Abstract
Regulated gene expression is essential for a proper progression through the cell cycle. The transcription factor NF-Y has a fundamental function in transcriptional regulation of cell cycle genes, particularly of G2/M genes. In order to investigate common and distinct functions of NF-Y subunits in cell cycle regulation, NF-YA, NF-YB and NF-YC have been silenced by shRNAs in HCT116 cells. NF-YA loss led to a delay in S-phase progression, DNA damage and apoptosis: we showed the activation of the replication checkpoint, through the recruitment of Δp53 and of the replication proteins PCNA and Mcm7 to chromatin. Differently, NF-YB depletion impaired cells from exiting G2/M, but did not interfere with S-phase progression. Gene expression analysis of NF-YA and NF-YB inactivated cells highlighted a common set of hit genes, as well as a plethora of uncommon genes, unveiling a different effect of NF-Y subunits loss on NF-Y binding to its target genes. Chromatin extracts and ChIP analysis showed that NF-YA depletion was more effective than NF-YB in hitting NF-Y recruitment to CCAAT-promoters. Our data suggest a critical role of NF-Y expression, highlighting that the lack of the single subunits are differently perceived by the cells, which activate diverse cell cycle blocks and signaling pathways.
INTRODUCTION
The histone-like transcription factor NF-Y consists of three subunits: NF-YA, NF-YB and NF-YC, which are all necessary for DNA binding of CCAAT boxes (1). NF-YB and NF-YC evolutionarily conserved core regions comprise a histone-fold motif, through which they interact with each other and with NF-YA. While NF-YB and NF-YC are ubiquitously expressed and their levels are relatively constant during the different phases of the cell cycle, NF-YA levels fluctuate throughout the cell cycle (2,3). Its expression increases at the onset of S phase and is reduced in IMR-90 fibroblasts after serum deprivation and in human monocytes (4,5). Although mRNA levels of NF-YA are relatively constant in growing and differentiated cells (2,6), it was recently reported that the ubiquitin–proteasome pathway and the acetylation status regulate NF-YA expression and consequently the functional activity of NF-Y (7). NF-YA is expressed as either a long- or short-form splice variant. The short isoform lacks 28 amino acids within the glutamine-rich domain, but maintains the C-terminal NF-YB/NF-YC interaction and DNA binding domains (8). The expression levels of the two isoforms vary in different cell types and a switch in their relative abundance was recently observed during THE differentiation of mouse ES cells (3,6–9).
The NF-Y complex is a fundamental player in the regulation of cell proliferation, supporting basal transcription of numerous cell cycle genes (10–12). In silico studies identified NF-Y as a common transcription factor associated to the regulatory module controlling cell cycle-dependent transcription of G2/M genes (13,14). In vivo, chromatin immunoprecipitation (ChIP) experiments demonstrated that the transcriptional activity of NF-Y during the cell cycle depends upon its timely regulated binding to cell cycle promoters (2,12,15). Additionally, NF-Y plays a pivotal role in the DNA-damage response, mediating the p53-dependent repression of G2/M genes. p53 negatively regulates NF-Y activity by decreasing the Cdk2-dependent phosphorylation of NF-YA. Moreover, a direct interaction of p53 with NF-Y has been observed on multiple CCAAT promoters (16–19).
ChIP on CHIP analysis highlights a bi-functional activity of NF-Y, which activates or represses transcription depending on the cellular and/or chromatin contexts (20,21). The function of NF-Y in gene transcription and cell cycle regulation has been investigated by expressing NF-YA dominant-negative mutants (NF-YA/DN) or by conditional inactivation of NF-YA. Stable expression of NF-YA/DN in mouse fibroblasts delays S-phase progression (22). A truncated NF-YA/Bdbd mutant, lacking the C-terminal transcription activation domain, leads to cell cycle arrest specifically at G2/M without apoptosis (23). Conditional inactivation of both NF-YA alleles in primary cultures of mouse embryonic fibroblasts (MEFs) completely blocks the progression of the cells into S phase and subsequently leads cells to apoptosis (24).
Our recent analysis of cell cycle progression upon NF-YB siRNA transfections highlights a different scenario from NF-YA deletion: NF-YB silenced cells show a massive delay in G2/M progression and only a small proportion of cells is impaired in progression to S phase (25). Microarray and reverse transcription polymerase chain reaction (RT-PCR) data indicate that genes of G2/M phase are repressed as well as genes for microtubule/cytoskeleton and spindle formation. We demonstrated that NF-YB knockdown activates p53 and its pro-apoptotic target genes, in the absence of DNA-damage.
In this study, we performed cellular and molecular analysis of HCT116 cells infected with shRNAs to specifically knock down NF-YA, NF-YB or NF-YC. Our data (i) highlight different phenotypes and transcriptional landscapes associated to each subunit silencing and (ii) suggest that NF-YA expression is fundamental to protect cells from DNA-damage induced apoptosis.
MATERIALS AND METHODS
Cell lines
The human colon carcinoma HCT116 and HCT116 p53−/− were generously provided by Bert Volgestein (Johns Hopkins University School of Medicine, Baltimore, MD). The cells were maintained as previously described (18,25,26). Human cervical carcinoma Hela and human colorectal carcinoma LoVo cell lines were maintained in DMEM supplemented with 10% FBS.
Short hairpin RNA
pLKO.1 non-target short hairpin RNA (shRNA) (SHC002), shRNA-NF-YA, targeting exon 6 (CCATCGTCTATCAACCAGTTA), shRNA-NF-YB targeting ex 5 (GCTATGTCTACTTTAGGCTTT) and shNF-YC targeting exon 5 (GCCTGGATTCACACAGAAGAT) were designed by Sigma-Aldrich. Puromycin resistance cassette was replaced with EGFP cassette. For lentivirus production, the shRNA vector plasmid (20 µg) and second-generation packaging plasmids (5 µg of pMD2-VSVG and 15 µg of pCMVΔR8.91) were cotransfected into HEK293T cells. Lentivirus-containing supernatant was collected 24 h after transfection, centrifuged at 3000 rpm to remove cell debris for 5 min, 0.45 µm-filtered and frozen at −80°C until use. The titer of lentiviral shRNA particles was determined by use of Fluorescent Activated Cell Sorter (FACS) counting of transduced GFP-positive cells. HCT116 cells were infected by spinoculation (1800 rpm for 45 min at 30°C) at MOI∼5. Forty-eight hours post-infection cells were harvested for cell cycle analysis, total extracts and RNA extraction.
Flow cytometric cell cycle analysis
Cells were harvested after shRNA infection and DNA distribution analysis of Propidium-Iodide-stained cells was performed by an Epics cytofluorimeter (Beckman Coulter). Apoptotic cells were identified by FACS using Annexin V-FITC conjugate (Bender MedSystems) following the protocol of the manufacturer. Replicative DNA content was analyzed using bivariate flow cytometry. Cells were pulsed with 20 µM BrdU for 30 min at 37°C. After washing with PBS1×/Tween-20 0.5%, cells were treated with 2 N HCl for 30 min at room temperature, followed by the addition of 0.1 M borate buffer, pH 8.5. Cells were washed and incubated with mouse anti-BrdU antibody (#317902 BioLegend) for 1 h at 4°C. After washing, cells were incubated with FITC anti-mouse (#F0313 Dako) for 1 h at 4°C. Finally, cells were counter stained with PI solution (propidium iodide 25 µg/ml, Na–Citrate 3.4 mM, NaCl 9.65 mM, NP-40 0.03%) for 15 min, and then analyzed by cytofluorimeter.
Indirect fluorescence staining was performed using anti-phospho H2A.X (Upstate #05-636) and mouse anti-FITC (#F0313 Dako). Briefly, cells were harvested, washed twice with PBS 1×, fixed in 1% formaldehyde for 10 min at 37°C, and post-fixed with 90% ethanol. After permeabilization with 0.25% Triton X-100 in PBS 1× for 5 min, cells were stained with primary antibody (1:25) o.n. at 4°C, and secondary antibody (1 : 50) for additional 2 h at 4°C. Cells were then treated with RNAse A for 40 min, incubated with propidium iodide (30 µg/µl) for additional 30 min at 4°C in the dark and analyzed with cytometer.
