Abstract
Type II toxin–antitoxin (TA) systems are generally composed of two genes organized in an operon, encoding a labile antitoxin and a stable toxin. They were first discovered on plasmids where they contribute to plasmid stability by a phenomenon denoted as ‘addiction’, and subsequently in bacterial chromosomes. To discover novel families of antitoxins and toxins, we developed a bioinformatics approach based on the ‘guilt by association’ principle. Extensive experimental validation in Escherichia coli of predicted antitoxins and toxins increased significantly the number of validated systems and defined novel toxin and antitoxin families. Our data suggest that toxin families as well as antitoxin families originate from distinct ancestors that were assembled multiple times during evolution. Toxin and antitoxin families found on plasmids tend to be promiscuous and widespread, indicating that TA systems move through horizontal gene transfer. We propose that due to their addictive properties, TA systems are likely to be maintained in chromosomes even though they do not necessarily confer an advantage to their bacterial hosts. Therefore, addiction might play a major role in the evolutionary success of TA systems both on mobile genetic elements and in bacterial chromosomes.
INTRODUCTION
Toxin–antitoxin (TA) systems are small genetic modules found on bacterial mobile genetic elements as well as in bacterial chromosomes. They appear to be specific of the eu-bacterial and archae-bacterial worlds as no homologous sequences are detected in eukaryotic genomes (except for the PIN toxin domain which is present in eukaryotes) (1). Toxins are always proteins but, based on the nature of the antitoxin and its mode of action, TA systems are currently divided into three classes. Antitoxins of type I and III systems are small RNAs that inhibit either toxin expression (type I) or activity (type III) (2,3). Antitoxins of type II systems are proteins that inactivate toxins by protein–protein complex formation (4). Type I and II systems were discovered in the mid-1980s on bacterial plasmids while type III was discovered only recently. For type I and II systems, it became rapidly clear that these systems were dedicated to plasmid maintenance. In addition to several mechanisms that prevent plasmid-free cells production, plasmids have evolved subtle molecular systems that act by killing plasmid-free daughter bacteria after plasmid loss. Type I and II systems are responsible for this phenomenon denoted as post-segregational killing or addiction (5,6). Addiction relies on the differential stability of the two components, antitoxins being less stable than toxins. In daughter bacteria that do not inherit a plasmid copy, the antitoxin and toxin pool is not replenished. Since the antitoxin is labile, the toxin is released from inhibition, leading to plasmid-free cell death. The cell is thus addicted to antitoxin production and in extenso to the presence of the TA genes (5). Therefore, type I and II systems contribute to an apparent plasmid stabilization at the population level. An extension of this function for type II systems is the capacity of plasmid-encoded systems to outcompete plasmids of the same incompatibility group devoid of TA systems (7).
Bacterial genome sequencing and database mining revealed that homologs of type I and II systems are found in bacterial chromosomes; the occurrence of type II systems being surprisingly high (8–12). Analysis of their distribution provided information regarding their evolution. While type I systems appear to have evolved by lineage specific-duplication (13), type II systems are thought to move from one genome to another by horizontal gene transfer (9,14). The number of type II systems varies greatly not only from one bacterial species to another, but also between isolates from the same species (9,15). The function(s) of chromosomally encoded type I and II systems remain(s) unclear (2,16,17). Although it has been proposed that type II systems might serve as stress response modules, convincing data are still lacking (18). Recent studies have shown that type II systems are involved in the stabilization of large genomic fragments (19) and of integrative conjugative elements (20), indicating that some TA systems might have retained their addictive properties.
Genetic organization of known type II TA systems is quite conserved. Typically, these systems comprise two small genes organized in an operon, the upstream gene encoding the antitoxin. General features such as small size of both components (31–204 amino acid for antitoxins and 41–206 amino acid for toxins) and short intergenic regions separating the two genes (−20 to +30 nt) are also well conserved (9). In general, antitoxins are composed of two domains, an amino-terminal domain responsible for DNA binding and a carboxy-terminal domain responsible for toxin interaction. The antitoxin–toxin protein complex, in which the toxin is inactive, is also responsible for negative autoregulation. Toxins typically are small, stable proteins that inhibit either replication by interacting with DNA-gyrase, or translation by cleaving messenger RNAs (mRNAs) or inhibiting elongation.
Classification of the type II TA systems is based on the amino acid sequence similarity of the toxins, each toxin family being associated with a specific antitoxin family. Thus, type II systems are currently divided into 10 families (8,9). However, a few examples of ‘hybrid’ associations of a toxin from one family and an antitoxin belonging to another family have recently been characterized (21,22). Furthermore, novel putative families of toxins and antitoxins (i.e. not homologous to the 10 families) have been predicted by bioinformatics approaches recently and some were found to be associated with known toxin or antitoxin families, indicating that ‘hybrid’ systems might be more common than originally thought (12,23,24). In fact, one such prediction in Escherichia coli K-12 was recently validated experimentally (8). Thus, predictions are that the type II TA systems are much more abundant and diversified than what is currently described. To evaluate this diversity, a bioinformatics approach was developed to explore prokaryotic genomes and to identify novel toxins and antitoxins. From our predictions, 18 antitoxin and 23 toxin sequences originating from different bacterial species were validated experimentally in E. coli, significantly increasing the number of families of type II antitoxins and toxins. Therefore, we propose to refer to antitoxin and toxin families independently instead of referring to TA system families.
MATERIALS AND METHODS
Bioinformatics approach
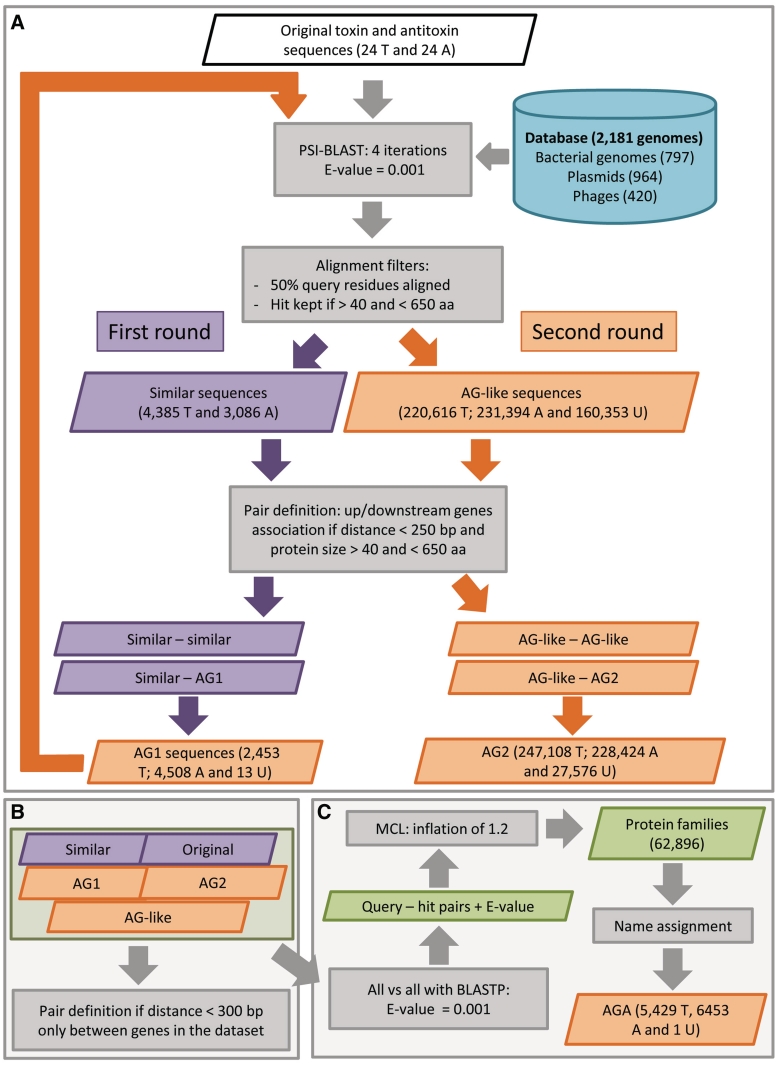
This approach is described in details in Supplementary ‘Materials and Methods’ section and summarized in Figure 1.
