Abstract
According to the model of translation initiation in eukaryotes, the 40S ribosomal subunit binds to capped 5′-end of mRNA and subsequently migrates along 5′-UTR in searching for initiation codon. However, it remains unclear whether the migration is the result of a random one-dimensional diffusion, or it is an energy-driven unidirectional movement. To address this issue, the method of continuous monitoring of protein synthesis in situ was used for high precision measurements of the times required for translation of mRNA with 5′-UTRs of different lengths and structures in mammalian and plant cell-free systems. For the first time, the relationship between the scanning time and the 5′-UTR length was determined and their linear correlation was experimentally demonstrated. The conclusion is made that the ribosome migration is an unidirectional motion with the rate being virtually independent of a particular mRNA sequence and secondary structure.
INTRODUCTION
The generally accepted paradigm for initiation of translation of eukaryotic capped mRNAs is based on the ribosomal scanning model first described by Kozak (1). This model proposes that an initiating 43S ribosomal particle binds to the cap structure at the 5′-end of the mRNA and migrates along the 5′-proximal non-coding nucleotide sequence [5′-untranslated region (5′-UTR)] until it encounters an AUG triplet which serves as the initiation codon. The process of searching for initiation codon was designated as scanning of the untranslated region by the initiating ribosomal particle. It was experimentally determined that the scanning process requires ATP (2). Over the last three decades, the original Kozak model has been tested and somewhat revised and in no case has the model been shown to be in principle conflict with experimental data (3–10) [see (11,12) for the reviews], although some alternative models have been also proposed (13).
On the other hand, a great deal of information has been obtained about initiation factors that are required for the scanning process and interact with scanning ribosomal particles [see (14–16) for the reviews]. Several initiation factors form a relatively stable complex with the scanning 40S ribosomal particle. They include eukaryotic initiation factors (eIF) eIF1, eIF1A, eIF2 with bound GTP and initiator Met-tRNAi and the large multi-subunit protein eIF3. All together, these factors and the 40S ribosomal subunit form the so-called 43S ribosomal initiation complex. Another large heterotrimeric protein complex, eIF4F, is involved in recruitment of the 43S complex to capped mRNAs. The complex eIF4F interacts via its largest subunit, eIF4G, with 40S-bound eIF3 and contributes to binding of the 43S complex with the 5′-located cap structure via its eIF4E subunit. The third subunit of eIF4F is eIF4A, which has ATP-dependent helicase and RNA-dependent ATPase activities (17,18). After the cap structure is recognized, the 43S complex starts its migration along the 5′-UTR of the mRNA.
Despite this detailed understanding of the composition of the scanning ribosomal complex and the compatibility of the scanning model with accumulated experimental data on translation initiation, the most important physical characteristics of the ribosome movement during the scanning process, such as the migration rate of scanning ribosomal particle and its quantitative dependence on 5′-UTR length and structure, have not been unequivocally addressed. Moreover, it remains unsolved what is the main function of ATP in the scanning process. Two alternative mechanisms of the search for the initiation codon are possible. It can be a diffusional wandering of the ribosomal particle along 5′-UTR and in this case ATP may be needed only for the unwinding of secondary structure barriers on the way of the scanning ribosomal complex. Alternatively, a specific ATP-dependent mechanism of the ordered unidirectional movement from the cap structure towards the start of the coding sequence can be the necessary attribute of the scanning process. The parameter that allows deciding between the two alternatives is the relationship between the length of 5′-UTR and the time required to find the initiation codon, i.e. the scanning time. In the case of the diffusional wandering the scanning time should be proportional to the squared length of the UTR, whereas in the case of the unidirectional movement, the relationship must be linear.
In the current study, we have performed the precision measurements of the aforementioned dependencies. The unique method of continuous in situ monitoring of luciferase activity proposed previously (19,20) was used. Mathematical analysis of the smooth kinetic curves that reflect the accumulation of active enzyme in real time allowed us to measure the duration of full-translation round with previously unattainable accuracy. Two in vitro translation systems based on the cell lysates of higher eukaryotes were used, namely S30 extracts of wheat germs and mammalian Krebs-2 ascites cells. We showed that increasing in 5′-UTR length resulted in increased translation time and that the time increment depended linearly on the length of the 5′-UTR, regardless of its primary and secondary structure. At the same time, the transit time (the combined time of elongation and termination) did not depend on the length of 5′-UTR. Also, we were able to discriminate between the initiation and elongation phases of translation due to the selective influence of K+ concentration on the rate of elongation. Thereby, the established linear dependence between scanning time and 5′-UTR length can be considered as the first quantitative experimental confirmation of net-unidirectional movement of the scanning ribosomal complex along the polynucleotide chain.
MATERIALS AND METHODS
Plasmid constructions
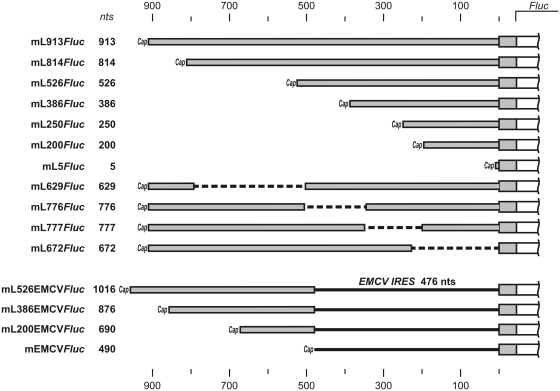
To prepare pL913Fluc, a fragment containing the first 952 nt of the human LINE-1 cDNA attached to the firefly luciferase coding region was obtained by PCR from plasmid pRluc-L15′-UTR-Fluc (21) using primers 5′-GTAATACGACTCACTATAGGGCCCGGCGGAGGAGCCAAG-3′ and 5′-CCCCGACTCTAGAATTACACGG-3′. After digestion with XbaI, the fragment was inserted into the Ecl136II–XbaI sites of the pUC19 vector, resulting in pL913Fluc. The pL913Fluc derivatives lacking different portions of the LINE-1 5′-UTR were prepared from the pL913Fluc parent plasmid by digestion with specific restriction endonucleases (Supplementary Data). The digested plasmids were blunt-ended using Klenow fragment or T4 DNA polymerase and re-ligated using T4 DNA ligase. To prepare pL200Fluc, a PCR fragment obtained from pL913Fluc using 5′-CCCCGAGCAGCCTAACTGG-3′ and 5′-CCAGCTGCGTTTTAGAGTTTCCAG-3′ primers was digested with BstXI and inserted into pL913Fluc from which the Bsp120I–BstXI fragment had been removed. The portion of the LINE-1 5′-UTR region contained in each of the constructs is schematically illustrated in Figure 1.
Figure 1.
Schematic representation of mRNAs containing various 5′-UTR sequences upstream of the luciferase coding region. mL913Fluc includes a 913 nt long 5′-UTR consisting of the full-length leader sequence of the human LINE-1 retrotransposon mRNA. The other 10 mRNAs have various deletions in the 5′-UTR, but all contain the same 45 nt 5′-terminal portion of the LINE-1 ORF fused to the Fluc coding sequence and the same 52 nt 3'-UTR (see Materials and methods). The length of the 5′-UTRs in each mRNA is indicated in nucleotides (nts). See Supplementary Figure S1 for 5′-UTRs secondary structure. mL526EMCVFluc, mL386EMCVFluc, mL200EMCVFluc and mL5EMCVFluc were prepared by the insertion of 488 nt EMCV IRES fragments into 5′-UTRs of mL526Fluc, mL386Fluc, mL200Fluc and mL5Fluc constructs, respectively.