Immunoblotting
Total extracts were prepared by resuspending the cell pellet in lysis buffer (50 mM Tris–HCl pH 8.0, 120 mM NaCl, 0.5% NP-40, 1 mM EDTA, protease and phosphatase inhibitors). Chromatin enriched extracts were prepared as described by Mendez and Stillman (27). For immunoblotting, an equivalent amount of cellular extracts were resolved by SDS–PAGE, electrotransferred to PVDF membrane, and immunoblotted with the following antibodies 1:1000 in TBS1X-BSA 1 mg/ml: anti-NF-YA (sc-10779 Santa-Cruz), anti-NF-YB (Pab 001 GeneSpin), anti-p53 DO-1 (sc-126 Santa Cruz), anti-phospho H2A.X (clone JBW301) (#05-636 Millipore), anti-phospho-p53 (Ser15) (#9284 Cell Signaling), anti-CycB1 (V152) (GTX72347 Genetex), anti-cleaved Casp7 and Casp9 (#9491 and #9501 Cell Signaling), anti-PARP1 (sc-8007 Santa Cruz), anti-PCNA (sc-56 Santa Cruz), anti-Mcm7 (sc-9966 Santa Cruz), anti-H2A (#39111 Active Motif), anti-Cdc6 (sc-9964 Santa Cruz), anti-Cdt1 (sc-28262 Santa Cruz), anti-phosphoThr68 Chk2 (#2661 Cell Signaling), anti-tubulin (T6074 Sigma Aldrich) and anti-actin (I-19) (sc-1616 Santa Cruz).
RT-PCR analysis
RNA was extracted using the RNeasy kit (Qiagen, Hilden, Germany), according to the manufacturer's protocol, from HCT116 infected cells. For cDNA synthesis, 3 µg of RNA was retrotranscribed with a Moloney murine leukemia virus reverse transcriptase (Finzymes). Semiquantitative PCRs were performed in a linear range of each amplification product. PCR oligonucleotides were previously described (25) or listed in Supplementary Data.
ChIP
Chip were performed as previously described (18,25,26). Chromatin was prepared upon 48 h shCTR, shNF-YA and sh-NF-YB infection. Quantitative real time PCR was performed using SYBR green IQ reagent (Biorad) in the iCycler machine (Biorad). Primers were designed to amplify genomic regions of 100–150-bp size. The relative sample enrichment was calculated with the following formula: 2ΔCtx − 2ΔCtb, where ΔCtx = Ct input − Ct sample and ΔCtb = Ct input − Ct control Ab.
Gene expression profiling and data analysis
Total cellular RNAs from HCT116 cells were isolated using RNeasy Kit (Qiagen, Germany) from shRNA infected cells. For each sample, 500 ng of total RNA were synthesized to biotinylated cRNA using the Illumina RNA Amplification Kit (Ambion Inc., Austin, TX, USA). Synthesis, HumanHT12 v 3.0 Expression BeadChip ILLUMINA arrays hybridization (Illumina Inc., San Diego, CA, USA), staining and scanning were performed according to the standard protocol supplied by Illumina.
For data analysis, the intensity files were loaded into the Illumina BeadStudio v. 3.1.3.0 software for quality control and gene expression analysis. First, the quantile normalization algorithm was applied on the data set to correct systematic errors. Background was subtracted. For differential expression analysis, technical replicates of each sample were grouped together and genes with a detection of P-value < 0.01, corresponding to a false-positive rate of 1%, were considered as detected. Differently expressed genes were selected with differential score cutoff set at ±40 and genes with fold difference <1.5 or >1.5 were further analyzed (Supplementary Tables S1 and S2). Gene Ontology (GO) enrichment P-values were computed according to the hypergeometric distribution (28). Only terms with P < 1.00E-05 GO were displayed and the terms have been ranked by decreasing P-value.
RESULTS
NF-Y subunits inactivation results in decreased proliferation
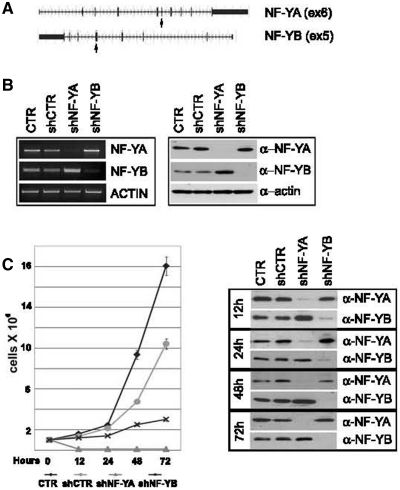
To understand the biological functions of NF-Y subunits, we knocked down the expression of NF-YA and NF-YB in HCT116 cells by lentiviral delivery of shRNAs. A non-target shRNA was used as control vector (shCTR). Of the five different shRNAs tested for NF-YA and NF-YB inactivation, we selected shNF-YA targeting exon 6 and sh-NF-YB targeting exon 5 (Figure 1A). Expression levels of NF-Y subunits were confirmed by western blot and RT-PCR analysis: NF-YA and NF-YB were completely depleted by 48 h after infection with shRNAs (Figure 1B). We noted that NF-Y inactivated cells grew more slowly than control cells, particularly a high percentage of cell death was observed in cell culture medium upon shNF-YA infection. The effect on cell proliferation was investigated through the estimation of growth rate. At 48 h post-infection, cells were trypsinized and equal numbers of cells were seeded into new culture dishes. Cell growth was significantly suppressed at 12, 24, 48 and 72 h post-seeding in NF-Y inactivated cells, particularly upon NF-YA loss (Figure 1C, left panel). Western blot showed the stability of NF-YA and NF-YB knock-down up to 72 h post-seeding (Figure 1C, right panel).
Figure 1.
(A) shRNAs were designed on human NF-YA and NF-YB genes, targeting exons 6 and 5, respectively (arrows). (B) Transcription (left panel) and protein levels (right panel) of NF-YA and NF-YB subunits after 48 h of shRNA lentiviral infection of HCT116 cells. (C) Left panel: growth rate of control and shRNA-infected HCT116 cells. Forty eight hours post-infection cells were trypsinized and equal number of cells were seeded into new culture dishes (time = 0 h). Viable cells were estimated after 12, 24, 48 and 72 h post-seeding. Right panel: western blot analysis of total extracts of control and shRNA infected cells. Actin was used as loading control.