Figure 1.
Association by guilt bioinformatics approach. Detection of ‘associated by guilt’ (AG) sequences (A). ‘Similar’ sequences (purple box) to the 48 ‘original’ sequences (white box) were detected by PSI-BLAST searches using the indicated parameters (gray boxes) and a database composed of 2181 genome sequences (blue cylinder). First round: AG toxins and antitoxins denoted as AG1 (orange box) were identified by paring of the ‘similar’ sequences using the indicated parameters (gray box). Number and type of sequences are indicated (T for toxin, A for antitoxin and U for unassigned). Second round: AG1 sequences were used as query for a second round of detection using the same parameters as in the initial steps to identify AG-like sequences (orange box). AG2 sequences were identified by pairing the AG-like sequences. Number and type of sequences are indicated. Pair definition (B). An additional round of pair definition was performed using sequences detected in (A) using the indicated parameters (gray box). Protein families (C). Protein families (green box) were generated by grouping all the proteins present in the dataset using the Markov clustering algorithm (MCL) with the indicated parameters (gray boxes). Names of ‘original’ or ‘similar’ sequences were propagated to the AG proteins within the same protein family. These sequences were then defined as ‘associated by guilt and annotated’ (AGA).
Bacterial strains, plasmids and media
Bacterial strains
The following E. coli strains were used: MC1061 (F− araD139, Δ(ara-leu)7696, galE15, galK16, Δ(lac)X74, rpsL (Strr), hsdR2 (rK−mK+), mcrA mcrB1) (25), MG1655 (rph1 ilvG rfb-50) (26), DJ624 Δara (MG1655 lacX74 mal::lacIq) (27) and DJ624λsfiA ::lacZ (this work).
Plasmids
The following plasmids were used and their relevant characteristics are indicated. The pBAD33 vector (p15A, Cmr, pBAD promoter) (28), the pBAD33-yoeB plasmid (pBAD33 derivative containing the yoeB toxin gene under the control of the pBAD promoter) (29), the pKK223-3 vector (ColE1, Ampr, pTac promoter) (30) and the pLac-staby vector (DelphiGenetics, Belgium) were used in this work.
Media
Luria-Bertani liquid and agar medium (LB) (Invitrogen) as well as M9 minimal liquid and agar medium (KH2PO4 (22 mM), Na2HPO4 (42 mM), NH4Cl (19 mM), MgSO4 (1 mM), CaCl2 (0.1 mM), NaCl (9 mM) supplemented with casamino acids [0.2% except for methionine incorporation (0.05%)] and carbon sources [glycerol, glucose or arabinose (1%)].
Antibiotics
Antibiotics were added at the following concentrations: chloramphenicol, 20 μg/ml; ampicillin, 100 μg/ml.
Experimental validation of novel toxins and antitoxins sequences
The experimental validation of the putative novel toxins and antitoxins families relies on a simple assay: expression of the putative toxins should cause growth inhibition while co-expression with their putative cognate antitoxins should restore normal growth. To that end, antitoxin and toxin candidate genes were cloned in compatible expression vectors under the control of inducible promoters.
Toxins cloning
Predicted toxin genes were cloned in the pBAD33 vector, under the control of the arabinose-inducible pBAD promoter. The predicted toxin coding sequences (CDS) were amplified from bacterial genomic DNA using the appropriate start and stop primers (Supplementary Table S1). Start primers carry a canonical Shine–Dalgarno (SD) sequence. Polymerase chain reaction (PCR) products were digested by XbaI and PstI, except the sauT3COL, nspT5PC7120, speT2TIGR4, speT1TIGR4 amplification products that were digested by SacI and KpnI; and ccrT3CB15 that was digested by XbaI and SalI. Digested products were ligated into the pBAD33 vector cleaved with the appropriate restriction enzymes. Toxins that did not confer an unambiguous growth inhibition phenotype when expressed from the pBAD33 vector were cloned in the pKK223-3 vector under the control of the pTac promoter. The PCR products were digested by EcoRI and PstI and cloned in the pKK223-3 vector cleaved with the same restriction enzymes.
Antitoxins cloning
Predicted antitoxin genes were cloned in the pLac-staby vector and/or in the pKK223-3 vector, which places the antitoxin genes under the control of the pLac and pTac promoters, respectively. Expression from both promoters is induced by isopropyl β-d-1-thiogalactopyranoside (IPTG) addition. Predicted CDSs were amplified from genomic DNA using the appropriate start and stop primers (Supplementary Table S1) and PCR products were ligated in the pLac-staby vector. The predicted antitoxins that failed to counteract the growth inhibition of their cognate toxins were cloned in the pKK223-3 vector under pTac control. The PCR products were digested by EcoRI and PstI and cloned in the pKK223-3 vector cleaved with the same restriction enzymes. To test the activity of antitoxins associated with toxins cloned in the pKK223-3 vector, the cognate antitoxin genes were cloned in the pBAD33 vector. PCR products were digested by XbaI and PstI and cloned in the pBAD33 digested with the same restriction enzymes.
Killing/rescue assay
The DJ624Δara strain was transformed by toxin-encoded plasmids and/or by antitoxin-encoded plasmids and the control vectors. Transformants were grown overnight in M9 liquid medium containing glucose (1%) and the appropriate antibiotics. Overnight cultures were diluted 100-fold in M9 medium containing glucose (1%) and the appropriate antibiotics and grown at 37°C to an OD 600 nm of 0.2. Dilutions (100–106) were plated on M9 plates containing either glucose (1%) or arabinose (1%) and IPTG (1 mM) and the appropriate antibiotics. Plates were incubated overnight at 37°C.
SOS induction
The DJ624λsfiA::lacZ strain containing the pBAD33, pBAD-parERK2, pBAD-ccrT4CB15 or pBAD-atuT1c58 plasmids was grown in M9 medium supplemented with glucose (1%) and chloramphenicol to an OD 600 nm of 0.1. Cultures were centrifuged and pellets resuspended in M9 medium containing arabinose (1%) and chloramphenicol. After 120 min of induction, aliquots were taken to perform β-galactosidase assays as described in ref. 31.
35S-methionine incorporation
The DJ624Δara strain containing the pBAD33 vector and its derivates containing the experimentally validated toxin genes and appropriate controls were grown in M9 medium supplemented with glycerol (1%) at 37°C. Toxin over-expression was induced at an OD 600 nm of 0.1 by adding arabinose (1%). After 180 min of induction, 1 ml aliquots were taken and 5 µCi of 35S-methionine was added. After 2 min of incubation at 37°C, 500 µl aliquots were removed and added to tubes containing 5 ml of cold 10% TCA and left 20 min on ice. Samples were then filtered on nitrocellulose filters (0.45 µm) saturated with the non-labeled precursor using a glass funnel. Filters were then washed in 10% TCA and air-dried. Filter-retained material was counted in 10 ml of scintillation liquid (Ready Protein +) in a liquid scintillation counter (Beckman). Translation rate was normalized to the 35S-methionine incorporated by the control strain (pBAD33 vector).
