Abstract
The type II poly(A)-binding protein PABP2/PABPN1 functions in general mRNA metabolism by promoting poly(A) tail formation in mammals and flies. It also participates in poly(A) tail shortening of specific mRNAs in flies, and snoRNA biogenesis in yeast. We have identified Caenorhabditis elegans pabp-2 as a genetic interaction partner of the let-7 miRNA, a widely conserved regulator of animal stem cell fates. Depletion of PABP-2 by RNAi suppresses loss of let-7 activity, and, in let-7 wild-type animals, leads to precocious differentiation of seam cells. This is not due to an effect on let-7 biogenesis and activity, which remain unaltered. Rather, PABP-2 levels are developmentally regulated in a let-7-dependent manner. Moreover, using RNAi PABP-2 can be depleted by >80% without significantly impairing larval viability, mRNA levels or global translation. Thus, it unexpectedly appears that the bulk of PABP-2 is dispensable for general mRNA metabolism in the larva and may instead have more restricted, developmental functions. This observation may be relevant to our understanding of why the phenotypes associated with human PABP2 mutation in oculopharyngeal muscular dystrophy (OPMD) seem to selectively affect only muscle cells.
INTRODUCTION
MicroRNAs (miRNAs) are small, non-coding RNAs that post-transcriptionally regulate gene expression in animals, plants and protozoa. Incorporated into a multi-subunit miRNA-induced silencing complex (miRISC), miRNAs serve as guide molecules to provide the specificity in target mRNA recognition by an antisense mechanism. Binding of miRISC ultimately prevents protein accumulation by target mRNA destabilization and/or translational repression, which may involve target mRNA deadenylation [reviewed by (1)].
The let-7 miRNA is phylogenetically conserved in bilaterian animals, with a remarkable 100% sequence identity of the mature miRNA in Caenorhabditis elegans and humans (2,3). let-7 was originally identified as a heterochronic gene in C. elegans (4). The genes of the heterochronic pathway (Figure 1A) control temporal patterning during post-embryonic development, i.e. they direct the developmental stage-specific execution of cell fates (5). Thus, loss of let-7 function causes a defect in the larval-to-adult (L/A) transition such that cells reiterate larval stage four (L4) cell fates in adult animals, ultimately leading to lethality by vulval bursting (4). For instance, the stem cell-like seam cells would normally exit the cell cycle and terminally differentiate at the L/A transition but continue to divide and fail to differentiate in let-7 mutant animals. In contrast to this retarded heterochronic phenotype, over-expression of let-7 or depletion of some of its targets such as lin-41, leads to the opposite, precocious phenotype, where seam cells differentiate prematurely at the L3-to-L4 molt (referred to as ‘L3 molt’ hereafter) (4,6). These functions of C. elegans let-7 in regulating temporal cell fates by controlling cell proliferation and differentiation are mirrored by mammalian let-7, which acts as a tumour suppressor and regulator of stem cells by repressing stem cell self-renewal and promoting differentiation (7).
Figure 1.
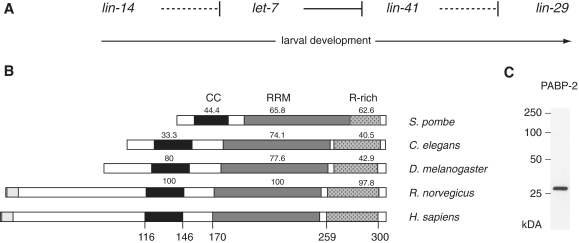
Conservation of eukaryotic PABP2. (A) Schematic depiction of the heterochronic pathway, which temporally regulates seam cell division and differentiation. For clarity, only those heterochronic genes investigated in this study are depicted. Solid lines represent direct repression of downstream genes, dashed lines indicate genetic interactions for which repression has not been shown to be direct (regulation of lin-29 by lin-41) or is assumed to be indirect (lin-14 versus let-7). (B) schematic representation of PABP2 protein in different species. The predicted coiled-coil (CC; black) region, RNA recognition motif (RRM; grey) and arginine-rich region (R-rich; dotted) are indicated. Human and rat proteins bear N-terminal extensions that contain poly-alanine tracts (light grey) that are expanded in disease. Numbers above domains indicate the degree of identity of the amino acid sequence to the corresponding human domains, numbers below the human sequence correspond to the amino acid positions. (B) Western blot using a polyclonal rat antibody reveals PABP-2 as a single band at ∼27 kDa.
We previously identified pabp-2, the C. elegans orthologue of the type II poly(A)-binding protein PABP2/PABPN1, in a reverse genetic screen for suppressors of let-7 loss-of-function lethality (8). Despite their shared name, type II or nuclear poly(A)-binding proteins are structurally and functionally unrelated to type I or cytoplasmic poly(A)-binding proteins, which have recently been reported to interact with miRISC (9–11). Mammalian PABP2 was initially identified as an enhancer of nuclear polyadenylation (12). In vitro, the poly(A) polymerase (PAP), the cleavage and polyadenylation specificity factor (CPSF) and PABP2 are both necessary and sufficient for faithful and efficient pre-mRNA polyadenylation (12,13). As poly(A) tail-length determines both the stability and the translation efficiency of an mRNA (14–16), PABP2 is thus likely to be of major importance to general mRNA metabolism.
CPSF and PABP2 cooperatively stimulate PAP in a process that involves CPSF binding to the polyadenylation signal AAUAAA, positioned ∼20 nt upstream of the cleavage site and PABP2 covering the growing poly(A) tail (13,17,18). Formation of a tight, spherical PABP2 particle is thought to fold back the growing poly(A) tail to maintain the contact between CPSF and PAP. Once the poly(A) tail has reached a critical length, no further PABP2 can be accommodated, and processivity of poly(A) tail synthesis is disrupted. Thus, PABP2 not only promotes polyadenylation, but also appears to act as a molecular ruler that defines the ultimate poly(A) tail length (17).