NF-YA and NF-YB depletion triggers different cell cycle blocks
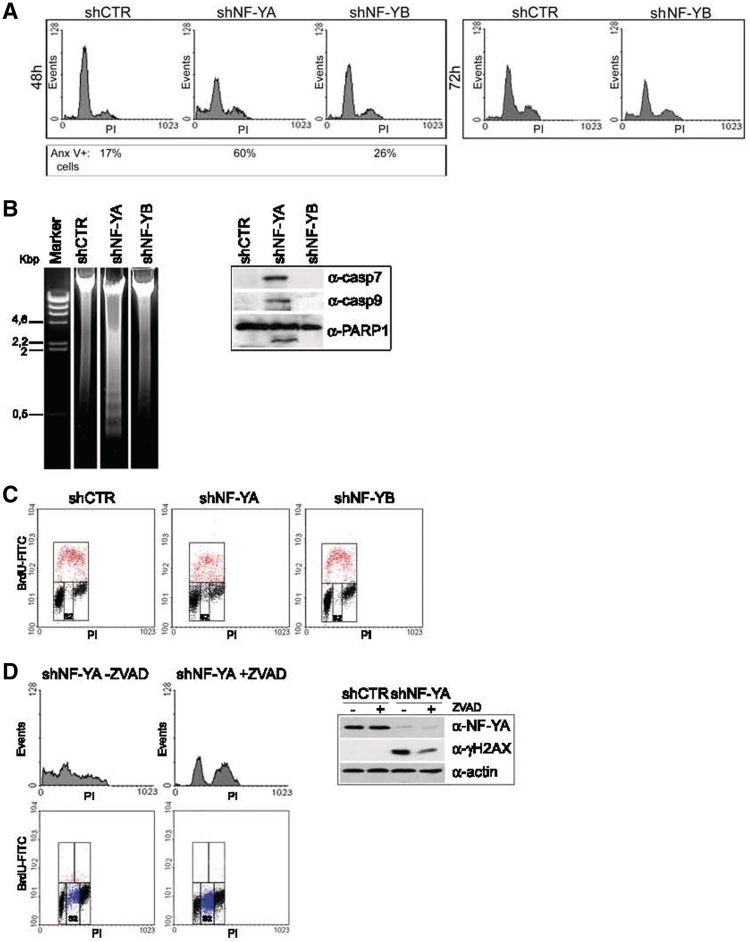
The reduced cell doublings in NF-Y inactivated populations could be due to apoptosis, cell cycle arrest, or both. To discriminate between these two possibilities, we performed a PI-cytometric analysis of the cells at 48 h post-infection (Figure 2A, left panel). The relative content of DNA indicated that the distribution of cells throughout the cell cycle was differently affected by NF-YA or NF-YB silencing. An increase of cells with an sub-G1 DNA content was observed only after shNF-YA (24% upon NF-YA and 5% upon NF-YB inactivation versus 4% in control cells). Compared to shCTR, shNF-YA expression increased S phase (from 7.6% to 15.5%), while G1 events decreased. NF-YB knock-down induced only a slight increase of G2/M population (from 16% to 23%).
Figure 2.
(A) PI/monoparametric FACS analysis of HCT116 cells after 48 h of infection with shCTR, shNF-YA and shNF-YB (left panel) and 72 h of infection with shCTR and shNF-YB (right panel). The percentage of Annexin V-positive cells is indicated under each panel. (B) Left panel: DNA agarose gel electrophoresis showing the typical DNA ladder pattern of apoptosis after 48 h of shNF-YA infection. Right panel: expression analysis of cleaved caspase 7 and −9 and PARP1 in NF-YA or NF-YB silenced cells. (C) BrdU/PI cytofluorimetric analysis of HCT116 cells 48 h post-infection with shRNAs. Red dots represent BrdU-positive cells. (D) Left panel: PI (upper panel) and BrdU/PI (lower panel) cytoflurimetric analysis of NF-YA inactivated cells untreated or treated with the pan-caspase inhibitor ZVAD. Blue dots represent non-cycling S cells (S2). Right panel: NF-YA and γH2AX expression analysis of total extracts of shCTR and shNF-YA cells, untreated or treated with ZVAD. Actin was used as loading control.
Although no sub-G1 events were detected upon 48 h from shNF-YB infection, cytofluorimetric analysis revealed an increase of apoptotic AnnexinV-positive cells (Figure 2A, left panel). Indeed, albeit to a lesser degree compared to shNF-YA, 72 h post-infection also in shNF-YB cells we identified sub-G1 events (from 2% in control cells to 11% in shNF-YB cells) (Figure 2A, right panel). Moreover, we observed that apoptosis activation was concurrent to a slight decrease of the G2/M population (from 28% in shCTR cells to 23% in shNF-YB cells), reproducing a phenotype similar to siRNA-mediated NF-YB silencing (25).
The significant increase of apoptotic cells upon 48 h from infection of shNF-YA was not ascribable to a different level of knock-down compared to shNF-YB. Dose–response experiments were performed with shNF-YA and shNF-YB: after 48 h, although at higher doses of MOI, no sub-G1 events were detected in shNF-YB cells. Conversely, shNF-YA cells underwent apoptosis despite a residual NF-YA expression (Supplementary Figure S1A).
A similar alteration of the cell cycle was observed after NF-YA and NF-YB silencing in p53-positive human colorectal carcinoma LoVo cells (data not shown). Interestingly, sub-G1 events also raised upon NF-YA knock-down in cervical carcinoma Hela cells, in which p53 is degraded by E6 espression (from ∼10% in shCTR to 35% in shNF-YA cells) (Supplementary Figure S1B, left panel). This result can be ascribed to the restored expression of p53 in shNF-YA cells, as demonstrated by western blot analysis (Supplementary Figure S1B, right panel). As observed for HCT116 cells, a G2/M increase was evident only in shNF-YB cells (from 11% to 20%).
As the histone-fold NF-YC subunit dimerizes with NF-YB and shares the activation domain with NF-YA, we wondered which cell cycle effect could be triggered by silencing NF-YC. At 48 h from the infection, the inactivation of all the decribed NF-YC isoforms (29) led to a slight increase of G2/M population and Annexin-V positive cells, without accumulating sub-G1 events (Supplementary Figure S1C), similarly to shNF-YB cells (Figure 2A).
The detection of a DNA ladder pattern highlighted that late stages of apoptosis occurred upon NF-YA loss (Figure 2B, left panel). Moreover, NF-YA depletion resulted in the activation of caspases, among which caspase −9 and −7, which triggered the proteolytic cleavage of PARP1 (Figure 2B, right panel and Supplementary Figure S1B).
The progression through the different phases of the cell cycle was analyzed in asynchronous cells by bivariate FACS analysis (Figure 2C). Compared to control cells, we detected an increase of G2/M events both in shNF-YA and shNF-YB cells (from 15% to 23.8% and 24.9%, respectively). NF-YA inactivation didn't result in any change of replicative events, although monoparametric analysis showed the doubling of cells in S phase (Figure 2A). This inconsistency could be explained by the increase of BrdU negative cells with an S-phase DNA content (S2), not observed in shCTR and shNF-YB cells (from 2% in shCTR and shNF-YB to 12% in shNF-YA cells). The S2 population could represent non-cycling S cells.
It has been reported that H2AX is phosphorylated (γH2AX) during apoptotic DNA fragmentation in mouse and human cells (30,31). Aditionally, γH2AX is a well-known marker of DNA damage, whose recruitment at DNA damaged sites seems to be fundamental for an efficient DNA repair (32,33). To discriminate between these two possibilities, apoptosis induced by NF-YA loss was inhibited by pre-treating cells with the pan-caspase inhibitor ZVAD. Although the percentage of apoptotic cells was completely abolished, H2AX phosphorylation was reduced but still sharp (Figure 2D, upper panel). Interestingly, the inhibition of apoptosis strongly increased the percentage of S2 cells from 12% to 40% (Figure 2D, lower panel).
These results suggested that S2 cells are addressed to apoptosis and that γH2AX activation is likely associated to DNA damage, triggered by NF-YA loss.
