Abstract
Ras proteins control many aspects of eukaryotic cell homeostasis by switching between active (GTP-bound) and inactive (GDP-bound) conformations, a reaction catalyzed by GTPase exchange factors (GEF) and GTPase activating proteins (GAP) regulators, respectively. Here, we show that the complexity, measured as number of genes, of the canonical Ras switch genetic system (including Ras, RasGEF, RasGAP and RapGAP families) from 24 eukaryotic organisms is correlated with their genome size and is inversely correlated to their evolutionary distances from humans. Moreover, different gene subfamilies within the Ras switch have contributed unevenly to the module’s expansion and speciation processes during eukaryote evolution. The Ras system remarkably reduced its genetic expansion after the split of the Euteleostomi clade and presently looks practically crystallized in mammals. Supporting evidence points to gene duplication as the predominant mechanism generating functional diversity in the Ras system, stressing the leading role of gene duplication in the Ras family expansion. Domain fusion and alternative splicing are significant sources of functional diversity in the GAP and GEF families but their contribution is limited in the Ras family. An evolutionary model of the Ras system expansion is proposed suggesting an inherent ‘decision making’ topology with the GEF input signal integrated by a homologous molecular mechanism and bifurcation in GAP signaling propagation.
INTRODUCTION
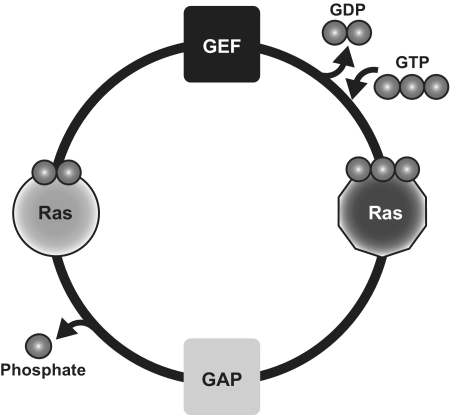
Ras proteins regulate many eukaryotic cellular processes, including cell growth, differentiation and survival (1,2). Ras signaling plays a major role in the homeostasis of multiple cellular pathways and alteration of its functionality leads to different pathologies, with cancer being the most typical. For example, 20% of all human tumors contain oncogenic mutations of Ras (3,4). The Ras switch works by alternating the activation state of the small guanine nucleotide binding protein Ras (active: bound to GTP; inactive: bound to GDP). Guanine nucleotide exchange factors (GEF) promote GDP release from Ras and favor GTP binding, whereas GTPase activator proteins (GAP) enhance the intrinsic GTPase activity of Ras that converts GTP into GDP (Figure 1).
Figure 1.
In the canonical cycle, Ras is bound to GDP (symbolized by two phosphate groups) during the basal state. GEF catalyzes the exchange between GDP and GTP, activating Ras that can bind to Ras effectors and perform the biological activity. GAP enhances the intrinsic GTPase activity of Ras, which returns to the inactive state. Disruption of any of these steps may lead to several pathologies, including cancer.
Ras is part of a large super-family of small GTPases that includes Rho, Rab and Arf (5). In this work, we focused on the Ras family of proteins and its regulators, and define the canonical Ras switch as composed by three different catalytic activities: Ras, GEF and GAP. GEF and GAP activities have been related to a variety of non-homologous sequence domains (6–8). We focused on families that perform Ras-specific regulation, including one GEF family (RasGEF) and two structurally differing GAP families (RapGAP and RasGAP) (9).
The Ras switch regulatory module has been studied from an evolutionary perspective before, but always with the different gene families in isolation and with a limited number of species (5,10–13). In this study, we consider the evolution of the canonical Ras module, composed by the gene families Ras, RasGEF, RasGAP and RapGAP, in 24 eukaryotic organisms with special emphasis on the Metazoan kingdom.
MATERIALS AND METHODS
Genome sequence and phylogenetic distance
Protein sequences for 24 organisms were included: 18 were downloaded from the EnsEMBL project (version 54) (14) and the other 6 from specialized databases (Supplementary Table S1). For EnsEMBL sequences, only translations from protein-coding genes were used and putative pseudogenes were excluded from the analysis. Divergence times were compiled from different sources, including fossil records and molecular distance estimations (15–17). When different estimations were obtained the average value was used (see Supplementary Figure S10 for a comparison between the two main sources of divergence time estimations and the average value used in this study).