RESULTS
Detecting ‘similar’ and potentially novel toxins and antitoxins
We collected the antitoxin and toxin sequences of type II systems that were experimentally validated and characterized at the time we started this work. In addition to the relBE, parDE, vapBC, mazEF, phd-doc, higAB, hipBA, omega-epsilon-zeta and ccd systems and their homologs, sequences of the vapXD system were added to our data set even though this system is poorly documented (32). We also included sequences of the parDEDL933-parE3, paaA2-parE2 (33) and yafNO systems that were identified by our procedure and tested as ‘proof of concept’. Note that yafNO characterization was published during the course of this work (34). The hicAB system was not included since it was not validated at the time we started this analysis (8). Our query data set was therefore composed of 24 sequences of antitoxin and 24 sequences of toxin that were denoted as ‘original’ sequences (Supplementary Table S2). We performed a comprehensive PSI-BLAST search (see Supplementary ‘Materials and Methods’ section) on 2181 prokaryotic genomes and detected more than 10 000 sequences. These sequences were grouped in eight toxin super-families and nine of antitoxin (Supplementary Tables S3 and S4).
Based on the conservation of type II systems organization, we developed an exploratory bioinformatics approach based on the ‘association by guilt’ of orphan toxins or antitoxins (i.e. not paired with ‘original’ antitoxins or toxins) to discover novel sequences potentially showing antitoxin or toxin activities (Figure 1 and see Supplementary ‘Materials and Methods’ section for details). These sequences were grouped into protein families based on their similarities (see Supplementary ‘Materials and Methods’ section for the detailed procedure and parameters). As a result, we distinguished two types of predicted antitoxin or toxin sequences: those belonging to a family composed of proteins of unknown function which were called AG (for ‘associated by guilt’), and those ‘associated by guilt’ but present in a family containing at least one ‘original’ sequence or one sequence presenting similarity with ‘original’ sequences (denoted as ‘similar’, see Supplementary ‘Materials and Methods’ section for details). Super-family names of these ‘original’ and/or ‘similar’ sequences were transferred to these AG sequences and they were denoted consequently ‘associated by guilt and annotated’ (AGA).
Experimental validation of predicted toxins and antitoxins
Our bioinformatics exploratory procedure led to the prediction of a large number of novel AG and AGA toxins and antitoxins located upstream and/or downstream of orphan ‘similar’ and originating from numerous bacterial species.
Experimental validation of these AG and AGA sequences was carried out in E. coli using a simple Kill/Rescue assay, in which expression of the toxin gene is expected to inhibit cell growth while co-expression of the toxin and antitoxin genes restores growth.
The toxin and antitoxin sequences were cloned in compatible vectors, under the control of different inducers (see ‘Materials and Methods’ section). These sequences were placed in the same genetic context, i.e. RBS and ATG as start codon, to achieve a comparable level of expression, although we cannot rule out that some genes are less expressed than others as we did not measure expression levels.
Predicted toxins
Twenty-three AG toxins were experimentally tested. Eight of them are paired with sequences ‘similar’ to ‘original’ antitoxins (Supplementary Table S5). Expression of the putative BceT5E33L, SpyT210270 and LmoT1EGD-e toxins did inhibit E. coli growth. Among the upstream open reading frames (ORFs) predicted to be antitoxins, only BceA5E33L, which is associated with the BceT5E33L toxin, turned out to be a functional antitoxin belonging to the HigA super-family of antitoxins. For the SpyT210270 toxin, the antitoxin activity of the downstream SpyA210270 ORF, although not predicted by our approach, was tested. Co-expression of SpyA210270 relieved SpyT210270-mediated cell growth inhibition. In this case, the antitoxin gene is located downstream of the toxin gene. For the third toxin that inhibits E. coli growth, LmoT1EGD-e, none of the flanking ORFs showed antitoxin activity.
Seven AG toxins associated with AGA antitoxins belonging to the HigA super-family were tested (Supplementary Table S6). Expression of four of them (SmeT11021, MavT1K10, SpyT19429 and SpyT310270) inhibited E. coli growth. However, only two of the predicted antitoxins (SmeA11021 and MavA1K10) relieved the growth inhibition mediated by their cognate toxins. For the SpyT19429 and SpyT310270 toxins, we were unable to detect antitoxin activity for their flanking ORFs.
Eight AG toxins associated with eight AG antitoxins were experimentally tested (Supplementary Table S7). Expression of BceT1E33L, SpyT110270, SpyT1M1 and EcoT1EDL933 resulted in E. coli growth inhibition and co-expression of their paired antitoxins relieved growth inhibition (Figure 2).
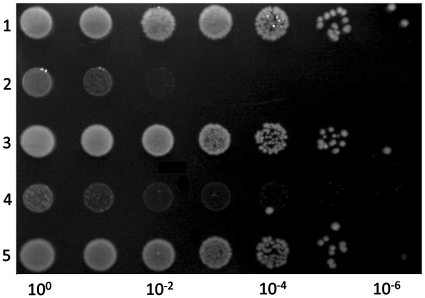
Figure 2.
The ecoA1-ecoT1EDL933 and the nspA5-nspT5PC7120 pairs constitute TA systems. The DJ624Δara strain containing the pBAD33 and pLac-staby plasmids (1), the pBAD33-ecoT1EDL933 and pLac-staby plasmids (2), the pBAD33-ecoT1EDL933 and pLac-staby-ecoA1EDL933 plasmids (3), the pBAD33-nspT5PC7120 and pLac-staby plasmids (4) or the pBAD33-nspT5PC7120 and pLac-staby- nspA5PC7120 plasmids (5) were grown in log phase in M9 medium supplemented with glucose (1%) and appropriate antibiotics. Serial dilutions (as indicated) were spotted on LB plates containing arabinose (1%) and IPTG (1 mM). Plates were incubated overnight at 37°C.
One predicted pair (MGAS10270 Spy0568 and Spy0569) presented a particular phenotype. This pair is composed of sequences similar to the E. coli hicAB system (8). HicA was shown to be a toxin cleaving mRNAs and HicB was shown to be its antitoxin. Our prediction was opposite: Spy0568, the S. pyogenes HicB homolog was predicted to be a toxin and Spy0569, the HicA homolog, the antitoxin. Surprisingly, expression of both proteins was toxic for E. coli (data not shown). We therefore renamed Spy0568 as SpyT410270 and Spy0569 as SpyT510270. The flanking ORFs being located on the complementary strand, we did not check their antitoxic activity.
Predicted antitoxins
Eleven AG antitoxins were experimentally tested. Eight predicted antitoxins are associated with sequences ‘similar’ to ‘original’ toxins (Supplementary Table S8). Expression of these toxins inhibited E. coli growth except for the RPC4130 ORF. The seven cognate antitoxins relieved the toxicity mediated by their associated toxins.
Three of these antitoxins (CcrA1CB15, SpeA2TIGR4 and CcrA4CB15) were validated by other groups during the course of this work, although their annotation turned out to be incorrect (9,35). The CcrA1CB15 antitoxin is associated with a sequence similar to ParE and was described as being a ParDRK2-like antitoxin in Caulobacter crescentus (36). We were unable to detect any sequence similarity between this sequence and the ParDRK2 antitoxin. The second case concerns the SpeA2TIGR4 antitoxin, which is associated with a toxin from the RelE family in Streptococcus pneumoniae. This ORF was named RelB2spn in a previous work (35). However, again, we were unable to detect any sequence similarity between this sequence and RelBK-12. The last case concerns the ccrA4CB15-ccrT4CB15 system that was annotated as relBE2 in C. crescentus (9) and experimentally analyzed (36). The CcrT4CB15 toxin belongs to the ParE/RelE super-family. We showed that expression of this toxin induces the SOS response, although less efficiently than ParERK2 (Figure 3), confirming our predictions and indicating that CcrT4CB15 is a ParE-like toxin. The associated CcrA4CB15 antitoxin, however, does not show any similarities to ‘original’ antitoxins. Thus, these three antitoxins represent novel sequences associated with ‘similar ‘toxins that present a growth inhibition phenotype do not show any similarities with ‘original’ antitoxins and were not described previously. The AtuA1c58 antitoxin is associated with AtuT1c58, which belongs to the ParE/RelE toxin super-family. Expression of this toxin induces the SOS system, indicating that AtuT1c58 is a ParE-like toxin (Figure 3). The NspA5PC7120 and AfuA2DSM4304 antitoxins are paired with toxins from the VapC super-family and the SpeA3TIGR4 antitoxin is associated with a toxin belonging to the Doc super-family.