Replication defects are associated to NF-YA loss
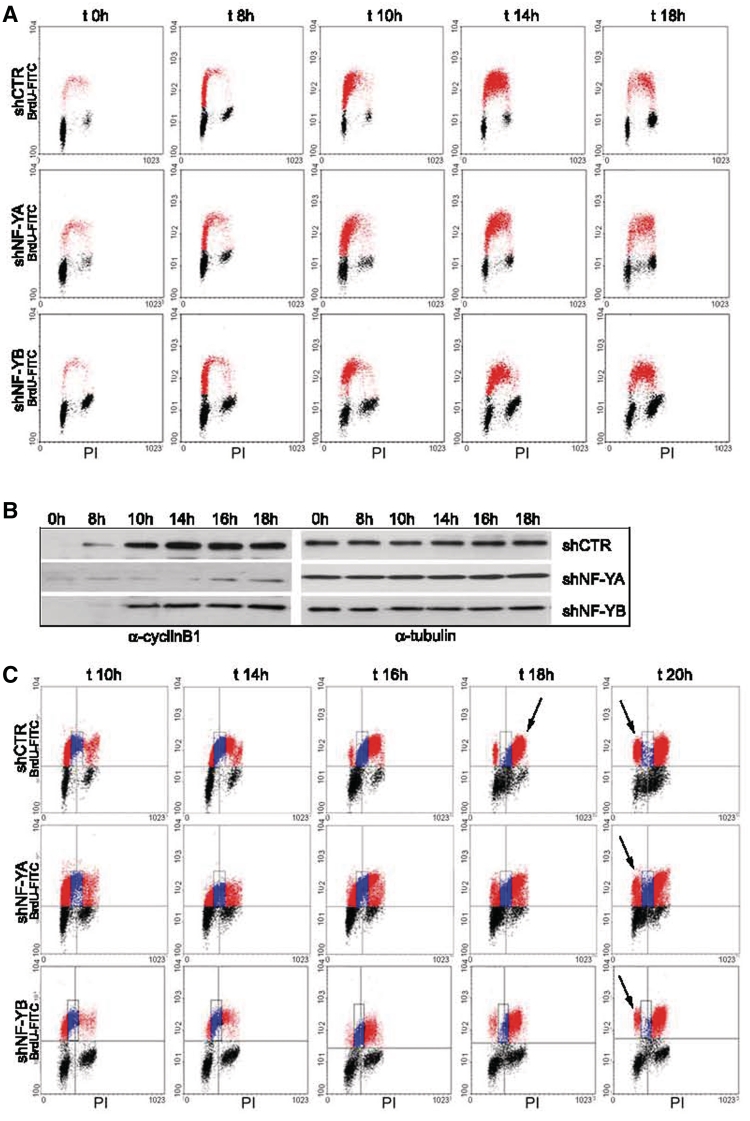
Cell cycle progression was studied in synchronous populations. Infected HCT116 cells were starved in serum-free for 30 h and then released in 10% FCS. The cells were harvested at different times and bivariate FACS analysis was performed (Figure 3A). At time 0 h, 85% of shCTR and 78% of shNF-YA cells were in G0/G1 phase. The proportion of cells passing through S phase was normalized to starting G0 cells. No relevant differences could be detected between control and shNF-YA cells up to 14 h, when BrdU-positive cells were mainly retained in early-S (55% in early-S and 34% in late-S in shCTR, and 57% in early-S and 28% in late-S in shNF-YA cells). Conversely, after 18 h, a dissimilarity was observed in the distribution of BrdU-positive cells. Control cells were mainly accumulated in late-S (15% of cells in early-S and 30% in late-S) and a G2/M increase was clearly observed. Following NF-YA inactivation, BrdU-positive cells were equally distributed between early and late-S (27% both in early- and late-S). Consequently, fewer cells entered the G2/M phase at 18 h compared to shCTR cells (24% in shNF-YA and 35% in shCTR cells). Moreover, it was evident the increase of the S2 population (from 1% of control cells to 11% after NF-YA loss). Western blot showed a time-course increase of cyclinB1 expression in control cells, clearly impaired upon NF-YA inactivation (Figure 3B). As to NF-YB inactivation, although serum deprivation, at time 0 h a significant percentage of cells was still retained in G2/M (15% compared to 5% of shCTR cells). When normalized to the starting G0 population, cells were only slightly delayed in entering the early S phase. Once entered, shNF-YB cells were not slowed down in passing to late S and G2/M. The kinetics of cell cycle progression was further supported by a reduced but still consistent expression of cyclinB1. Lower levels of cyclinB1 were expected, considering that few NF-Y molecules can still transactivate the CCNB1 CCAAT-gene.
Figure 3.
(A) Time-course BrdU/PI cytofluorimetric analysis at time 0, 8, 10, 14 and 18 h post-release from serum starvation of shRNA infected cells. (B) CyclinB1 expression analysis of total extracts of shCTR, shNF-YA and shNF-YB cells at the indicated times after release from serum starvation. Tubulin was used as loading control. (C) BrdU pulse-chase biparametric FACS analysis of synchronized G0 cells upon NF-YA and NF-YB inactivation versus control cells.
The effect of NF-Y abrogation on DNA replication was investigated by BrdU pulse-chase experiments on synchronized G0 cells. After 10 h from the release, shNF-YA and shNF-YB cells were pulsed with BrdU for 30 min, after which the BrdU was washed away and the cells were allowed to grow in normal medium for different time points. We followed the BrdU-positive cells as they passed through S (Figure 3C, blue dots), G2/M and returned to G1 phase (red dots). shCTR BrdU-positive cells completed S phase in 6–8 h (from 10 to 16–18 h) and clearly entered the G2 phase at 16 h, growing to 90% at time 18 h (Figure 3C, first panel t = 18 h, arrow). At 20 h, ∼25% of cells moved to next G1 phase (last panel t = 20 h, arrow). Upon NF-YA inactivation, it was evident that more than 30% of cells were still in S phase at 20 h (middle panel, blue dots) and fewer cells reached the G2/M and entered the next G1 phase. The duration of S phase upon NF-YB inactivation was similar to shCTR (Figure 3C, third panel). The main defect observed was associated to the prolonged G2/M, which halved the percentage of cells moving to the next G1 at 20 h (from 25% in shCTR to 12% in shNF-YB).
These results suggest that NF-Y could have an important role in DNA replication and G2/M progression.
NF-YA-induced DNA-damage and p53 activation are coupled with S-phase defects
γH2AX staining upon NF-YA inactivation was maintained although apoptosis inhibition (Figure 2D). It is possible that H2AX phosphorylation is an indication of early DNA-damage that culminates in the S-phase arrest. To investigate this hypothesis, we analyzed γH2AX staining in shNF-YA cells released from serum starvation (Supplementary Figure S2A). γH2AX quantification respect to total H2A highlighted a sharp increase at 16 and 18 h from the release, corresponding to defects in cell cycle progression (Figure 3A). Bivariate distributions representing cellular DNA content versus γH2AX of asynchronous shNF-YA cells showed both 2N and 4N γH2AX positive cells (Supplementary Figure S2B). These data hint that DNA damage doesn't occur uniquely in S population, although it is possible that S damaged cells are more prone to apoptosis (Figure 2D). In response to DNA damage, H2AX has been reported to be phosphorylated by ATM, ATR, which is mainly associated to replication stress (34), as well as DNA-PK, observed during apoptotic DNA fragmentation (30). To inhibit ATM/ATR activation, we co-treated shNF-YA cells with non-toxic dose of caffeine (1 µM) and we analyzed cell cycle distribution by cytofluorimetric analysis (Supplementary Figure S2C). Caffeine didn't completely inhibit apoptosis, but decreased the percentage of sub-G1 cells of ∼25% (from ∼40% without caffeine to 30% upon caffeine treatment), suggesting the contribution of ATM/ATR to apoptosis activation.
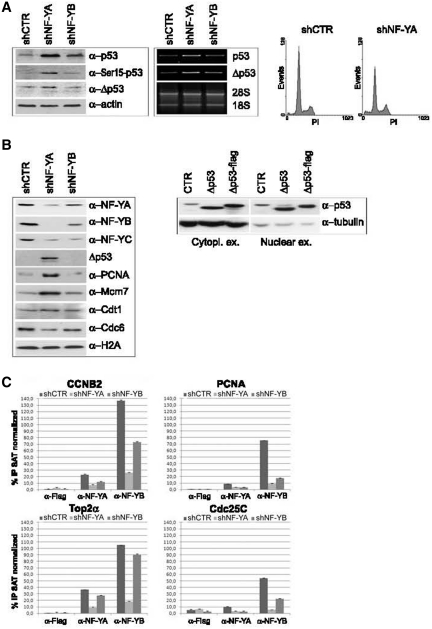
One of the main signal transducer of DNA damage is p53. Western blot analysis of shCTR extracts highlighted a slight p53 expression, which increased after NF-Y depletion (Figure 4A, left and middle panels). As shown in Figure 4A (right panel), a less severe activation of apoptosis was observed upon NF-YA loss in HCT116 p53−/− cells (from 2.8% in shCTR to 11% in shNF-YA cells), hinting that p53 plays a role in apoptosis induced in shNF-YA cells.