Constructs pL200EMCVFluc, pL386EMCVFluc and pL526EMCVFluc carrying the fragment of Encephalomyocarditis virus (EMCV) IRES (373–848) located adjacent to Fluc initiation codon were performed as follows: PCR fragment obtained from pTE1 (22) using primers 5′-CGAGCCACAAAGGTGGGCCCGGAAACCTGG-3′ and 5′-CACGCCATCTTTGTGGCCATATTATCATCG-3′ was digested with BstXI and ligated into BstXI sites of pL200Fluc, pL386Fluc and pL526Fluc, respectively. The construct pEMCVFluc was prepared by the substitution of LINE-1 leader in pL913Fluc by the same EMCV IRES fragment (Figure 1 for the schematic representation of the constructs).
The stable GC-rich stem–loop (6) was inserted into LINE-1 5′-UTR by coligation of PCR fragments containing the 5′- and 3′-halves of the hairpin into pL913Fluc between NheI and Bsp119I sites (Supplementary Data for details).
In vitro transcription
To generate mRNA for use in in vitro translation, plasmids (pL913Fluc and derivatives) were linearized with HindIII and transcribed in vitro according to Pokrovskaya and Gurevich (23) with minor modifications. Reaction mixtures contained 50 µg/ml linearized plasmid, 200 U/ml T7 RNA polymerase, 200 U/ml RNase inhibitor, 100 µg/ml Ac-BSA, 2 mM each ATP, GTP and CTP, 0.3 mM GTP and 6 mM m7GpppG or ApppG in 200 mM HEPES–KOH buffer, pH 8.0 with 12 mM MgCl2 and 40 mM DTT. Reactions (50 μl volume) were incubated at 38°C for 1 h. The mRNA was extracted from the reactions with phenol/chlorophorm/NaOAc, pH 5.0, passed through a Sephadex 50 mini-spin column (Pharmacia), ethanol precipitated and washed. The pellets of RNA were dried under vacuum and dissolved in DEPC-treated water. These mRNA concentrations were calculated from UV absorbance and were named after the corresponding template plasmids, but with the prefix ‘m’ instead of ‘p’ (i.e. mL913Fluc, mL5Fluc, etc.).
In vitro translation
Wheat germ cell-free translation system
Wheat germ extract (WGE) was prepared from germs of wheat, sort Kazakhstanskaya 4, basically according to the protocol of Erickson and Blobel (24) with a modification in the germ washing procedure (25). The detailed protocol is described in (26). The concentration of the obtained WGE was 246 OD260/ml. The final translation mixture contained 20% v/v WGE, 100 µg/ml creatine phosphokinase, 500 U/ml RNase inhibitor, 50 mg/ml yeast total tRNA, 0.1 mM each amino acid, 1 mM ATP, 0.6 mM GTP and 16 mM creatine phosphate in 20 mM HEPES–KOH buffer pH 7.6 with 1.25 mM Mg(OAc)2, 2.5 mM DTT, 0.25 mM spermidine and 0.1 mM luciferin. The total K+ concentration in the translation system was 35 mM.
Krebs-2 cell-free translation system
Whole-cell extracts were prepared from mouse Krebs-2 ascites cells as described by Dmitriev et al. (27) (see Supplementary Data for details). The final translation mixture contained 50% v/v Krebs-2 extract, 100 µg/ml creatine phosphokinase, 500 U/ml RNase inhibitor, 50 mg/ml calf total tRNA, 25 μM each amino acid, 1 mM ATP, 0.2 mM GTP and 8 mM creatine phosphate in 20 mM HEPES–KOH buffer pH 7.6 with 0.6 mM Mg(OAc)2, 100 mM KOAc, 1 mM DTT, 0.5 mM spermidine and 0.1 mM luciferin. As shown earlier, the capped luciferase mRNA remains practically intact in Krebs-2 translation system for at least 1 h (28).
For both types of cell-free translation systems, reaction components were mixed on ice, adjusted to 80% of the final volume and incubated for 2 min at 25°C (WGE) or 30°C (Krebs-2). A quantity of 2 µl of preheated 5-fold concentrated mRNA were diluted with 8 μl of the prepared reaction mixture and immediately put into the temperature-controlled cell of a Chemilum-12 multichannel luminometer. The intensity of light emission generated through luciferase activity was measured continuously by collecting the streaming data on the computer as a kinetic curve.
Calculation of the full-translation time
Calculations of full-translation time (the time between the start of the reaction and the appearance of luciferase activity) were made using the IgorPro 6.0 program (WaveMetrics, Inc.). The portions of the original kinetic luminescence curves L(t) corresponding to the first 15 min of the reaction were fitted by the numerical solutions of a parameterized differential equation system describing normal Gauss distribution:
| (1) |
where parameter tsyn—the position of the Gaussian peak—corresponds to the full-translation time. The precision of tsyn determination was figured out from the curve approximation quality.
Adjustment of K+ concentration during cell-free translation
In microcentrifuge tubes, 20 μl of 20 mM KOAc were placed and dried under vacuum. Wheat germ translation system mixtures (150µl) containing 25 nM of a given mRNA template were set-up as described above and incubated at 25°C. At regular 30 s intervals after the start of the translation reaction, 10 μl portions of the translation reaction mixture were added to the preheated tubes with dried KOAc, vortexed, briefly spun down and immediately put into the luminometer cell for continuous recording of luciferase activity.
Transit time evaluation
Ten microlitres of WGE translation reaction mixtures with certain mRNAs were set up as described above and incubated 4 min at 25°C. After that, 0.5 μl of 60 μM edeine was added and reaction tubes were put into the luminometer cell for continuous recording of luciferase activity. In control reactions edeine was substituted by 0.5 μl of deionized water. The moment of the system response to edeine addition was determined from the point of divergence between the edeine treated and the control time course curves. Transit time was accepted to be equal to the delay between edeine addition and the response observed.
Cell culture and transfection procedures
Human embryonic kidney HEK293T cells were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and transfected with target and reference (Rluc) mRNAs using Unifectin-56, as described previously (21,27). After 2 h of incubation, cells were harvested and luciferase activities were analyzed with the Dual Luciferase Assay kit (Promega).
RESULTS
Construction of luciferase-encoding mRNAs with 5′-UTRs of different lengths
To investigate the dependence of full-translation time (the time from the start of the mRNA scanning to the release of active protein) on the length of the 5′-UTR, we generated a series of recombinant Fluc-mRNAs that were identical in their coding sequences but differed in lengths of their 5′-UTRs. The longest 5′-UTR used was the full-length natural leader sequence of the human LINE-1 retrotransposon. This 900 nt leader was shown to provide efficient cap-dependent translation initiation in mammalian cell-free translation systems and to be scanned by the ribosomal initiation complex in searching for the initiation codon, with no indications of IRES or shunting mechanisms (21). We demonstrated that this leader directs strictly cap-dependent translation also in a wheat germ cell-free translation system (see Supplementary Figure S2).
Ten different deletion mutants of the full-length LINE-1 5′-UTR were constructed in such a way that the deletions randomly covered the entire 900 nt sequence of the original leader (Figure 1). Six of the mutants were produced by sequential 5′-truncations. The other four mutants resulted from internal deletions more or less evenly distributed along the entire leader; as a result, we got Fluc-mRNAs with 5′-UTRs of approximately similar lengths but different sequences that allowed testing the effect of mRNA primary and secondary structure on ribosomal scanning rate. The full-length LINE-1 5′-UTR sequence and the 10 shortened sequences were used to construct plasmids in which the 5′-UTR was followed by a 45 nt 5′-terminal portion of the LINE-1 ORF fused to the Fluc coding sequence. Corresponding mRNAs were synthesized from each plasmid using in vitro transcription in the presence of excess m7GpppG as a substrate for co-transcriptional capping.