Sequence detection and analysis
Protein family members for Ras, GAP and GEF were identified using Hidden Markov Model (HMM) profiles from the Pfam database (version 23) of protein domains. Domain searches were performed with the software package HMMER (version 2.3.2) (18). Pfam models for domains with identifiers Ras (PF00071), Rap_GAP (PF02145), RasGAP (PF00616) and RasGEF (PF00617) were used. In this work, we refer to Ras, RapGAP, RasGAP and RasGEF for both the domains and the gene families indistinctly. The models were used to search the gene translations, and sequences were collected on the basis of an E-value cutoff of 1E-02 (18). Ras human sequences obtained from Uniprot were used to identify and separate the Ras family sequences from the other families (Rho, Rab, Arf, etc.). For each organism, the detected sequences were combined with the human ones and an alignment and phylogenetic tree were constructed (see below). The branches containing the human orthologs were then selected. Sequences were filtered to keep one sequence per gene; therefore, multiple transcript were disregarded (the gene with the longest transcript was selected). Linear regression model fits and graphics were performed using the statistical software R (19).
Alignments and phylogenetic trees
Multiple sequence alignments were generated with ClustalW (20), and phylogenetic trees were constructed using the Neighbor-Joining (NJ) method implemented in the software Quicktree (21). Tree topology reliability was assessed with the bootstrap method using 1000 replications. TreeDyn (22) was used to visualize and annotate the trees. Trees were annotated with the organism name, gene id, symbol and the quality of the current gene model (only for EnsEMBL genes). Color squares are used to summarize clades: mammals (violet), Sauria (birds and lizard, orange), fish (blue), insects (green) and fungi (yellow). Outgroups Monosiga brevicollis (choanoflagellate) and Dictiostelium discoideum (slime mold) are colored in gray, whereas all other intermediate species are in pink. The domain composition for each sequence was computed (see below) and presence of the domain is indicated with black squares (gray squares mean the domain is not present). Bootstrap percentage values are indicated at each node, with values above 80% colored in red.
Gene classification into subfamilies
Sequences were classified into comprehensive subfamilies when possible, accounting for known annotations, presence of protein domains, and bootstrap support. The subfamilies were supported in most cases by high bootstrap values. For each subfamily, we counted the number of genes.
Exon, splice variants, domain and architecture distributions
The number of exons per gene was obtained for EnsEMBL genomes with the EnsEMBL Perl API and the distribution of exons was computed. Domains were detected with the program hmmpfam (HMMER) against the Pfam database (version 23). The domain architecture was computed for non-overlapping domains. Domains and architectures were separated by species and the frequencies computed. The splice variant sequences in humans were based on data from EnsEMBL version 41 in the ASTD database (23).
Similarity of Ras module domains and architecture profiles between species
Ras, RasGEF, RasGAP and RAPGAP domain and architecture occurrence matrices were joined into two matrices, respectively, for performing profile similarity measures (Supplementary Material). The Euclidean distance (ED) was used to measure the distance between pairs of profiles.
RESULTS AND DISCUSSION
Ras switch module genetic expansion in eukaryotes
Sequences for each gene family were detected by using HMM profiles. A broad spectrum of the eukaryote phylogeny was covered, with 24 organisms (Figure 2 and Supplementary Table S1) including 18 species in the Metazoan kingdom, three fungi, M. brevicollis (an unicellular protist considered the closest relative to the Metazoans) and D. discoideum (slime mold, out-group of the fungi/metazoan clade). The plant Arabidopsis thaliana was included as a negative control since plants have Rho and Rab homologs but not Ras (24).
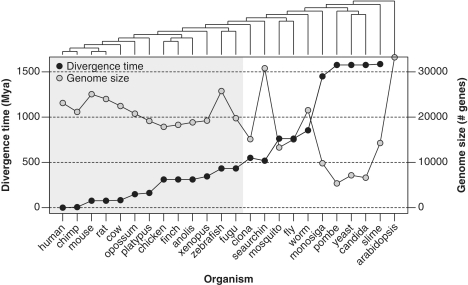
Figure 2.