Consistent with the function assigned to PABP2 in vitro, depletion of PABP2 in cultured mouse myoblasts led to a shortening of mRNA poly(A) tails (19). In Drosophila, PABP2 was shown to be essential for viability, and a transgene bearing a point mutation that prevents PAP stimulation was unable to rescue the lethality of a null allele (20). In contrast, deletion of pabp2 in Schizosaccharomyces pombe was tolerated and, unexpectedly, caused hyperadenylation of bulk mRNA (21). Moreover, fission yeast Pab2 was found to participate in the processing of 3′-extended small nucleolar (sno)RNAs (22). Finally, despite a nuclear steady-state localization, PABP2 shuttles between nucleus and cytoplasm, consistent with additional cytoplasmic roles (23). Indeed, cytoplasmic PABP2 functions to shorten the poly(A) tails of oscar and cyclinB mRNAs in Droshophila embryos, establishing an essential developmental function (20). Taken together, although strong evidence supports an important role of PABP2 in general mRNA metabolism, these functions might not be generally conserved across eukaryotes, and PABP2 might have been recruited for additional or alternative functions in different organisms.
Little is known about C. elegans PABP-2. Like its mammalian counterpart, PABP-2 contains a putative coiled-coil region, a single RNA recognition motif (RRM), and a C-terminal arginine-rich domain (Figure 1B). However, like its Drosophila and S. pombe orthologues, C. elegans PABP-2 lacks a region of homology to the N-terminus of mammalian PABP2. In human PABP2, this region includes a polyalanine tract, the expansion of which causes oculopharyngeal muscular dystrophy (OPMD), a late-onset, progressive disease (24).
Here, we demonstrate that in C. elegans, depletion of PABP-2 not only rescues loss of let-7 function, but also causes precocious seam cell differentiation. Surprisingly, efficient depletion of PABP-2 leaves global translation and mRNA levels largely unaffected, while causing accumulation of the LIN-29 transcription factor, the most downstream effector gene known in the heterochronic pathway. Moreover, PABP-2 concentration decreases during animal development in a let-7-dependent manner, although PABP-2 is unlikely to be a direct let-7 target. Our results support the idea that the bulk of PABP-2 in C. elegans larvae is not required for general mRNA metabolism but may play more specialized roles in development. Given the tissue-specificity of phenotypes seen upon PAPB2 mutation in human OMPD, such non-canonical functions of PABP2 may deserve more detailed study also in other animals, including humans.
MATERIALS AND METHODS
Caenorhabditis elegans strains and handling
Strains were maintained and cultured as described (25). Wild-type N2, MT7626: let-7(n2853) and VT516 lin-29(n546)/mnC1 dpy-10(e128) unc-52(e444)II strains were provided by CGC. him-5; [ajm-1::gfp/MH27::GFP; rol-6] (26) was used to visualize seam cells. MT19756: nIs408[lin-29b::mCherry] contains an integrated array (nEx1681) formed by the injection of PCR product (50 ng/µl) containing the lin-29B locus (LG II sequence 11 917 298–11 927 996), which has mCherry inserted in place of the stop codon, and a plasmid carrying ttx-3::gfp (40 ng/µl). The lin-29b::mCherry reporter rescues the Pvl, alae, molting and seam cell division defects of the putative null allele lin-29(n836) (David T. Harris and H. Robert Horvitz, unpublished data). maIs105[col-19::gfp]; let-7(n2853) was provided by Frank Slack and Ryusuke Niwa. HW761: lin-29(n546)/mnC1 dpy-10(e128) unc-52(e444)II; him-5; [ajm-1::gfp/MH27::GFP; rol-6] was used to visualize seam cells in a lin-29(lf) background and was established by crossing VT516 with him-5; [ajm-1::gfp/MH27::GFP; rol-6] males. HW758 nIs408[lin-29b/mCherry] I; him-5; [ajm-1::gfp/MH27::GFP; rol-6], used to visualize lin-29b expression and seam cell fusion in the same animals, was established by crossing MT19756 with him-5; [ajm-1::gfp/MH27::GFP; rol-6] males.
RNAi by feeding synchronized L1 larvae on RNAi plates at 25°C was performed as described (27). Unless indicated otherwise, animals for molecular studies were harvested at the L4 stage, when let-7 levels are high (4). RNAi feeding constructs from published RNAi libraries (28,29) were used against daf-12, hbl-1, lin-41, lin-14, pab-1, eif-3.e and pabp-2.
To assess brood sizes, wild-type animals were grown at 25°C on L4440 (mock(RNAi)) control or pabp-2(RNAi) feeding plates. Gravid adults were singled and transferred onto OP50 plates for further growth at 25°C. The number of progeny was counted 24 and 72 h after transfer to OP50 plates. Since egg laying in control animals was essentially complete by 24 h, with animals producing fewer than seven progeny within the following 48 h, the analysis was restricted to the first 24 h.
lin-29 epistasis
lin-29 epistasis was tested using HW761 lin-29(n546)/mnC1 dpy-10(e128) unc-52(e444)II; him-5; [ajm-1::gfp/MH27::GFP; rol-6) animals. mnC1 homozygotes arrest in late larval development and eventually die. Hence, young adults segregate into 1/3 lin-29(n546) homozygotes and 2/3 lin-29(n546)/mnC1 heterozygotes, which was confirmed by the frequency of lin-29(lf) phenotypes including protruding vulva and sterility. Synchronized HW761 L1 larvae were grown on RNAi feeding plates at 25°C until young adult stage. Failure of terminal seam cell differentiation was assessed by fluorescence microscopy. The reverse epistasis experiment, the suppression of pabp-2(RNAi) phenotypes in lin-29(lf) animals could not be tested. At the L3 molt, when we examined precocious cell fusion, lin-29(n546) homozygous, lin-29(n546)/mnC1 heterozygous and mnC1 homozygous animals were all indistinguishable, so that only one-fourth of the animals would have the desired lin-29(n546) genotype. Thus, the maximum possible reduction of precocious seam cell fusion falls within the variability of our results.
