Abstract
Human nuclear RNase P is required for transcription and processing of tRNA. This catalytic RNP has an H1 RNA moiety associated with ten distinct protein subunits. Five (Rpp20, Rpp21, Rpp25, Rpp29 and Pop5) out of eight of these protein subunits, prepared in refolded recombinant forms, bind to H1 RNA in vitro. Rpp20 and Rpp25 bind jointly to H1 RNA, even though each protein can interact independently with this transcript. Nuclease footprinting analysis reveals that Rpp20 and Rpp25 recognize overlapping regions in the P2 and P3 domains of H1 RNA. Rpp21 and Rpp29, which are sufficient for reconstitution of the endonucleolytic activity, bind to separate regions in the catalytic domain of H1 RNA. Common themes and discrepancies in the RNA-protein interactions between human nuclear RNase P and its related yeast and archaeal counterparts provide a rationale for the assembly of the fully active form of this enzyme.
INTRODUCTION
RNase P is an ubiquitous ribonucleoprotein (RNP) enzyme that is involved in the biogenesis of tRNA (1,2). The RNA subunit of RNase P is the catalytic moiety of the enzyme (3–6). In human, nuclear RNase P is composed of H1 RNA and at least ten distinct protein subunits, termed Rpp14, Rpp20, Rpp21, Rpp25, Rpp29, Rpp30, Rpp38, Rpp40, Pop5 and Pop1 (7–11). These proteins have homologs in nuclear RNase P of Saccharomyces cerevisiae (12) and other eukaryotes (13). Rpp21, Rpp29, Rpp30, Pop5 and Rpp38 have functional homologs in archaea (14,15), an indication that the RNP core of archaeal/eukaryal RNase P is made up in part of highly conserved proteins.
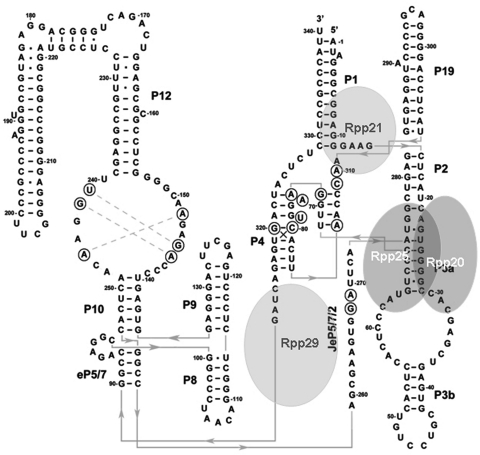
H1 RNA can be folded into a two-dimensional structure that consists of four potential co-axial helices (Figure 1). This proposed structure is based on models of eukaryal RNase P RNAs (16) and the crystal structures of bacterial counterparts (17,18). H1 RNA has two prominent structural features, the P3 and P4 domains (Figure 1) (16). The P3 domain is a helix–loop–helix RNA shown to be functional in nucleolar localization and its equivalent in yeast constitutes a major docking site for binding of various proteins (19–21). The P4 domain is a conserved pseudoknot critical for H1 RNA function in precursor tRNA cleavage (22).
Figure 1.
H1 RNA structure and its interaction with protein subunits. A proposed secondary structure of H1 RNA was deduced from the model of eukaryal RNase P RNA (16) and the crystal structures of bacterial RNase P RNAs (17,18). Four major coaxial helices are seen: P2–P3–P19, P1–P4-JeP5/7/2, P8–P9, eP5/7-P10–P12. The orientation of the extended part of P12 (positions 178–221) relative to the co-axial helix is unknown. Bases in circles are conserved. The approximate binding sites of the protein subunits Rpp20, Rpp25, Rpp21 and Rpp29 in H1 RNA are based on the nuclease footprinting analyses described in this study.
The endonucleolytic activity of human RNase P has been reconstituted in vitro (22). Thus, H1 RNA with recombinant Rpp21 and Rpp29 proteins, prepared in denatured forms, has endonucleolytic activity in a reaction buffer that contains 10 mM Mg2+ ions at pH 7.5 (22; this study). However, H1 RNA itself can remove the 5′ leader of precursor tRNA in the presence of high concentrations of Mg2+ ions at pH 6 (5). Reconstitution of the full activity of related archaeal RNase P RNPs has been obtained using recombinant Rpp21, Rpp29, Rpp30 and Pop5 proteins (15,23–28). A fifth protein subunit, Rpp38, also known as L7Ae, enhances the optimal reaction temperature of the reconstituted enzyme and increases its catalysis (15,26). Archaeal Rpp30/Pop5 and Rpp21/Rpp29 form heterodimers (28–30). Rpp30/Pop5 binds to the catalytic domain of its corresponding RNA moiety and acts on enhancing the catalytic efficiency of the reconstituted RNP, whereas the Rpp21/Rpp29 heterodimer recognizes the specificity domain and increases enzyme affinity (28–30).
Yeast three-hybrid genetic screens and UV crosslinking analysis using purified RNase P reveal that Rpp21, Rpp29, Rpp30 and Rpp38 interact with H1 RNA (31). Moreover, RNA–protein interaction studies using GST-fused recombinant proteins demonstrate that Rpp20, Rpp21, Rpp25, Rpp29, Rpp38 and Pop1, but not Rpp14 and Pop5, bind to H1 RNA in pull-down assays (32). Streptavidin pull-down experiments show that Rpp20, which was translated in vitro, does not interact with a biotinylated P3 RNA fragment, whereas Rpp25 weakly interacts with this piece of RNA (33). Similarly, Rpp20 does not interact with P3 RNA of human MRP RNA, while Rpp25 displays negligible binding affinity (34). Nonetheless, these two proteins form heterodimers that synergistically bind to this P3 RNA domain (33,34).
In S. cerevisiae, three-hybrid screens demonstrate that Pop1 and Pop4 (Rpp29) interact with Rpr1 RNA of nuclear RNase P (35), while filter binding assays show that recombinant Pop1, Pop3, Pop4, Pop6, Pop7 and Snm1 proteins bind to MRP RNA (36); Pop1 recognizes the P3 domain of Rpr1 RNA (19). Biochemical and structural studies establish that Pop6 and Pop7, the homologs of Rpp25 and Rpp20, contact the large internal loop and stem of the P3 domain of the MRP RNA and Rpr1 RNA (20,21). As is the case with its human counterpart, Pop6 binds to the P3 domain of these RNA transcripts only in the presence of Pop7, with which it forms a heterodimer (20,34).