Figure 4.
(A) Left panel: p53, Phospho-Ser15 p53 and Δp53 expression analysis of HCT116 total extracts 48 h post-infection. Actin was used as loading control. Middle panel: RT-PCR analysis of p53 and Δp53 mRNA transcripts. Right panel: monoparametric cell cycle analysis of HCT116 p53−/− cells after shCTR and shNF-YA infection for 48 h. (B) Left panel: expression analysis of chromatin enriched extracts with the indicated antibodies. Right panel: p53 and Δp53 expresison analysis of cytoplasmic and nuclear extracts from HCT116 cells transiently transfected with Δp53 and Δp53-Flag. (C) ChIP analysis of NF-Y binding to the indicated CCAAT-promoters in shCTR, shNF-YA and shNF-YB cells. The enrichment was calculated as percentage IP recovery (%IP) of NF-YA and NF-YB over input normalized to satellite DNA. The input signal consists of 20% of the amount of chromatin used with each immunoprecipitation.
We then checked for p53 phosphorylation at Ser15, a critical event in its activation following DNA-damage. Ser15-p53 phosphorylation was observed in HCT116 shNF-YA cells (Figure 4A, left panel). ATM and ATR directly phosphorylate p53 on Ser15 and activate other kinases, such as Chk1, Chk2 and Plk3, which in turn mediate the phosphorylation of p53 on additional residues. Western blot analysis of phospho-Chk2 (Thr68) showed its overexpression upon NF-YA loss (Supplementary Figure S2C, right panel).
In addition to its post-translational modifications, it was reported that alternative splicing of p53 has a role in promoter selectivity of target genes. In particular, a novel human p53 isoform (Δp53) has been found to be associated to intra-S checkpoint activation (35). Western blot analysis in HCT116 cells with DO-1 antibody, recognizing the N-terminus domain of p53, detected a lower migrating p53 isoform, whose molecular weight is consistent with Δp53. A slight increase of Δp53 was observed in shNF-YA cells, both as protein and mRNA levels (Figure 4A, left and middle panels). While Rohaly et al. showed Δp53 binding to DNA in damaged S-phase cells (35), Chan et al. (36) demonstrated that Δp53 is excluded from the nucleus, due to the deletion of a nuclear localization signal, and only slightly accumulated in the nucleus when it is coexpressed with p53. Chromatin-enriched extracts showed Δp53 accumulation after NF-YA silencing, corroborating its possible activity in the attenuation of S phase progression (Figure 4B, left panel). Differential enriched extracts from HCT116 cells transiently transfected with Δp53 showed that its overexpression led to both cytoplasmic and nuclear localization in this cellular context (Figure 4B, right panel), consistently with its possible recruitment to chromatin.
In addition, higher levels of chromatin-bound PCNA and of the replication helicase Mcm7 were found in shNF-YA cells, compared to shCTR and shNF-YB cells. Conversely, Cdt1 and Cdc6, which are required for the assembly of the pre-RC and loading of the MCM complex, didn't increase their expression levels (Figure 4B). Besides its role in the initiation of DNA replication, PCNA has been demonstrated to be an important component of multiple DNA repair (37), while an excess of Mcm2-7 is required for human cells to survive replicative stress (38). These data suggested that NF-YA loss can trigger the activation of the S-checkpoint, differently from NF-YB abrogation.
As expected, the amount of both NF-YA and NF-YB associated with chromatin decreased upon NF-Y subunits inactivation (Figure 4B, left panel). Interestingly, NF-YA depletion was more efficacious in hitting NF-Y binding to DNA than NF-YB loss. To better investigate NF-Y chromatin-recruitment, we performed ChIP assays with anti-NF-YA and anti-NF-YB antibodies (Figure 4C). Real-time qPCR further supported the hypothesis that NF-YA inactivation impaired NF-Y recruitment on CCAAT-promoters more strongly than NF-YB depletion.
Changes in gene expression upon NF-YA and NF-YB silencing
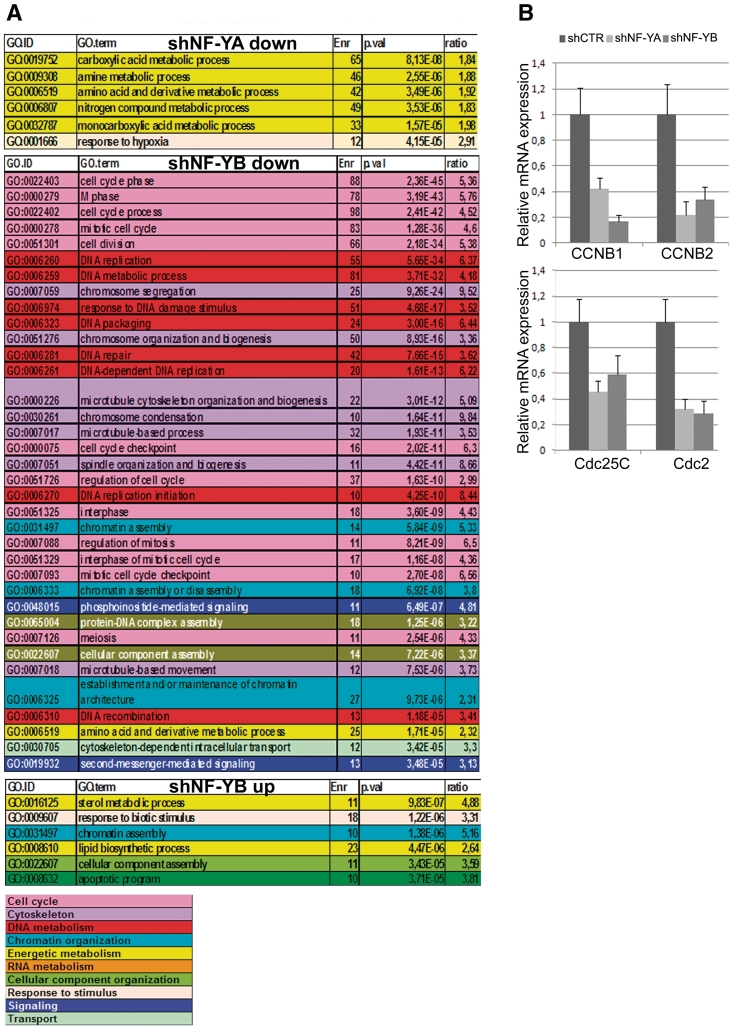
Expression profiling analysis of NF-Y inactivated cells was performed with the Illumina platform, which targets more than 37 000 annotated genes. Using relatively stringent criteria and employing the hypergeometric distribution to compute an enrichment P-value, we found that in shNF-YA cells 1195 and 1412 genes were up- and down-regulated, respectively, while transcription levels of only 515 and 685 genes increased and decreased following NF-YB depletion (Supplementary Tables S1 and S2). GO enrichment analysis of the shNF-YA repressed genes identified metabolic process as the statistically most important term retrieved (Figure 5A). No functional categories with P-value < 1.00E−05 were found among shNF-YA up-regulated genes. Conversely, NF-YB repressed genes were statistically retrieved in cell cycle, microtubule/cytoskeleton and DNA-metabolism functions, as already observed upon NF-YB siRNA transfection (25). NF-YB inactivation led to the up-regulation of relatively few GO terms, among which were those of lipid and sterol metabolism: this is somewhat surprising, since NF-Y is known to cooperate with SREBP1 and SREBP2 to activate many of these genes (39,40).
Figure 5.
(A) Gene Ontology (GO) terms displaying a statistically significant over-representation in the sample sets (down- and up-regulated) retrieved from microarray expression analysis of NF-YA and NF-YB silenced versus shCTR cells. Terms have been pruned and grouped according to manually determined macro-categories identified by different colors. (B) mRNA expression analysis of CCNB1, CCNB2, Cdc25C and Cdc2 by real time RT-PCR. mRNA transcripts have been normalized versus TBP and β-actin expression levels.
