Abstract
Epigenetic DNA methylation is involved in many biological processes. An epigenetic status can be altered by gain or loss of a DNA methyltransferase gene or its activity. Repair of DNA damage can also remove DNA methylation. In response to such alterations, DNA endonucleases that sense DNA methylation can act and may cause cell death. Here, we explored the possibility that McrBC, a methylation-dependent DNase of Escherichia coli, cleaves DNA at a replication fork. First, we found that in vivo restriction by McrBC of bacteriophage carrying a foreign DNA methyltransferase gene is increased in the absence of homologous recombination. This suggests that some cleavage events are repaired by recombination and must take place during or after replication. Next, we demonstrated that the enzyme can cleave a model DNA replication fork in vitro. Cleavage of a fork required methylation on both arms and removed one, the other or both of the arms. Most cleavage events removed the methylated sites from the fork. This result suggests that acquisition of even rarely occurring modification patterns will be recognized and rejected efficiently by modification-dependent restriction systems that recognize two sites. This process might serve to maintain an epigenetic status along the genome through programmed cell death.
INTRODUCTION
Epigenetic DNA methylation is involved in many aspects of biological processes. [In this article, we consider any heritable information superimposed to the DNA sequence to be epigenetic (1)]. In eukaryotes, 5-methyl-cytosine plays primarily regulatory roles in chromatin organization, gene expression and genome maintenance, and its disturbance is related to human diseases (2). In prokaryotes, DNA methylation serves at least two roles. One role is regulation of gene expression, DNA repair and coordination of replication and division, as in Enteric bacteria (Dam) and Caulobacter relatives (CcrM). This role is associated with persistence of the gene in a given taxon. Another role is distinction of self from non-self, in which the ‘self’ methylation pattern is enforced by cleavage of DNA with foreign patterns. This role for methylation is associated with sporadic gene distribution within and between taxa. Both unmethylated and methylated sequences can be cleavage targets. Unmethylated sequences are recognized by Type I–III restriction endonucleases (such as EcoK, EcoRI and PvuII and EcoP15) and both DNA strands are cleaved if the sequence is not protected by cognate methylation. On the other hand, methylated sequences are recognized by Type IV restriction endonucleases, typically with weaker sequence specificity.
Three different types of methylation are found: N6-methyladenine, N4-methylcytosine and 5-methylcytosine. All can be found associated with sequence-specific DNAses (restriction endonucleases) in the sporadically distributed ‘self-identity’ role in bacteria. So far only N6-methyl-adenine has been associated with cell cycle regulation, transcriptional regulation and host–pathogen interaction (3,4). In some cases, the roles are blurred, as with adenine-specific DNA methylation by several Type III restriction-modification systems, which though variable and sporadically distributed (5), nevertheless are involved in phase variable gene expression (6). 5-methyl-cytosine or N4-methyl-cytosine DNA modification is used by several restriction-modification systems for control of their own transcription (7,8). Epigenetic DNA methylation, N6-methyl-adenine for prokaryotes and 5-methyl-cytosine for eukaryotes, is also involved in silencing of selfish genetic elements and other aspects of intragenomic conflicts (9). Dcm, a persistent DNA 5-methyl-cytosine methyltransferase in Escherichia coli and related bacteria, defends its host genome from attack by specific Type II restriction-modification systems (10) (see below).
Changes in DNA methylation state can cause cell death in many organisms although their underlying mechanisms and their biological significance likely vary. DNA methylation level can be decreased through DNA replication following loss of a methyltransferase gene or by removal of methylated bases during repair of damaged DNA (9,11–15). Some Type II restriction-modification systems enforce maintenance of specific DNA methylation by restriction-mediated killing of cell lineages that have lost the modification gene and thus methylation activity (16–18). Reduced methylation leads to Type I restriction endonuclease-mediated cell death under DNA damage repair and other conditions (12,14,15). On the other hand, acquisition of a specific DNA methylation pattern also causes cell death. For example, cell death upon exogenous expression of methyltransferases has been reported in eukaryotes. Expression of mouse DNA methyltransferases induces lethality in a fly and a frog (19,20). Though the underlying mechanisms that trigger cell death and the biological significance of the lethality in these heterologous eukaryotic systems remain unclear, a prokaryotic model has provided a simple case with an elucidated mechanism and probable biological significance (21). A methylated DNA-specific DNase (McrBC of Escherichia coli) causes cell death upon entry and expression of a cytosine methyltransferase gene (21). [Mcr stands for modified cytosine restriction (22).] When the methylation system enters the cell and begins to methylate the host genome, McrBC senses the epigenetic change, triggering cell death through chromosomal cleavage. Intact (unmethylated) genomes with mcrBC genes would survive in neighboring clonal cells. This presumably accounts for the incompatibility of wild-type E. coli K-12 with many modification methyltransferases (22) and may be generalizable to other modification-dependent restriction systems (23–25) that are known to induce DNA damage in the presence of genes for target DNA methyltransferases. This can be interpreted as a mechanism to maintain epigenetic integrity of a genome (9,21). The DNA methyltransferases represent potential threats to such epigenomic integrity of prokaryotic genomes because of their frequent horizontal transfer (26–29).
Some biochemical characteristics of the McrBC protein have been revealed. McrBC enzyme consists of two subunits, McrB and McrC. The N-terminal domain of McrB specifically binds to a methylated recognition site (30), while its C-terminal domain has GTPase activity (31). McrC is the endonuclease subunit, with a PD.(D/E)xK motif shared by many DNases (32,33). Efficient cleavage of a linear DNA by the enzyme requires two recognition sites on it (34,35): the composite recognition sequence of E. coli McrBC can be represented as RmC N40–2000 RmC, where R is A or G (34,36). Methylation does not need to be on the same DNA strand, so the two methylated sites do not need to be in a particular orientation (34). DNA double-strand cleavage occurs between the two sites, preferably at ∼30 bp inward from one of the sites, through interaction of two McrBC complexes on the DNA (34). The interaction is facilitated through translocation of the enzyme complexes along the DNA (35). The enzyme can cleave a linear substrate with a single modified element (RmC) if an obstacle, such as a bound protein, is also present (35).