Polyribosome preparation and analysis
Polyribosome preparations were performed by sucrose density gradient ultracentrifugation as described (30). For each gradient fraction, 400 ng of RNA was reverse transcribed using the ImProm-II Reverse Transcription System (Promega) according to the manufacturer's recommendations. Random hexamer primers were used to avoid a bias against short poly(A) tails, which may occur as a consequence of miRNA action (1) or PABP-2 depletion.
Quantitative PCR reactions were performed in technical duplicate using the ABsoluteTM QPCR SYBRs Green ROX Mix (Thermo Fisher Scientific) on an ABI Prism 7000 real-time thermal cycler. Relative transcript levels were calculated using the 2[−ΔΔC(T)] method (31). (For primer pairs see Supplementary Data.) The relative transcript levels were corrected for the total amount of RNA extracted from each fraction and mapped as percentage of the sum of all fractions. Reverse transcription of total RNA was performed on aliquots of the same samples that were used for polysome profiling using unequal duplicates of 400 and 800 ng of RNA input. Repetition of one experiment using oligo(dT)-priming of reverse transcription instead of random hexamers did not change the results. The fold-change in transcript levels between pabp-2 and mock(RNAi) derived from total RNA or from the sum of all fractions also yielded comparable results, confirming the robustness of the assay.
Northern blotting
RNA samples were separated on TBE Urea PAGE gels and transferred to Hybond Nx membrane (GE Healthcare). Chemical cross-linking with EDC was performed according to the method described in (32). Antisense DNA oligonucleotides were 5′-labelled using T4 polynucleotide kinase (PNK) and [y-32P]ATP. (See Supplementary Data for oligonucleotide sequences). Radioactive signals were detected using a Storage Phosphor Screen and a Typhoon 9400 scanner and quantified with Imagequant TL software (all GE Healthcare).
Antibodies and western blotting
SDS–PAGE and western blotting was performed according to standard protocols (33). To obtain an antibody against PABP-2, recombinant GST-PABP-2 was expressed in Escherichia coli, purified on glutathione sepharose 4B (GE Healthcare), released by thrombin cleavage and gel extracted. Polyclonal antibodies against PABP-2 were raised in rats by Charles River Laboratories (Kisslegg, Germany) and unprocessed immune serum was used 1:500 to detect PABP-2 as a single band. Actin was detected by monoclonal mouse anti-actin MAB1501 (Millipore, 1:1000 dilution). Horseradish peroxidase-conjugated anti-mouse (NA931V, GE Healthcare) or anti-rat (112-035-003, Jackson Labs) secondary antibodies were used for signal detection by ECL (GE healthcare); bands were quantified in ImageJ (34).
Nomarski and fluorescent imaging
Microscopy images were acquired using an Axioplan microscope (axio imager Z1, Zeiss) equipped with a CCD camera (AxioCam Mrm, Zeiss). Adobe Photoshop software was used to crop images or to adjust levels, leaving gamma unaltered.
RESULTS
RNAi-mediated knockdown of pabp-2 suppresses let-7(n2853) lethality
The temperature sensitive let-7(n2853) allele harbours a point mutation in the seed region of the mature let-7 miRNA that impairs target mRNA silencing (4,35). As a consequence, mutant animals die by bursting through the vulva at the L/A transition when grown at 20°C or above (Figure 2A and C). To identify let-7 interaction partners, we previously screened an RNAi-by-feeding library of approximately 2400 genes on C. elegans chromosome I for suppression of let-7-associated lethality (8). In the course of this screen, we identified pabp-2, encoding the type II poly(A)-binding protein PABP-2, as a potent suppressor. Almost 60% of synchronized let-7(n2853) L1 larvae reach the adult stage when grown on bacteria expressing a double-stranded RNA against the genomic region of pabp-2 at 25°C (Figure 2B and C). In let-7 wild-type animals, RNAi-mediated knockdown of pabp-2 led to a >10-fold reduction in brood size and fully penetrant early larval arrest of viable progeny (Figure 2D). Double mutant let-7(n2853); pabp-2(RNAi) animals similarly often bore dead embryos and rare viable progeny underwent early larval arrest (Figure 2B and E).
Figure 2.
RNAi-mediated knockdown of pabp-2 suppresses let-7(n2853) lethality. (A) let-7(n2853) animals die at the L/A transition by bursting through the vulva (white arrowhead) when grown at 25°C, (B) let-7(n2853); pabp-2(RNAi) animals survive into adulthood as indicated by the presence of embryos (asterisks), although most embryos die in utero, (C) Suppression of let-7(n2853)-mediated vulval bursting upon pabp-2(RNAi) or daf-12(RNAi), which served as a positive control. In this and subsequent figures, ‘mock(RNAi)’ denotes control animals that were fed bacteria carrying the insertless L4440 RNAi vector. n ≥ 4 independent trials with ≥70 animals each, (D) Number of viable progeny during the first 24 h of egg laying at 25°C; n = 10 animals each. (E) Arrested F1 progeny of a let-7(n2853); pabp-2(RNAi) mother. The number of cells in the gonad (black arrowhead) indicates that progeny arrest at late L1/early L2 stage. (F) At late L4, PABP-2 protein levels were reduced by >80% in pabp-2(RNAi) relative to mock(RNAi) animals. Non-adjacent lanes of the same blot are shown. Error bars denote SEM. Scale bars are 20 μm.
RNAi-mediated depletion of PABP-2 was mirrored in homozygous pabp-2(ok1121) mutant progeny, deleted for pabp-2, which similarly died in utero of their heterozygous (balanced) mothers or underwent early larval arrest (data not shown). Further reflecting specificity, three additional RNAi constructs against exons 2 and 3 as well as the entire coding region of pabp-2, also suppressed let-7 lethality, albeit to variable extents (data not shown).