To better define RNA–protein interactions in human RNase P, we have purified eight of its protein subunits in refolded recombinant forms and tested the properties of these polypeptides in binding to H1 RNA by gel shift assays and nuclease mapping analysis. Our findings reveal that the intermolecular interactions between H1 RNA and its cognate protein subunits exhibit common themes when compared with those shown for the archaeal and other eukaryal counterparts but some differences are observed.
MATERIALS AND METHODS
Preparation of recombinant protein subunits of human RNase P
Rpp14, Rpp20, Rpp21, Rpp25, Rpp29, Rpp30, Rpp38, Rpp40 and hPop5 were overexpressed as histidine-tagged proteins in Escherichia coli and were purified as previously described (8) but in refolded and soluble forms (Demma, M., Alfiya-Mor, N. and Wesolowski D., unpublished data). Briefly, each overexpressed recombinant protein was dissolved in 6 M urea before it was loaded onto a nickel-charged resin column (8). The columns with the bound proteins were sequentially washed with buffers that contained 4, 2 and 0 M urea to allow gradual protein folding. Elution of the proteins was done with an imidazole gradient in a buffer containing 20 mM Tris–HCl, pH 7.5, 50 mM NaCl and 2 mM MgCl2. DTT was added to the eluted fractions at a final concentration of 2 mM. Substantial amounts of Pop5 and Rpp29 were lost during the refolding stages (data not shown). In some cases, Pop5 and Rpp14 were purified on Sephacryl S100 gel filtration columns (Figure 2C). The purity (>95%) of the polypeptides was determined by SDS–PAGE followed by Coomassie blue staining or Ponceau red dye staining of equal concentrations of proteins transferred to nitrocellulose membranes. The membranes were subjected to Northwestern blot analysis as described previously (37).
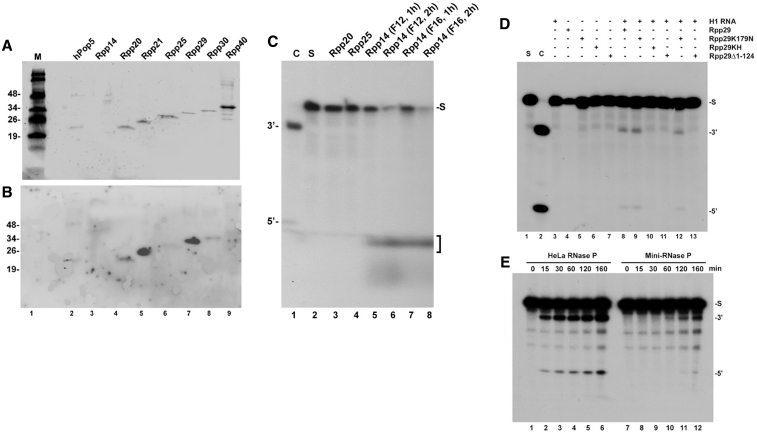
Figure 2.
Preparation of recombinant proteins and reconstitution of RNase P activity. (A) Refolded recombinant Pop5, Rpp14, Rpp20, Rpp21, Rpp25, Rpp29, Rpp30 and Rpp40 proteins (1 pmol each) were separated in SDS–PAGE, transferred to nitrocellulose membrane and stained with Ponceau red dye (37). Rpp14 was not stained using this method (lane 3). A protein size marker in kDa is shown. (B) Northwestern blot analysis of the membrane seen in (A) was performed using internally 32P-labeled H1 RNA as previously described (37). (C) 3′-5′ exoribonucleolytic activity of recombinant Rpp14 using labeled pSupS1 as substrate (lane 2; 0.1 pmol). Fractions F12 and F16 that correspond to the peak of Rpp14 eluted from a Sephacryl S-100 gel filtration column were examined in 1× Exo Buffer (10 mM Tris HC1, pH 8.0, 15 mM Na2HPO4, 50 mM KCl and 1 U of rRNasin) for 1 and 2 h as described by Jiang and Altman (38). The RNA products were analyzed in a denaturing 8% polyacrylamide gel and the degraded RNA (mononucleotides) are indicated by a bracket. Lane 1 shows pSupS1 cleaved by a DEAE-purified HeLa RNase P and used as size marker. (D) H1 RNA (1 pmol) was incubated with Rpp21 (1 pmol) in the presence of Rpp29 (2 pmol) or Rpp29K179N, Rpp29KH or Rpp29Δ1–124 (37) for 2 h on ice. Cleavage of pSupS1 (lane 1; 0.1 pmol) was then examined for 5 h in 2× MRP/1× TNET buffer (22; see ‘Materials and Methods’ section). Cleavage products were analyzed in denaturing 8% polyacrylamide gel. The positions of the substrate (S), tRNA (3′) and 5′ leader sequence (5′) are indicated. In lane 12, H1 RNA activity was reconstituted in the presence of Rpp21 and Rpp29 at a molar ratio of 1:1:1. Control for cleavage by a freshly prepared DEAE-purified HeLa nuclear RNase P (∼3 × 10−7 U) is shown (C; lane 2). (E) Kinetic of processing of pSupS1 by 1:100 diluted DEAE-purified RNase P versus reconstituted RNase P (also called mini-RNase P, which is composed of H1 RNA, Rpp21 and Rpp29).
Gel shift assays
An internally 32P-labeled H1 RNA (5–10 × 103 cpm) and excess amounts (0.5–1 µg) of poly(I:C) RNA were incubated in 100 µl total volumes with recombinant proteins in 1× MK buffer (10 mM Tris–HCl, pH 7.5, 150 mM KCl, 0–10 mM MgCl2, 1-5 mM DTT and 5% glycerol) for 15 min on ice and then for 20 min at room temperature. For binding of Rpp21 or Rpp29, incubation was in 2× TNET buffer (37) for 30 min on ice and 10 min at room temperature. 10 µl of 10× DNA loading dye (50% glycerol, 0.05% bromophenol blue and 0.05% xylene cyanol) were added, tubes were centrifuged for 5 min at 5000 rpm and protein–RNA complexes were separated in native 4–5% polyacrylamide gels (40:0.5 of acrylamide/bis), which contained 1 mM MgCl2 and 2% glycerol, in 1× TBE running buffer. These gels have low resolution capacity of RNA–protein complexes. H1 RNA appears in two separate bands, which most likely represent two conformers, even though it was extracted as a single band from denaturing gels. Nonetheless, the two bands of H1 RNA are shifted to a single complex in the presence of interacting protein, an inference that these conformers are equally bound by the protein. Bands were visualized by autoradiography and quantitated. K1/2 expresses the protein concentration at which 50% binding to H1 RNA was achieved. The gel shift assays have been repeated 3–5 times. Reproducible results were obtained.