A large-scale analysis of protein–protein interaction in E. coli revealed that the methyltransferase subunit of EcoKI interacts with DnaB helicase, a component of DNA replication machinery (37). This association might promote maintenance methylation after passage of the replication fork. In addition, a different Type I restriction endonuclease, EcoR124I, cleaves a model DNA replication fork at the branch point (38). Such an association between restriction and DNA replication was also suggested by an in vivo observation: the alleviation of restriction by homologous recombination functions even when only a single genome of a DNA bacteriophage enters a cell (39). Homologous recombination requires two copies of homologous DNA; therefore, this result implies that, at least for a subpopulation of the restricted phage, two copies of phage DNA were formed and subsequently suffered restriction cleavage, which was then followed by recombinational repair.
Since many aspects of McrBC action resemble those of Types I and III enzymes, we were encouraged to test it for action on DNA surrounding a DNA replication fork. We found that the McrBC enzyme indeed shows cleavage activity on a model DNA replication fork with methylation.
MATERIALS AND METHODS
Bacteria, bacteriophages and plasmids
The bacterial strains, all derivatives of E. coli K-12, the bacteriophage lambda and P1 strains, and the plasmids are listed in Table 1. The oligonucleotides are listed in Supplementary Table S1.
Table 1.
Bacteria, bacteriophages and plasmids
| Bacteria | Genotype | Source/Reference |
|---|---|---|
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB) [F′ traD36 proAB+ lacIqZΔM15] | Laboratory collection (University of Tokyo) |
| DH5alpha MCR | F− λ− ϕ80 dlacZ ΔM15Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 phoA supE44 thi-1 gyrA96 relA1 Δ(mrr-hsdRMS-mcrBC) | S. Ohta (53) |
| AB1157 | thr-1 leu-6 thi-1 lacY1 galK2 ara-14 xyl-5 mtl-1 proA2 his-4 argE3 str-31 tsx-33 supE44 | (54) |
| JC5519 | As AB1157 but recB21 recC22 | (55) |
| JC8679 | F−thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 glnV44(AS) galK2(Oc) λ−sbcA23 his-60 recC22 relA1 recB21 rpsL31(strR) xylA5 mtl-1 argE3(Oc) thi-1 | (56) |
| JC8691 | As AB1157 but recB21 recC22 sbcA23 | (56) |
| BMF1 | As JC8679 but ΔmcrB::Km | (21) |
| BNH3480 | As AB1157 but ΔmcrB::Km | Constructed by transduction with BMF1 as the donor. |
| Bacteriophage | Comments | Source/reference |
| P1vir | For transduction | Laboratory collection (University of Tokyo) |
| LIK891 | Bam10 ΔB int+ Δ(red-gam) imm21 nin5 shn60 | (57) |
| LEF1 | The same as LIK891 but carrying a pvuIIM gene | (21) |
| Plasmids | Properties | Source/reference |
| pMC63 | Two M.FnuDII sites 63 bp apart, Cmr | (36) |
| pMC0 | Single M.FnuDII site, Cmr | (36) |
| pBR322::fnuDIIM | Ampr | Laboratory collection (New England Biolabs) (58) |
| pACYC177 | Ampr | Laboratory collection (University of Tokyo) (59) |
| pME63 | Figure 2 | This work |
| pMap63 | Figure 2 | This work |
| pKI2 | pACYC177::fnuDIIM, Ampr | This work |
To construct pME63, an oligo DNA (mcGap1-cis) was inserted into BglII-SphI-cleaved pMC63. To construct pMap63, another oligo DNA (mcGap1) was similarly inserted into pMC63. To construct pKI2, a fragment containing fnuDIIM was amplified by PCR from pBR322::fnuDIIM with the primers, M.FnuDII-F and M.FnuDII-R. It was cut with BamHI and XhoI and ligated to BamHI–XhoI-cleaved pACYC177.
Bacteriophage infection
Bacterial strains were grown at 37°C to stationary phase in L broth with appropriate antibiotic selection. Then, the culture was diluted 1/50 and grown in tryptone broth (1.0% Bacto-tryptone and 0.5% NaCl), supplemented with 0.2% maltose, 10 mM MgSO4 and 10 µg/ml vitamin B1, to log phase. The culture (2 × 108 cells/ml) was used as a host for measuring plaque formation efficiency of unmodified lambda phage (LIK891) at a multiplicity of infection of less than 0.1 with serial dilutions. After 10 min of incubation at room temperature, the infected culture was mixed with molten top agar and poured onto tryptone plates (1.0% Bacto-tryptone, 0.5% NaCl and 1% agar) for overnight incubation at 37°C. The plaque forming efficiency of each phage lysate (unmodified lambda phage and lambda::pvuIIM) was calculated as the ratio of the titer on the strain being measured to the average titer of the lysate on the recBC sbcA mcrB::Km strain.
Forked DNA with methylation
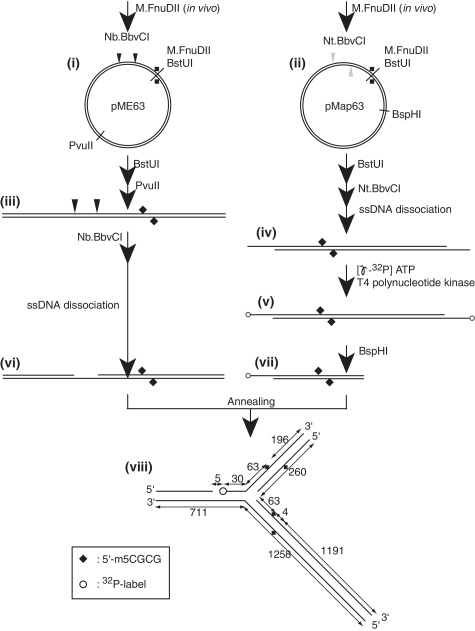
The substrates for the cleavage assay were prepared by annealing two DNA fragments with complementary single-strand regions as previously reported (38) with some modifications (see Figure 2 below). The two starting plasmids, pME63 and pMap63, were methylated in vivo by propagating in bacterial cells (DH5alpha_Mcr) carrying a plasmid (pKI2) producing M.FnuDII. To eliminate potential unmethylated plasmid molecules, the plasmid preparations were treated with BstUI (New England Biolabs), which recognizes the same sequence as M.FnuDII but cannot cleave DNAs modified by M.FnuDII [see Figure 2(i), (ii) below]. The pME63 was linearized with PvuII (New England Biolabs) [see Figure 2(iii) below]. Then it was treated with a nicking enzyme, Nb.BbvCI (New England Biolabs), while pMap63 was nicked with Nt.BbvCI (New England Biolabs). The resulting short-DNA strands were dissociated by incubation at 78°C for 10 min and 37°C for 10 min in the presence of the complementary single-strand oligonucleotide gap1-cis-C [for Figure 2(vi) below] or gap1 [for Figure 2(iv) below] as described before (38). The 5′-ends of intermediate (iv) were labeled with [γ-32P]ATP (Perkin Elmer) and T4 polynucleotide kinase (TaKaRa) (v), followed by cleavage with BspHI (New England Biolabs) and recovery of the left half for removal of one of the end labels (vii). To prepare the fork DNA, the two gapped DNAs {(vi), (vii)} were annealed by mixing in equimolar amounts and incubating at 75°C for 10 min. The switch of the incubator was turned off. When the temperature reached 37°C, that temperature was maintained for 10 min. About 65–80% of the two parts were found annealed (judged by gel electrophoresis). The resulting solution was directly subjected to the cleavage assay. The resulting eM63(++) [see Figure 2(viii) below] is a long-branched DNA with two methylated sites, both located 63 bp away from the branch point. To prepare an unlabeled substrate, unlabeled structure (vii) was prepared by BstUI, Nt.BbvCI and BspHI treatments, followed by single-strand DNA dissociation and removal.