In situ monitoring of luciferase activity after translation start: precision measurement of the full-translation time
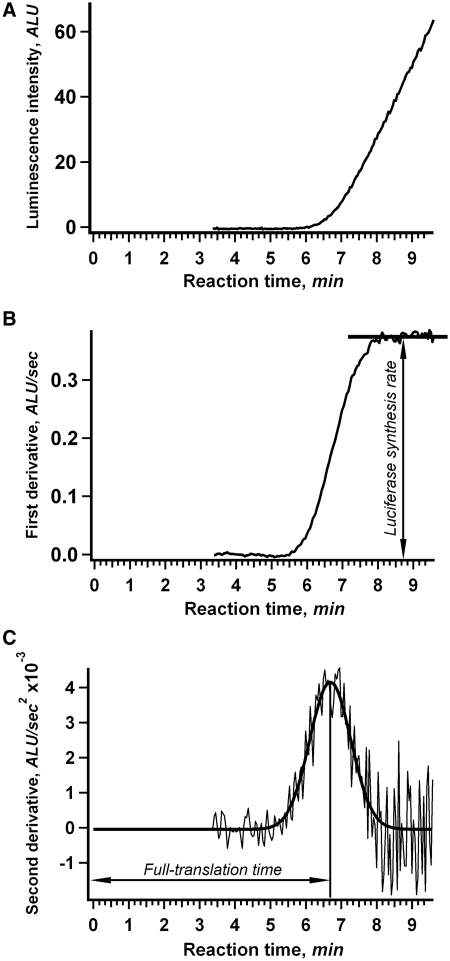
The 11 different mRNAs containing LINE-1-derived 5′-UTRs and the same Fluc coding sequence (Figure 1) were used as templates for in vitro translation with the continuous in situ recording of luciferase activity. Figure 2 shows representative results obtained for translation of the mL200Fluc mRNA (containing the 5′-UTR of a moderate length, ∼200 nt) in a wheat germ cell-free translation system. As shown in Figure 2A, the continuous measurement of luciferase activity over time produces a time course of translation that is a smooth curve with a high signal-to-noise ratio suitable for use in complex numerical analyses (see below). The delay in signal appearance after the start of translation reflects the fact that the first active (full-length) molecules of luciferase cannot appear in the reaction mixture until the first ribosomes finish translating the entire mRNA. The useful property of firefly luciferase is that its N-terminal amino acid residues are critical for the enzyme stability (29), so that any leaky scanning of cognate AUG and artificial initiation at downstream inframe methionine codons would yield inactive truncated proteins undetectable by the method. As shown previously, folding of the firefly luciferase occurs co-translationally and the full-length protein displays its activity immediately upon translation termination (19). Also, it should be mentioned that in the wheat germ system used, the elongation rate was found to be constant for at least the first 30 min of the cell-free translation reaction (20). Therefore, the observed time lag between the start of the reaction and the appearance of active luciferase can be interpreted as the time required for synthesis of a complete protein molecule during a full-translation round consisting of initiation (including scanning), elongation and termination. This time will be referred to as ‘full-translation time’.
Figure 2.
Kinetics of in vitro translation as revealed by continuous in situ measurement of luciferase activity. (A) Time course of the synthesis of active luciferase in a cell-free system. mL200Fluc mRNA (25 nM) was translated in a wheat germ cell-free translation system at 25°C. Luminescence was measured in situ in the luminometer cell. (B) First derivative of the curve shown in (A). The plateau level indicates the steady-state rate of luciferase synthesis. (C) Second derivative of the curve shown in (A). The bold line represents the Gauss function fit of the data. The marked position of the Gaussian peak corresponds to the average full-translation time which represents the combined durations of initiation, elongation and termination.
It is noteworthy that the kink at the initial rise of the kinetic curve that reflects the appearance of active luciferase is smooth rather than sharp (e.g. Figure 2A). This reflects the statistical spread in synthesis time for discrete molecules of luciferase within the reaction. For normal statistical distributions, the second derivative of the kinetic curve must fit the Gauss function. An example of the numerical differentiation and Gaussian fitting is presented in Figure 2B and C. The position of the Gauss distribution peak indicates the average time for synthesis of full-length luciferase in a given translation round (average full-translation time). In the example shown in Figure 2C for translation of the mL200Fluc mRNA, the average full-translation time was 6 min 40 s. In practice, we used the inversed procedure in order to diminish the effect of noise and increase the computational stability: the numerical solution of the corresponding differential equation system was fitted to the original kinetic data (see Materials and methods).
The reliability of the measured parameters depends both on the signal-to-noise ratio and on the reproducibility of the experiments. The signal-to-noise ratio automatically follows from the curve fitting procedure and was found to be acceptable in our experiments. Reproducibility of the results was estimated by analyzing three independent data sets. On the whole, the accuracy of our measurements of full-translation time (the delay in the appearance of the active protein after the start of translation) could be estimated in the range of 2–10 s depending on mRNA construct, translation system and reaction conditions or between 0.3% and 1.5% of the measured value.
Full-translation time increases linearly with increasing 5′-UTR length
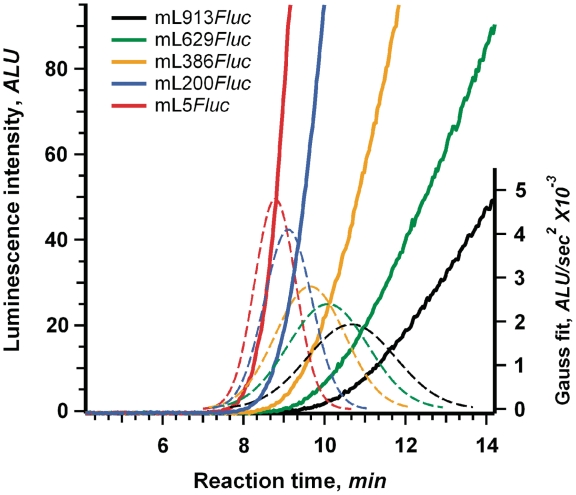
In order to determine the impact of 5′-UTR length on full-translation time, we translated each of the 11 mRNAs shown in Figure 1 both in the wheat germ cell-free translation system and in the cell-free translation system based on extracts of mouse Krebs-2 ascites cells. The kinetic curves for several representative mRNAs translated in the wheat germ system are presented in Figure 3. The data shown are for five mRNAs with 5′-UTRs ranging from 5 nt (mL5Fluc) to 913 nt (mL913Fluc). As expected, mRNAs with longer 5′-UTRs displayed a longer delay in the appearance of luciferase activity after translation start, i.e. the increased full-translation time, as compared to mRNAs with shorter 5′-UTRs. Besides, translation of mRNAs with long 5′-UTRs resulted in lower productivity of the translation systems (Supplementary Figure S3). This could be the result of a significant off-rate of scanning ribosomal particles during their movement along extended 5′-UTRs. The problem of reduced processivity of ribosomal scanning in cell-free systems was noted previously (11,30).
Figure 3.
Determination of full-translation time for mRNAs with different length 5′-UTRs. Time course of luciferase synthesis in the wheat germ system (effective K+ concentration is 35 mM) translating the indicated mRNAs. Dashed lines represent the Gaussian fitting of the second derivatives of the corresponding kinetic curves (see legend to Figure 2).