Figure 3.
Overexpression of the CcrT4CB15 and AtuT1C58 toxins induce the SOS system in E. coli. The DJ624λsfiA::lacZ strain containing the pBAD33 (1), pBAD-parERK2 (2), pBAD-ccrT4CB15 (3) or pBAD-atuT1c58 (4) plasmids were grown in M9 medium. After induction of toxin expression by arabinose addition, samples were taken to perform β-galactosidase assays as described in ‘Materials and Methods’ section.
Three AG antitoxins associated with AGA toxins were experimentally tested (Supplementary Table S6). Expression of these three toxins (NspT1PC7120, NspT2PC7120 and NeuT1C91) resulted in E. coli growth inhibition and co-expression of their associated antitoxins relieved this inhibition (Figure 2). Two of these toxins belong to the VapC super-family and the other to the ParE/RelE super-family.
Thus, this approach led to the validation of 23 toxins and 18 antitoxins. For 5 of the 23 toxins, we were unable to identify the cognate antitoxin, suggesting that they might be solitary toxins or that expression levels of some of the antitoxins might be too low to rescue the toxic phenotype. Eleven of the 34 predicted toxins did not affect E. coli growth; it is possible that, as mentioned above, expression levels are too low; these sequences might be false-positives (not related to toxins) or they might be ‘true’ toxins but only for their natural host and not for E. coli.
Toxin activities
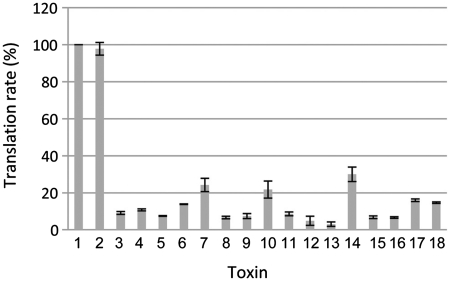
To gain insights into the mode of toxicity of the novel validated toxins, several tests were performed. Cell morphology was observed by microscopy and SOS induction, transcription as well as translation rates were measured in E. coli overexpressing the active toxins. In addition to the novel toxins, we also tested the AGA NspT1PC7120 and NspT5PC7120 toxins belonging to the VapC super-family in order to confirm this annotation. Neither cell morphology, SOS induction nor transcription rates were affected by toxin overexpression (data not shown and Supplementary Figure S1). However, expression of these toxins drastically reduced 35S-methionine incorporation (from 10% to 30% as compared to the control vector) (Figure 4). These data show that all of the tested toxins affect translation. In addition, preliminary data seem to indicate that expression of some of these toxins induces mRNA cleavage in E. coli (data not shown).
Figure 4.
Overexpression of the novel toxins inhibits translation in E. coli. The DJ624Δara strain containing the pBAD33 (1), pBAD-parERK2 (2), pBAD-yoeB (3), pBAD-ecoT1EDL933 (4), pBAD-mavT1K10 (5), pBAD-spyT110270 (6), pBAD-spyT210270 (7), pBAD-bceT1E33L (8), pBAD-smeT11021 (9), pBAD-spyT1M1 (10), pBAD-bceT5E33L (11), pBAD-nspT1PC7120 (12), pBAD-nspT2PC7120 (13), pBAD-spyT410270 (14), pBAD-spyT510270 (15), pBAD-spyT310270 (16), pBAD-spyT19429 (17) and pBAD-lmoT1EGD-e (18) were grown in M9 medium. After induction of toxin expression by arabinose addition, cultures were labeled with 35S-methionine. Translation rate was measured as described in ‘Materials and Methods’ section.
Evolutionary relationships between novel and ‘original’ super-families
Six of the 23 toxins validated experimentally define four novel super-families (Table 1). These super-families were denoted as Gin (for growth inhibition).
Table 1.
Occurrence of 12 toxin super-families in 2181 prokaryotic genomes
| Toxin super-families (n = 7034) |
Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RelE/ParE (n = 2057) | CcdB/MazF (n = 645) | Zeta (n = 1434) | Doc (n = 859) | YafO (n = 25) | VapD (n = 29) | HipA (n = 421) | VapC (n = 1267) | GinA (n = 101) | GinB (n = 98) | GinC (n = 89) | GinD (n = 9) | 12 | |
| ‘Original’ sequences | RelEK-12, RelE307, HigB, PasB, StbE, YoeB, Txe, YafQ, ParERK2, ParE1, ParE3 | CcdBF, CcdBO157, MazF, YdcE, PemK, ChpBK | Zeta | Doc | YafO | VapD | HipA | VapC, MvpT | – | – | – | – | 24 |
| Validated ‘similar’ sequences | CcrT1CB15, AtuT1C58, CcrT4CB15, SpeT2TIGR4 | NT | NT | SpeT3TIGR4 | NT | NT | NT | AfuT2DSM4304, NspT5PC7120 | – | YgjNa | – | – | 7 |
| Validated AGA sequences | NeuT1C91 | NT | NT | NT | NT | NT | NT | NspT2PC7120, NspT1PC7120 | – | – | – | – | 3 |
| Validated AG sequences | EcoT1EDL933, MavT1K10 | NT | NT | NT | NT | NT | NT | NT | SpyT110270, SpyT210270, BceT1E33L | SmeT11021 | SpyT1M1 | BceT5E33L | 8 |
The Zeta super-family might contain a significant number of false-positive sequences that were selected on the basis of ATP-binding domain found in the ‘original’ Zeta sequence.
n represents the number of sequences; NT for not tested.
aIndicates that the YgjN toxin was validated during the course of this work (37).
Three of the novel toxins constitute the GinA super-family (SpyT110270, SpyT210270 and BceT1E33L). The GinB super-family is composed of the SmeT11021 novel toxin as well as the YgjN toxin, which was identified in E. coli during the course of this work (37). This toxin was considered as a homolog of RelE (37). However, while we detected sequence similarity between SmeT11021 and YgjN (data not shown, E-value: 10e-7), we were unable to detect any sequence similarity between SmeT11021 and the members of the ParE/RelE super-family. The GinC and GinD super-families are composed of SpyT1M1 and BceT5E33L, respectively. Regarding the solitary toxins, they define four novel super-families (data not shown).
Twelve of the 18 antitoxins that were experimentally validated define 11 novel antitoxin super-families that are unrelated to the ‘original’ super-families. They were named Fiz for full toxin neutralization (Table 2).
Table 2.