Figure 2.
Preparation of a long-branched DNA with methylation. The two plasmids were modified in vivo by M.FnuDII to generate 5′-m5CGCG, a McrBC recognition sequence {(i), (ii)}. Potential unmethylated plasmids were eliminated by cleavage with BstUI (5′-CGCG). pME63 was cleaved with PvuII and then with nicking endonuclease Nb.BbvCI (iii), while pMap63 was treated with nicking endonuclease Nt.BbvCI (iv). The resulting short single strands were dissociated by heating and removed by annealing with a complementary single-strand oligo DNA. The 5′-ends of intermediate (iv) were labeled with 32P (v), followed by BspHI cleavage for removal of one of the radio-labels (vii). The two DNAs with complementary single-strand regions {(vi), (vii)} were annealed to form a branched structure {viii, eM63(++)} as detailed in ‘Materials and Methods’ section. Open circle, 32P label at 5′-end; filled diamond, DNA methylation.
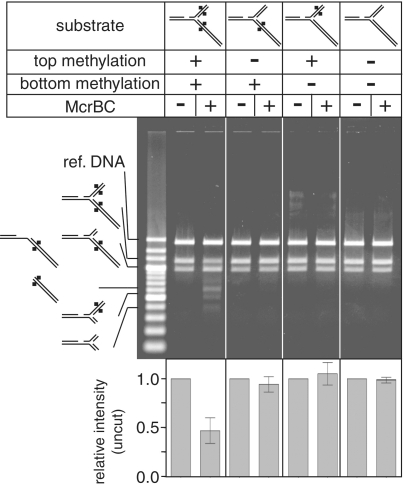
To make branched DNAs with methylation only in a single arm, or one without methylation (see Figure 5 below), plasmids without in vivo methylation were used as starting materials for the not methylated part. The procedure was the same as above except for omission of the BstUI treatments.
Figure 5.
Fork cleavage requires two methylated arms. Branched DNA substrates with different methylation patterns were challenged with the McrBC enzyme at 37°C for 30 min. A reference DNA was added after the reaction was stopped to normalize, if any, uneven staining through the gel. This was followed by agarose gel electrophoresis, and the products were visualized with ethidium bromide. Ref. DNA, the reference DNA. Quantification results are provided as mean ± SD from three independent reactions.
A linear two-site substrate (l.pMC63) was prepared by linearizing methylated pMC63 as previously reported (36). Methylation was by growth in vivo with a compatible plasmid bearing fnuDIIM.
Purification of McrB and McrC
Component proteins were purified separately from expression hosts described by Sutherland et al. (34). A modification of purification strategy 2 was followed, resulting in single protein bands on a polyacrylamide gel. Their storage was at −20°C in 500 mM NaCl, 10 mM Tris (pH 7.5), 0.1 mM EDTA, 1 mM DTT, 50% glycerol.
Cleavage assay
The reaction was performed with 1.3–1.6 nM of the substrates (estimated from the annealing efficiency judged by electrophoretic separation of annealing products, and taking into account incomplete annealing), 10 mM Tris–HCl (pH 7.9), 1 mM GTP, 10 mM MgCl2, 1 mM Dithiothreitol at 37°C, for 30 min with 10 U of McrBC. The total reaction volume was 15 μl. The cleavage activity with a preferred substrate, a linear duplex with two recognition sites, reached a plateau with 10 U of the McrBC enzyme under this condition (Supplementary Figure S1). The reaction was started with addition of GTP and stopped by addition of sodiumdodecylsulfate to 0.1%. The resulting solution was subjected to 1% agarose gel electrophoresis, followed by visualization with ethidium bromide and UV light. Where appropriate, the gel was dried and its 32P-signal was detected by an imaging analyzer, FLA5100 (Fuji-Film, Japan). In the quantitative experiments with ethidium bromide stain (see Figure 5 below, Supplementary Figure S1), 25 ng of a reference DNA was added after the reaction stop for normalization.
The contrast of presented figures was adjusted as follows. Raw data were imported to Photoshop CS version 8.0 (Adobe), and the image mode was changed to gray-scale. The settings of adjustment curves were then changed to set the end points in a linear relationship. These settings were (input, output) = (50, 0) and (190, 255) for Figure 3A left side (see below); (30, 0) and (190, 255) for Figure 3A right side (see below); (80, 30) and (130, 255) for Figure 5 (see below). These adjustments were independent of the quantification results. Quantitation employed analysis by the software package Image Gauge version 4.22 (Fuji-Film), and the graphs were prepared by the GraphPad PRISM version 4.03 (GraphPad softwear).
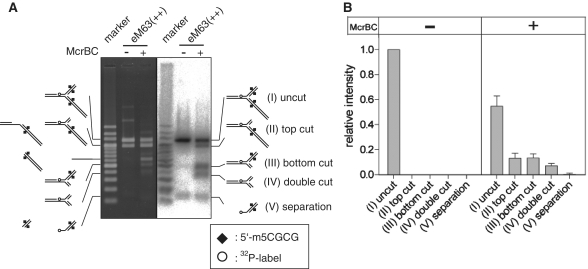
Figure 3.