Nuclease footprinting analysis
H1 RNA (60 pmol), transcribed in vitro, was dephosphorylated with calf alkaline phosphatase and then 5′-end labeled by incubation with 50 µCi of [γ-32P]ATP (3000 Ci/mmol) and T4 polynucleotide kinase. Labeled RNA was separated in a 5% polyacrylamide/8 M urea gel and the full-length H1 RNA band, 340-nt in length, was extracted. The labeled H1 RNA (1 × 104 cpm; ∼0.125 ng) was incubated without or with recombinant protein(s) at concentrations indicated in each experiment in a binding buffer (see above) that contained 100–500 ng of unlabeled H1 RNA. After incubation for 20 min on ice, the partial digestion reaction was in a final volume of 100 µl by addition of 1 U RNase T2 (Invitrogen) or RNase T1 (Sigma) for 5–30 min at 25°C. Proteins were then removed by Proteinase K/SDS and RNA was extracted by phenol, ethanol precipitated, resuspended in a loading buffer and analyzed in an 8% polyacrylamide sequencing gel.
Assay for RNase P activity
Cleavage of the 5′ leader of Schizosaccharomyces pombe precursor tRNASer (pSupS1) or E. coli precursor tRNATyr by purified or reconstituted RNase P was performed in 60 µl of 1× MRP buffer (20 mM Tris–HCl, pH 7.5; 10 mM MgCl2, 40 mM KCl, 20 μg/ml bovine serum albumin, 0.2 U of rRNasin, 1 mM DTT and 1 μg of poly I:C RNA) and 1× TNET buffer [20 mM Tris–HCl, pH 7.5; 35 mM NaCl, 0.1 mM Na2EDTA (disodium salt), 0.01% Triton X-100 and 1 mM β-mercaptoethanol) at 37°C (22). After pre-incubation of H1 RNA (1-2 pmol) with freshly prepared Rpp21 and Rpp29 (1–4 pmol) for 30 min on ice, labeled precursor tRNA (0.035–0.07 pmol) was added per assay and processing was performed at 37°C. The substrate was uniformly labeled with α[32P]GTP (3,000 mCi/mmol, Amersham). Time of incubation was as indicated in each experiment. The cleavage reactions were stopped by adding urea dye and products were separated in an 8% polyacrylamide/7 M urea gel. Bands were visualized by autoradiography and quantitated. A DEAE-purified HeLa nuclear RNase P was used as positive control for substrate cleavage. Freshly prepared DEAE-purified RNase P has a specific activity of ∼15 × 105 U/mg protein (7,8). One unit is defined as the amount of enzyme that is required to process 1 µmol of pSupS1 in 1 min at 37°C. This activity declines with time of storage of the enzyme.
RESULTS
Preparation of functional recombinant protein subunits of human RNase P in refolded states
A Ponceau red dye staining of recombinant Rpp14, Rpp20, Rpp21, Rpp25, Rpp29, Rpp30, Rpp40 and Pop5 proteins (8) revealed that seven of these proteins were purified to homogeneity (Figure 2A, lanes 2 and 4–9; see ‘Materials and Methods’ section). Rpp14 was not stained by this dye (Figure 2A, lane 3) but was detectable by silver staining or Western blot analysis using its corresponding antibodies (Alfiya-Mor, N. and N. Jarrous, data not shown).
To check if the polypeptides were functional, we tested Rpp14 for its known 3′–5′ exoribonucleolytic activity on an internally 32P-labeled precursor tRNASer (pSupS1)(38), whereas Rpp21 and Rpp29 were assessed in the activation of the 5′-endonucleolytic activity of H1 RNA (22) using pSupS1 as substrate. These ribonucleolytic activities of the individual subunits are relevant to RNase P function (22,38). Rpp30 and Rpp40 were not examined in any assay since these proteins are not known to exhibit any particular function. As expected, Rpp14 degraded pSupS1 in the absence of Mg2+ ions (Figure 2C, lanes 5–8; 38). Rpp21 and Rpp29 mediated the correct cleavage of the 5′ leader of pSupS1 by H1 RNA (Figure 2D, lane 8). H1 RNA, Rpp21 or Rpp29 alone were inactive (Figure 2D, lanes 3 and 4), an indication that these three subunits are needed for the cleavage reaction. This reconstituted activity was <0.01% of that exhibited by a DEAE-purified HeLa nuclear RNase P (Figure 2D, lane 2 versus 8) and <1% of the activity exhibited by 1:100 diluted enzyme (Figure 2E, lanes 1–6 versus 7–12). A slight increase in substrate cleavage was observed using Rpp29K179N (Figure 2D, lane 8 versus 9), a mutant protein shown previously to activate the M1 RNA subunit of E. coli RNase P (37). Lysine 179 is a conserved residue critical for protein function (37). Rpp29K179N worked optimally with H1 RNA and Rpp21 at a molar ratio of 2:1:1, respectively, while a less activity was obtained at an equimolar ratio of these subunits (Figure 2D, lane 9 versus 12), assuming that these subunits were equally soluble and functional. The truncated Rpp29Δ1–124 protein, which lacks the first 124 amino acids of Rpp29, and the mutant Rpp29KH (37) were both deficient in reconstitution of RNase P activity (Figure 2D, lanes 10 and 11).
These results confirm that Rpp21 and Rpp29 are functional in activation of H1 RNA, which mediates the removal of the 5′ leader of precursor tRNA (22). This result has been first reported in 2003 (22), and since then the assay has been repeated numerous times. For unknown reasons, not every preparation of H1 RNA was able to reconstitute the endonucleolytic activity of RNase P. About one of four independent preparations of H1 RNA synthesized in vitro was functional. Nonetheless, reconstitution assays performed with a functional preparation of H1 RNA were all successful.
Interaction of Rpp20, Rpp25, Rpp21, Rpp29 and Pop5 with H1 RNA
A rapid screen for potential interactions between H1 RNA and the eight recombinant protein subunits described above was performed by Northwestern blot analysis using a labeled H1 RNA as probe (37; see ‘Materials and Methods’ section). This analysis revealed binding of H1 RNA to Rpp21 and Rpp29 in a hybridization buffer that contained 200 mM NaCl (Figure 2B, lanes 5 and 7). Positive binding signals of this transcript with Rpp20, Rpp25, Pop5 and Rpp30, but not with Rpp14 and Rpp40, were also detected (Figure 2B).