To verify PABP-2 depletion directly and to examine the extent of knockdown, we generated a rat polyclonal antibody against PABP-2 recombinantly expressed in E. coli. When tested on whole animal lysates, this antibody recognized a single band of an apparent size of ∼27 kDa (Figure 1C), slightly above the predicted size but consistent with the migration pattern observed for the recombinant protein (data not shown). When examined in mid-L4 stage animals, we found that pabp-2(RNAi) reduced PABP-2 protein levels by >80% relative to animals exposed to mock RNAi (Figure 2F).
Depletion of PABP-2 causes precocious seam cell fusion
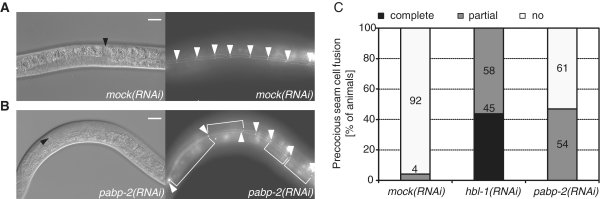
To ascertain that the suppression of the let-7(n2853) lethality reflected a heterochronic function of PABP-2, we examined seam cell differentiation. Terminal differentiation of seam cells at the L/A transition in wild-type animals involves their fusion into a syncytium. In let-7(n2853) animals, seam cells fail to terminally differentiate at the L/A transition whereas overexpression of let-7, or depletion of some of its targets such as lin-41 or hbl-1, causes seam cells to fuse precociously, at the L3 molt (4,6,42,43) (Figure 3C). Whereas only 4% of wild-type animals exposed to mock RNAi displayed seam cell fusion at this stage, this number was increased to 47% of animals on pabp-2 RNAi (Figure 3). Thus pabp-2(RNAi) causes heterochronic defects opposite to let-7(n2853).
Figure 3.
Depletion of PABP-2 causes precocious seam cell fusion. Synchronized him-5; [ajm-1::gfp/MH27::GFP; rol-6] L1-stage larvae were exposed to RNAi as indicated and examined for precocious seam cell fusion upon reaching L4 stage. (A and B) Photomicrographs of animals grown on mock and pabp-2(RNAi), respectively. Arrowheads in the Nomarski micrographs (left panels) point to the distal tips of the gonads. Arrowheads in the GFP micrographs (right panels) indicate the cell–cell junctions between seam cells in absence of cell fusion visualized by the expression of AJM-1::GFP. (C) The penetrance of precocious seam cell fusion from three independent experiments. The numbers on the bars indicate the absolute number of animals assigned to the respective category. Scale bars are 20 μm.
pabp-2(RNAi) promotes LIN-29 activity
To situate pabp-2 more clearly in the heterochronic pathway, we examined its relation to lin-29, the most downstream heterochronic gene known to regulate seam cell differentiation (5). In wild-type animals, this zinc finger transcription factor is upregulated during the L4 stage to drive transcription of direct targets such as the adult cuticular collagen col-19 (36,37).
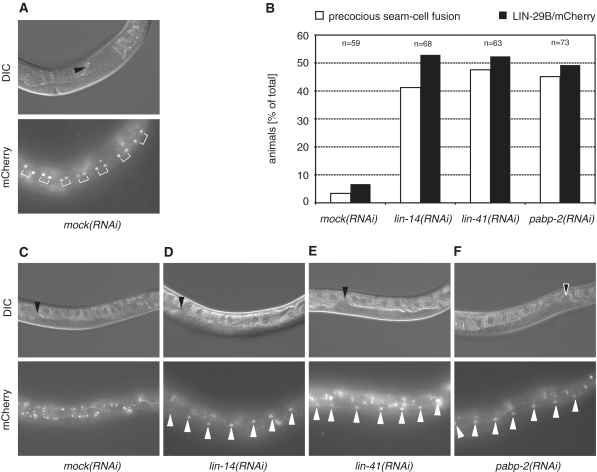
Using a functional lin-29b::mCherry fusion gene (see ‘Materials and Methods’ section), LIN-29 first becomes visible in wild-type seam cells in L4 stage animals, prior to seam cell fusion (Figure 4A). As reported for endogenous LIN-29 (36), accumulation of LIN-29/mCherry occurs precociously at the L3 molt upon RNAi-mediated depletion of the early acting heterochronic gene lin-14 (53% of animals) or the late acting lin-41 (52% of animals) (Figure 4B–E). RNAi against pabp-2 caused a similar precocious LIN-29B/mCherry accumulation (50%; Figure 4B and F). Knockdown of all three genes also caused comparable levels of precocious seam cell fusion (Figure 4B).
Figure 4.
LIN-29/mCherry accumulates precociously in pabp-2(RNAi) animals. Strain HW761 expressing lin-29b/mCherry was exposed to RNAi by feeding as synchronized L1-stage larvae. (A and C–F) Photomicrographs of animals grown on the indicated RNAi. Upper panels show Nomarski micrographs and lower panels show fluorescence micrographs. Arrowheads in the Nomarski micrographs point to the distal tip of the gonad, white arrowheads in the fluorescence micrographs point to seam cell nuclei. (A) Mock-treated L4 animal accumulating LIN-29B at mid L4. The brackets indicate the nuclei of the daughter cells of the final seam cell division. (B) Percentage of animals exhibiting precocious seam cell fusion (white bars) and LIN-29B accumulation in seam cell nuclei at late L3. The number of animals examined is indicated above the bars. (C–F) Late L3 stage animals exposed to (C) mock RNAi, (D) lin-14(RNAi), (E) lin-41(RNAi) and (F) pabp-2(RNAi).