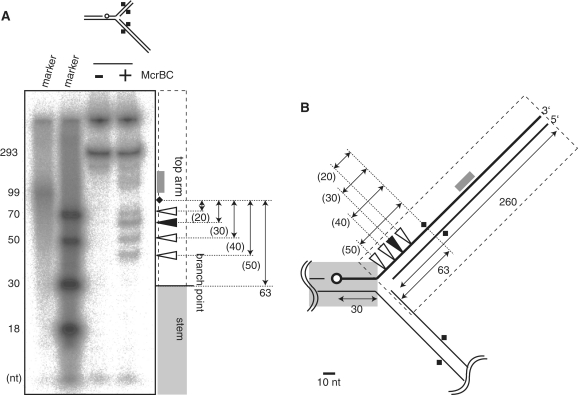
Cleavage of a model DNA replication fork by McrBC. (A) The substrate eM63++ (Figure 2A) was cleaved with McrBC, leaving products that had lost a part of the top arm, bottom arm or both. The starting material contains equal amounts of structures Precursor (vi) and Fork (viii) of Figure 2: two ethidium-stained bands in the untreated reaction (left panel, middle lane), only one of which carries 32P-label (right panel, middle lane). A portion of the material corresponds to Precursor (vii) in Figure 2; this is too faint to see by ethidium, but is visible at the bottom of the gel in the autoradiogram. This substrate was incubated with McrBC at 37°C for 30 min, followed by agarose gel electrophoresis. Precursor structure (vi) comigrates with product structure (II, top cut). Left, ethidium bromide stain; Right 32P signal. (B) Quantification of the 32P signal. Mean + SD from three independent reactions. Marker, 32P-labeled 200 bp ladder (TaKaRa).
Determination of the cleavage sites
The model fork substrate eM63(++) labeled with 32P was digested as described above, followed by heat denaturation at 95°C for 5 min. The products were subjected to electrophoresis through 12% polyacrylamide gel with 6 M urea. The marker ssDNA was prepared by end-labeling oligo DNAs of the indicated length (Supplementary Table S1) with 32P. The 32P-signal was detected by an imaging analyzer, FLA5100 (Fuji-Film, Japan).
RESULTS
Relation between methyl-specific DNase action and DNA replication in vivo
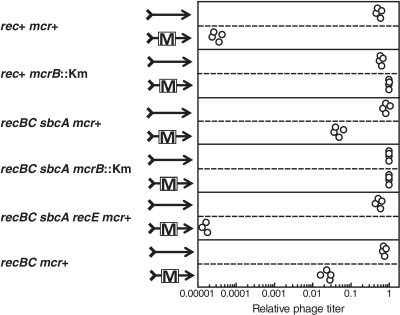
When an incoming DNA does not have appropriate DNA methylation pattern, it will be cleaved by restriction systems in the host cell. Such cleaved DNA can be subject to homologous recombination to repair the break. An relationship between DNA replication and restriction have been reported under conditions where only a single bacteriophage genome can enter a cell (39). For a Type I and some Type III restriction systems, phage restriction was alleviated by activation of the RecET homologous recombination pathway contributed by the Rac prophage (39,40) (Rec stands for recombination.) This suggested that the phage genome breakage could be repaired through the RecET pathway of homologous recombination. Because homologous recombination requires at least two copies of very similar DNAs, these results suggested that the DNA of the alleviated population of the phage was not cleaved by the restriction endonuclease until after replication of the region. In order to examine if McrBC-mediated restriction has a similar relation with homologous recombination machinery, bacteriophage lambda with or without the PvuII cytosine-N4-methyltransferase gene (which creates susceptible sites) was used to infect E. coli strains, with various genotypes with respect to homologous recombination, under the condition of single infection (plaque formation in the presence of excess bacterial cells within soft agar) (Figure 1). The bacteriophage carrying the methyltransferase gene was severely restricted in the wild type rec+ strain. This restriction is dependent on both McrBC activity (for cleavage) and M.PvuII activity (to create the modified substrate; Figure 1, 1st line pair, rec+ mcr+, and 2nd line pair, rec+ mcr−) as previously reported (21). The severe restriction was dramatically alleviated in a strain with an activated RecET pathway, in which RecE, RecT and Lar are active but do not compete with ExoV (Figure 1, recBC sbcA mcr+). The residual restriction is still dependent on McrB function (Figure 1, 4th line pair, recBC sbcA mcrB−). This restriction alleviation was dependent on the recE gene that encodes exonuclease VIII, an essential component of the pathway (Figure 1, recBC sbcA recE mcr+).
Figure 1.
McrBC-mediated restriction of a methyltransferase-carrying phage is alleviated by RecET-mediated homologous recombination. Lambda phage (arrow) with or without pvuIIM gene (box) on its genome (see ‘Materials and Methods’ section) was allowed to infect various E. coli strains under conditions of single infection (plaque assay). The average titer of phage without pvuIIM in the recBC sbcA mcrB::Km strain was taken as unity. Strains are: rec+ mcr+, AB1157; rec+ mcrB::Km, BNH3480; recBC sbcA mcr+, JC8679; recBC sbcA mcrB::Km, BMF1; recBC sbcA recE mcr+, BIK784; recBC mcr+, JC5519.
In the wild-type cell, the major homologous recombination pathway is mediated by RecBCD enzyme. The processive exonuclease activity of RecBCD degrades linear phage genome fragments after restriction cleavage, destroying the potential recombination substrate (41). As expected, the severity of this restriction is alleviated but not abolished by mutational inactivation of the RecBC enzyme (Figure 1, 6th line pair, recBC mcr+, versus 1st line pair, rec+ mcr+). Because RecBCD pathway and RecET pathway can repair chromosomal cleavage equally well, these effects on phage restriction are unlikely to be explained by chromosomal damage repair.
These results indicate that the bacteriophage genome restricted by McrBC can be repaired by homologous recombination machinery of RecET pathway. Because repair by homologous recombination needs multiple sets of homologous DNAs and because we have a condition of single DNA infection of a cell, this result suggests that a substantial fraction of restriction cleavage by the McrBC enzyme occurs after replication or during replication of the phage genome under our conditions.
A model DNA replication fork with epigenetic methylation on long branches
Although the above in vivo observation does not provide evidence for an association between McrBC-mediated cleavage and DNA replication, it reminded us of similar observations with Types I and III restrictions (39). Those observations led us to demonstrate that Type I restriction endonuclease cleaves a model DNA replication fork in vitro (38), proposed occurrence of such cleavage of DNA replication fork in vivo, and discussed its likely biological significance (9,38). Because there is similarity between McrBC and Types I and III systems (9), we naturally thought of a possibility of McrBC-mediated cleavage of methylated DNA around a DNA replication fork. Encouraged by these observations and considerations, we tested for McrBC action, if any, on a model DNA replication fork in vitro.