Occurrence of 20 antitoxin super-families in 2181 prokaryotic genomes
| Antitoxin superfamilies (n = 10 829) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phd (n = 955) | RelB (n = 304) | PasA (n = 656) | VapB (n = 665) | HigA (n = 3351) | VapX (n = 10) | CcdA (n = 74) | Epsilon (n = 11) | ParD (n = 10) | FizA (n = 1962) | ||
| ‘Original’ sequences | Phd, YefM, Axe, PasB, StbD, YafN, RelB307 | RelBK-12, DinJ, Paa1 | PasA, YdcD, ParDEDL933 | VapB, MvpA, MazE, ChpBI, PemI | HigA, HipB | VapX | CcdAF | Epsilon | ParDRK2 | – | |
| Validated ‘similar’ sequences | NT | NT | NT | NT |
|
NT | NT | NT | NT | – | |
| Validated AGA sequences | NT | NT | NT | NT |
|
NT | NT | NT | NT | – | |
| Validated AG sequences | NT | NT | SpeA2TIGR4 | SpeA3TIGR4 | EcoA1EDL933 | NT | NT | NT | NT | SpyA21070 | |
| Antitoxin superfamilies (n = 10 829) |
Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FizB (n = 1542) | FizC (n = 1121) | FizD (n = 103) | FizE (n = 26) | FizF (n = 10) | FizG (n = 9) | FizH (n = 8) | FizI (n = 6) | FizJ (n = 5) | FizK (n = 1) | 20 | |
| ‘Original’ sequences | – | – | – | – | – | – | – | – | – | – | 24 |
| Validated ‘similar’ sequences | – | – | – | – | – | – | – | – | – | – | 1 |
| Validated AGA sequences | – | – | – | – | – | – | – | – | – | – | 2 |
| Validated AG sequences | CcrA1CB15 | SpyA1M1 | NeuA1C91 | AtuA1C58 | SpyA110270 | NspA2PC7120, NspA5PC7120 | AfuA2DSM4304 | CcrA4CB15 | BceA133L | NspA1PC7120 | 15 |
The HigA super-family might contain a significant number of false-positive sequences that were selected on the basis of the HTH-XRE domain found in the ‘original’ HigA sequence.
n represents the number of sequences; NT for not tested.
aIndicates that the YgjM antitoxin was validated during the course of this work (37).
Association between antitoxin and toxin super-families
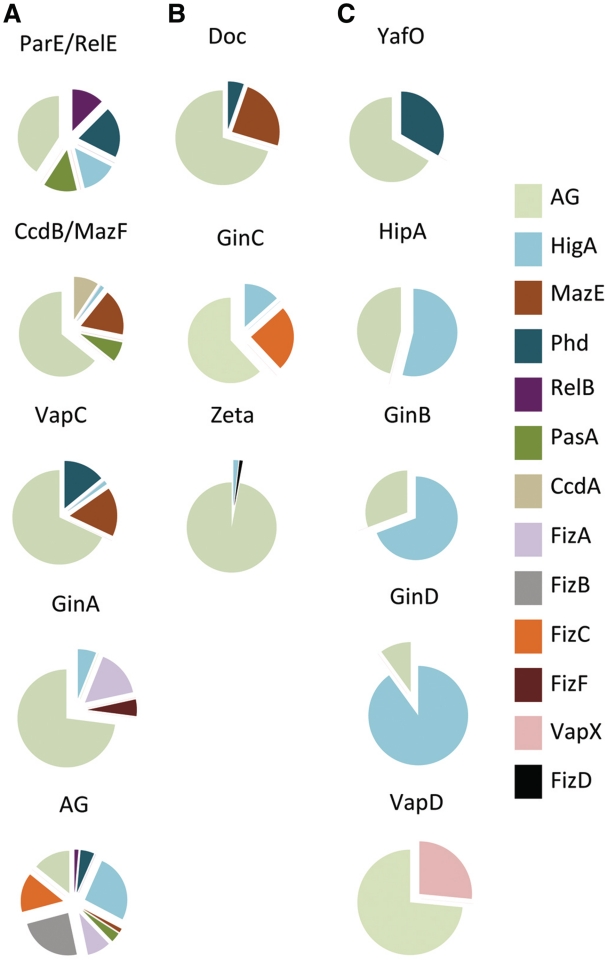
In general, a large number of predicted toxin sequences are associated with antitoxin AG sequences (Figure 5). Three main categories can be defined: toxin super-families that have multiple partners (>4) such as the large ParE/RelE, VapC and CcdB/MazF super-families and the smaller GinA super-family (Figure 5A). The second category is composed of Doc, GinC and Zeta super-families. Sequences belonging to these super-families are associated with three different antitoxin super-families (Figure 5B). In the last category, toxin super-families are associated with AG sequences and a second super-family, Phd for the YafO super-family and HigA for the HipA, GinB and GinD super-families.
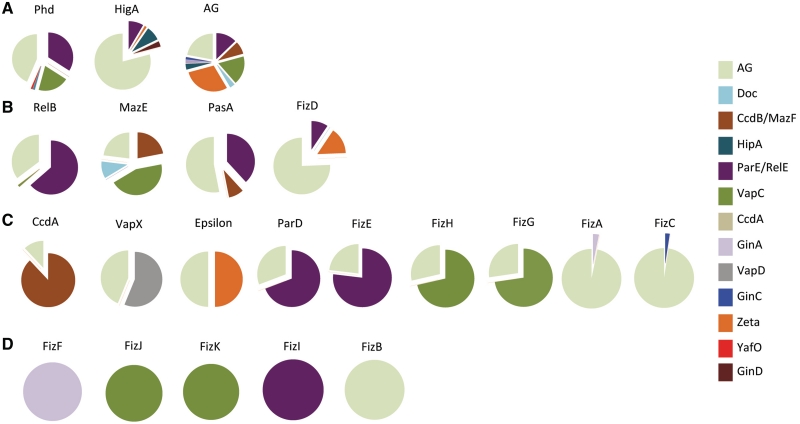
Figure 5.
Associations of toxin super-families. The 12 toxin super-families are indicated above the pie chart. Each section of the pie chart represents the relative abundance of antitoxin sequences belonging to a given super-family associated with toxin sequences. Each antitoxin super-family is represented by a specific color. In (A), toxin super-families that are associated with multiple antitoxin super-families (>4). In (B), toxin super-families that are associated with three different antitoxin super-families. In (C), toxin super-families that are associated with AG sequences and another antitoxin super-family. Associations occurring at less than 1% were not considered for clarity.
As observed for the predicted toxins, a large number of predicted antitoxins are associated with AG sequences (Figure 6). Categories similar to those defined for toxin super-families can be defined. Three super-families of antitoxin are promiscuous and associate with five or more toxin families i.e. the large PhD and HigA super-families (Figure 6A). The second category groups super-families that associated with three or four different toxin super-families such as the RelB and FizD super-families (Figure 6B). Most of the antitoxin super-families are quite restrictive in their associations (Figure 6C). They are associated with AG sequences and a second toxin super-family. Five of the novel super-families associate exclusively with a specific toxin super-family (FizB, FizF, FizI, FizJ and FizK) (Figure 6D).
Figure 6.
Associations of antitoxin super-families. The 20 antitoxin super-families are indicated above the pie chart. Each section of the pie chart represents the relative abundance of toxin sequences belonging to a given super-family associated with antitoxin sequences. Each toxin super-family is represented by a specific color. In (A), antitoxin super-families that are associated with multiple toxin super-families (five or more). In (B), antitoxin super-families that are associated with three or four different toxin super-families. In (C), antitoxin super-families that are associated with AG sequences and another toxin super-family. In (D), antitoxin super-families that are associated with a specific toxin super-family or to AG sequences. Associations occurring at <1% were not considered for clarity.
