Mammalian cellular differentiation and development depend on stable, somatically heritable epigenetic switches. A good example is X chromosome inactivation, the random silencing of either the paternal or maternal X chromosome but not both. Once silenced, genes on the inactive X (Xi) remain genetically silent in all progeny cells, even though identical genes on the active X (Xa) in the same nucleus are expressed. In most human cells, reactivation of a silent gene on the Xi is below the level of detection (1). Methylation changes are frequent in cancer cells and difficult to distinguish from mutations, leading to the burgeoning field of cancer epigenetics (2, 3). Epigenetics, the study of changes in gene function that do not depend on changes in primary DNA sequence (4), depends on stable, heritable marking of DNA or chromatin. Because heritability is such a key feature, important questions are: What is the fidelity with which an epigenetic state is transmitted from one cell generation to the next? How frequent are mistakes? Are there epigenetic repair mechanisms? This Commentary will discuss three articles relevant to these questions: (i) an important study of methylation fidelity by Laird et al. (5) reported in this issue of PNAS, (ii) a recent study of de novo methylation by Chen et al. (6), and (iii) a kinetic treatment of dynamic, stochastic DNA methylation (7).
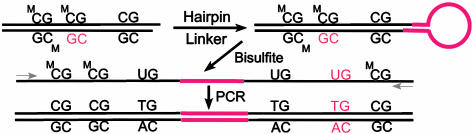
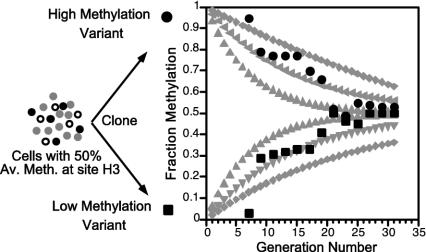
Cytosine DNA methylation was the first epigenetic mark correctly identified (8, 9) and its inheritance mechanism at least superficially understood (10). 5-Methylcytosine (mC) is found mainly in symmetrical CpG dyads (mCG/GmC), which are transiently converted to hemimethylated sites (mCG/GC) by DNA replication but then converted back to symmetrically methylated sites by a DNA methyltransferase with high specificity for hemimethylated CpG sites. The method of Laird et al. (5) (Fig. 1) extends the bisulfite method for determining cytosine methylation. Bisulfite treatment of single-stranded DNA deaminates C, changing it to U, but does not deaminate mC. PCR after bisulfite treatment converts U to T, and subsequent cloning and DNA sequencing gives the position of mC in individual molecules (11). Numerous studies using the bisulfite method have clearly established that methylation patterns often show molecule-to-molecule variation (12-14), but, nevertheless, the methylation pattern error rate as calculated by Ushijima et al. (15) is very low. Molecule-to-molecule variation but stable average methylation of specific sites (16) and stable patterns over larger regions (15) need to be explained. The simple, yet elegant, method introduced by Laird et al. (5) is to use hairpin ligation to covalently join the complementary strands of individual DNA molecules, thereby allowing one to identify and quantitate hemimethylated sites, even in repetitive sequences. Both the gain and loss of methylation can be measured. Laird et al. use their method to study 22 CpG sites in the CpG island of the human FMR gene in normal lymphocytes. This gene is X-linked, and the CpG island is methylated only on the Xi. They find that substantial de novo methylation takes place on the Xi, up to 16% per replication event. Overall their results are consistent with and lend credence to the kinetic or stochastic model for methylation used by Riggs and collaborators (7) to analyze and understand clone-to-clone variation in methylation patterns of the human X-linked PGK promoter and a site 3 kb upstream of mouse Igf2 (Fig. 2). The stochastic methylation model assumes that for each CpG dyad in each DNA molecule there is a certain efficiency (probability) of methylation maintenance (Em), maintenance failure (1 – Em), or de novo methylation (Ed). The theoretical curves in Fig. 2 are based on the solution of the differential equations dMi/dt = Mi·Emi + Edi·U and dUi/dt = (1 – Edi)·Ui + (1 – Emi)·Mi, where Mi and Ui are, respectively, the number of methylated and unmethylated molecules of a specific CpG dyad (7). A clear prediction of this treatment is that clones initially with low methylation at a site caused by stochastic fluctuation in the population will gain methylation, whereas high methylation clones will lose methylation at the same rate until an equilibrium is reached (see Fig. 2). At equilibrium, percent methylation (%M) is given by: %M = 100 Ed/(1 – Ed – Em). This treatment predicts that methylation at most sites in cell lines, and in tissue cell types, will be stochastically variable to a certain extent, with each site depending on site-specific probabilities Emi and Edi. This model easily explains stable partial methylation in tissues and cell lines, as is observed (16). By using the above equation for %M at equilibrium and their estimates of Ed and Em obtained by the hairpin-bisulfite method, Laird et al. (5) calculated for the FMR region an expected methylation level of 81% at equilibrium, in excellent agreement with the experimentally observed level of 80.5% in normal lymphocytes.