As a model replication fork, we prepared a Y-shaped DNA with long branches (Figure 2) as we did for a Type I restriction endonuclease (38). Because previous reports have shown that distance between their two recognition sites on a linear DNA substrate affects DNA cleavage activity of McrBC, with an optimum distance of 63 bp (36), our model fork was designed to carry methylation sites 63 bp away from the branch point [Figure 2(viii)]. A forked substrate with two methylated sites {eM63(++)} was prepared through nicking enzyme treatment to generate a dissociable single strand, dissociation of the strand leading to gap formation, and intermolecular annealing at the gaps as illustrated in Figure 2. It was labeled with 32P at the stem region.
A methyl-specific endonuclease can cleave a model DNA replication fork
When this Y form was treated with McrBC, a decrease of the substrate and appearance of new DNA species were observed in electrophoresis (Figure 3). We assigned a structure to each of these products, based on mobility, presence of label and sizes and labels of the denaturation products (Figures 3 and 4, Supplementary Data). The products that retained branched structure had lost the top arm (II), the bottom arm (III), or both (IV) (Figure 3A). Quantification revealed that the cleavages of the top arm (II) and the bottom arm (III) were equally efficient, while double cleavage (IV) was less efficient (Figure 3B). This is similar to the result obtained with linear substrates in which cleavage appeared to occur ∼30 bp from one or the other of the two collaborating modified elements (36). A possible product that can be produced by strand separation (V) was not shown to increase above the level in the starting preparation under this condition.
Figure 4.
Mapping cleavage sites. (A) A model replication fork (eM63++) cleaved by McrBC was subjected to polyacrylamide gel electrophoresis under denaturatingconditions (6M urea). (B) Cleavage sites. Open circle, 32P-label at 5′-end; Square, DNA methylation; Filled triangle, major product; white triangle, minor product; gray box, products corresponding to the smear. There are two labeled bands in the untreated lane, corresponding to the strand participating in the fork (293 nt), and not denaturated materials stacked in the well.
To our best knowledge, this is the first demonstration that a methyl-specific DNase can cleave a branched DNA, a model DNA replication fork.
Cleavage occurs near the branch point
In order to map cleavage sites on the model fork, products from the 32P-labeled substrate were subjected to polyacrylamide gel electrophoresis under a denaturing condition (Figure 4A). The result indicates that the majority of the cleavage events occurred between the methylated site and the branch point (Figure 4, triangles). The cleavage closest to the branch point was only ∼10 bp away from it. The predominant cleavage was ∼30 bp from the modified base as previously reported (35,36,42). As found also by Pieper et al. (42) there is a 10-bp cadence in cleavage position. We also found some cleavage in the distal region from the methylation site (Figure 4, gray box).
Cleavage requires two methylated arms
Next we examined whether methylation of both the arms is needed for the fork cleavage by the McrBC enzyme. In the case of a Type I restriction endonuclease, recognition of only one of the two arms is sufficient for cleavage at the branch point (38) (the branch point substitutes for the collision that otherwise depends on a second translocating complex). We prepared model replication forks carrying methylation in only one of the arms, or no methylation (see ‘Materials and Methods’ section). Unexpectedly, no cleavage by McrBC was observed with the substrates carrying only one methylation (Figure 5). These results indicate that the cleavage requires two methylated arms. Possible mechanism of the fork cleavage will be considered in Discussion together with its biological significance.
DISCUSSION
Cleavage of DNA replication fork by McrBC
In the present study, we showed that phage restriction by the McrBC enzyme is alleviated by homologous recombination under the single infection condition, and that the McrBC enzyme can cleave a branched DNA with two methylated recognition sites in vitro. Most features of this cleavage are compatible with observations made on linear substrates: two modified elements are required (34); cleavage occurs preferentially ∼30 bp from one modified element or the other (36,42); cleavages also occur further away with a spacing compatible with a 10 bp cadence (42).
In contrast to the enzyme's behavior on linear substrates, we did observe a significant population of products migrating in the position expected for a product in which both arms were cleaved off. With linear substrates, double-strand cleavage occurs near one modified element or the other but not both (36,42). Activity on a single-element substrate has been observed only when a physical block to translocation was also present (35). With this in mind, we considered that the doubly cut arms might result from sequential double-strand cleavage events, one mediated by collaboration between two elements and the second mediated by a single element combined with fork interaction in place of a translocation block. However, a substrate with a single modified element was refractory to cleavage, a result not compatible with sequential double-strand cleavage events (Figure 5).
At least four possible explanations for the doubly cut products come to mind. First, a concerted model: some population of substrate molecules is conformed so that four strands at once can be cleaved by collaborating complexes. This might be accomplished by a supercomplex with four McrC subunits. Second, a modified sequential model: a conformation might exist that allows two elements to collaborate in cleaving one strand near each site in one event, with the resulting nicked product competent as a substrate for a second event of the same kind. Third, a nicks-around-the-branch-point model: the first nicked product in the ‘modified sequential model’ might leave two arms with double-strand breaks at the discontinuous region of the branch point. The cut at 10 nt from the branch point (50 nt from the modified element) (Figure 4) can leave a product holding the arm with only 7 base pairs (or 10 base pairs for the bottom arm), which would be dissociated spontaneously or through a short branch migration. Fourth, a fork-migration model. In this, we envision a single cleavage event on a structure in which fork migration had resulted in a four-armed structure [the standard ‘chicken foot’ fork regression model (43)]. Cleavage mediated by interaction of two of the four arms to remove one arm, followed by re-migration to form a three-armed structure again, could make a structure with the same conformation as a double-cut substrate but with differently disposed sequence.