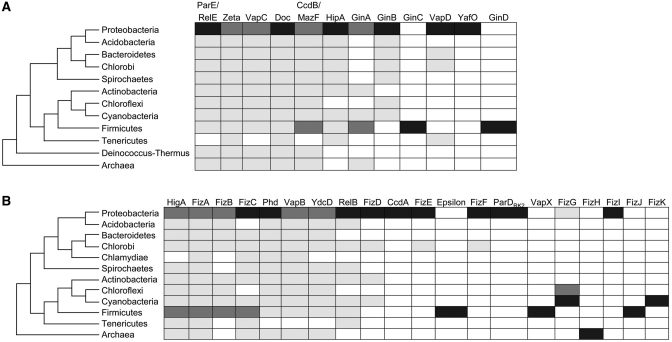
Phyletic and genomic distribution of the toxin and antitoxin super-families
Phyletic distribution varies from one super-family to the other. The ParE/RelE, Zeta, VapC, Doc, CcdB/MazF, HipA and GinB super-families of toxins are widely distributed in bacterial phyla, although GinB is not detected in Firmicutes (Figure 7A). At least 70% of their members are detected in Proteobacteria, Firmicutes (except GinB), Cyanobacteria and Actinobacteria. The remaining sequences are detected in various phyla, with all of them except HipA and GinB being present in Archae. The GinA and VapD super-families, on the other hand, are restricted to a smaller number of phyla. Note that the VapD sequences are neither detected in Firmicutes nor in Archae. The GinC, YafO and GinD are specific to a single phylum. Most of the toxin sequences are located in chromosomes (Supplementary Figure S2A), except for the VapD sequences that are detected on plasmids (40%) and GinA sequences on phages (30%). The GinC, YafO and GinD sequences are detected neither on plasmids nor on phages.
Figure 7.
Phyletic distribution of toxin and antitoxin super-families. Sequences of toxin super-families (A) and antitoxin super-families (B) are detected in the bacterial phyla indicated at the left of the figure in different proportions: not detected (white), 0.1–20% (gray), 20.1–60% (dark gray), above 60.1% (black).
As observed for the toxin super-families, phyletic distribution of the antitoxin super-families varies from one super-family to the other (Figure 7B). The HigA, FizA, FizB, FizC, Phd, VapB, PasA and RelB super-families appear widely distributed in bacterial phyla. At least 70 % of the members of these super-families are found in Proteobacteria, Firmicutes and Cyanobacteria (except for RelB sequences). The remaining 30% varies depending on the family, with members of HigA, FizA, Phd, VapB and PasA super-families being detected in Archae. The other super-families appear to be much more restricted. A majority of the FizG sequences are detected in Cyanobacteria. Sequences of FizE and FizF super-families are distributed in two phyla: Proteobacteria and Green sulfur bacteria. Members of the CcdA, ParD and FizI super-families are exclusively found in Proteobacteria, while those of VapX and Epsilon are restricted to Firmicutes and FizH to Archae. The only representative of FizK super-family is found in Cyanobacteria.
Regarding their genomic location (Supplementary Figure S2B), more than 85% of the sequences are located on chromosomes except for the RelB, FizF, CcdA and ParD super-families with 20–40% of the sequences being located on plasmids. The Epsilon super-family represents an exception with its 11 representative sequences being found only on plasmids.
Genome size and number of type II TA systems are not correlated
Figure 8 shows that there is no significant correlation between the number of predicted toxin and antitoxin sequences and the total number of ORFs predicted in a given genome. Even the smallest genomes of some of the intracellular obligate species seem to contain predicted toxin and antitoxin sequences (Table 3). While Buchnera and Chlamydia/Chlamydophyla genomes are devoid of predicted toxin and antitoxin sequences, some Rickettsias (such as R. bellii OSU 85-389, R. bellii RML369-C, R. akari or R. felis) contain more than 20 predicted sequences, representing between 1.8% and 2.6% of the total number of ORFs. Interestingly, the number of predicted toxins and antitoxins in the Rickettsia genus is quite variable (0–36). Variability is also found within the Wolbachia genus (0–6 predicted sequences). For the 20 bacterial isolates with the highest content of toxin and antitoxin predicted sequences, it varies from 37 to 97 per genome, representing from 0.8% to 2.5% with respect to the total number of ORFs (Supplementary Table S9). These bacteria belong to four different phyla (Proteobacteria, Cyanobacteria, Green sulfur and Actinobacteria). High genome plasticity appears to be a common theme for most of these strains (described for 12 over 20, e.g. Microcystis aeruginosa, Gleobacter violaceum and Nitrosomanas europeae). They also tend to have a versatile metabolism (6 over 20, e.g. Nitrosomanas europeae, Rhodopseudomas palustris and Azoarcus sp.) or a particular metabolism (4 over 20, e.g. Geobacter uraniireducens, Pelodictyon phaeoclathratiforme and Caulobacter sp.). Two isolates are symbionts (Verminephrobacter eiseniae and Photorabdus luminescens) and another is part of phototrophic bacterial consortia (C. chlorochromatii). Five of the strains belong to the Mycobacterium tuberculosis complex and are pathogenic (or deriving from a pathogenic strain in the case of M. tuberculosis H37Ra).
Figure 8.
No correlation between the total number of CDS and the number of predicted antitoxin and toxin sequences. Only ‘original’ and ‘similar’ antitoxin and toxin sequences were considered.
Table 3.
Predicted antitoxin and toxin sequences in intracellular obligate species
| Bacterial species | Phyla | CDS in genome | Predicted A and T (%) |
|---|---|---|---|
| Buchnera aphidicola str. APS | Proteobacteria | 564 | 0 |
| Buchnera aphidicola str. Bp | Proteobacteria | 504 | 0 |
| Buchnera aphidicola str. Cc | Proteobacteria | 357 | 0 |
| Buchnera aphidicola str. Sg | Proteobacteria | 546 | 0 |
| Chlamydia muridarum Nigg | Chlamydiae | 904 | 0 |
| Chlamydia trachomatis 434/Bu | Chlamydiae | 874 | 0 |
| Chlamydia trachomatis A/HAR-13 | Chlamydiae | 911 | 0 |
| Chlamydia trachomatis D/UW-3/CX | Chlamydiae | 895 | 0 |
| Chlamydia trachomatis L2b/UCH-1/proctitis | Chlamydiae | 874 | 0 |
| Chlamydophila abortus S26/3 | Chlamydiae | 932 | 0 |
| Chlamydophila caviae GPIC | Chlamydiae | 998 | 0 |
| Chlamydophila felis Fe/C-56 | Chlamydiae | 1005 | 0 |
| Chlamydophila pneumoniae AR39 | Chlamydiae | 1112 | 0 |
| Chlamydophila pneumoniae CWL029 | Chlamydiae | 1052 | 0 |
| Chlamydophila pneumoniae J138 | Chlamydiae | 1069 | 0 |
| Chlamydophila pneumoniae TW-183 | Chlamydiae | 1113 | 0 |
| Rickettsia akari str. Hartford | Proteobacteria | 1259 | 23 (1.8) |
| Rickettsia bellii OSU 85-389 | Proteobacteria | 1476 | 32 (2.2) |
| Rickettsia bellii RML369-C | Proteobacteria | 1429 | 26 (1.8) |
| Rickettsia canadensis str. McKiel | Proteobacteria | 1093 | 1 (0.09) |
| Rickettsia conorii str. Malish 7 | Proteobacteria | 1374 | 13 (0.9) |
| Rickettsia felis URRWXCal2 | Proteobacteria | 1400 | 36 (2.6) |
| Rickettsia massiliae MTU5 | Proteobacteria | 968 | 16 (1.7) |
| Rickettsia prowazekii str. Madrid E | Proteobacteria | 835 | 0 |
| Rickettsia rickettsii str. 'Sheila Smith' | Proteobacteria | 1345 | 14 (1.0) |
| Rickettsia rickettsii str. Iowa | Proteobacteria | 1384 | 13 (0.9) |
| Rickettsia typhi str. Wilmington | Proteobacteria | 838 | 0 |
| Mycobacterium leprae TN | Actinobacteria | 1605 | 1 (0.06) |
| Wigglesworthia glossinidia | Proteobacteria | 611 | 0 |
| Wolbachia endosymbiont of Drosophila melanogaster | Proteobacteria | 1195 | 6 (0.5) |
| Wolbachia endosymbiont strain TRS of Brugia malayi | Proteobacteria | 805 | 0 |
| Wolbachia pipientis | Proteobacteria | 1275 | 2 (0.2) |
For each genome, the total number of CDS is indicated.