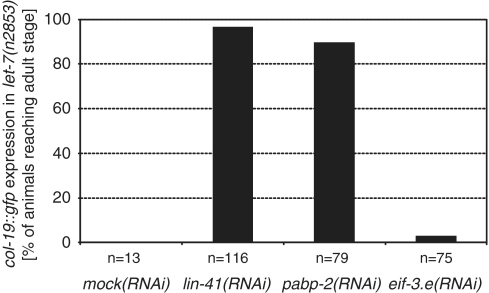
To examine whether altered LIN-29 accumulation was functionally relevant and able to explain the rescue of let-7 mutant animals, we examined activation of the LIN-29 target col-19 (37) using a col-19::gfp reporter (38). This cuticular collagen is expressed in adults but not in larvae (39), and fails to be activated in let-7(n2853) mutant adults, where LIN-29 levels remain low (4). Consistent with restored function of LIN-29, let-7(n2853); pabp-2(RNAi) animals displayed highly penetrant (90%) expression of col-19::gfp, similar to what was observed with control let-7(n2853); lin-41(RNAi) animals (Figure 5). This effect is specific and not an indirect consequence of restored animal viability, since escaping let-7(n2853) adults on mock(RNAi) failed to activate col-19 expression, as did let-7(n2853) animals exposed to eif-3.e(RNAi), a potent suppressor of let-7(n2853) vulval bursting [Figure 5; M. Rausch and M. Ecsedi, unpublished data; (8)].
Figure 5.
pabp-2(RNAi) restores expression of the LIN-29 target col-19 in let-7(n2853) animals. Expression of col-19::gfp in let-7(n2853) adults. Note that only animals that bypassed let-7(n2853) lethality at the L/A transition could be scored. (Survival of let-7(n2853) animals in this assay: mock(RNAi): 5%; lin-41(RNAi): 94%; pabp-2(RNAi): 55%; eif-3.e(RNAi): 76%). The total number of animals examined in three biological replicates is indicated.
Finally, we wished to examine how lin-29 and pabp-2 interact genetically. Since technical reasons prevented us from examining whether loss of lin-29 suppressed the precocious seam cell phenotype of pabp-2(RNAi) (see ‘Materials and Methods’ section), we investigated whether the retarded seam cell fusion phenotype seen in lin-29(n546) null mutant animals (37) was suppressed by pabp-2(RNAi). As expected (see ‘Material and Methods’ section), approximately one-third (31%) of adult animals derived from balanced lin-29(n546)/mnC1 heterozygous hermaphrodites displayed unfused seam cells on mock RNAi. This number remained unchanged when the two control genes lin-14 and lin-41 were depleted by RNAi, consistent with their function upstream of lin-29 in the heterochronic pathway (5) (Figure 1A). Similarly, pabp-2(RNAi) was unable to suppress the seam cell fusion defect in lin-29(n546) mutant animals. Thus, taken together, the expression and genetic interaction data support a model where papb-2 functions upstream of, and at least in part through, lin-29.
PABP-2 protein is overexpressed in late L4-stage let-7(n2853) animals
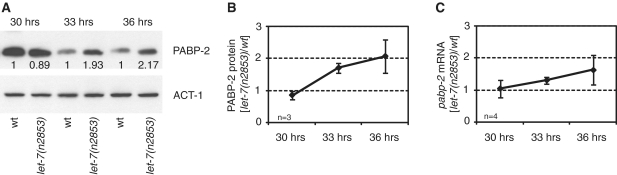
The genetic interaction between pabp-2 and let-7 might be explained by repression of pabp-2 expression by let-7. However, sequence analysis using RNAhybrid (40) revealed that the entire pabp-2 locus was devoid of sequences enabling let-7:pabp-2 mRNA duplex formation. Still, to examine whether let-7 might indirectly regulate PABP-2 levels, we assessed its levels by western blotting. In contrast to the apparent house-keeping function of PABP2 in mRNA metabolism, but consistent with a function in temporal patterning, we found a substantial decline of PAPB-2 protein levels in wild-type animals during the L4 stage such that we observed an ∼5-fold signal decrease between 30 and 33 h after initiating growth of synchronized L1 larvae at 25°C (Figure 6A). Moreover, PABP-2 levels were comparable in wild-type and let-7(n2853) mutant animals at 30 h, but ∼2-fold elevated in let-7(n2853) at 33 and 36 h (Figure 6A and B). A similar trend was observed when examining pabp-2 mRNA levels (Figure 6C), suggesting that loss of let-7 activity affects PABP-2 levels through increased transcription or stability of the papb-2 mRNA, Thus, PAPB-2 levels are not only developmentally regulated, but this regulation is also mediated, in part, by let-7.
Figure 6.
pabp-2 is over-expressed in let-7(n2853) starting late L4. (A–C) Expression of PABP-2 protein and pabp-2 mRNA in wild-type and let-7(n2853) animals at 30, 33 and 36 h of postembryonic development (corresponding to mid-L4 to late L4/young adult stage). Numbers below the PABP-2 bands in (A) indicate PABP-2 signal by western blotting normalized to actin signal, with wild-type signal set to one for each time point. (B) Protein quantification normalized to PABP-2 signal in wild-type animals for each time-point; shown is the average of three independent trials. Note that in wild-type animals, PABP-2 signal declines ∼5-fold over the time course. (C) mRNA quantification by RT–qPCR.