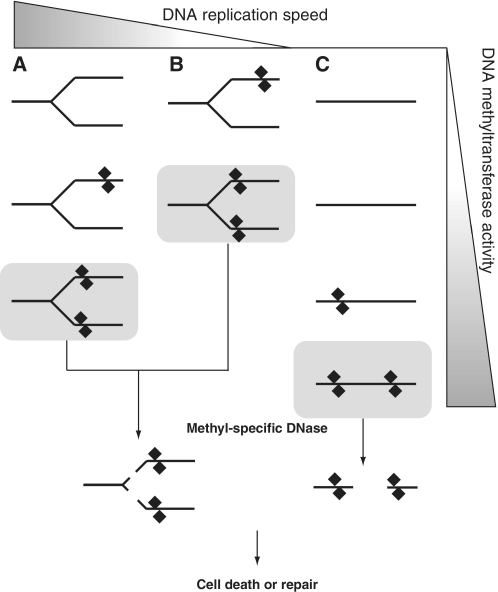
From the results obtained, we consider biological significance of the possible cleavage at a DNA replication fork. The first question is why the DNase needs two methylated arms for cleavage. One idea is that it would provide a sensitive response to acquired epigenetic DNA methylation (Figure 6). It is possible that the genomic region around DNA replication fork is more accessible to DNA methyltransfersases than the other regions not undergoing replication. For example, in E. coli, the chromosome is condensed with nucleoid-associated proteins (NAPs) such as H-NS and StpA (44–48). A methyltransferase being established in the cell, during the accumulation phase of expression may begin by modifying these regions (Figure 6A and B), rather than the other regions that would require higher concentration of the enzyme for the modification (Figure 6C). When the modification activity reaches a level that can provide two of the modified sister DNAs, the DNase may manifest the cleavage activity, followed by cell death or repair of the cleaved DNA. Slower progression of the fork would give the methyltransferase time to modify both the duplexes and, consequently, make the fork more susceptible to the cleavage (Figure 6B). When the modification activity was increased because of accumulation of the methyltransferase, it would cause chromosomal cleavage by the DNase even in the regions outside the replication forks (Figure 6C).
Figure 6.
DNA replication fork as a target for DNA methylation and methyl-specific DNase action (a hypothesis). (A) When a methyltransferase gene is established or switched on in a cell, DNA around the replication fork is expected to be methylated earlier than elsewhere because of its higher accessibility to the methyltransferases. Although methyl-specific DNases would not cleave a replication fork with DNA methylation only in one of the two daughter duplexes near the branch point, they would cleave a fork with DNA methylation in both the arms, which represents a higher level of DNA methylation. (B) Slower replication might increase the probability of occurrence of such two-arm methylation around a fork and, therefore, the DNase-mediated cleavage. (C) Unreplicating regions might be less sensitive to the methylation. The DNase-mediated cleavage there might require higher methyltransferase activity.
On the other hand, modification ahead of the fork would be converted after fork passage to a structure hemimethylated on both arms. McrBC will act on hemi-modified sites. Thus, immediately following fork passage, a symmetrically modified site would be sensitive to cleavage. This would be true even for very rare sites. It is of note that McrBC cleaves very poorly linear DNAs on which two recognition sites are spaced more than 3000 bp apart (36). Thus, fork passage would provide an opportunity to eliminate the new modification that might otherwise be insensitive to McrBC surveillance. This property would characterize any enzyme that preferentially cleaves two-site substrates and capable of cleaving hemi-modified sites. The recently characterized family of Type IV enzymes exemplified by MspJI (49) should behave this way, for example.
Such a response to an invading epigenetic system would decrease possibility of its establishment and spread to the siblings (9). It may be similar to abortion of virus multiplication through programmed death.
Branched DNA cleavage activity of McrBC (Type IV) and a Type I restriction endonuclease
We earlier reported that a Type I restriction endonuclease EcoR124 cleaves model DNA replication forks (38). Type I restriction endonucleases and Type IV restriction endonucleases, including the McrBC, have related biochemical activities (50). They likely translocate along DNA and cleave the DNA when they collide with another translocating enzyme molecule (35,50–52).
However, their fork cleavage reactions are different in products, positions and mechanisms.
EcoR124I cleavage needed an active recognition site only on one arm (38). However, McrBC needs active recognition sites on both arms (Figure 5).
The cleavage position by McrBC was observed 10–40 bp from the branch point (Figure 4), so the position of cleavage appears to be directed by the position of modification. In contrast, EcoR124I cleaved in the vicinity of the branch point, ∼5 bp (0–8 bp) from the branch point (38), so the position of cleavage appeared to be directed by the position of the translocation block.
Although the McrBC generated a product that lost both of the arms (Figure 3A), the EcoR124I did not generate such a product (38).
In addition, the McrBC did not show any DNA strand separation activity in the tested condition (Figure 3B), though the EcoR124I did show a strand separation activity (38).
The fork cleavage activity of EcoR124I is a DNA structure-dependent activity. Its likely action is as follows: from an active recognition site, it starts translocating along DNA. When it encounters a branch structure, it cleaves DNA there (38). McrBC activates DNA cleavage activity when it interacts with Lac repressor protein bound to a DNA (35). This result suggested that ‘road blocks’ on the DNA can promote cleavage. Our results, however, suggest that McrBC may cleave the branched DNA with a different mechanism. Probably, the cleavage may be triggered by an interaction, across the branch point, between two enzyme complexes translocating from the two different recognition sites. More detailed biochemical characterization would be required to examine this possibility.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Japan Society for the Promotion of Science (JSPS) (Grants-in-Aid for Scientific Research); the global Center of Excellence program ‘Genome Information Big Bang’ from Ministry of Education, Culture, Sports, Science and Technology (MEXT) (to I.K.); JSPS, Naito foundation, Kato foundation, Takeda foundation, Sumitomo foundation and ‘Grants-in-aid for Scientific Research (to N.H.); The Japan Science Society, Sasakawa Scientific Research Grant (to K.I.). Funding for open access charge: University of Tokyo.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank people who provided bacterial strains listed in Table 1. We thank Rich Roberts and anonymous reviewers for discussions of biological implications.