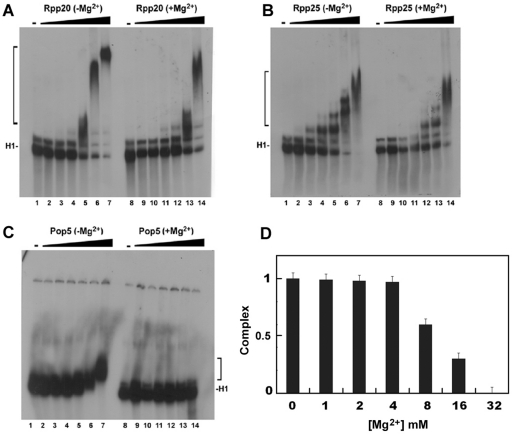
Gel shift assays corroborated the above findings that Rpp20, Rpp25 and Pop5 interact with H1 RNA (Figures 3A–C; see ‘Materials and Methods’ section). Rpp20 and Rpp25 bind to H1 RNA in the form of monomers and multimers, thus generating small and large complexes (Figure 3A and C). The large complexes were generated as a result of protein-protein interaction (self-oligomerization) at high protein concentrations (see below). A total of 50% binding of H1 RNA by Rpp20 and Rpp25, expressed as K1/2, was obtained at 0.5 and 0.23 µM, respectively (Figure 3E and F). These assays were performed in a buffer that contained 2 mM MgCl2, 150 mM KCl and excess amounts of poly I:C, which was used as non-specific competitor (see ‘Materials and Methods’ section). Similarly, Rpp21 and Rpp29 bound to H1 RNA but formed large complexes that remained in the gel wells (Figure 4A). These complexes could be released by the addition of unlabeled H1 RNA used as a specific competitor (data not shown). The calculated K1/2 for binding of Rpp21 and Rpp29 to H1 RNA were ∼0.5 and 0.14 µM, respectively (Figure 4B). Thus, these two proteins exhibit moderate binding affinity to naked H1 RNA. By contrast, Rpp14, Rpp30 or Rpp40 did not interact with this transcript (Supplementary Figure S1).
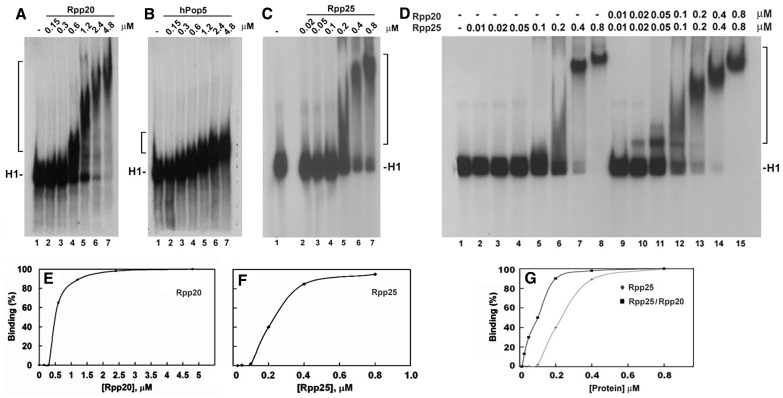
Figure 3.
Rpp20, Rpp25 and Pop5 bind to H1 RNA in gel shift assays. (A–C) Increasing concentrations (in µM) of Rpp20, Pop5 and Rpp25 were incubated with 32P-labeled H1 RNA (5000 cpm) and the formation of complexes was analyzed in 4% native gel and visualized by autoradiography (upper panel; see ‘Materials and Methods’ section). Rpp20 and Rpp25 form monomers and oligomers thus generating large complexes. H1 RNA appears as two bands in native gels (see C, lane 1) but both bands were shifted as one complex in the presence of Rpp25. (D) A gel shift assay using H1 RNA (lane 1) and increasing concentrations of Rpp25 alone (lanes 2–8) or in combination with Rpp20 added at indicated equimolar ratios (lanes 9–15). Proteins were pre-incubated for 30 min before addition of H1 RNA. Complexes are indicated by a bracket. (E and F) Quantitation of H1 RNA–Rpp20 and H1 RNA–Rpp25 complexes, which include single and multiple proteins bound to H1 RNA (37). The K1/2 was calculated as the protein concentration at which 50% binding of protein to H1 RNA was seen. (G) Quantitation of Rpp25/H1 RNA and Rpp20/Rpp25/H1 RNA complexes seen in (D).
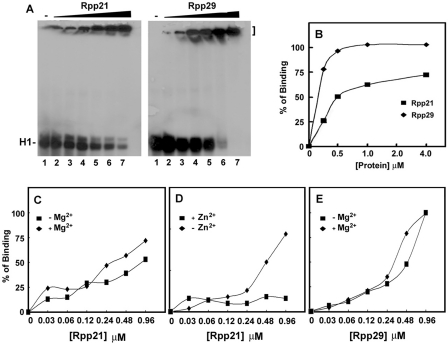
Figure 4.
Binding of recombinant Rpp21 and Rpp29 to H1 RNA. (A) Gel shift assays using increasing concentrations (0.03-0.92 µM) of Rpp21 and Rpp29 were done as described in Figure 3. These two soluble proteins formed complexes with H1 RNA that were trapped in the gel wells. (B) Gel shift assays were done as in panel A but protein concentrations were as indicated and percentage of binding to H1 RNA was quantitated and plotted. (C–E) Binding of H1 RNA by Rpp21 and Rpp29 in the presence or absence of 10 mM MgCl2 or ZnCl2 in the binding buffer.
The combination of increasing concentrations of Rpp20 with Rpp25, which were pre-incubated together, increased their binding affinity to H1 RNA, when compared with the affinity exhibited by Rpp25 (Figure 3D, lanes 2–8 versus 9–15; Supplementary Table S1) or by Rpp20 (Figure 3A and Supplementary Table S1) alone. These two proteins bind jointly to H1 RNA with 50% binding seen at protein concentration of ∼0.1 µM of each protein (Figure 3G and Supplementary Table S1). Rpp20 and Rpp25 functioned optimally at equimolar ratios (data not shown), thus supporting the concept that they heterodimerize (34) as soluble and functional proteins.
Effect of Mg2+ ions on H1 RNA–protein interactions
Mg2+ ions are essential for substrate cleavage by H1 RNA (5) and by human RNase P at concentrations of ∼20–30 mM (22). This divalent ion is also critical for H1 RNA folding (5). However, higher concentrations of Mg2+ ions impede the activity of RNase P (data not shown). We tested the effect of Mg2+ ions on the binding of H1 RNA by Rpp20, Rpp21, Rpp25, Rpp29 and Pop5 (Supplementary Table S1). The presence of 10 mM Mg2+ ions impeded the binding of Rpp20 and Rpp25 to H1 RNA (Figure 5A, lane 5 versus 12 and 5B, lane 4 versus 11; Supplementary Table S1) and inhibited the interaction of Pop5 with this transcript (Figure 5C, lane 7 versus 14). In contrast, binding of H1 RNA by Rpp21 was enhanced in the presence of Mg2+ ions (Figure 4C; Supplementary Table S1), while the interaction of Rpp29 with this RNA was optimal at 2 mM Mg2+ ions (Figure 4E; Supplementary Table S1). Severe inhibition of complex formation of Rpp20/Rpp25 with H1 RNA was seen at 32 mM of this divalent ion (Figure 5D), a concentration that is favorable for the activity of human RNase P (22). The presence of Zn2+ ions had a strong inhibitory effect on binding of H1 RNA by Rpp21 (Figure 4D; Supplementary Table S1), which has a potential zinc metal-binding motif (11).