Fig. 1.
The hairpin-bisulfite PCR method for determining cytosine methylation in complementary strands.
Fig. 2.
Methylation modeling and comparison with experimental data. Modeling (gray curves) was done by using the equation in figure 4 of Pfeifer et al. (7) and various Em, Ed values: 0.90, 0.10 (outer curves); 0.95, 0.05 (middle curves); and 0.97, 0.03 (inner curves). Two curves were calculated for each set of parameters, one starting with an unmethylated site (M = 0, U = 1) and one starting with a methylated site (M = 1, U = 0). The experimental data were obtained by the authors by using mouse cell line BML-2, which was known to be 50% methylated at a specific HpaII site (H3), located near the Igf2 gene but not imprinted (13). Seventeen clones were analyzed for fraction methylation of H3 beginning at generation seven after cloning by using a PCR-based assay with an internal standard (21). As predicted by the stochastic model, the methylation level was quite variable when first assayed soon after cloning. Shown are data from one low methylation clone (▪) and one high methylation clone (•). All 17 clones returned to near 50% methylation by the 30th generation.
The gain and loss of methylation can be measured by hairpin-bisulfite PCR.
The first mammalian cytosine DNA methyltransferase to be identified (Dnmt1) is almost certainly the main maintenance DNA methyltransferase; it is part of replication foci and has a strong preference for hemimethylated sites (17). More recently, other DNA methyltransferases have been studied, and two of them, Dnmt3a and Dnmt3b, have de novo methylation activity. Chen et al. (6) demonstrate that deletion of both Dnmt3a and Dnmt3b from embryonic stem cells results in a loss of genomewide methylation, which can be restored by reintroduction of either Dnmt3a or Dnmt3b but not Dnmt1. Deletion of Dnmt1 results in a rapid, ≈90% reduction of DNA methylation by the first passage, but deletion of Dnmt3a and Dnmt3b results in a very slow loss of methylation, taking 5 months and 70 generations for 90% reduction of global methylation. Taking this many generations with Ed = 0 caused by the deletions is consistent with an average Em of 0.95, similar to that found for Hpa site 3 in Igf2 (Fig. 2) and the FMR CpG island (5). It seems clear that de novo methylation is required for the maintenance of methylation patterns, not just in the initial establishment of the patterns (5-7).
We thus are beginning to get quantitative data on methylation fidelity. Maintenance provided by Dnmt1 is good (average Em ≈95), but are the measured maintenance efficiencies adequate for stable switches? For the X-linked PGK promoter and the FMR region studied, the answer is probably yes. Both regions have multiple methylated sites (60 and 22, respectively) and a relatively high rate of de novo methylation on the Xi. Methylation-loss lesions will not accumulate; the rate of methylation ”repair” will exceed methylation loss, and a methylation-silenced gene on the Xi will not escape (7, 18). A clear distinction thus must be made between site-specific methylation fidelity and epigenetic fidelity (18), which often will depend on multiple sites. Also, although precise measurements still need to be made, it is likely that for many sites maintenance is much better than average with Em ≥ 0.99 (7).
A different question remains unanswered and important. What keeps CpG islands on the Xa unmethylated? De novo methylation of active unmethylated promoters on the Xa has so far not been detected, and each group speculates that de novo methylation on the Xa is much less than on the Xi (5, 7, 18). Similarly, autosomal CpG islands must also be protected from de novo methylation (15). A search for endogenous inhibitors of Dnmt3a and Dnmt3b could be very informative. Site-specific, active demethylation is also a likely possibility, at least in some cases (19). A large number of factors are likely to affect Ed and Em values. Some of these are chromatin accessibility, chromatin remodeling proteins, methylation density, repetitive DNA, histone modifications, site-specific proteins, and small interfering RNA. Some sites will be essentially fully methylated and some will be completely unmethylated, but most will be partially methylated; few will have exactly the same Em and Ed values. Noteworthy are experiments by Han et al. (20) showing that in mammalian cells the lac repressor, when bound to the lac operator in the absence of inducer, reduces %M of episomal and integrated lac operator sites by protection from de novo methylation. Conceptually, the establishment, maintenance, and change of specific methylation patterns in the genome can be understood. Although the details will be complicated, the general concept has emerged that %M at specific sites, and methylation patterns over larger regions, will be determined by site-specific Em and Ed parameters, which in turn are controlled by local chromatin fine structure, ancillary factors, and various Dnmt activities. The new hairpin-bisulfite technique will be a powerful tool aiding progress toward understanding these important details of methylation-dependent epigenetics.