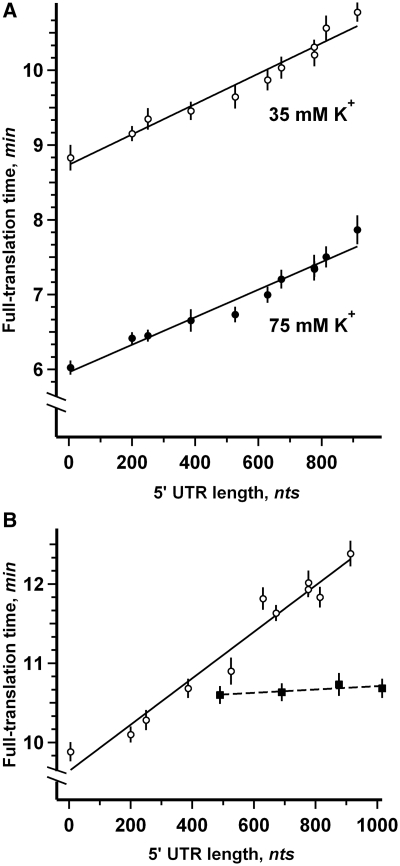
Figure 4 shows the dependence of full-translation time on 5′-UTR length as determined for all 11 mRNAs in the wheat germ (Figure 4A) and Krebs-2 (Figure 4B) cell-free translation systems (see Supplementary Table S1 for numerical values). Remarkably, for all the cases analyzed, the data show that the full-translation time displayed positive correlation with 5′-UTR length and that the relationship was close to linear. As expected, the maximum delay in the appearance of luciferase activity after translation start was observed with the full-length 913 nt leader in mL913Fluc; the difference with the values obtained for virtually leaderless sequence (mL5Fluc) was ∼2 and 2.5 min in the wheat germ and Krebs-2 systems, respectively.
Figure 4.
Linear dependence of full-translation time on 5′-UTR length. (A) Fluc mRNAs with 5′-UTRs of different lengths were translated in the wheat germ cell-free system at 25°C with effective K+ concentration adjusted to either 35 mM (open symbols) or 75 mM (solid symbols). Error bars denote the sum of the standard errors derived from the curve fitting procedure and the dispersion of data obtained in three independent experiments. The lines represent the least squares approximation of data points. (B) The same mRNAs were translated in the Krebs-2 cell-free system at 30°C (open symbols). Four Fluc mRNAs with 5′-UTRs of different length with the EMCV IRES sequence inserted adjacent to AUG codon were translated in the same system (solid symbols).
Quite different situation was observed in the case of translation of luciferase mRNAs with 5′-UTRs consisting of LINE-1 leader fragments of different length followed by EMCV IRES sequence located adjacent to AUG codon (Figure 4B). Here the full-translation time was virtually constant and did not depend on the overall length of 5′-UTR. The result implies that in this case the alternative IRES-dependent initiation mechanism was dominant despite the fact that the mRNAs were capped. The direct binding of ribosomal subunits to the specific IRES structure was in no way affected by the preceding 5′-UTR sequence. This is a good proof of the absence of any unspecific effect of the lengthened 5′-UTR on the translation process. Therefore, it can be asserted that the linear increase of the translation time with the increase of 5′-UTR length is due to the cap-dependent initiation process that involves scanning as its essential stage.
The effect of increased 5′-UTR length on full-translation time is due to increased scanning time
As our set of mRNAs had identical coding sequences it is likely that the observed dependence of full-translation time on 5′-UTR length reflects the increase of the scanning time rather than some differences in the elongation stage. To test this directly, we made use of the finding first reported by Cahn and Lubin (31) that the elongation stage of eukaryotic translation is particularly sensitive to potassium ion (K+) concentration in the surrounding medium, both in vivo and in vitro. The reduction of K+ concentration was considered to inhibit the elongation stage of translation more or less specifically (32). In agreement with the literature, we found that increasing the K+ concentration in the wheat germ cell-free translation system resulted in shorter translation time for all mRNAs tested (Figure 4A), but had a negligible influence on the dependence between full-translation time and 5′-UTR length (as reflected by the parallelism of the lines approximating high and low K+ data sets in Figure 4A).
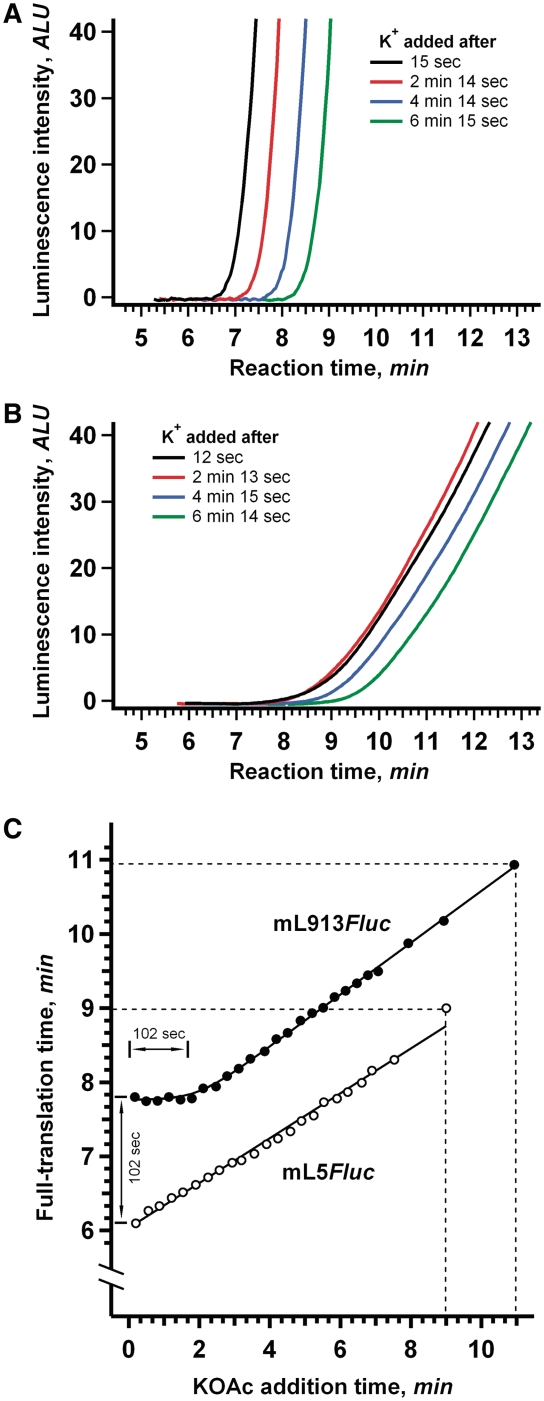
In our experiments on discrimination of initiation and elongation stages, in vitro translation was initiated at a lower K+ concentration (35 mM) and then portions of the translation mixture were supplemented with additional K+ (∼75 mM final concentration) at 30 s intervals as the reaction proceeded. Based on the assumption that the increased (K+) selectively stimulates elongation, with no effect on scanning rate, the following two predictions can be made: (i) When K+ is added at any time during the scanning phase, it should affect the whole subsequent elongation stage identically and, therefore, result in the identical decrease of full-translation times. In other words, a plateau should be observed in the full-translation time versus K+ addition time dependence plot. In the case of the leaderless mRNA (mL5Fluc) where no scanning phase is expected, this plateau should be absent and (ii) After the scanning phase, i.e. during elongation, the later K+ is added, the less decrease of the full-translation time should be observed. This dependence should be close to linear and its slope should be the same for all mRNAs, irrespective of the lengths of their 5′-UTRs.
The results of our experiments were found to be in full conformity with the predictions made. Figure 5 illustrates the dependence of full-translation time on the time of K+ addition for the two mRNA constructs with the most different 5′-UTR lengths (mL913Fluc with a 913 nt leader and mL5Fluc with a 5 nt leader). For the mRNA with the shortest 5′-UTR the dependence was linear throughout the translation process. A very different situation was observed for the mRNA with the longest (913 nt) 5′-UTR: K+ additions made at any time during the first 2 min of the translation reaction had identical effect on full-translation time (Figure 5C). Therefore, during translation of mL913Fluc, the elongation phase was preceded by a phase insensitive to K+ concentration; this phase is to be attributed to the initiation stage. Moreover, the observed duration of this phase (∼100 s, see Figure 5C) is in agreement with the difference between full-translation time measurements for the virtually leaderless mRNA (mL5Fluc) and the full-length leader mRNA (mL913Fluc). This result confirmed our assumption that the effect of 5′-UTR length on full-translation time is due to its effect on the initiation stage of translation, most probably on the scanning phase, but not on the elongation stage.