Figure 5.
Effect of Mg2+ ions on the interaction of Rpp20, Rpp25 and Pop5 with H1 RNA. (A–C) Gel shift assays were performed as describe in Figures 3 and 4 in the presence or absence of 10 mM MgCl2 in the binding buffer. Protein concentrations for Rpp20 were 0, 0.12, 0.24, 0.48, 0.96, 1.92 and 3.84 µM, for Rpp25 were 0, 0.03, 0.06, 0.12, 0.24, 0.48 and 0.96 µM and for Pop5 were 0, 0.125, 0.25, 0.5, 1, 2 and 4 µM. (D) Effect of increasing concentrations of MgCl2 on binding of Rpp20/Rpp25 to H1 RNA. The two proteins were added at equimolar ratio, 0.8 µM each. The ternary complexes were quantitated and plotted.
These findings reveal profound effects of high concentrations of a divalent ion on the intermolecular interaction between RNA and protein subunits of RNase P. These effects could be direct or indirect through alteration of RNA folding.
Rpp20, Rpp25, Rpp21 and Rpp29 bind to the catalytic domain of H1 RNA
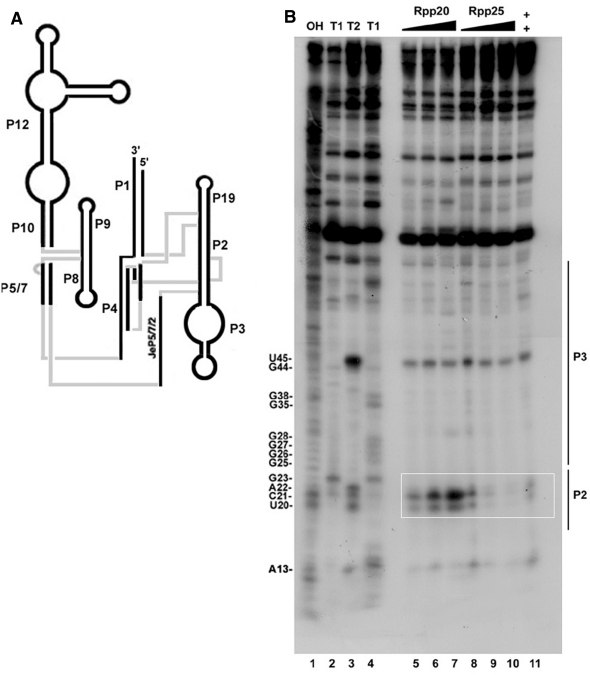
Footprinting cleavage analysis using RNase T2 revealed that binding of Rpp20 to H1 RNA protected this transcript at several positions in the P2 and P3 domains (Figure 6B, lanes 5–7 versus 8–10). The most prominent positions protected from nuclease attacks were U45, which is located in the apical loop of P3, and the A22 and U20 residues positioned in P2 (Figure 6B, lane 7 versus 8–10). C21 became more sensitive to cleavage in the presence of this protein (Figure 6B, lane 7 versus 9). Cleavage of A22 and U20 indicates that they are not base paired in P2, as is predicted by the two-dimensional model of H1 RNA (Figure 1). In a longer exposure of the film, we noticed that the sequence that spans positions U45 to A22 was shielded from nuclease attacks at several positions in the presence of Rpp20 (Figure 6B, lane 3 versus 5–7). In contrast, binding of Rpp25 to H1 RNA was more confined, as manifested by protection of U20, C21 and A22 from nuclease cleavages (Figure 6B, lanes 3 versus 8–10). As expected, the presence of both Rpp20 and Rpp25 primarily protected U20, C21 and A22 (Figure 6B, lane 3 versus 7 and 11). Footprinting analysis using RNase T1 was not informative, as this nuclease exclusively attacks guanosine residues in single stranded regions in refolded H1 RNA that seem not to be protected by Rpp20 and Rpp25 (data not shown).
Figure 6.
RNase T2 footprint analysis of H1 RNA bound by Rpp20 and Rpp25. (A) A model of H1 RNA structure. (B) RNase T2 footprint analysis of 5′-end 32P-labeled H1 RNA bound by 0.5, 1 and 2 µM of Rpp20 (lanes 5–7) or Rpp25 (lanes 8–10) or both proteins at a final concentration of 1 µM each (lane 11). Alkali ladder (lane 1) and partial digestion of H1 RNA using 0.4 units of RNase T1 (lane 2) or RNase T2 without protein (lane 3) were used as size markers. Lane 4 shows RNase T1 cleavage of H1 RNA that produced G ladder. Positions of nucleotides and the approximate location of the P2 and P3 domains are shown. The overlapping region protected by Rpp20 and Rpp25 is boxed.
The nuclease mapping analyses show that Rpp20 and Rpp25 recognize the P2 and P3 domains, both of which are found on the same co-axial helix in H1 RNA (Figure 6A). The observation that similar patterns of protection were seen in the presence of low and high concentration of Rpp20 and Rpp25 suggests that the large complexes formed with H1 RNA (Figure 3A and C, lanes 6 and 7) are generated as a result of protein–protein interaction (self-oligomerization) and not due to RNA–protein interactions with distinct sites in this transcript. The protection of U45 by these two proteins may be due to alteration in H1 RNA structure when bound by these proteins, even though direct contact of Rpp20/Rpp25 with the apical loop of P3 is not excluded (39). The binding of Rpp20 and Rpp25 to the P3 domain of H1 RNA agrees with the mapping analyses that demonstrate that the yeast homologs Pop7 and Pop6 contact the equivalent P3 domain in the MRP RNA (20,21).