let-7 biogenesis occurs normally in PABP-2 depleted animals
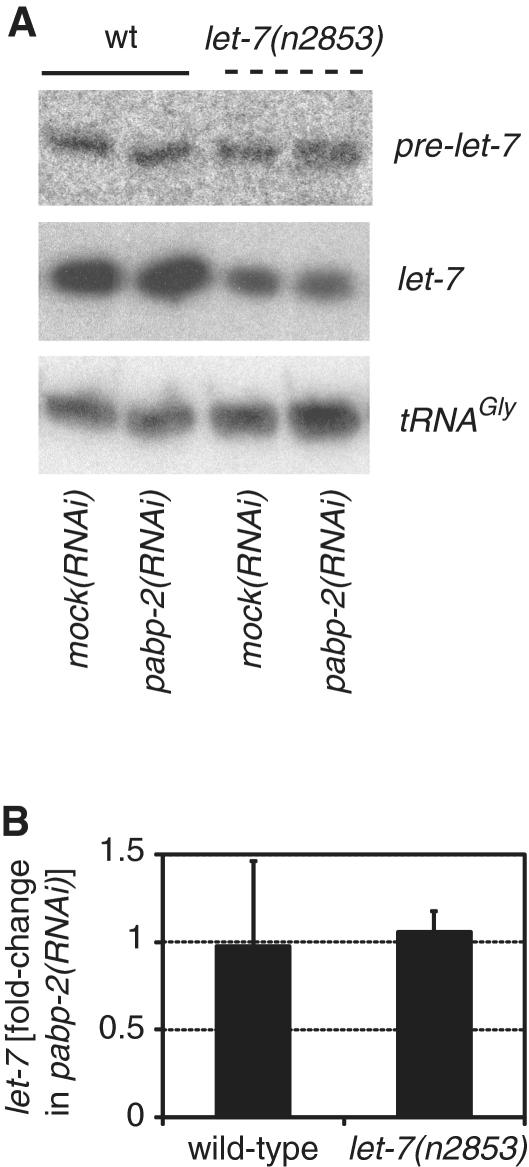
MicroRNAs of the let-7 family are components of several double negative regulatory loops, where factors that are repressed by the miRNAs themselves repress let-7 biogenesis [reviewed in (7)]. The fact that depletion of pabp-2 mirrored let-7 overexpression together with the known RNA-binding activity of PABP2 proteins and their role in the biogenesis of different RNA species thus prompted us to ask whether PABP-2 might also normally inhibit let-7 function, perhaps by interfering with its biogenesis. Hence, we used northern blotting to study whether pabp-2(RNAi) enhanced let-7 biogenesis. Consistent with a previous report (4), levels of mature let-7 were ∼3-fold reduced in let-7(n2853) animals relative to wild-type animals, whereas pre-let-7 levels were largely unaffected or only modestly increased (Figure 7A). However, depletion of PABP-2 affected neither pre-let-7 nor mature let-7 levels regardless of whether wild-type or let-7(n2853) mutant animals were investigated (Figure 7). Thus, PABP2 does not play a significant role in let-7 biogenesis.
Figure 7.
let-7 biogenesis occurs normally in PABP-2-depleted animals. (A) Northern blots using total RNA from synchronized late L4 animals grown on mock or pabp-2(RNAi). Oligonucleotides specific for pre-let-7, let-7 or tRNAGly(TCC) were used. The experiment was performed on biological triplicates, a representative example is shown. (B) Relative fold-change of mature let-7 levels in pabp-2(RNAi) versus mock(RNAi) in wild-type and let-7(n2853) animals. n = 3, error bars represent SEM.
Depletion of PABP-2 does not affect let-7 target mRNA stability
Caenorhabditis elegans miRNAs regulate their cognate targets by target mRNA destabilization (30,41) and/or translational repression at the initiation step (30). To test whether depletion of PABP-2 affected let-7 target gene silencing, we first determined the mRNA levels of the let-7 target genes daf-12, lin-41 and hbl-1, all three of which function in seam cell temporal patterning (4,6,27,42,43). We further included lin-14 and lin-28, two targets of the lin-4 miRNA (44,45), to examine potentially more general roles of pabp-2 in miRNA function, as well as act-1 and tbb-1 as non-miRNA regulated reference genes.
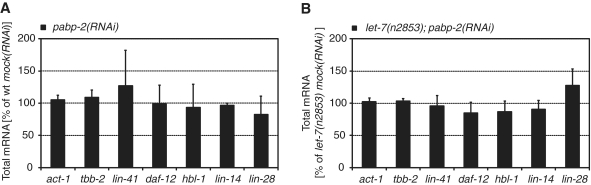
We extracted total RNA of L4-stage wild-type and let-7(n2853) animals grown on pabp-2(RNAi) or mock(RNAi). As the expression levels of let-7 target genes are typically low at this stage, we used quantitative reverse transcription–PCR (qRT–PCR) to detect transcripts. The expression levels of act-1 (actin) and tbb-2 (β-tubulin) were comparable in pabp-2(RNAi) and mock(RNAi), suggesting that the depletion of PABP-2 did not substantially affect overall mRNA levels. Moreover, the levels of the miRNA target genes lin-41, daf-12, hbl-1, lin-14 and lin-28 did not show any statistically significant changes, regardless of whether pabp-2(RNAi) was performed on wild-type or let-7(n2853) mutant animals (Figure 8). We conclude that PABP-2 depletion does not affect let-7-mediated mRNA degradation.
Figure 8.
Depletion of PABP-2 does not affect let-7 target mRNA stability. (A and B) Analysis of total mRNA levels in wild-type or let-7(n2853) mutant animals by qRT-PCR. mRNA levels in animals exposed to pabp-2(RNAi) are given as percentage of the mRNA levels observed in mock(RNAi) animals.
Depletion of PABP-2 has only minor effects on translation efficiency
Since miRNA-mediated mRNA degradation and translational repression may be independent mechanisms of target mRNA silencing, we next performed polysome profiling on animals exposed to pabp-2(RNAi) to establish the translation initiation efficiency of miRNA targets. To this end, we used sucrose-density gradient ultracentrifugation to fractionate whole animal lysates from L4-stage wild-type and let-7(n2853) animals grown on either pabp-2(RNAi) or mock(RNAi) feeding plates. We then performed qRT–PCR to analyse the distribution of transcripts across the fractions.