See companion article on page 204.
References
- 1.Gartler, S. M. & Goldman, M. A. (1994) Dev. Genet. 15, 504-514. [DOI] [PubMed] [Google Scholar]
- 2.Jones, P. A. & Baylin, S. B. (2002) Nat. Rev. Genet. 3, 415-428. [DOI] [PubMed] [Google Scholar]
- 3.Clark, S. J. & Melki, J. (2002) Oncogene 21, 5380-5387. [DOI] [PubMed] [Google Scholar]
- 4.Russo, E., Martienssen, R. & Riggs, A. D. (1996) Epigenetic Mechanisms of Gene Regulation (Cold Spring Harbor Lab. Press, Plainview, NY).
- 5.Laird, C. D., Pleasant, N. D., Clark, A. D., Sneeden, J. L., Hassan, K. M. A., Manley, N. C., Vary, J. C., Jr., Morgan, T., Hansen, R. S. & Stöger, R. (2004) Proc. Natl. Acad. Sci. USA 101, 204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, T., Ueda, Y., Dodge, J. E., Wang, Z. & Li, E. (2003) Mol. Cell. Biol. 23, 5594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfeifer, G. P., Steigerwald, S. D., Hansen, R. S., Gartler, S. M. & Riggs, A. D. (1990) Proc. Natl. Acad. Sci. USA 87, 8252-8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riggs, A. D. (1975) Cytogenet. Cell. Genet. 14, 9-25. [DOI] [PubMed] [Google Scholar]
- 9.Holliday, R. & Pugh, J. E. (1975) Science 187, 226-232. [PubMed] [Google Scholar]
- 10.Bird, A. (2002) Genes Dev. 16, 6-21. [DOI] [PubMed] [Google Scholar]
- 11.Frommer, M., McDonald, L. E., Millar, D. S., Collis, C. M., Watt, F., Grigg, G. W., Molloy, P. L. & Paul, C. L. (1992) Proc. Natl. Acad. Sci. USA 89, 1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoger, R., Kajimura, T. M., Brown, W. T. & Laird, C. D. (1997) Hum. Mol. Genet. 6, 1791-1801. [DOI] [PubMed] [Google Scholar]
- 13.Feil, R., Walter, J., Allen, N. D. & Reik, W. (1994) Development (Cambridge, U.K.) 120, 2933-2943. [DOI] [PubMed] [Google Scholar]
- 14.Millar, D. S., Ow, K. K., Paul, C. L., Russell, P. J., Molloy, P. L. & Clark, S. J. (1999) Oncogene 18, 1313-1324. [DOI] [PubMed] [Google Scholar]
- 15.Ushijima, T., Watanabe, N., Okochi, E., Kaneda, A., Sugimura, T. & Miyamoto, K. (2003) Genome Res. 13, 868-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turker, M. S., Swisshelm, K., Smith, A. C. & Martin, G. M. (1989) J. Biol. Chem. 264, 11632-11636. [PubMed] [Google Scholar]
- 17.Leonhardt, H., Page, A. W., Weier, H. U. & Bestor, T. H. (1992) Cell 71, 865-873. [DOI] [PubMed] [Google Scholar]
- 18.Riggs, A. D., Xiong, Z., Wang, L. & LeBon, J. M. (1998) Novartis Found. Symp. 214, 214-225. [DOI] [PubMed] [Google Scholar]
- 19.Bruniquel, D. & Schwartz, R. H. (2003) Nat. Immunol. 4, 235-240. [DOI] [PubMed] [Google Scholar]
- 20.Han, L., Lin, I. G. & Hsieh, C. L. (2001) Mol. Cell. Biol. 21, 3416-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singer-Sam, J., LeBon, J. M., Tanguay, R. L. & Riggs, A. D. (1990) Nucleic Acids Res. 18, 687. [DOI] [PMC free article] [PubMed] [Google Scholar]