Promoter sequences of genes differentially expressed were analyzed with the Pscan algorithm (41) using the new NF-Y-PSFM matrix (positional sequence frequency matrix) (42), to identify whether the NF-Y binding site motif was over-represented in shNF-Y modulated genes. The CCAAT box showed a significant enrichment with P-value 4.91E−13 and 7.76E−19 in promoter sets of genes down-regulated after NF-YA or NF-YB loss, respectively. This analysis confirmed the direct involvement of NF-Y in the transcriptional regulation of down-regulated sets of genes. On the other hand, the CCAAT module was not enriched in up-regulated genes, suggesting an indirect effect.
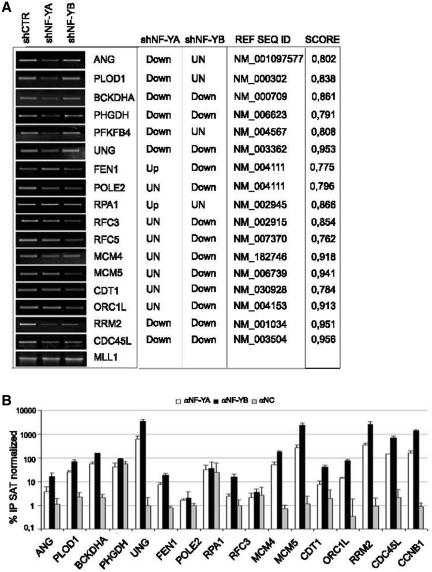
Semiquantitative and real time RT-PCRs were performed to validate the profiling analysis (Figure 6A and Supplementary Figure S3). With the exclusion of POLE2 and RPA1, predicted to be unmodified and up-regulated upon NF-YA loss, respectively, and MCM4, predicted to be down-regulated in shNF-YB cells, qRT-PCR analysis matched with the microarray profiles. The NF-Y matrix (42) and the Pscan algorithm (41) were used to scan the promoters sequence of these possible NF-Y targets. The CCAAT scores give an estimate of the likelihood of the sequences to represent a site recognized by NF-Y (Figure 6A, right panel). To investigate the binding of NF-Y to the predicted target genes, ChIP analysis was performed with anti NF-YA and NF-YB antibodies in HCT116 cells. As showed in Figure 6B, NF-Y was bound to 13 of the 16 analyzed genes, suggesting that NF-Y is directly involved in their transcriptional regulation. CCNB1 promoter, a well-known NF-Y target, was used as positive control (Figure 6B).
Figure 6.
(A) Semiquantitive RT-PCR validation of the profiling analysis of the indicated transcripts after NF-YA and NF-YB silencing compared to control cells. mRNA transcripts have been normalized versus MLL1 expression levels. The predicted changes of the expression levels from Illumina platform (up, down or unchanged), the Ref Seq ID and the estimated CCAAT score are indicated in the table. (B) ChIP analysis of NF-YA and NF-YB binding to the indicated genes in HCT116 cells. The enrichment was calculated as percentage IP recovery (%IP) of NF-YA and NF-YB over input normalized to satellite DNA.
By real time RT-PCR we further analyzed cell cycle G2/M CCAAT-regulated genes, which were retrieved as down-regulated genes only upon NF-YB loss. As shown in Figure 5B, both NF-YA and NF-YB inactivation led to CCNB1, CCNB2, Cdc25C and Cdc2 transcriptional decrease. Changes in gene expression correlated with decreased recruitment of NF-Y on gene regulatory regions observed by ChIP analysis (Figure 4C).
Thus, although NF-YA loss impaired the transcription of a wide panel of CCAAT-genes, the metabolic regulatory genes might be mainly affected. Alterations in gene expression of these genes could likely contribute to trigger apoptosis.
DISCUSSION
The role of NF-Y in controlling cell growth and apoptosis has been mainly attributed to its activity in gene transcription. We previously demonstrated that transient NF-YB inactivation by siRNA led to a delay in G2/M transition induced by the down-regulation of G2/M CCAAT-genes (25). To dissect various functions of NF-Y subunits in vivo, we have utilized shRNAs targeting NF-YA, NF-YB and NF-YC in HCT116 cells. The comparison between the two subunits inactivation has led to new insights into the function of NF-Y into the cell.
The estimation of growth rate of NF-YA and NF-YB inactivated cells (Figure 1) has questioned the role of the two subunits in the cell cycle. Although NF-Y transcriptional activity needs the association of all the subunits, our data suggest that each NF-Y component could have specific functions. Interestingly, western blot analysis highlighted that NF-YA depletion affects the expression of NF-YB subunit (Figure 1). It is possible that NF-YA and the stable heterodimer NF-YB/NF-YC could have distinct functions in the cell. This hypothesis is further supported by the results gained by NF-YC inactivation (Supplementary Figure S1C), showing an effect on the cell cycle comparable to NF-YB loss.
Silencing NF-YA expression affects S-phase progression
NF-YA depleted cells had a very severe phenotype, with a strong increase of cell death which is reverted by caspase inhibition (Figure 2D). Moreover, we highlighted the presence of non-cycling S cells (S2 fraction), which increased as apoptosis was suppressed. BrdU incorporation didn't raise upon treatment of shNF-YA cells with ZVAD. On the other hand, the increase of S2 cells suggests that the main cause of apoptosis activation was not the inefficient BrdU incorporation, rather the presence of replicative defects. The analysis of cell cycle progression in synchronized cells pointed out a delay in completing DNA replication. BrdU pulse-chase experiment on synchronized G0 cells clearly identified a fraction of cells delayed in clearing S-phase (Figure 3C).
The increase of γH2AX staining in NF-YA inactivated cells, still detected after apoptosis inhibition, suggests that DNA-damage occurs upon NF-YA loss. NF-YA depletion could originate spontaneous DSBs in S-phase cells and NF-Y itself could be fundamental for the intra-S damage checkpoint. By the analysis of chromatin enriched extracts, we observed the increase of PCNA and Mcm7 binding to DNA (Figure 4), which is consistent with a response to DNA replication defects. In addition to replication proteins, PCNA interacts with a variety of proteins involved in DNA-repair, such as nucleases and polymerases implicated in NER, BER, DSBs repair and translesion synthesis (43). An excess of Mcm2–7 loading onto chromatin has been shown to license dormant origins, in order to maintain DNA replication rates despite the increased checkpoint activity (38). This pathway has an important function in maintaining genetic stability in mammalian cells. The stabilization of p53 protein by the phosphorylation of Ser15 supports the activation of a DNA-damage pathway. Moreover, its splice variants Δp53, which is recruited to chromatin of NF-YA inactivated cells, is suggestive of the activation of intra-S checkpoint. Δp53 has been shown to bind and transactivate only p53 target genes involved in cell cycle arrest, such as p21, indicating a differential transcriptional activity in damaged S phase cells. Given the increase of apoptosis in NF-YA inactivated cells, it's clear that p53 apoptotic target genes are transactivated too. Indeed, p53 depleted cells were clearly impaired in apoptosis induction (Figure 4A). Therefore, we suppose that a complex regulatory network is involved in controlling S phase progression through Δp53; and cell death through phosphorylated p53, which has also been shown to bind sites of DNA-damage (44). If the origin of DSBs can delay S-phase progression or if S-phase accumulated cells are more sensitive to DNA-damage remains unknown.