Figure 5.
The effect of K+ addition during the translation reaction on full-translation time. (A) The K+ concentration in the wheat germ system translating 25 nM mL5Fluc mRNA (5 nt 5′-UTR) was increased from 35 mM to 75 mM at the indicated times after the start of the translation reaction. (B) The same as (A), but in the system translating mL913Fluc mRNA (913 nt 5′-UTR). (C) Dependence of full-translation time on the time of K+ addition after the start of the translation reaction. At the indicated time points, the final concentration of K+ was increased from 35 mM to 75 mM in the wheat germ system translating 25 nM mL5Fluc mRNA (open symbols) or mL913Fluc mRNA (solid symbols). Dashed lines indicate the translation times measured without extra K+ addition.
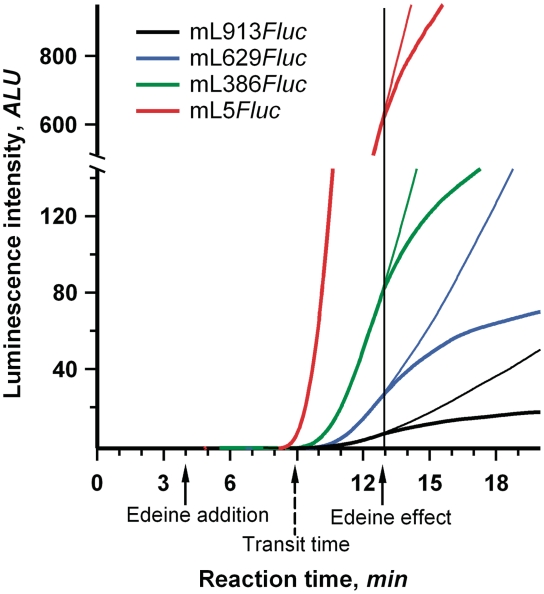
In addition, the negligible effect of 5′-UTR length on elongation can be checked by direct measurement of the time required for a translating ribosome to pass over the coding sequence of mRNA (the so-called transit time). The transit time can be defined as the combined time of elongation and termination and does not depend on the duration of initiation. Hence, any process specifically affecting the initiation stage of translation should not influence the transit time. The commonly used methodology for the transit time evaluation involves pulse-chase experiments with radioactive amino acids (33,34). Unfortunately, the low precision of the method hardly allows analyzing the time differences in a seconds scale. We employed the alternative technique based on the inhibition of translation with initiation-specific antibiotic. Edeine, linear oligopeptide antibiotic, affects the late step in eukaryotic translation initiation process, allowing 40S ribosomal subunits to bind to mRNA and to scan 5′-UTR, but preventing recruitment of the 60S ribosomal subunits and formation of the 80 S initiation complexes (35,36). At low concentrations (<5 µM), it has no discernible effect on elongation in wheat germ translation system (37). Addition of edeine in the course of translation reaction immediately prevents entering of new ribosomes into elongation. Evidently, the inhibitory effect of the antibiotic is reflected on a time course curve only after mRNA-bound ribosomes have read over the full coding sequence and terminated translation. This delay of the response to edeine addition coincides with the transit time.
We added edeine to 3 µM concentration in 4 min after the start of the reaction in wheat germ systems translating mRNAs with different length 5′-UTRs 913, 629, 386 and 5 nt. In spite of the fact that the values of full-translation time were quite different for the mRNAs used, the response to the antibiotic addition occurred virtually at the same moment (Figure 6). This implies that the transit time was the same in all the cases and, as expected, the increased length of 5′-UTR had no effect on the duration of elongation and termination. It was also found that the transit time coincided with the full-translation time of leaderless mRNA (5 nt long 5′-UTR). Apparently, the duration of initiation in the absence of the extended leader was very small, at least under the value of experimental error (5–10 s). Thus, the full-initiation time was proved to be virtually equivalent to the time required for the 5′-UTR scanning.
Figure 6.
Evaluation of transit time for Fluc mRNAs with 5′-UTRs of different length. A quantity of 0.5 μl of 60 μM edeine (bold line) or the same volume of mQ (fine line) was added to the wheat germ system translating 25 nM of the indicated mRNAs. Vertical line points up the equivalence of the moments of the systems response to the edeine addition. Dashed arrow indicates the estimated value of transit time.
The scanning rate is of the same order of magnitude as the movement rate of translating ribosome
The rate of ribosomal scanning can be determined from the slope of the linear dependencies between full-translation time and 5′-UTR length presented in Figure 4. These calculations gave scanning rates of ∼6 and 8 nt/s for the Krebs-2 (30°C) and wheat germ (25°C) cell-free translation systems, respectively. These results are in reasonable agreement with the estimate of ∼10 nt/s made earlier in a yeast cell-free translation system (30). Previously, it was shown that the full-translation time of leaderless mRNAs corresponded well with their transit time, which is a measure solely of the duration of elongation (20). Therefore, we estimated the rate of translating ribosome movement during elongation from the full-translation time of mL5Fluc (5 nt leader). The rate of elongation was found to be 3 nt/s in the Krebs-2 system (30°C) and 3 or 4.5 nt/s (depending on the K+ concentration) in the wheat germ system (25°C). Hence, the movement rates of scanning and translating ribosomes are within the same order of magnitude. This implies that initiation of translation is a relatively time-consuming process that can contribute significantly to the overall duration of protein synthesis.
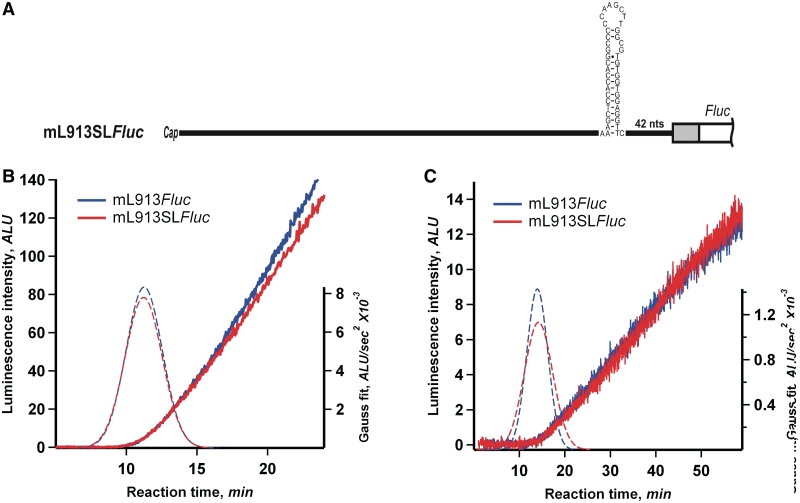
The scanning rate does not respond to differences in secondary structure of 5′-UTR
During the scanning process, the ribosomal initiation complex is supposed to encounter various local secondary structure elements, such as double-helical regions. Surprisingly, our experiments (Figure 4) have demonstrated that the scanning rate is virtually the same with different 5′-UTRs which possess various content and stability of secondary structure. At the same time, according to our estimations, no stem–loops or other double-helical elements with calculated thermodynamic stability greater than −20 kcal/mol can be predicted in the 5′-UTRs used in the experiments (Supplementary Figure S1). In order to test a possible effect of a secondary structure element of high stability on in vitro translation directly, we inserted the synthetic stem–loop (hairpin) with the calculated thermodynamic stability of about −30 kcal/mol, which was earlier used by Kozak (6), into the supposedly unstructured 3′-part of mL913Fluc 5′-UTR, 42 nt upstream of the initiation codon (Figure 7A). As seen in Figure 7B and C, translation of this mRNA in both wheat germ and ascite cell-free systems revealed no significant change of full-translation time and the rate of protein synthesis, as compared with the original mL913Fluc. Similar experiments with mL777Fluc and mL386Fluc demonstrated analogous results (see Supplementary Data and Supplementary Figure S4). Transfection of the two mRNAs into the cultured human embryonic kidney (HEK293) cells led to similar yields of active luciferase (Supplementary Data and Supplementary Figure S5). Therefore, the addition of a relatively stable stem-loop element into 5′-UTR had a weak, if any, effect on the rate and processivity of scanning. These results are in full agreement with the experiments of Kozak, where the insertion of an individual stem–loop with the predicted thermodynamic stability of ΔG = −30 kcal/mol in the middle of the 5′-UTR did not inhibit the initiation of translation in mammalian COS cells (6).
