxylases involved in J biosynthesis in genomic DNA of African trypanosomes. Nucleic Acids Res. 2009;37:1452–1462. doi: 10.1093/nar/gkn1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dipaolo C, Kieft R, Cross M, Sabatini R. Regulation of Trypanosome DNA glycosylation by a SWI2/SNF2-like protein. Mol. Cell. 2005;17:441–451. doi: 10.1016/j.molcel.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 21.Kieft R, Brand V, Ekanayake DK, Sweeney K, DiPaolo C, Reznikoff WS, Sabatini R. JBP2, a SWI2/SNF2-like protein, regulates de novo telomeric DNA glycosylation in bloodstream form Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;156:24–31. doi: 10.1016/j.molbiopara.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vainio S, Genest PA, ter Riet B, van Luenen H, Borst P. Evidence that J-binding protein 2 is a thymidine hydroxylase catalyzing the first step in the biosynthesis of DNA base J. Mol. Biochem. Parasitol. 2009;164:157–161. doi: 10.1016/j.molbiopara.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grover RK, Pond SJ, Cui Q, Subramaniam P, Case DA, Millar DP, Wentworth P., Jr O-glycoside orientation is an essential aspect of base J recognition by the kinetoplastid DNA-binding protein JBP1. Angew. Chem. Int. Ed. Engl. 2007;46:2839–2843. doi: 10.1002/anie.200604635. [DOI] [PubMed] [Google Scholar]
- 27.Sabatini R, Meeuwenoord N, van Boom JH, Borst P. Site-specific interactions of JBP with base and sugar moieties in duplex J-DNA. Evidence for both major and minor groove contacts. J. Biol. Chem. 2002;277:28150–28156. doi: 10.1074/jbc.M201487200. [DOI] [PubMed] [Google Scholar]
- 28.Muller M, Weigand JE, Weichenrieder O, Suess B. Thermodynamic characterization of an engineered tetracycline-binding riboswitch. Nucleic Acids Res. 2006;34:2607–2617. doi: 10.1093/nar/gkl347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman J, Egan D, Walter TS, Meged R, Berry I, Ben Jelloul M, Sussman JL, Stuart DI, Perrakis A. Towards rationalization of crystallization screening for small- to medium-sized academic laboratories: the PACT/JCSG+ strategy. Acta Crystallogr. D Biol. Crystallogr. 2005;61:1426–1431. doi: 10.1107/S0907444905024984. [DOI] [PubMed] [Google Scholar]
- 30.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 31.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 32.Grosse-Kunstleve RW, Adams PD. Substructure search procedures for macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1966–1973. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- 33.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skubak P, Ness S, Pannu NS. Extending the resolution and phase-quality limits in automated model building with iterative refinement. Acta Crystallogr. D Biol. Crystallogr. 2005;61:1626–1635. doi: 10.1107/S0907444905032233. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 39.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 41.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 42.Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 2006;78:1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- 43.Pascal BD, Chalmers MJ, Busby SA, Mader CC, Southern MR, Tsinoremas NF, Griffin PR. The Deuterator: software for the determination of backbone amide deuterium levels from H/D exchange MS data. BMC Bioinformatics. 2007;8:156. doi: 10.1186/1471-2105-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mooij WT, Cohen SX, Joosten K, Murshudov GN, Perrakis A. “Conditional restraints”: Restraining the free atoms in ARP/wARP. Structure. 2009;17:183–189. doi: 10.1016/j.str.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 46.Svergun DI, Barberato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 47.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 49.Shen A, Higgins DE, Panne D. Recognition of AT-rich DNA binding sites by the MogR repressor. Structure. 2009;17:769–777. doi: 10.1016/j.str.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cruz AK, Titus R, Beverley SM. Plasticity in chromosome number and testing of essential genes in Leishmania by targeting. Proc. Natl Acad. Sci. USA. 1993;90:1599–1603. doi: 10.1073/pnas.90.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dossin Fde M, Dufour A, Dusch E, Siqueira-Neto JL, Moraes CB, Yang GS, Cano MI, Genovesio A, Freitas-Junior LH. Automated nuclear analysis of Leishmania major telomeric clusters reveals changes in their organization during the parasite's life cycle. PLoS ONE. 2008;3:e2313. doi: 10.1371/journal.pone.0002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robertson AB, Dahl JA, Vagbo CB, Tripathi P, Krokan HE, Klungland A. A novel method for the efficient and selective identification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2011;39:e55. doi: 10.1093/nar/gkr051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Genest PA, Ter Riet B, Cijsouw T, van Luenen HG, Borst P. Telomeric localization of the modified DNA base J in the genome of the protozoan parasite Leishmania. Nucleic Acids Res. 2007;35:2116–2124. doi: 10.1093/nar/gkm050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.