Previously, conditional inactivation of NF-YA in MEFs resulted in the inhibition of S-phase entry and progression, and finally led to cell death (24). The effect of NF-YA inactivation on cell proliferation has been mainly ascribed to changes in expression of multiple genes controlling the G1 to S progression (24). GO enrichment analysis of the shNF-YA repressed genes didn't identify down-regulation of genes possibly involved in G1 phase or DNA replication. Conversely, only the category of metabolic processes emerged as the statistically most important terms retrieved. The role of NF-Y in metabolic pathways has been recently identified (39,45,46). Genome wide analysis of the occupancy of SREBP1, key transcriptional regulator of cholesterol and fatty acid metabolism, highlighted that cholesterol biosynthesis, lipid metabolism and aminoacid activation genes were preferentially regulated by the combination of SREBP1 and NF-Y binding sites (39). More recently, it has been described that SREBP-2- and NF-Y-mediated regulation of FDPS gene transcription modulates cell proliferation (40).
NF-YB loss affects G2/M phase progression
The main defect observed in shNF-YB cells was associated to the prolonged G2/M, which halved the percentage of cells moving to the next G1 at 20 h post-release from serum starvation (Figure 3). A similar effect was observed upon the expression of the NF-YA/Bdbd mutant, which lacks the transcription activation domain but still binds NF-YB/NF-YC and the CCAAT-box (23). Bdbd expression inhibited the entry of cells into mitosis and arrested cells at G2 phase. NF-YB loss doesn't impair NF-Y binding as evidently as NF-YA depletion. NF-Y recruitment is reduced but not completely abolished. This reflects the condition induced by NF-YA/Bdbd: NF-Y is still bound to CCAAT-boxes, but is no more able to activate the transcription. In particular, the transcription of G2/M genes is affected by NF-YB loss (Figure 4). The analysis of microarray data gained by shNF-YB infection perfectly overlaps with our previous results of siRNA NF-YB inactivated cells (25). Cell cycle, microtubule/cytoskeleton and G2/M retrieved categories were expected, given the established role of NF-Y in controlling cell cycle regulated genes.
The comparison between NF-YA and NF-YB depletion strongly highlights that the lack of the single subunits are differently perceived by the cell, which activate diverse cell cycle blocks. The loss of NF-YA, which is the only subunit that can address NF-Y to the CCAAT-box, strongly affects NF-Y binding, thus leading to no occupancy of CCAAT boxes. This could induce some type of genome damage, which may delay cells in completing S phase and finally leads to cell death. Conversely, NF-YB depletion only reduces NF-Y binding to its target genes and the main consequence observed is the repression of CCAAT-genes transcription, particularly of G2/M genes, which show high affinity for NF-Y. Finally, this leads to a delay in exiting the G2/M phase.
This study suggests a new role of CCAAT-box occupancy by NF-Y beyond its transcriptional activity, which could play an important function in protecting cells from DNA damage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Associazione Italiana per la Ricerca sul Cancro-MFAG (grants number 6192 to C.I. and IG-8586 to A.W.); Italian Ministry for Education, University and Research (grant PRIN 2008CJ4SYW_004); European Union (CRESCENDO I.P., contract LSHM-CT2005-018652). Funding for open access charge: Associazione Italiana per la Ricerca sul Cancro-MFAG.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Mantovani for stimulating discussion and V. Zappavigna for α-Cdc6 and α-Cdt1 antibodies. We sincerely thank Irena Dornreiter (Universität Hamburg, Germany) for the kind gift of Δp53 expression vectors (26). While this manuscript was under review, Gurtner et al. reported that unrestricted NF-Y activity leads to a p53- and E2F1-dependent apoptosis (47). Their results support that misregulation of NF-Y, both caused by NF-YA overexpression and NF-YA loss, has a central role in controlling cell proliferation and apoptosis.
REFERENCES
- 1.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 2.Bolognese F, Wasner M, Dohna CL, Gurtner A, Ronchi A, Muller H, Manni I, Mossner J, Piaggio G, Mantovani R, et al. The cyclin B2 promoter depends on NF-Y, a trimer whose CCAAT-binding activity is cell-cycle regulated. Oncogene. 1999;18:1845–1853. doi: 10.1038/sj.onc.1202494. [DOI] [PubMed] [Google Scholar]
- 3.Gurtner A, Manni I, Fuschi P, Mantovani R, Guadagni F, Sacchi A, Piaggio G. Requirement for down-regulation of the CCAAT-binding activity of the NF-Y transcription factor during skeletal muscle differentiation. Mol. Biol. Cell. 2003;14:2706–2715. doi: 10.1091/mbc.E02-09-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang ZF, Huang DY. Regulation of thymidine kinase expression during cellular senescence. J. Biomed. Sci. 2001;8:176–183. doi: 10.1159/000054030. [DOI] [PubMed] [Google Scholar]
- 5.Marziali G, Perrotti E, Ilari R, Testa U, Coccia EM, Battistini A. Transcriptional regulation of the ferritin heavy-chain gene: the activity of the CCAAT binding factor NF-Y is modulated in heme-treated Friend leukemia cells and during monocyte-to-macrophage differentiation. Mol. Cell Biol. 1997;17:1387–1395. doi: 10.1128/mcb.17.3.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farina A, Manni I, Fontemaggi G, Tiainen M, Cenciarelli C, Bellorini M, Mantovani R, Sacchi A, Piaggio G. Down-regulation of cyclin B1 gene transcription in terminally differentiated skeletal muscle cells is associated with loss of functional CCAAT-binding NF-Y complex. Oncogene. 1999;18:2818–2827. doi: 10.1038/sj.onc.1202472. [DOI] [PubMed] [Google Scholar]
- 7.Gurtner A, Fuschi P, Magi F, Colussi C, Gaetano C, Dobbelstein M, Sacchi A, Piaggio G. NF-Y dependent epigenetic modifications discriminate between proliferating and postmitotic tissue. PLoS One. 2008;3:e2047. doi: 10.1371/journal.pone.0002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li XY, Hooft van Huijsduijnen R, Mantovani R, Benoist C, Mathis D. Intron-exon organization of the NF-Y genes. Tissue-specific splicing modifies an activation domain. J. Biol. Chem. 1992;267:8984–8990. [PubMed] [Google Scholar]
- 9.Grskovic M, Chaivorapol C, Gaspar-Maia A, Li H, Ramalho-Santos M. Systematic identification of cis-regulatory sequences active in mouse and human embryonic stem cells. PLoS Genet. 2007;3:e145. doi: 10.1371/journal.pgen.0030145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kabe Y, Yamada J, Uga H, Yamaguchi Y, Wada T, Handa H. NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol. Cell Biol. 2005;25:512–522. doi: 10.1128/MCB.25.1.512-522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasner M, Haugwitz U, Reinhard W, Tschop K, Spiesbach K, Lorenz J, Mössner J, Engeland K. Three CCAAT-boxes and a single cell cycle genes homology region (CHR) are the major regulating sites for transcription from the human cyclin B2 promoter. Gene. 2003;312:225–237. doi: 10.1016/s0378-1119(03)00618-8. [DOI] [PubMed] [Google Scholar]
- 12.Salsi V, Caretti G, Wasner M, Reinhard W, Haugwitz U, Engeland K, Mantovani R. Interactions between p300 and multiple NF-Y trimers govern cyclin B2 promoter function. J. Biol. Chem. 2003;278:6642–6650. doi: 10.1074/jbc.M210065200. [DOI] [PubMed] [Google Scholar]
- 13.Linhart C, Elkon R, Shiloh Y, Shamir R. Deciphering transcriptional regulatory elements that encode specific cell cycle phasing by comparative genomics analysis. Cell Cycle. 2005;4:1788–1797. doi: 10.4161/cc.4.12.2173. [DOI] [PubMed] [Google Scholar]