The total number of ‘similar’ and AGA sequences belonging to the super-families described in Supplementary Tables S2 and S3 were considered with the exception of those belonging to the Zeta and HigA super-families as many false-positive sequences are suspected.
The percentage of predicted sequences with respect to the total number of CDS in the genome is indicated between brackets.
A for antitoxin, T for toxin.
DISCUSSION
Discovering novel toxin and antitoxin sequences
The ‘association by guilt’ approach was already used to predict novel TA systems by Makarova et al. (24). The approach we used in this work allowed the identification of sequence pairs presenting unusual features such as a total absence of similarity with the ‘original’ sequences, a larger size for both the antitoxins and toxins (>200 amino acid) and larger intergenic distances (up to 300 bp). Comparison between their data and ours indicates that our approach predicted their sequences (data not shown) but, more importantly, confirmed that there are many more toxins and antitoxin families not related to ‘original’ ones or simply uncharacterized yet, than previously thought and that remain to be experimentally studied. The successful experimental validation of some of our predictions is a direct proof of the results presented in the two works.
Activities of type II toxins
Quite unexpectedly, the novel toxins identified in this work are all translation inhibitors. We cannot exclude that a bias might have been introduced by the experimental validation of foreign genes in E. coli. Toxins targeting specific pathways might not be toxic for E. coli and therefore not validated by our Killing/rescue assay. The counter-argument is that if these genes are involved in stable maintenance of mobile genetic elements and move through horizontal transfer, toxins should target conserved mechanisms to ensure their function and maintenance in different bacterial hosts. ‘Original’ toxins inhibiting translation use a variety of mechanisms from cleavage of free-RNAs, cleavage of mRNAs during translation, inhibition of elongation by 30S binding and by EF-Tu phosphorylation (38–40), and it remains to be shown whether novel toxins will extend this list of activities. Mechanisms of action of these toxins are currently being investigated in our laboratory.
Origin and evolution of type II TA systems
Evolution of the TA systems remains largely unknown. While some authors proposed that these systems might derive from a common ancestor (41), other groups are in favor of several ancestors (10,12). Our data indicate that the validated toxins (both ‘original’ and those discovered in this analysis) constitute 12 unrelated super-families. No structural homologs could be detected for the novel toxins in the PDBsum structure database except for SpyT1M1. The structure of its homolog is unrelated to type II toxins. This indicates that the novel toxins do not share any evolutionary relationship with ‘original’ super-families, although they are functionally related. However, only the resolution of their three-dimensional structures will unambiguously answer this question since similar structures have been detected for toxins that do not show significant sequence similarity (42). Subsequent divergence is observed for toxins within the ParE/RelE and CcdB/MazF super-families as they acquired different activities (DNA-gyrase inhibitors and mRNA-interferases).
For the antitoxins, our data indicate that they form 20 unrelated super-families. Further analyses will be required to define whether or not these antitoxins have a DNA-binding domain. Retracing antitoxin evolution might be complex since antitoxins are composed in general of two independent domains, an amino-terminal DNA-binding domain and a carboxy-terminal toxin-binding domain. DNA-binding domains might be recruited from or by other types of activities totally unrelated to antitoxins. A variation on the theme is observed in the case of three-component systems. In these systems [epsilon-omega-zeta (43) and paaR1-paaA1-parE3 (33)], DNA-binding and antitoxin activities are encoded by two separated polypeptides. Molecular mechanisms underlying transcriptional regulation of three-component systems remain to be determined.
Abundance and diversity
Another level of diversity is observed for antitoxin and toxin associations. Sequences of the RelE/ParE, CcdB/MazF and VapC super-families are paired with antitoxin sequences of the RelB, Phd, MazE and PasA super-families, supporting the idea that antitoxins and toxins were assembled multiple times as suggested previously (12). Sequences belonging to these super-families are abundant and present on plasmids. Other super-families are less promiscuous in their association. The reason is unclear, although super-families such as GinC, GinD, YafO, FizH or FizI are less represented and tend to be phyla- and chromosome-specific (although we cannot exclude that some are located on genomic islands), which might reduce their possibilities of association. However, although CcdA, Epsilon and ParD antitoxin sequences are present on plasmids, and therefore should be prone to horizontal gene transfer, they are phyla-specific and not abundant. Thus, what makes the evolutionary success of a given antitoxin or toxin super-family is still largely unknown. We have to keep in mind the biases introduced by the fact that a majority of the fully sequenced genomes available at NCBI are of Proteobacteria (745/1566 genomes) and that genomic islands within chromosomes remain difficult to identify.
Nevertheless, in general, these entities are abundant in bacterial chromosomes. Some genomes contain as much as 97 predicted antitoxin and toxin sequences (Supplementary Table S9), representing 1.5% with respect to the total number of CDSs. An even higher percentage is observed in some obligate intracellular bacterial genomes (Table 3). Predicted antitoxin and toxin sequences in R. belii and R. felis represent as much as 2.2% to 2.6%, respectively. Interestingly, these strains also contain a high number of repeat sequences (3.7% and 4.4%, respectively) and other genes indicative of genome plasticity (e.g. phage-related genes, transposases) (44,45). In contrast, other strains of the Rickettsia genus such as R. typhi and R. prowezaki, appear to be devoid of predicted antitoxin and toxin sequences (Table 3). Interestingly, both strains are closely related in the Rickettsia phylogeny and have undergone massive reductive evolution (a general phenomenon observed in obligate intracellular bacteria) as compared to R. felis and R. bellii (46). The absence of TA systems in some obligate intracellular bacteria might be due to massive gene loss. The comparison of M. leprae and its free-living relative M. tuberculosis complex strains supports this idea (one predicted sequence versus 43–45) (Table 3 and Supplementary Table S9). A contrario, a high content of TA systems might reflect an intense gene flux (Supplementary Table S9). Interestingly, R. felis carries a putative conjugative plasmid (44) and might still be prone to horizontal gene transfer. This could possibly explain the presence of predicted toxin and antitoxin sequences in this strain.
As a conclusion, we propose that toxin and antitoxin genes have emerged and assembled multiple times during evolution. Associated with mobile genetic elements, TA systems promote their stability and propagate efficiently within the bacterial world. Because of their addictive properties, they integrate stably in bacterial chromosomes without providing necessarily selective advantages to their hosts. Interestingly, most of the toxin families are translation inhibitors, suggesting that this activity might have been selected rather than more detrimental ones such as DNA-gyrase inhibitors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fonds de la Recherche Scientifique (FRSM-3.4530.04); European Union (STREP CRAB, LSHM- CT-2005-019023); Fondation Van Buuren and Fonds Jean Brachet; the FRIA (to D.G. and J.G.); and European Spatial Agency (EXANAM ESA-AO-07-Concordia to R.L.). Funding for open access charge: FNRS (FRSM-3.4530.04).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to the scientific community for kindly providing us with genomic DNA and bacterial strains and to the reviewers for their helpful comments. LVM thanks Marie Deghorain, Nadim Majdalani and Pierre Smeesters for critical reading of the manuscript and Susan Gottesman, Michael Maurizi, Didier Mazel and Gisela Storz for discussions.