The nuclease mapping patterns of H1 RNA bound by Rpp21 and Rpp29 were rather distinct from those obtained with Rpp20 and Rpp25. Thus, RNase T2 footprinting analysis showed that Rpp21 protected in a dose-dependent manner the cleavage of A13, which is located in the junction that links P1 with P2 (Figure 7B, lanes 5–7 versus 4 and Supplementary Figure S2, lanes 3–5 versus 1). Cleavage of positions A22, C21 and U20 in P2 was also reduced (Figures 7B and Supplementary Figure S3). No additional cleavage sites, particularly those in the specificity domain of H1 RNA, were protected by Rpp21 (Figure 7B and Supplementary Figure S2; S-domain). These results are in accordance with the observation that endogenous Rpp21 could be crosslinked to an RNA fragment that is composed of the first 74 nt of H1 RNA (31). In contrast to Rpp21, which mainly protected A13, Rpp29 shielded A85, which was one of the foremost residues protected from nuclease attacks (Figure 7C, lanes 1–3 versus 4–7 and 8–10; Supplementary Figure S3, lane 5 versus 1). The residue A85 is located in the junction that links P4 with eP5/7 (Figure 1). A decrease in the cleavage of the region spanning positions A85 to A13, which corresponds to the P2 and P3 domains, was evident in the presence of Rpp29 (Figure 7C, lanes 1–3 versus 4–7 and 8–10; Supplementary Figure S3, lanes 3–5 and lanes 7–10 versus lanes 1 and 2). Rpp29 did not affect the cleavage pattern of the specificity domain (Figure 7C and Supplementary Figure S3), even after fine resolution of this region by longer runs in sequencing gels (data not shown). As expected, the combination of Rpp21 and Rpp29 increased the protection of the entire region from A85 to A13 in the catalytic domain of H1 RNA, whereas no alteration in the nuclease sensitivity pattern was seen in the specificity domain (Supplementary Figure S2, lanes 7–8 versus lane 1).
Figure 7.
RNase T2 footprint analysis of H1 RNA bound by Rpp21 and Rpp29. (A) A model of H1 RNA structure. (B) The 5′-end 32P-labeled H1 RNA was incubated without or with Rpp21 at final concentrations of 0.5, 1 and 2 µM (lane 4 versus 5–7) in a binding buffer for 20 min and then digested with 1 U of RNase T2 for 5 min at 25°C. The specificity (S) domain is indicated. Positions of key nucleotides and RNA size markers (lanes 1 and 2) are shown. Position of A13 protected by Rpp21 is boxed. (C) Digestion of 5′-end-labeled H1 RNA with RNase T2 for the indicated times in the absence (lanes 1–3) or presence of increasing concentrations of Rpp29 (lanes 4–7). In lanes 8–10, identical concentrations of Rpp29 were used but digestion with the nuclease was for the indicated times. Position of A85 protected by Rpp29 is boxed.
The nuclease mapping analyses demonstrate that Rpp20 and Rpp25 bind to the P2/P3 domain (Figure 1), while Rpp21 and Rpp29 mainly interact with two distinct junctions in the catalytic domain of H1 RNA (Figure 1).
DISCUSSION
We have studied intermolecular interactions between the RNA and protein subunits of human nuclear RNase P using defined recombinant components. Five proteins, Rpp20, Rpp25, Rpp21, Rpp29 and Pop5, bind directly to H1 RNA in vitro. We have also demarcated the binding regions of Rpp20, Rpp21, Rpp25 and Rpp29 in this catalytic RNA. Other protein subunits of RNase P, such as Rpp14, Rpp30 and Rpp40, exhibit no binding capability to H1 RNA and therefore they may assemble with the RNP through protein–protein interactions. Elucidation of these RNA–protein interactions could be helpful in understanding the assembly process of human RNase P and the development of efficient reconstitution systems for this nuclear RNP complex.
The two Alba-like protein subunits, Rpp20 and Rpp25, recognize regions in the P3 and P2 domain in H1 RNA. This result is consistent with findings that show that these two conserved proteins bind to the P3 domain of human MRP RNA (33,34,39) and that the yeast homologs, Pop7 and Pop6, bind as heterodimers to the equivalent P3 domain in the MRP RNA with comparable affinities (20,21). Accordingly, these evolutionary related proteins exhibit common properties in binding to this functional domain in eukaryal RNase P and RNase MRP RNAs. Moreover, Rpp20 and Rpp25 and their yeast homologs heterodimerize and bind jointly to their cognate RNAs. However, while Rpp25 can bind to H1 RNA (this study), the yeast counterpart Pop6 binds to its Rpr1 RNA only in the presence of Pop7, with which it forms heterodimers (20,34). In the case of the P3 RNA of human MRP RNA, Rpp20 does not bind to this RNA fragment and Rpp25 recognizes this RNA with an insignificant affinity, Kd = 1 mM (34). However, the combination of Rpp20 and Rpp25 elicits their heterodimerization and the resulting heterodimer binds to the P3 RNA with high affinity, which was 1000-fold higher than that obtained with Rpp25 alone, thus demonstrating a synergistic effect (34). In the case of H1 RNA, however, Rpp20 and Rpp25 individually bind to this transcript with moderate affinity, while the combination of both proteins enhances further their binding affinity. The final binding affinities of Rpp20 and Rpp25 to either H1 RNA or P3 RNA are comparable (33,34, this study).
The ability of Rpp20 or Rpp25 to recognize H1 RNA seems to be valid also for binding to DNA, as both proteins belong to a superfamily of chromatin binding proteins. This conclusion is supported by observation that demonstrates that Rpp20 binds to chromatin of rRNA genes at early G1 phase in synchronized human cells, while Rpp25 occupies these genetic loci at late G1 or early S phase (40). The dual ability of Rpp20 and Rpp25 in binding to RNA and DNA implicates these subunits in structural roles, such as in the assembly of RNase P and its connection to chromatin (Dehtiar et al., manuscript submitted for publication). It has been noted that many chromatin binding proteins form dimers or higher order oligomers essential for their function in chromatin structure and function in gene expression (41). The P3 domain of H1 RNA, to which Rpp20 and Rpp25 bind, has been shown to be critical for nucleolar localization of this transcript (11,32,33). The differences in the nucleic acid binding properties between Rpp20 and Rpp25 and their yeast homologs could reflect on the mode of assembly of their corresponding RNP complexes and by this means on their biological roles.