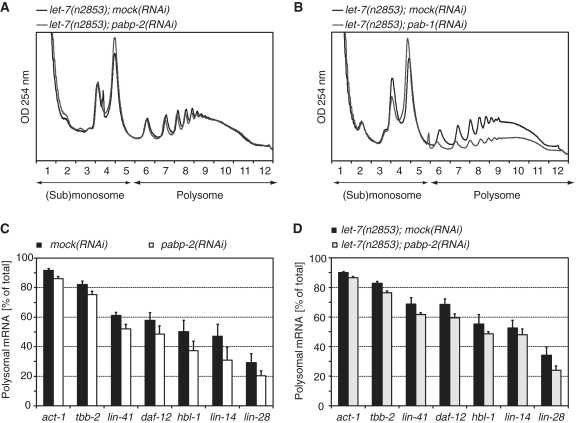
Surprisingly, given the importance of poly(A) length in controlling mRNA translation and the suspected role of PABP-2 in determining polyadenylation globally, the UV-absorbance gradient profiles of lysates of pabp-2(RNAi) and mock(RNAi) were essentially the same (Figure 9A). A small increase was only seen in the 80S peak, which represents mRNAs bound by one ribosome and, possibly, also free ribosomes, but the relative amount of RNA recovered from polysomal fractions did not change significantly (Supplementary Figure S1A). In contrast, when we knocked down pab-1, one of two C. elegans orthologues of the type I poly(A)-binding protein, a considerable depletion in polyribosomes resulted (Figure 9B). Thus, unlike pab-1, knockdown of pabp-2 does not appear to have a general effect on translation.
Figure 9.
Depletion of PABP-2 has only minor effects on translation efficiency. (A) Typical polysome profiles of let-7(n2853) animals grown on mock(RNAi) or pabp-2(RNAi). Fractions 1–5 comprise the (sub)-monosome, fractions 6–12 the polysomes. (B) RNAi against pab-1, the C. elegans orthologue of the human or yeast cytoplasmic (type I) poly(A)-binding protein restrains translation. (C and D) Polysomal fractions of mRNAs are plotted as percentage of the total (=monosomal + polysomal fractions) in late L4 wild-type (C) and let-7(n2853) (D) animals grown on either mock(RNAi) or pabp-2(RNAi). Numbers are the averages of ≥3 independent experiments. Error bars represent SEM.
When we specifically examined the miRNA target genes lin-41, daf-12, hbl-1, lin-14 and lin-28 from the polysomal fractions in wild-type and let-7(n2853) animals, we found them to be consistently, though modestly, depleted from polysomes upon PABP-2 knockdown. However, the control genes act-1 and tbb-2 were depleted as well, albeit to a lower extent (Figure 9C and D). Although we cannot exclude that this moderate inhibition of translation in pabp-2(RNAi) animals may account for the rescue of let-7(n2853) lethality, it thus appears that PABP-2 has no major and specific function in miRNA-mediated translational repression.
DISCUSSION
PABP2 functions as a general mRNA metabolic factor in mammals, but also plays specific developmental roles in fly embryogenesis. We have revealed a heterochronic function of C. elegans PAPB-2 by demonstrating its genetic interaction with the heterochronic let-7 miRNA, its temporally regulated expression, its responsiveness to let-7 levels and its role in the prevention of precocious seam cell fusion. The genetic interaction between let-7 and pabp-2 and the fact that pabp-2 lacks obvious miRNA binding sites suggested that pabp-2 is either a negative regulator of let-7 biogenesis or let-7 activity; or that pabp-2 antagonizes the developmental role of let-7 in seam cell and vulva development without a direct molecular interaction. The first two scenarios appeared particularly appealing considering that PAPB-2 is an RNA binding protein involved in polyadenylation, because the let-7 primary miRNA is polyadenylated (46), and because mRNA deadenylation is considered to be one of the mechanisms through which miRNAs silence their target mRNAs (1).
However, the data we present here argue against a specific function of PAPB-2 in either process, as let-7 levels were unaffected by PAPB-2 depletion, as were the levels of let-7 target mRNAs. In contrast, we observed a mild depletion of let-7 target mRNAs from polysomes upon pabp-2(RNAi), but this effect was also observed for control genes and thus non-specific. Formally, we cannot rule out that such a general translational repression rescues let-7(n2853) lethality by sufficiently reducing translation of the key let-7 targets daf-12 and lin-41. However, we consider this unlikely because the extent of translational repression observed across four independent experiments was variable, whereas the suppression of let-7(n2853) lethality was not, i.e. there was little correlation between these two read-outs. Therefore, we consider it more likely that PABP-2 functions downstream of, or in parallel to, let-7 in the heterochronic pathway. Given that PABP-2 accumulates to inappropriate levels in the let-7(n2853) mutant, we favour the former possibility. An attractive possibility that remains to be explored is that PAPB-2 functions together with LIN-41.
Restoration of expression of the LIN-29 target gene col-19 in let-7(n2853); pabp-2(RNAi) double mutant animals, precocious LIN-29/mCherry accumulation in pabp-2(RNAi) single mutant animals and failure to suppress the retarded seam cell phenotype of the lin-29(n546) mutation further suggests that papb-2 acts upstream of lin-29. With regard to LIN-29/mCherry accumulation we note with interest that we failed to observe good correlation of precocious LIN-29/mCherry accumulation and precocious seam cell fusion at the L3 molt, i.e. we observed fused seam cells without detectable LIN-29/mCherry accumulation, as well as unfused seam cells displaying strong LIN-29/mCherry signal (data not shown). In contrast, in wild-type seam cells, fusion during the L4 stage was preceded by LIN-29/mCherry accumulation. Although we cannot rule out that our reporter might not fully capture all the details of endogenous LIN-29 accumulation, it will be interesting to examine whether LIN-29 activity is subject to regulation, e.g. through post-translational modification or the requirement of co-factors that are themselves subject to regulation. In accord with the notion of additional layers of regulation, precocious accumulation of LIN-29 was previously found to be insufficient to drive seam cell differentiation in early lin-41(RNAi) larvae (6).