REFERENCES
- 1.Arcus VL, Rainey PB, Turner SJ. The PIN-domain toxin–antitoxin array in mycobacteria. Trends Microbiol. 2005;13:360–365. doi: 10.1016/j.tim.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 2008;72:579–589. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blower TR, Pei XY, Short FL, Fineran PC, Humphreys DP, Luisi BF, Salmond GP. A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat. Struct. Mol. Biol. 2011;18:185–90. doi: 10.1038/nsmb.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin–antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 5.Yarmolinsky MB. Programmed cell death in bacterial populations. Science. 1995;267:836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]
- 6.Gerdes K, Rasmussen PB, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl Acad. Sci. USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper TF, Heinemann JA. Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. Proc. Natl Acad. Sci. USA. 2000;97:12643–12648. doi: 10.1073/pnas.220077897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jorgensen MG, Pandey DP, Jaskolska M, Gerdes K. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 2009;191:1191–1199. doi: 10.1128/JB.01013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guglielmini J, Szpirer C, Milinkovitch MC. Automated discovery and phylogenetic analysis of new toxin–antitoxin systems. BMC Microbiol. 2008;8:104. doi: 10.1186/1471-2180-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin–antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anantharaman V, Aravind L. New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 2003;4:R81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. Abundance of type I toxin–antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010;38:3743–3759. doi: 10.1093/nar/gkq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mine N, Guglielmini J, Wilbaux M, Van Melderen L. The decay of the chromosomally encoded ccdO157 toxin–antitoxin system in the Escherichia coli species. Genetics. 2009;181:1557–1566. doi: 10.1534/genetics.108.095190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magnuson RD. Hypothetical functions of toxin–antitoxin systems. J. Bacteriol. 2007;189:6089–6092. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Melderen L, Saavedra De Bast M. Bacterial toxin–antitoxin systems: more than selfish entities? PLoS Genet. 2009;5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Melderen L. Toxin–antitoxin systems: why so many, what for? Curr. Opin. Microbiol. 2011;13:781–785. doi: 10.1016/j.mib.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Szekeres S, Dauti M, Wilde C, Mazel D, Rowe-Magnus DA. Chromosomal toxin–antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol. Microbiol. 2007;63:1588–1605. doi: 10.1111/j.1365-2958.2007.05613.x. [DOI] [PubMed] [Google Scholar]
- 20.Wozniak RA, Waldor MK. A toxin–antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009;5:e1000439. doi: 10.1371/journal.pgen.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grady R, Hayes F. Axe-Txe, a broad-spectrum proteic toxin–antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 2003;47:1419–1432. doi: 10.1046/j.1365-2958.2003.03387.x. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt O, Schuenemann VJ, Hand NJ, Silhavy TJ, Martin J, Lupas AN, Djuranovic S. prlF and yhaV encode a new toxin–antitoxin system in Escherichia coli. J. Mol. Biol. 2007;372:894–905. doi: 10.1016/j.jmb.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makarova KS, Grishin NV, Koonin EV. The HicAB cassette, a putative novel, RNA-targeting toxin–antitoxin system in archaea and bacteria. Bioinformatics. 2006;22:2581–2584. doi: 10.1093/bioinformatics/btl418. [DOI] [PubMed] [Google Scholar]
- 24.Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of type 2 toxin–antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct. 2009;4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 26.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 27.Vanderpool CK, Gottesman S. The novel transcription factor SgrR coordinates the response to glucose-phosphate stress. J. Bacteriol. 2007;189:2238–2248. doi: 10.1128/JB.01689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christensen SK, Maenhaut-Michel G, Mine N, Gottesman S, Gerdes K, Van Melderen L. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol. Microbiol. 2004;51:1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- 30.Brosius J, Holy A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc. Natl Acad. Sci. USA. 1984;81:6929–6933. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilbaux M, Mine N, Guerout AM, Mazel D, Van Melderen L. Functional interactions between coexisting toxin-antitoxin systems of the ccd family in Escherichia coli O157:H7. J. Bacteriol. 2007;189:2712–2719. doi: 10.1128/JB.01679-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daines DA, Jarisch J, Smith AL. Identification and characterization of a nontypeable Haemophilus influenzae putative toxin–antitoxin locus. BMC Microbiol. 2004;4:30. doi: 10.1186/1471-2180-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallez R, Geeraerts D, Sterckx Y, Mine N, Loris R, Van Melderen L. New toxins homologous to ParE belonging to three-component toxin–antitoxin systems in Escherichia coli O157:H7. Mol. Microbiol. 2010;76:719–732. doi: 10.1111/j.1365-2958.2010.07129.x. [DOI] [PubMed] [Google Scholar]
- 34.Singletary LA, Gibson JL, Tanner EJ, McKenzie GJ, Lee PL, Gonzalez C, Rosenberg SM. An SOS-regulated type 2 toxin–antitoxin system. J. Bacteriol. 2009;191:7456–7465. doi: 10.1128/JB.00963-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nieto C, Pellicer T, Balsa D, Christensen SK, Gerdes K, Espinosa M. The chromosomal relBE2 toxin-antitoxin locus of Streptococcus pneumoniae: characterization and use of a bioluminescence resonance energy transfer assay to detect toxin-antitoxin interaction. Mol. Microbiol. 2006;59:1280–1296. doi: 10.1111/j.1365-2958.2006.05027.x. [DOI] [PubMed] [Google Scholar]
- 36.Fiebig A, Castro Rojas CM, Siegal-Gaskins D, Crosson S. Interaction specificity, toxicity and regulation of a paralogous set of ParE/RelE-family toxin–antitoxin systems. Mol. Microbiol. 2010;77:236–251. doi: 10.1111/j.1365-2958.2010.07207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christensen-Dalsgaard M, Jorgensen MG, Gerdes K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 2010;75:333–348. doi: 10.1111/j.1365-2958.2009.06969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science. 2009;323:396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu M, Zhang Y, Inouye M, Woychik NA. Bacterial addiction module toxin Doc inhibits translation elongation through its association with the 30S ribosomal subunit. Proc. Natl Acad. Sci. USA. 2008;105:5885–5890. doi: 10.1073/pnas.0711949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Condon C. Shutdown decay of mRNA. Mol. Microbiol. 2006;61:573–583. doi: 10.1111/j.1365-2958.2006.05270.x. [DOI] [PubMed] [Google Scholar]
- 41.Gerdes K. Toxin–antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 2000;182:561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown BL, Grigoriu S, Kim Y, Arruda JM, Davenport A, Wood TK, Peti W, Page R. Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 2009;5:e1000706. doi: 10.1371/journal.ppat.1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de la Hoz AB, Ayora S, Sitkiewicz I, Fernandez S, Pankiewicz R, Alonso JC, Ceglowski P. Plasmid copy-number control and better-than-random segregation genes of pSM19035 share a common regulator. Proc. Natl Acad. Sci. USA. 2000;97:728–733. doi: 10.1073/pnas.97.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogata H, La Scola B, Audic S, Renesto P, Blanc G, Robert C, Fournier PE, Claverie JM, Raoult D. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2006;2:e76. doi: 10.1371/journal.pgen.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogata H, Renesto P, Audic S, Robert C, Blanc G, Fournier PE, Parinello H, Claverie JM, Raoult D. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 2005;3:e248. doi: 10.1371/journal.pbio.0030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogata H, Audic S, Renesto-Audiffren P, Fournier PE, Barbe V, Samson D, Roux V, Cossart P, Weissenbach J, Claverie JM, et al. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science. 2001;293:2093–2098. doi: 10.1126/science.1061471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.