- 14.Elkon R, Linhart C, Sharan R, Shamir R, Shiloh Y. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caretti G, Salsi V, Vecchi C, Imbriano C, Mantovani R. Dynamic recruitment of NF-Y and histone acetyltransferases on cell-cycle promoters. J. Biol. Chem. 2003;278:30435–30440. doi: 10.1074/jbc.M304606200. [DOI] [PubMed] [Google Scholar]
- 16.Allen KA, Williams AO, Isaacs RJ, Stowell KM. Down-regulation of human topoisomerase IIalpha correlates with altered expression of transcriptional regulators NF-YA and Sp1. Anticancer Drugs. 2004;15:357–362. doi: 10.1097/00001813-200404000-00008. [DOI] [PubMed] [Google Scholar]
- 17.Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M, Dobbelstein M, Del Sal G, Piaggio G, Mantovani R. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol. Cell Biol. 2005;25:3737–3751. doi: 10.1128/MCB.25.9.3737-3751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basile V, Mantovani R, Imbriano C. DNA damage promotes histone deacetylase 4 nuclear localization and repression of G2/M promoters, via p53 C-terminal lysines. J. Biol. Chem. 2006;281:2347–2357. doi: 10.1074/jbc.M507712200. [DOI] [PubMed] [Google Scholar]
- 19.Yun J, Chae HD, Choi TS, Kim EH, Bang YJ, Chung J, Choi KS, Mantovani R, Shin DY. Cdk2-dependent phosphorylation of the NF-Y transcription factor and its involvement in the p53-p21 signaling pathway. J. Biol. Chem. 2003;278:36966–36972. doi: 10.1074/jbc.M305178200. [DOI] [PubMed] [Google Scholar]
- 20.Ceribelli M, Dolfini D, Merico D, Gatta R, Vigano AM, Pavesi G, Mantovani R. The histone-like NF-Y is a bifunctional transcription factor. Mol. Cell Biol. 2008;28:2047–2058. doi: 10.1128/MCB.01861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Testa A, Donati G, Yan P, Romani F, Huang TH, Vigano MA, Mantovani R. Chromatin immunoprecipitation (ChIP) on chip experiments uncover a widespread distribution of NF-Y binding CCAAT sites outside of core promoters. J. Biol. Chem. 2005;280:13606–13615. doi: 10.1074/jbc.M414039200. [DOI] [PubMed] [Google Scholar]
- 22.Hu Q, Maity SN. Stable expression of a dominant negative mutant of CCAAT binding factor/NF-Y in mouse fibroblast cells resulting in retardation of cell growth and inhibition of transcription of various cellular genes. J. Biol. Chem. 2000;275:4435–4444. doi: 10.1074/jbc.275.6.4435. [DOI] [PubMed] [Google Scholar]
- 23.Hu Q, Lu JF, Luo R, Sen S, Maity SN. Inhibition of CBF/NF-Y mediated transcription activation arrests cells at G2/M phase and suppresses expression of genes activated at G2/M phase of the cell cycle. Nucleic Acids Res. 2006;34:6272–6285. doi: 10.1093/nar/gkl801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhattacharya A, Deng JM, Zhang Z, Behringer R, de Crombrugghe B, Maity SN. The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 2003;63:8167–8172. [PubMed] [Google Scholar]
- 25.Benatti P, Basile V, Merico D, Fantoni LI, Tagliafico E, Imbriano C. A balance between NF-Y and p53 governs the pro- and anti-apoptotic transcriptional response. Nucleic Acids Res. 2008;36:1415–1428. doi: 10.1093/nar/gkm1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basile V, Ferrari E, Lazzari S, Belluti S, Pignedoli F, Imbriano C. Curcumin derivatives: molecular basis of their anti-cancer activity. Biochem. Pharmacol. 2009;78:1305–1315. doi: 10.1016/j.bcp.2009.06.105. [DOI] [PubMed] [Google Scholar]
- 27.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ceribelli M, Benatti P, Imbriano C, Mantovani R. NF-YC complexity is generated by dual promoters and alternative splicing. J. Biol. Chem. 2009;284:34189–34200. doi: 10.1074/jbc.M109.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee B, Kessinger C, Kobayashi J, Chen BP, Chen DJ, Chatterjee A, Burma S. DNA-PK phosphorylates histone H2AX during apoptotic DNA fragmentation in mammalian cells. DNA Repair. 2006;5:575–590. doi: 10.1016/j.dnarep.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Rogakou EP, Nieves-Neira W, Boon C, Pommier Y, Bonner WM. Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at Ser139. J. Biol. Chem. 2000;275:9390–9395. doi: 10.1074/jbc.275.13.9390. [DOI] [PubMed] [Google Scholar]
- 32.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA\double strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Downey M, Durocher D. γH2AX as a checkpoint maintenance signal. Cell Cycle. 2006;5:1376–1381. doi: 10.4161/cc.5.13.2899. [DOI] [PubMed] [Google Scholar]
- 34.Ward IM, Minn K, Chen J. UV-induced ataxia-telangiectasia-mutated and Rad3-related (ATR) activation requirs replication stress. J. Biol. Chem. 2004;279:9677–9680. doi: 10.1074/jbc.C300554200. [DOI] [PubMed] [Google Scholar]
- 35.Rohaly G, Chemnitz J, Dehde S, Nunez AM, Heukeshoven J, Deppert W, Dornreiter I. A novel human p53 isoform is an essential element of the ATR-intra-S phase checkpoint. Cell. 2005;122:21–32. doi: 10.1016/j.cell.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 36.Chan WM, Poon RYC. The p53 isoform Δ53 lacks intrinsic transcriptional activity and reveals the critical role of nuclear import in dominant-negative activity. Cancer Res. 2007;67:1959–1969. doi: 10.1158/0008-5472.CAN-06-3602. [DOI] [PubMed] [Google Scholar]
- 37.Szuts D, Christov C, Kitching L, Krude T. Distinct populations of human PCNA are required for initiation of chromosomal DNA replication and concurrent DNA repair. Exp. Cell Res. 2005;311:240–250. doi: 10.1016/j.yexcr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4:e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishimoto K, Tachibana K, Hanano I, Yamasaki D, Nakamura H, Kawai M, Urano Y, Tanaka T, Hamakubo T, Sakai J, et al. Sterol-regulatory-element-binding protein 2 and nuclear factor Y control human farnesyl diphosphate synthase expression and affect cell proliferation in hepatoblastoma cells. Biochem. J. 2010;429:347–357. doi: 10.1042/BJ20091511. [DOI] [PubMed] [Google Scholar]
- 41.Zambelli F, Pesole G, Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009;37:W247–W252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dolfini D, Zambelli F, Pavesi G, Mantovani R. A perspective of promoter architecture from the CCAAT box. Cell Cycle. 2009;8:4127–4137. doi: 10.4161/cc.8.24.10240. [DOI] [PubMed] [Google Scholar]
- 43.Warbrick E. The puzzle of PCNA's many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 44.Al Rashid ST, Dellaire G, Cuddihy A, Jalali F, Vaid M, Coackley C, Folkard M, Xu Y, Chen BP, Chen DJ, et al. Evidence for the direct binding of phosphorylated p53 to sites of DNA breaks in vivo. Cancer Res. 2005;65:10810–10821. doi: 10.1158/0008-5472.CAN-05-0729. [DOI] [PubMed] [Google Scholar]
- 45.Pallai R, Simpkins H, Chen J, Parekh HK. The CCAAT box binding transcription factor, nuclear factor-Y (NF-Y) regulates transcription of human aldo-keto reductase 1C1 (AKR1C1) gene. Gene. 2010;459:11–23. doi: 10.1016/j.gene.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schiavoni G, Bennati AM, Castelli M, Fazia MA, Beccari T, Servillo G, Roberti R. Activation of TM7SF2 promoter by SREBP-2 depends on a new sterol regulatory element, a GC-box, and an inverted CCAAT-box. Biochim. Biophys. Acta. 2010;1801:587–592. doi: 10.1016/j.bbalip.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Gurtner A, Fuschi P, Martelli F, Manni I, Artuso S, Simonte G, Ambrosino V, Antonini A, Folgiero V, Falcioni R, et al. Transcription factor NF-Y induces apoptosis in cells expressing wild-type p53 through E2F1 upregulation and p53 activation. Cancer Res. 2010;70:9711–9720. doi: 10.1158/0008-5472.CAN-10-0721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.