Figure 7.
Addition of stable secondary structure element to LINE-1 5′-UTR. (A) mL913SLFluc was prepared by the insertion of GC-rich hairpin (Kozak 1986) with the calculated thermodynamic stability of about −30 kcal/mol into mL913Fluc, 42 nt downstream the AUG codon. (B) Time course of luciferase synthesis in the wheat germ system translating mL913Fluc and mL913SLFluc. The positions of Gaussian peaks (dashed lines) correspond to the average full-translation time values (see legend to Figure 2). (C) The same mRNAs were translated in the Krebs-2 cell-free system at 30°C.
DISCUSSION
The fact that the scanning has been shown to be ATP dependent (2) is usually explained as the necessity of ATP for RNA helicases required to melt hairpins of structured 5′-UTRs and clear the way for the moving particles. However, it should be emphasized that any unidirectional movement requires energy by itself, regardless of the problem of secondary structure unwinding. An energy-independent unidirectional movement, even along unstructured (or melted) mRNA regions, is in violation of the second law of thermodynamics. Therefore, the directional movement of the scanning ribosomal particle implies the presence of a special energy-dependent mechanism that restricts the backward diffusional shifts.
The alternative possibility is that the scanning ribosomal complex searches for the initiation codon through the ATP-independent diffusional mechanism, the so-called phaseless wandering. This mechanism was proposed and then confirmed in the case of the movement of prokaryotic ribosomal particles through intercistronic UTRs of polycistronic mRNAs (38,39). The diffusional mechanism was also recently proposed for eukaryotic ribosomal particles initiating translation on poly(A) leaders of poxviral mRNAs (40). In principle, one could not exclude the possibility of such a random wandering mechanism in the case of the eukaryotic cap-dependent initiation; while the motion itself being energy independent, ATP could be required for helicases to provide the unfolding of local secondary structure barriers in 5′-UTR. Hence, the unidirectionality of the scanning process during cap-dependent initiation arises as a principal problem to be experimentally solved.
The two alternatives described above can be distinguished by measuring the relationship between the length of 5′-UTR and the time required for its scanning. In the case of the random wandering, the scanning time should be proportional to the squared length of the UTR, whereas in the case of the unidirectional movement the relationship must be linear. Earlier the importance of this issue was adequately understood by specialists, but technical means to solve the problem were insufficient. Nevertheless, some ‘attempts to measure the lag time between addition of mRNA to a (prewarmed) cell-free translation system and the first initiation events suggested that the lag was more closely related to the length of the 5′-UTR than to the square of the length (S. Grünert and R.J. Jackson, unpublished data)’ [cited from (11)].
In this work, we approached the problem by precise determination of the dependency of the scanning time on the length of the 5′-UTR sequence. The initiation time was analyzed as a part of the overall duration of the translation epicycle. Previously, it was reported that the moment of the appearance of active translation product in cell-free system corresponds to the full time required for the translation of single mRNA molecule (20,30). In conventional laboratory practice, the translation process is studied by means of collecting aliquots from the translation mixture at particular time points. However, as the differences in translation time for mRNAs with different length 5′-UTRs are comparable with the time interval between collection of samples, the procedure of discrete sampling could not provide data suitable for rigorous quantitative analysis. To avoid this problem, we took advantage of a more precise technique based on continuous in situ monitoring of the appearance of the newly synthesized protein. This methodology produces smooth kinetic curves with high accuracy and reproducibility. The power of the method was previously demonstrated in studies of co-translational protein folding (19) and translation initiation rate during the course of polysome formation (20). Here, we applied this technique to measure the full-translation time of a set of mRNAs with 5′-UTRs of different lengths. For this purpose, a variety of truncated derivatives of the originally long 5′-UTR were fused with the same luciferase coding sequence and the full-translation time of each mRNA was measured. We found that the dispersion of translation time could be described by a normal Gauss distribution and, therefore, we were able to apply standard statistical analysis to our data. It was shown that the duration of translation can be determined with the mean deviation of about 5 s, i.e. ∼1% of the total value (6–13 min) and is just ∼5% of the maximum difference observed in translation times (∼100 s). Such a high precision has been previously unattainable in translation experiments and here allowed us to perform statistically reliable analysis of the dependence of translation time and, hence, scanning time on 5′-UTR length.
In our study, we employed in vitro translation systems based on two different eukaryotic cell extracts, both plant (wheat germs) and mammalian (mouse Krebs-2 ascites cells). This allowed us to use a long natural leader that was shown to manifest efficient cap-dependent translation initiation. We chose the 5′-UTR of human LINE-1 retrotransposon mRNA (900 nt) from among the variety of eukaryotic mRNA leaders of extreme length [see (41) for the review]. The leader was well characterized in terms of its structural characteristics and obeying the classical mechanism of cap-dependent initiation (21,42). In particular, it was shown that shortening of the LINE-1 leader by deletion of different length fragments did not significantly alter initiation efficiency (21). This allowed us to design a set of deletion mutants with 5′-UTR lengths covering the entire 900 nt range more or less evenly. The most remarkable result we obtained was a direct positive correlation between full-translation time and 5′-UTR length. As the coding sequence was identical in all the mRNAs used, the differences in full-translation time could be attributed only to the time required for initiation. Additional data attributing this effect of 5′-UTR length specifically to the initiation stage of translation was obtained through selective modulation of the elongation rate by altering K+ concentration and by direct measurements of transit time.
The human LINE-1 5′-UTR has been predicted to possess a developed secondary structure with multiple stem-loops unevenly scattered along its sequence (21). The deletions that we made to generate our set of 11 constructs were virtually random and the resulting truncated 5′-UTRs differed substantially in their secondary structure content and stability. Nevertheless, when full-translation time was plotted against 5′-UTR length, all the points fell close to the linear fit (Figure 4). As this plot reflected the dependence of the duration of initiation stage on 5′-UTR length (see the previous paragraph) and only the time of migration of the initiation complex along the 5′-UTR could correlate with the 5′-UTR length, we assumed the differences in translation time to be due to the differences in scanning time. This implies that the scanning time depended mainly on the length, rather than on the local primary or secondary structures of the leader sequence. To sum up, for the first time it was shown that the relationship between scanning time and 5′-UTR length is linear that made it possible to unambiguously conclude that the migration of the scanning ribosomal particle is unidirectional movement rather than stochastic random wandering.
As discussed above, the unidirectional character of the movement certainly points to the existence of a special energy-dependent mechanism. Such a mechanism can be realized at the molecular level in the form of an ATP-fed Brownian ratchet-and-pawl device (43), as proposed for other molecular moving machines, including myosin, dynein, kinesin, RNA polymerase, etc. [e.g. (44–46)], as well as translating ribosomes (47–49). The crucial point in such molecular nanomachines is the energy-dependent restriction of the backward diffusional shifts, while the forward diffusion is allowed (rectification of Brownian motion).