REFERENCES
- 1.Casadesus J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 3.Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marinus MG, Casadesus J. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 2009;33:488–503. doi: 10.1111/j.1574-6976.2008.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furuta Y, Abe K, Kobayashi I. Genome comparison and context analysis reveals putative mobile forms of restriction-modification systems and related rearrangements. Nucleic Acids Res. 2010;38:2428–2443. doi: 10.1093/nar/gkp1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srikhanta YN, Fox KL, Jennings MP. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat. Rev. Microbiol. 2010;8:196–206. doi: 10.1038/nrmicro2283. [DOI] [PubMed] [Google Scholar]
- 7.Beletskaya IV, Zakharova MV, Shlyapnikov MG, Semenova LM, Solonin AS. DNA methylation at the CfrBI site is involved in expression control in the CfrBI restriction-modification system. Nucleic Acids Res. 2000;28:3817–3822. doi: 10.1093/nar/28.19.3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Driscoll J, Fitzgerald GF, van Sinderen D. A dichotomous epigenetic mechanism governs expression of the LlaJI restriction/modification system. Mol. Microbiol. 2005;57:1532–1544. doi: 10.1111/j.1365-2958.2005.04769.x. [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa K, Fukuda E, Kobayashi I. Conflicts targeting epigenetic systems and their resolution by cell death: novel concepts for methyl-specific and other restriction systems. DNA Res. 2010;17:325–342. doi: 10.1093/dnares/dsq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi N, Naito Y, Handa N, Kobayashi I. A DNA methyltransferase can protect the genome from postdisturbance attack by a restriction-modification gene complex. J. Bacteriol. 2002;184:6100–6108. doi: 10.1128/JB.184.22.6100-6108.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivancic-Bacce I, Vlasic I, Cogelja-Cajo G, Brcic-Kostic K, Salaj-Smic E. Roles of PriA protein and double-strand DNA break repair functions in UV-induced restriction alleviation in Escherichia coli. Genetics. 2006;174:2137–2149. doi: 10.1534/genetics.106.063750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makovets S, Powell LM, Titheradge AJ, Blakely GW, Murray NE. Is modification sufficient to protect a bacterial chromosome from a resident restriction endonuclease? Mol. Microbiol. 2004;51:135–147. doi: 10.1046/j.1365-2958.2003.03801.x. [DOI] [PubMed] [Google Scholar]
- 13.Makovets S, Doronina VA, Murray NE. Regulation of endonuclease activity by proteolysis prevents breakage of unmodified bacterial chromosomes by type I restriction enzymes. Proc. Natl Acad. Sci. USA. 1999;96:9757–9762. doi: 10.1073/pnas.96.17.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cromie GA, Leach DR. Recombinational repair of chromosomal DNA double-strand breaks generated by a restriction endonuclease. Mol. Microbiol. 2001;41:873–883. doi: 10.1046/j.1365-2958.2001.02553.x. [DOI] [PubMed] [Google Scholar]
- 15.Blakely GW, Murray NE. Control of the endonuclease activity of type I restriction-modification systems is required to maintain chromosome integrity following homologous recombination. Mol. Microbiol. 2006;60:883–893. doi: 10.1111/j.1365-2958.2006.05144.x. [DOI] [PubMed] [Google Scholar]
- 16.Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995;267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 17.Handa N, Nakayama Y, Sadykov M, Kobayashi I. Experimental genome evolution: large-scale genome rearrangements associated with resistance to replacement of a chromosomal restriction-modification gene complex. Mol. Microbiol. 2001;40:932–940. doi: 10.1046/j.1365-2958.2001.02436.x. [DOI] [PubMed] [Google Scholar]
- 18.Kusano K, Naito T, Handa N, Kobayashi I. Restriction-modification systems as genomic parasites in competition for specific sequences. Proc. Natl Acad. Sci. USA. 1995;92:11095–11099. doi: 10.1073/pnas.92.24.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyko F, Ramsahoye BH, Kashevsky H, Tudor M, Mastrangelo MA, Orr-Weaver TL, Jaenisch R. Mammalian (cytosine-5) methyltransferases cause genomic DNA methylation and lethality in Drosophila. Nat. Genet. 1999;23:363–366. doi: 10.1038/15551. [DOI] [PubMed] [Google Scholar]
- 20.Kimura H, Suetake I, Tajima S. Exogenous expression of mouse Dnmt3 induces apoptosis in Xenopus early embryos. J. Biochem. 2002;131:933–941. doi: 10.1093/oxfordjournals.jbchem.a003184. [DOI] [PubMed] [Google Scholar]
- 21.Fukuda E, Kaminska KH, Bujnicki JM, Kobayashi I. Cell death upon epigenetic genome methylation: a novel function of methyl-specific deoxyribonucleases. Genome Biol. 2008;9:R163. doi: 10.1186/gb-2008-9-11-r163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raleigh EA, Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc. Natl Acad. Sci. USA. 1986;83:9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anton BP, Raleigh EA. Transposon-mediated linker insertion scanning mutagenesis of the Escherichia coli McrA endonuclease. J. Bacteriol. 2004;186:5699–5707. doi: 10.1128/JB.186.17.5699-5707.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heitman J, Model P. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J. Bacteriol. 1987;169:3243–3250. doi: 10.1128/jb.169.7.3243-3250.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waite-Rees PA, Keating CJ, Moran LS, Slatko BE, Hornstra LJ, Benner JS. Characterization and expression of the Escherichia coli Mrr restriction system. J. Bacteriol. 1991;173:5207–5219. doi: 10.1128/jb.173.16.5207-5219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bujnicki JM, Radlinska M. Molecular evolution of DNA-(cytosine-N4) methyltransferases: evidence for their polyphyletic origin. Nucleic Acids Res. 1999;27:4501–4509. doi: 10.1093/nar/27.22.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nobusato A, Uchiyama I, Kobayashi I. Diversity of restriction-modification gene homologues in Helicobacter pylori. Gene. 2000;259:89–98. doi: 10.1016/s0378-1119(00)00455-8. [DOI] [PubMed] [Google Scholar]
- 28.Nobusato A, Uchiyama I, Ohashi S, Kobayashi I. Insertion with long target duplication: a mechanism for gene mobility suggested from comparison of two related bacterial genomes. Gene. 2000;259:99–108. doi: 10.1016/s0378-1119(00)00456-x. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gast FU, Brinkmann T, Pieper U, Kruger T, Noyer-Weidner M, Pingoud A. The recognition of methylated DNA by the GTP-dependent restriction endonuclease McrBC resides in the N-terminal domain of McrB. Biol. Chem. 1997;378:975–982. doi: 10.1515/bchm.1997.378.9.975. [DOI] [PubMed] [Google Scholar]