Rpp21 and Rpp29 have the capability to reconstitute the endonucleolytic activity of H1 RNA in tRNA processing in vitro (Figure 2; ref. 22). These two conserved proteins act as cofactors in RNA-based catalysis by playing a key role in recognition of precursor tRNA substrates (22; see below). Nuclease footprinting analyses indicate that Rpp21 and Rpp29 contact the catalytic domain of H1 RNA, consistent with previous reports (31,32). Specifically, Rpp21 seems to interact with the junction that connects P1 with P2, a finding that agrees with other studies in which it has been shown that endogenous Rpp21 could be crosslinked to an RNA fragment that consists of the first 74 nt of H1 RNA (31). Rpp29 protects the junction that bridges the P4 with the specificity domain from nuclease attack (Figure 7 and Supplementary Figure S3). These two junctions link co-axial helices in H1 RNA (Figure 1) and thereby their recognition by Rpp21 and Rpp29 involves proper folding of H1 RNA. In fact, the proximity of binding of Rpp29 to P4, which is essential for the formation of the catalytic center of RNase P RNA (16), is consistent with this protein being a co-factor in catalysis. This role is corroborated by the ability of Rpp29 from human and Dictyostelium discoideum to associate and activate M1 RNA in vitro (37,42). UV crosslinking experiments and oligonucleotide interference assays reveal that Rpp29 recognizes precursor tRNA and not its mature tRNA form (37). Moreover, nuclease mapping analysis shows that Rpp29 protects position G(-5) in pSupS1 (Supplementary Figure S4), similarly to protein cofactors of bacterial RNase P that bind to the catalytic domain of the RNA (43,44) and contact the 5′ leader of precursor tRNA (44,45). Archaeal Rpp21 and Rpp29, which recognize the specificity domain of their cognate RNAs (30,46), also play a role in substrate recognition and cleavage site selection by reconstituted archaeal RNPs (47). Accordingly, the function of these two protein cofactors seems to have been preserved from archaea to humans, even though their arrangement in their corresponding RNPs may differ (48). In this context, it will be interesting to map the binding sites of the protein subunits associated with the Crenarchaeon Pyrobaculum aerophilum RNase P RNA shown to have a miniature specificity domain, if any (49).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Israel Science Foundation (grant no. 673/06); United States–Israel Binational Science Foundation (grant no. 2005/009 to N.J.). Funding for open access charge: Israel Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Venkat Gopalan, Ohio State University, Columbus, USA, and Leif Kirsebom, Uppsala University, Sweden, for their valuable comments on this article.
REFERENCES
- 1.Altman S. The road to RNase P. Nat. Struct. Biol. 2000;7:827–828. doi: 10.1038/79566. [DOI] [PubMed] [Google Scholar]
- 2.Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 2006;31:333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 4.Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. RNase P RNAs from some Archaea are catalytically active. Proc. Natl Acad. Sci. USA. 1999;96:7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kikovska E, Svärd SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc. Natl Acad. Sci. USA. 2007;104:2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willkomm DK, Hartmann RK. An important piece of the RNase P jigsaw solved. Trends Biochem. Sci. 2007;32:247–250. doi: 10.1016/j.tibs.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Eder PS, Kekuda R, Stolc V, Altman S. Characterization of two scleroderma autoimmune antigens that copurify with human ribonuclease P. Proc. Natl Acad. Sci. USA. 1997;94:1101–1106. doi: 10.1073/pnas.94.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarrous N, Altman S. Human ribonuclease P. Methods Enzymol. 2001;342:93–100. doi: 10.1016/s0076-6879(01)42538-9. [DOI] [PubMed] [Google Scholar]
- 9.van Eenennaam H, Lugtenberg D, Vogelzangs JHP, van Venrooij WJ, Pruijn GJM. hPop5, a protein subunit of the human RNase MRP and RNase P endoribonucleases. J. Biol. Chem. 2001;276:31635–31641. doi: 10.1074/jbc.M103399200. [DOI] [PubMed] [Google Scholar]
- 10.Guerrier-Takada C, Eder PS, Gopalan V, Altman S. Purification and characterization of Rpp25, an RNA-binding protein subunit of human ribonuclease P. RNA. 2002;8:290–295. doi: 10.1017/s1355838202027954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jarrous N. Human ribonuclease P: subunits, function, and intranuclear localization. RNA. 2002;8:1–7. doi: 10.1017/s1355838202011184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao S, Scott F, Fierke CA, Engelke DR. Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem. 2002;71:165–189. doi: 10.1146/annurev.biochem.71.110601.135352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartmann E, Hartmann RK. The enigma of ribonuclease P evolution. Trends Genet. 2003;19:561–569. doi: 10.1016/j.tig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Hall TA, Brown JW. Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA. 2002;8:296–306. doi: 10.1017/s1355838202028492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho IM, Lai LB, Susanti D, Mukhopadhyay B, Gopalan V. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc. Natl Acad. Sci. USA. 2010;107:14573–14578. doi: 10.1073/pnas.1005556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marquez SM, Chen JL, Evans D, Pace NR. Structure and function of eukaryotic ribonuclease P RNA. Mol. Cell. 2006;24:445–456. doi: 10.1016/j.molcel.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl Acad. Sci. USA. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 19.Ziehler WA, Morris J, Scott FH, Millikin C, Engelke DR. An essential protein-binding domain of nuclear RNase P RNA. RNA. 2001;7:565–575. doi: 10.1017/s1355838201001996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perederina A, Esakova O, Koc H, Schmitt ME, Krasilnikov AS. Specific binding of a Pop6/Pop7 heterodimer to the P3 stem of the yeast RNase MRP and RNase P RNAs. RNA. 2007;13:1648–1655. doi: 10.1261/rna.654407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perederina A, Esakova O, Quan C, Khanova E, Krasilnikov AS. Eukaryotic ribonucleases P/MRP: the crystal structure of the P3 domain. EMBO J. 2010;29:761–769. doi: 10.1038/emboj.2009.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mann H, Ben-Asouli Y, Schein A, Moussa S, Jarrous N. Eukaryotic RNase P: role of RNA and protein subunits of a primordial catalytic ribonucleoprotein in RNA-based catalysis. Mol. Cell. 2003;12:925–935. doi: 10.1016/s1097-2765(03)00357-5. [DOI] [PubMed] [Google Scholar]
- 23.Kouzuma Y, Mizoguchi M, Takagi H, Fukuhara H, Tsukamoto M, Numata T, Kimura M. Reconstitution of archaeal ribonuclease P from RNA and four protein components. Biochem. Biophys. Res. Commun. 2003;306:666–673. doi: 10.1016/s0006-291x(03)01034-9. [DOI] [PubMed] [Google Scholar]