When discussing possible mechanisms of 5′-UTR scanning it should be taken into account that random melting of secondary structure by ribosome-free ATP-dependent helicases can hardly support unidirectional movement of the scanning particle. A tempting model could be a helicase attached at the front of the scanning ribosomal particle (50), provided immediate refolding of helices occurred behind the ribosomal particle (in order to block its backward diffusion). The weakness of this model is the strict requirement for a developed, secondary structure along 5′-UTR. On the other hand, the initiation factor eIF4A, known as RNA helicase, ATP-dependent RNA-binding protein and RNA-dependent ATPase [(17) for the review], was reported to be attached at the rear of the scanning particle (51). The role of this protein in the ribosome-bound state could be the involvement in an ATP-dependent pawl, which temporarily fixes the scanning particle on mRNA to prevent the backward diffusional shifts, rather than the role of bona fide RNA helicase (52). According to this model, the scanning particle moves along more or less unfolded RNA chain due to Brownian motion, but the backward motion is periodically blocked by the ATP-dependent interaction of the particle-bound initiation factors eIF4A and eIF4B with RNA, these proteins play the roles of a spring and a pawl, respectively, in the Feynman's type ratchet mechanism. Anyhow, both the aforementioned models are principally consistent with the paradigm of ATP-dependent unidirectional movement.
The unidirectional movement at the molecular level does not necessarily imply the absence of stochasticity in the movement process. As all the spatial displacements of particles at the size level of ribosomes are caused by thermal (Brownian) motion, they must be stochastic. Typical patterns of net-unidirectional movements (‘biased Brownian movements’), with occasional backward steps, were demonstrated in direct experiments using single molecule detection techniques for the cases of kinesin walking along a microtubule fibril (53) and myosin heads stepping along actin microfilaments (46).
As already mentioned, neither full-size LINE-1 5′-UTR, nor any of the studied deletion mutants contain perfect stems or stem–loops with a stability greater than −20 kcal/mol (see ‘Results’ section). It is known that the insertion of a stable 13 bp GC-rich stem-loop (ΔG = −30 kcal/mol) in the middle of the 5′-UTR did not inhibit the initiation of translation in mammalian COS cells (6). Here, we demonstrated that the insertion of the same stable stem–loop in the unstructured part of LINE-1 leader had a weak effect, if any, on scanning rate both in vivo and in vitro. It is noteworthy that the initiation of translation in Saccharomyces cerevisiae was reported to be more sensitive to the stability of 5′-UTR secondary structure, as compared with mammalian systems (54,55). This can be explained by some peculiarities of the yeast translation apparatus: the same stem–loop structures were shown to be far less inhibitory in the reticulocyte lysate system (55). On the other hand, it should be noted that despite the closeness of experimental points to the linear fit, some deviations exceeded the values of the experimental error (Figure 4). This may indicate that local unique structures or nucleotide sequences within 5′-UTRs can exert some influence on the rate of scanning.
An important point to be also mentioned is that any reliable information about the real state of the secondary structure of mRNA in eukaryotic cells in vivo and in eukaryotic cell extracts is absent. There are all grounds to assert that the presence of various RNA helicases and RNA binding proteins should make the secondary structure of mRNA to be melted in a more or less degree or at least significantly deviated from the structures predicted for isolated RNAs. Hence, it cannot be excluded that hairpins and other local secondary structure elements of moderate stability does not really exist in mRNAs under cytosol conditions, but rather melted or half-melted.
The linear character of the time-to-length dependency allowed us to estimate the scanning rate. As mentioned in the ‘Results’ section, the rates of scanning determined under our experimental conditions were 6 nt/s at 30°C for Krebs ascite extract and 8 nt/s at 25°C for wheat germ extract. Earlier, using the conventional procedure of collecting aliquots from the translation mixture at particular time points, translation times of mRNAs with artificial low-structured 5′-UTRs of different lengths [multiple copies of GCN4 leader, (56)] were measured in yeast translation system (30). Although precise dependencies of the time on the length were not obtained, they managed to estimate an approximate average rate of the scanning, which was reported to be ∼10 nt/s at 26°C. Thus, despite the inevitable variations of this parameter on the source of cell extract, incubation temperature, ionic conditions, etc. the reported value was found to be similar to ours.
Taken together, our results provide experimental proof for several basic principles of the eukaryotic translation initiation. First, the initiation time is basically equivalent to the time required for the ribosomal particle to pass over the 5′-UTR, i.e. to the scanning time. Second, the movement of the ribosomal particle along the 5′-UTR occurs in a net-unidirectional manner. Third, the scanning 43S particles can overcome various sections of 5′-UTRs with an essentially constant rate more or less independently of the differences in their local structural elements.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Program of State Support of Leading Scientific Schools (NSh-8488.2010.4 to A.S.S.); Program for Basic Researches on Molecular and Cellular Biology of the Presidium of Russian Academy of Sciences (to A.S.S.); Russian Foundation for Basic Research (RFBR 07-04-01120 to S.E.D.); Grant of President of Russian Federation (MK-5309.2011.4 to S.E.D.) and Fogarty International Center (R03TW008217). Funding for open access charge: Fogarty International Center (award no. R03TW008217).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to A.A. Komar and A.F. Finkelstein for helpful discussions and to P. Baker for critically reading the manuscript.
REFERENCES
- 1.Kozak M. How do eucaryotic ribosomes select initiation regions in messenger mRNA? Cell. 1978;15:1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- 2.Kozak M. Role of ATP in binding and migration of 40S ribosomal subunits. Cell. 1980;22:459–467. doi: 10.1016/0092-8674(80)90356-6. [DOI] [PubMed] [Google Scholar]
- 3.Kozak M. Inability of circular mRNA to attach to eukaryotic ribosomes. Nature. 1979;280:82–85. doi: 10.1038/280082a0. [DOI] [PubMed] [Google Scholar]
- 4.Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1983;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984;12:3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl Acad. Sci. USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konarska M, Filipowicz W, Domdey H, Gross HJ. Binding of ribosomes to linear and circular forms of the 5′-terminal leader fragment of tobacco-mosaic-virus RNA. Eur. J. Biochem. 1981;114:221–227. doi: 10.1111/j.1432-1033.1981.tb05139.x. [DOI] [PubMed] [Google Scholar]
- 9.Cigan AM, Feng L, Donahue TF. tRNAi(met) functions in directing the scanning ribosome to the start site of translation. Science. 1988;242:93–97. doi: 10.1126/science.3051379. [DOI] [PubMed] [Google Scholar]
- 10.Jackson RJ. The ATP requirement for initiation of eukaryotic translation varies according to the mRNA species. Eur. J. Biochem. 1991;200:285–294. doi: 10.1111/j.1432-1033.1991.tb16184.x. [DOI] [PubMed] [Google Scholar]
- 11.Jackson RJ. A comparative view of initiation site selection mechanisms. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 127–183. [Google Scholar]