In vitro, mammalian PABP2 is required for pre-mRNA polyadenylation (12), supporting an important function of PAPB2 in general mRNA metabolism in vivo. Consistent with this notion, PABP2 is essential for embryonic viability in flies (20) and C. elegans (this study). Surprisingly however, we could deplete PABP-2 by >80% from C. elegans larvae without affecting their viability or mRNA stability, and with little if any effect on global translation, indicating that, in this situation, the bulk of PABP-2 is dispensable for general mRNA metabolism. Although unexpected in view of the above findings, we note that this mirrors, in an animal, the situation in yeast where pab2 can be deleted in S. pombe and does not exist in Saccharomyces cerevisiae. Moreover, siRNA-mediated depletion of mouse PABP2 from murine primary myoblasts resulted in a decrease of particularly long poly(A) tracts (∼300 nt), but seemed to have little effect on shorter poly(A) tracts (∼100 nt) (19). Finally, a polyalanine tract expansion in the N-terminus of human PAPB2 leads to OPMD, an adult-onset, progressive disease characterized by selective phenotypes restricted to a subset of muscle cells. Although redundant activities, residual PABP2 function, or, in the case of OPMD, toxicity of the accumulating mutant PABP2 may explain some of these phenomena, we would like to propose that functions of PABP2 that affect only selected mRNAs (20) may deserve equal consideration to general mRNA metabolic roles in future studies on PABP2 function. Clearly, the fact that C. elegans PABP-2 levels are developmentally regulated, in a let-7-dependent manner, supports functions beyond mere housekeeping activity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Novartis Research Foundation through the Friedrich Miescher Institute; the Swiss National Science Foundation (grant number 3100A0-114001); and the European Research Council (ERC Starting Independent Investigator Grant number 241985—‘miRTurn’). Howard Hughes Medical Institute to the laboratory of Dr. H. Robert Horvitz (to D.H.). Funding for open access charge: ERC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
The authors thank Dr. David Baillie, Dr. Ryusuke Niwa, Dr. Frank Slack and the Caenorhabditis Genetics Center (CGC) for strains used in this work, Magdalene Rausch and Matyas Ecsedi for sharing unpublished data and Dr. Rafal Ciosk and Dr. Witold Filipowicz for critical reading of this manuscript.
REFERENCES
- 1.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 2.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 3.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 5.Rougvie AE. Intrinsic and extrinsic regulators of developmental timing: from miRNAs to nutritional cues. Development. 2005;132:3787–3798. doi: 10.1242/dev.01972. [DOI] [PubMed] [Google Scholar]
- 6.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 7.Büssing I, Slack FJ, Großhans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Ding XC, Slack FJ, Großhans H. The let-7 microRNA interfaces extensively with the translation machinery to regulate cell differentiation. Cell Cycle. 2008;7:3083–3090. doi: 10.4161/cc.7.19.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zekri L, Huntzinger E, Heimstädt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell. Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walters RW, Bradrick SS, Gromeier M. Poly(A)-binding protein modulates mRNA susceptibility to cap-dependent miRNA-mediated repression. RNA. 2010;16:239–250. doi: 10.1261/rna.1795410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wahle E. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell. 1991;66:759–768. doi: 10.1016/0092-8674(91)90119-j. [DOI] [PubMed] [Google Scholar]
- 13.Bienroth S, Keller W, Wahle E. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 1993;12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beilharz TH, Preiss T. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA. 2007;13:982–997. doi: 10.1261/rna.569407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2009;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn U, Gundel M, Knoth A, Kerwitz Y, Rudel S, Wahle E. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 2009;284:22803–22814. doi: 10.1074/jbc.M109.018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wahle E. Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem. 1995;270:2800–2808. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- 19.Apponi LH, Leung SW, Williams KR, Valentini SR, Corbett AH, Pavlath GK. Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum. Mol. Genet. 2010;19:1058–1065. doi: 10.1093/hmg/ddp569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benoit B, Mitou G, Chartier A, Temme C, Zaessinger S, Wahle E, Busseau I, Simonelig M. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila. Dev. Cell. 2005;9:511–522. doi: 10.1016/j.devcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Perreault A, Lemieux C, Bachand F. Regulation of the nuclear poly(A)-binding protein by arginine methylation in fission yeast. J. Biol. Chem. 2007;282:7552–7562. doi: 10.1074/jbc.M610512200. [DOI] [PubMed] [Google Scholar]
- 22.Lemay JF, D'Amours A, Lemieux C, Lackner DH, St-Sauveur VG, Bahler J, Bachand F. The nuclear poly(A)-binding protein interacts with the exosome to promote synthesis of noncoding small nucleolar RNAs. Mol. Cell. 2010;37:34–45. doi: 10.1016/j.molcel.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 23.Lemay JF, Lemieux C, St-Andre O, Bachand F. Crossing the borders: Poly(A)-binding proteins working on both sides of the fence. RNA Biol. 2010;7:291–295. doi: 10.4161/rna.7.3.11649. [DOI] [PubMed] [Google Scholar]
- 24.Brais B, Bouchard JP, Xie YG, Rochefort DL, Chretien N, Tome FM, Lafreniere RG, Rommens JM, Uyama E, Nohira O, et al. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat. Genet. 1998;18:164–167. doi: 10.1038/ng0298-164. [DOI] [PubMed] [Google Scholar]
- 25.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohler WA, Simske JS, Williams-Masson EM, Hardin JD, White JG. Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr. Biol. 1998;8:1087–1090. doi: 10.1016/s0960-9822(98)70447-6. [DOI] [PubMed] [Google Scholar]
- 27.Großhans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev. Cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 28.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 29.Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding XC, Großhans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 2009;28:213–222. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 33.Schagger H. Tricine-SDS-PAGE. Nat. Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 34.Abramoff M, Magelhaes P, Ram S. Image Processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 35.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bettinger JC, Lee K, Rougvie AE. Stage-specific accumulation of the terminal differentiation factor LIN-29 during Caenorhabditis elegans development. Development. 1996;122:2517–2527. doi: 10.1242/dev.122.8.2517. [DOI] [PubMed] [Google Scholar]
- 37.Rougvie AE, Ambros V. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development. 1995;121:2491–2500. doi: 10.1242/dev.121.8.2491. [DOI] [PubMed] [Google Scholar]
- 38.Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z, Kirch S, Ambros V. The Caenorhabditis elegans heterochronic gene pathway controls stage-specific transcription of collagen genes. Development. 1995;121:2471–2478. doi: 10.1242/dev.121.8.2471. [DOI] [PubMed] [Google Scholar]
- 40.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 42.Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 43.Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 44.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 45.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 46.Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.