- 24.Boomershine WP, McElroy CA, Tsai HY, Wilson RC, Gopalan V, Foster MP. Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P. Proc. Natl Acad. Sci. USA. 2003;100:15398–15403. doi: 10.1073/pnas.2535887100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai HY, Pulukkunat DK, Woznick WK, Gopalan V. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc. Natl Acad. Sci. USA. 2006;103:16147–16152. doi: 10.1073/pnas.0608000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukuhara H, Kifusa M, Watanabe M, Terada A, Honda T, Numata T, Kakuta Y, Kimura M. A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 2006;343:956–964. doi: 10.1016/j.bbrc.2006.02.192. [DOI] [PubMed] [Google Scholar]
- 27.Terada A, Honda T, Fukuhara H, Hada K, Kimura M. Characterization of the archaeal ribonuclease P proteins from Pyrococcus horikoshii OT3. J. Biochem. 2006;140:293–298. doi: 10.1093/jb/mvj144. [DOI] [PubMed] [Google Scholar]
- 28.Pulukkunat DK, Gopalan V. Studies on Methanocaldococcus jannaschii RNase P reveal insights into the roles of RNA and protein cofactors in RNase P catalysis. Nucleic Acids Res. 2008;36:4172–4180. doi: 10.1093/nar/gkn360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honda T, Kakuta Y, Kimura K, Saho J, Kimura M. Structure of an archaeal homolog of the human protein complex Rpp21-Rpp29 that is a key core component for the assembly of active ribonuclease P. J. Mol. Biol. 2008;384:652–662. doi: 10.1016/j.jmb.2008.09.056. [DOI] [PubMed] [Google Scholar]
- 30.Xu Y, Amero CD, Pulukkunat DK, Gopalan V, Foster MP. Solution structure of an archaeal RNase P binary protein complex: formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions. J. Mol. Biol. 2009;393:1043–1055. doi: 10.1016/j.jmb.2009.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang T, Guerrier-Takada C, Altman S. Protein-RNA interactions in the subunits of human nuclear RNase P. RNA. 2001;7:937–941. doi: 10.1017/s1355838201010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welting TJ, van Venrooij WJ, Pruijn GJ. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004;32:2138–2146. doi: 10.1093/nar/gkh539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welting TJ, Peters FM, Hensen SM, van Doorn NL, Kikkert BJ, Raats JM, van Venrooij WJ, Pruijn GJ. Heterodimerization regulates RNase MRP/RNase P association, localization, and expression of Rpp20 and Rpp25. RNA. 2007;13:65–75. doi: 10.1261/rna.237807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hands-Taylor KL, Martino L, Tata R, Babon JJ, Bui TT, Drake AF, Beavil RL, Pruijn GJ, Brown PR, Conte MR. Heterodimerization of the human RNase P/MRP subunits Rpp20 and Rpp25 is a prerequisite for interaction with the P3 arm of RNase MRP RNA. Nucleic Acids Res. 2010;38:4052–4066. doi: 10.1093/nar/gkq141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houser-Scott F, Xiao S, Millikin CE, Zengel JM, Lindahl L, Engelke DR. Interactions among the protein and RNA subunits of Saccharomyces cerevisiae nuclear RNase P. Proc. Natl Acad. Sci. USA. 2002;99:2684–2689. doi: 10.1073/pnas.052586299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aspinall TV, Gordon JM, Bennett HJ, Karahalios P, Bukowski JP, Walker SC, Engelke DR, Avis JM. Interactions between subunits of Saccharomyces cerevisiae RNase MRP support a conserved eukaryotic RNase P/MRP architecture. Nucleic Acids Res. 2007;35:6439–6450. doi: 10.1093/nar/gkm553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharin E, Schein A, Mann H, Ben-Asouli Y, Jarrous N. RNase P: role of distinct protein cofactors in tRNA substrate recognition and RNA-based catalysis. Nucleic Acids Res. 2005;33:5120–5132. doi: 10.1093/nar/gki828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang T, Altman S. A protein subunit of human RNase P, Rpp14, and its interacting partner, OIP2, have 3′–>5′ exoribonuclease activity. Proc. Natl Acad. Sci. USA. 2002;99:5295–5300. doi: 10.1073/pnas.072083699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welting TJ, Mattijssen S, Peters FM, van Doorn NL, Dekkers L, van Venrooij WJ, Heus HA, Bonafé L, Pruijn GJ. Cartilage-hair hypoplasia-associated mutations in the RNase MRP P3 domain affect RNA folding and ribonucleoprotein assembly. Biochim. Biophys. Acta. 2008;1783:455–466. doi: 10.1016/j.bbamcr.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 40.Reiner R, Krasnov-Yoeli N, Dehtiar Y, Jarrous N. Function and assembly of a chromatin-associated RNase P that is required for efficient transcription by RNA polymerase I. PLoS One. 2008;3:e4072. doi: 10.1371/journal.pone.0004072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marianayagam NJ, Sunde M, Matthews JM. The power of two: protein dimerization in biology. Trends Biochem. Sci. 2004;29:618–625. doi: 10.1016/j.tibs.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 42.Stamatopoulou V, Toumpeki C, Tzakos A, Vourekas A, Drainas D. Domain architecture of the DRpp29 protein and its interaction with the RNA subunit of Dictyostelium discoideum RNase P. Biochemistry. 2010;49:10714–10727. doi: 10.1021/bi101297z. [DOI] [PubMed] [Google Scholar]
- 43.Buck AH, Kazantsev AV, Dalby AB, Pace NR. Structural perspective on the activation of RNase P RNA by protein. Nat. Struct. Mol. Biol. 2005;12:958–964. doi: 10.1038/nsmb1004. [DOI] [PubMed] [Google Scholar]
- 44.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragón A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784–789. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koutmou KS, Zahler NH, Kurz JC, Campbell FE, Harris ME, Fierke CA. Protein-precursor tRNA contact leads to sequence-specific recognition of 5′ leaders by bacterial ribonuclease P. J. Mol. Biol. 2010;396:195–208. doi: 10.1016/j.jmb.2009.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Honda T, Hara T, Nan J, Zhang X, Kimura M. Archaeal homologs of human RNase P protein pairs Pop5 with Rpp30 and Rpp21 with Rpp29 work on distinct functional domains of the RNA subunit. Biosci. Biotechnol. Biochem. 2010;74:266–273. doi: 10.1271/bbb.90550. [DOI] [PubMed] [Google Scholar]
- 47.Sinapah S, Wu S, Chen Y, Pettersson BM, Gopalan V, Kirsebom LA. Cleavage of model substrates by archaeal RNase P: role of protein cofactors in cleavage-site selection. Nucleic Acids Res. 2010;39:1105–1116. doi: 10.1093/nar/gkq732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jarrous N, Gopalan V. Archaeal/Eukaryal RNase P: subunits, functions and RNA diversification. Nucleic Acids Res. 2010;38:7885–7894. doi: 10.1093/nar/gkq701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai LB, Chan P, Cozen A, Bernick DL, Brown J, Gopalan V, Lowe T. Discovery of the elusive Pyrobaculum RNase P: an unexpected form of an ancient RNA. Proc. Natl Acad. Sci. USA. 2010;107:22493–22498. doi: 10.1073/pnas.1013969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.