- 12.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chappell SA, Edelman GM, Mauro VP. Ribosomal tethering and clustering as mechanisms for translation initiation. Proc. Natl Acad. Sci. USA. 2006;103:18077–18082. doi: 10.1073/pnas.0608212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q Rev. Biophys. 2004;3:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 16.Pestova TV, Lorsh JR, Hellen CUT. The mechanism of translation initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- 17.Rogers GW, Jr, Komar AA, Merrick WC. eIF4A: the godfather of the DEAD box helicases. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- 18.Marintchev A, Edmonds K, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolb VA, Makeyev EV, Spirin AS. Folding of firefly luciferase during translation in a cell-free system. EMBO J. 1994;13:3631–3637. doi: 10.1002/j.1460-2075.1994.tb06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alekhina OM, Vassilenko KS, Spirin AS. Translation of non-capped mRNAs in a eukaryotic cell-free system: acceleration of initiation rate in the course of polysome formation. Nucleic Acids Res. 2007;35:6547–6559. doi: 10.1093/nar/gkm725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dmitriev SE, Andreev DE, Terenin IM, Olovnikov IA, Prassolov VS, Merrick WC, and Shatsky IN. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated. Mol. Cell. Biol. 2007;27:4685–4697. doi: 10.1128/MCB.02138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evstafieva AG, Ugarova TY, Chernov BK, Shatsky IN. A complex RNA sequence determines the internal initiation of encephalomyocarditis virus RNA translation. Nucleic Acids Res. 1991;19:665–671. doi: 10.1093/nar/19.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pokrovskaya ID, Gurevich VV. In vitro transcription: preparative RNA yields in analytical scale reactions. Anal. Biochem. 1994;220:420–423. doi: 10.1006/abio.1994.1360. [DOI] [PubMed] [Google Scholar]
- 24.Erickson AH, Blobel G. Cell-free translation of messenger RNA in a wheat germ system. Methods Enzymol. 1983;96:38–50. doi: 10.1016/s0076-6879(83)96007-x. [DOI] [PubMed] [Google Scholar]
- 25.Madin K, Sawasaki T, Ogasawara T, Endo Y. A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: plants apparently contain a suicide system directed at ribosomes. Proc. Natl Acad. Sci. USA. 2000;97:559–564. doi: 10.1073/pnas.97.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirokov VA, Kommer A, Kolb VA, Spirin AS. Continuous-exchange protein-synthesizing systems. In: Grandi G, editor. Methods in Molecular Biology: In Vitro Transcription and Translation Protocols. 2nd edn. Vol. 375. Totowa, NJ: Humana Press; 2007. pp. 19–55. [DOI] [PubMed] [Google Scholar]
- 27.Dmitriev SE, Andreev DE, Adyanova ZV, Terenin IM, Shatsky IN. Efficient cap-dependent translation of mammalian mRNAs with long and highly structured 5′-untranslated regions in vitro and in vivo. Mol. Biol. 2009;43:108–113. [PubMed] [Google Scholar]
- 28.Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 29.Sung D, Kang H. The N-terminal amino acid sequences of the firefly luciferase are important for the stability of the enzyme. Photochem. Photobiol. 1998;68:749–753. doi: 10.1562/0031-8655(1998)068<0749:tntaas>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 30.Berthelot K, Muldoon M, Rajkowitsch L, Hughes J, McCarthy JEG. Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol. Microbiol. 2004;51:987–1001. doi: 10.1046/j.1365-2958.2003.03898.x. [DOI] [PubMed] [Google Scholar]
- 31.Cahn F, Lubin M. Inhibition of elongation steps of protein synthesis at reduced potassium concentrations in reticulocytes and reticulocyte lysate. J. Biol. Chem. 1978;253:7798–7803. [PubMed] [Google Scholar]
- 32.Spirin AS, Ryazanov AG. Regulation of elongation rate. In: Trachsel H, editor. Translation in Eukaryotes. Roca Raton, FL: CRC Press; 1991. pp. 325–350. [Google Scholar]
- 33.Wu RS, Warner JR. Cytoplasmic synthesis of nuclear proteins: kinetics of accumulation of radioactive proteins in various cell fractions after brief pulses. J. Cell Biol. 1971;51:643–652. doi: 10.1083/jcb.51.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sørensen MA, Pedersen S. Determination of the peptide elongation rate in vivo. In: Martin R, editor. Protein Synthesis, Methods and Protocols. Totowa, NJ: Humana Press; 1998. pp. 129–142. [DOI] [PubMed] [Google Scholar]
- 35.Hunt T. The control of globin synthesis in rabbit reticulocytes. Ann. NY Acad. Sci. 1974;241:223–231. doi: 10.1111/j.1749-6632.1974.tb21880.x. [DOI] [PubMed] [Google Scholar]
- 36.Kozak M, Shatkin AJ. Migration of 40S ribosomal subunits on messenger RNA in the presence of edeine. J. Biol. Chem. 1978;253:6568–6577. [PubMed] [Google Scholar]
- 37.Kopeina GS, Afonina ZA, Gromova KV, Shirokov VA, Vasiliev VD, Spirin AS. Step-wise formation of eukaryotic double-row polyribosomes and circular translation of polysomal mRNA. Nucleic Acids Res. 2008;36:2476–2488. doi: 10.1093/nar/gkm1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarabhai A, Brenner S. A mutant which reinitiates the polypeptide chain after chain termination. J. Mol. Biol. 1967;27:145–162. doi: 10.1016/0022-2836(67)90357-9. [DOI] [PubMed] [Google Scholar]
- 39.Adhin MR, van Duin J. Scanning model for translational reinitiation in eubacteria. J. Mol. Biol. 1990;213:811–818. doi: 10.1016/S0022-2836(05)80265-7. [DOI] [PubMed] [Google Scholar]
- 40.Shirokikh NE, Spirin AS. Poly(A) leader of eukaryotic mRNA bypasses the dependence of translation on initiation factors. Proc. Natl Acad. Sci. USA. 2008;105:10738–10743. doi: 10.1073/pnas.0804940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. reviews0004.1–0004.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andreev DE, Dmitriev SE, Terenin IM, Prassolov VS, Merrick WC, Shatsky IN. Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res. 2009;37:6135–6147. doi: 10.1093/nar/gkp665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feynman R, Leighton R, Sands M. The Feynman Lectures on Physics. Vol. 1. Reading, MA: Addison-Wesley Publishing Company; 1963. [Google Scholar]
- 44.Cordova NJ, Ermentrout B, Oster GF. Dynamics of single-motor molecules: The thermal ratchet model. Proc. Natl Acad. Sci. USA. 1992;89:339–343. doi: 10.1073/pnas.89.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gelles J, Landick R. RNA polymerase as a molecular motor. Cell. 1998;93:13–16. doi: 10.1016/s0092-8674(00)81140-x. [DOI] [PubMed] [Google Scholar]
- 46.Yanagida T, Ueda M, Murata T, Esaki S, Ishii Y. Brownian motion, fluctuation and life. Biosystems. 2007;88:228–242. doi: 10.1016/j.biosystems.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 47.Spirin AS. Ribosome as a molecular machine. FEBS Lett. 2002;514:2–10. doi: 10.1016/s0014-5793(02)02309-8. [DOI] [PubMed] [Google Scholar]
- 48.Spirin AS. The ribosome as an RNA-based molecular machine. RNA Biol. 2004;1:3–9. [PubMed] [Google Scholar]
- 49.Spirin AS. The ribosome as a conveying thermal ratchet machine. J. Biol. Chem. 2009;284:21103–21119. doi: 10.1074/jbc.X109.001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 52.Spirin AS. How does a scanning ribosomal particle move along the 5′-untranslated region of eukaryotic mRNA? Brownian ratchet model. Biochemistry. 2009;48:10688–10692. [Google Scholar]
- 53.Nishiyama M, Higuchi H, Ishii Y, Taniguchi Y, Yanagida T. Single molecule processes on the stepwise movement of ATP-driven molecular motors. Biosystems. 2003;71:145–156. doi: 10.1016/s0303-2647(03)00122-9. [DOI] [PubMed] [Google Scholar]
- 54.Oliveira CC, van den Heuvel JJ, McCarthy JE. Inhibition of translational initiation in Saccharomyces cerevisiae by secondary structure: the roles of the stability and position of stem-loops in the mRNA leader. Mol. Microbiol. 1993;3:521–532. doi: 10.1111/j.1365-2958.1993.tb01713.x. [DOI] [PubMed] [Google Scholar]
- 55.Vega Laso MR, Zhu D, Sagliocco F, Brown AJ, Tuite MF, McCarthy JE. Inhibition of translational initiation in the yeast Saccharomyces cerevisiae as a function of the stability and position of hairpin structures in the mRNA leader. J. Biol. Chem. 1993;268:6453–6462. [PubMed] [Google Scholar]
- 56.Hinnebusch AG. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J. Biol. Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.