- 31.Pieper U, Schweitzer T, Groll DH, Gast FU, Pingoud A. The GTP-binding domain of McrB: more than just a variation on a common theme? J. Mol. Biol. 1999;292:547–556. doi: 10.1006/jmbi.1999.3103. [DOI] [PubMed] [Google Scholar]
- 32.Pieper U, Pingoud A. A mutational analysis of the PD…D/EXK motif suggests that McrC harbors the catalytic center for DNA cleavage by the GTP-dependent restriction enzyme McrBC from Escherichia coli. Biochemistry. 2002;41:5236–5244. doi: 10.1021/bi0156862. [DOI] [PubMed] [Google Scholar]
- 33.Bujnicki JM, Rychlewski L. Grouping together highly diverged PD-(D/E)XK nucleases and identification of novel superfamily members using structure-guided alignment of sequence profiles. J. Mol. Microbiol. Biotechnol. 2001;3:69–72. [PubMed] [Google Scholar]
- 34.Sutherland E, Coe L, Raleigh EA. McrBC: a multisubunit GTP-dependent restriction endonuclease. J. Mol. Biol. 1992;225:327–348. doi: 10.1016/0022-2836(92)90925-a. [DOI] [PubMed] [Google Scholar]
- 35.Panne D, Raleigh EA, Bickle TA. The McrBC endonuclease translocates DNA in a reaction dependent on GTP hydrolysis. J. Mol. Biol. 1999;290:49–60. doi: 10.1006/jmbi.1999.2894. [DOI] [PubMed] [Google Scholar]
- 36.Stewart FJ, Raleigh EA. Dependence of McrBC cleavage on distance between recognition elements. Biol. Chem. 1998;379:611–616. [PubMed] [Google Scholar]
- 37.Arifuzzaman M, Maeda M, Itoh A, Nishikata K, Takita C, Saito R, Ara T, Nakahigashi K, Huang HC, Hirai A, et al. Large-scale identification of protein–protein interaction of Escherichia coli K-12. Genome Res. 2006;16:686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishikawa K, Handa N, Kobayashi I. Cleavage of a model DNA replication fork by a Type I restriction endonuclease. Nucleic Acids Res. 2009;37:3531–3544. doi: 10.1093/nar/gkp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Handa N, Kobayashi I. Type III restriction is alleviated by bacteriophage (RecE) homologous recombination function but enhanced by bacterial (RecBCD) function. J. Bacteriol. 2005;187:7362–7373. doi: 10.1128/JB.187.21.7362-7373.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kushner SR, Nagaishi H, Clark AJ. Isolation of exonuclease VIII: the enzyme associated with sbcA indirect suppressor. Proc. Natl Acad. Sci. USA. 1974;71:3593–3597. doi: 10.1073/pnas.71.9.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Handa N, Ichige A, Kusano K, Kobayashi I. Cellular responses to postsegregational killing by restriction-modification genes. J. Bacteriol. 2000;182:2218–2229. doi: 10.1128/jb.182.8.2218-2229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pieper U, Groll DH, Wunsch S, Gast FU, Speck C, Mucke N, Pingoud A. The GTP-dependent restriction enzyme McrBC from Escherichia coli forms high-molecular mass complexes with DNA and produces a cleavage pattern with a characteristic 10-base pair repeat. Biochemistry. 2002;41:5245–5254. doi: 10.1021/bi015687u. [DOI] [PubMed] [Google Scholar]
- 43.Michel B, Flores MJ, Viguera E, Grompone G, Seigneur M, Bidnenko V. Rescue of arrested replication forks by homologous recombination. Proc. Natl Acad. Sci. USA. 2001;98:8181–8188. doi: 10.1073/pnas.111008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dame RT. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 45.Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 46.Schroder O, Wagner R. The bacterial DNA-binding protein H-NS represses ribosomal RNA transcription by trapping RNA polymerase in the initiation complex. J. Mol. Biol. 2000;298:737–748. doi: 10.1006/jmbi.2000.3708. [DOI] [PubMed] [Google Scholar]
- 47.Lithgow JK, Haider F, Roberts IS, Green J. Alternate SlyA and H-NS nucleoprotein complexes control hlyE expression in Escherichia coli K-12. Mol. Microbiol. 2007;66:685–698. doi: 10.1111/j.1365-2958.2007.05950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoebel DM, Free A, Dorman CJ. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology. 2008;154:2533–2545. doi: 10.1099/mic.0.2008/020693-0. [DOI] [PubMed] [Google Scholar]
- 49.Zheng Y, Cohen-Karni D, Xu D, Chin HG, Wilson G, Pradhan S, Roberts RJ. A unique family of Mrr-like modification-dependent restriction endonucleases. Nucleic Acids Res. 2010;38:5527–5534. doi: 10.1093/nar/gkq327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dryden DTF, Murray NE, Rao DN. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 2001;29:3728–3741. doi: 10.1093/nar/29.18.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seidel R, van Noort J, van der Scheer C, Bloom JG, Dekker NH, Dutta CF, Blundell A, Robinson T, Firman K, Dekker C. Real-time observation of DNA translocation by the type I restriction modification enzyme EcoR124I. Nat. Struct. Mol. Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 52.Ellis DJ, Dryden DTF, Berge T, Edwardson JM, Henderson RM. Direct observation of DNA translocation and cleavage by the EcoKI endonuclease using atomic force microscopy. Nat. Struct. Biol. 1999;6:15–17. doi: 10.1038/4882. [DOI] [PubMed] [Google Scholar]
- 53.Grant SG, Jessee J, Bloom FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl Acad. Sci. USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bachmann BJ. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt FC, editor. Escherichia coli and Salmonella. Washington DC: American Society for Microbiology; 1987. pp. 1190–1219. [Google Scholar]
- 55.Willetts NS, Clark AJ. Characteristics of some multiply recombination-deficient strains of Escherichia coli. J. Bacteriol. 1969;100:231–239. doi: 10.1128/jb.100.1.231-239.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gillen JR, Willis DK, Clark AJ. Genetic analysis of the RecE pathway of genetic recombination in Escherichia coli K-12. J. Bacteriol. 1981;145:521–532. doi: 10.1128/jb.145.1.521-532.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Handa N, Ohashi S, Kusano K, Kobayashi I. Chi-star, a chi-related 11-mer sequence partially active in an E. coli recC1004 strain. Genes Cells. 1997;2:525–536. doi: 10.1046/j.1365-2443.1997.1410339.x. [DOI] [PubMed] [Google Scholar]
- 58.Lunnen KD, Barsomian JM, Camp RR, Card CO, Chen SZ, Croft R, Looney MC, Meda MM, Moran LS, Nwankwo DO, et al. Cloning type-II restriction and modification genes. Gene. 1988;74:25–32. doi: 10.1016/0378-1119(88)90242-9. [DOI] [PubMed] [Google Scholar]
- 59.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.