Phylogenetic relationship for all the species used in this work. In the left Y-axis it is represented the species divergence times (MYA) from human obtained from different sources (combined and averaged), including fossil and molecular distance estimations. In the right Y-axis, it is represented the species genome size, considered as the number of estimated protein-coding genes.
All the gene families in the Ras module are broadly distributed over all the species except Arabidopsis. This co-absence of all Ras module’s families in Arabidopsis, which contains other Ras homologs such as Rho and Rab genes, supports the modular consistency of the Ras switch regulatory system defined in this work (Figure 3). RapGAP orthologs are missing in the budding yeast Saccharomyces cerevisiae. However, the other two fungi, including another budding yeast (Candida), have RapGAP homologs, suggesting that this functionality was lost in S. cerevisiae after the split of the fungi clade. Interestingly, S. cerevisiae has more RasGAP genes than the other fungi, suggesting that compensation of the RapGAP function is performed by some of these extra proteins.
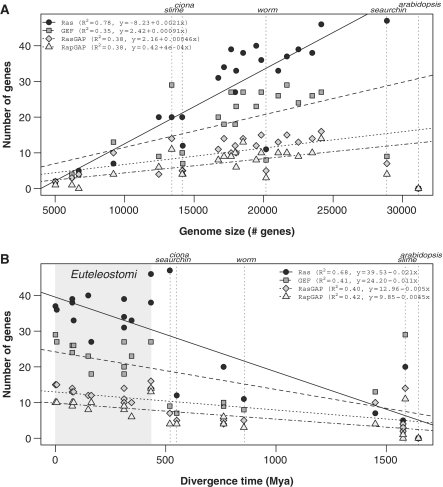
Figure 3.
(A) The number of detected genes versus the total number of protein-coding genes (genome size) for Ras, GEF, RasGAP and RapGAP. (B) The number of detected genes versus the phylogenetic distance to humans. The location of the Euteleostomi clade is highlighted with gray. Fitted linear models are for the entire data set except Arabidopsis, Ciona, Sea urchin and slime mold.
Plotting the number of sequences against the genome size, measured as the total number of protein coding genes, reveals a linear trend (Figure 3A and Supplementary Figure S1A). The regression models reflect highly significant correlation values (Ras: R2 = 0.78, P = 2.74E-08; RasGEF: R2 = 0.35, P = 2.78E-03; RasGAP: R2 = 0.38, P = 1.67E-03; RapGAP: R2 = 0.38, P = 1.68E-03). A few species (worm, sea urchin, Ciona and slime mold) diverge from this tendency. Removing them greatly improves the correlation with the linear model (Ras: R2 = 0.92, P = 9.26E-11; RasGEF: R2 = 0.86, P = 9.50E-09; RasGAP: R2 = 0.79, P = 4.33E-07; RapGAP: R2 = 0.82, P = 1.05E-07—compare regression lines in Figure 3A and Supplementary Figure S1A). This improvement in the correlation can be explained in part by the uncertainty in the total number of genes, which can significantly affect draft genomes like that of sea urchin and Ciona. However, for the slime mold, this divergence could be explained in terms of alternative evolutionary pathways (see below).
The slope of the linear models above is a measure of the expansion rate. Assuming that the expansion of these families is mainly due to gene duplication (25), the expansion rate would be a measure of the duplication rate. The fastest expanding family is Ras, with a duplication rate of 0.23% (slope 2.3E-03), followed by GEF, with a duplication rate of 0.14% (slope 1.4E-03). Both RasGAP and RapGAP have the smallest duplication rates (slopes of 6.8E-04E-04 and 6.0E-04, respectively). From a functional perspective, because RasGAP and RapGAP perform the same catalytic activity on Ras, we can calculate a combined duplication rate for GAP (slope of 1.3E-03) and arrive at a value closer to that of GEF.
Another measure, the Relative Duplication Rate (RDR), indicates how fast a gene family expands relative to the others. Taking Ras as the baseline, the RDR is 0.61 for GEF, 0.29 for RasGAP and 0.26 for RapGAP. If RasGAP and RapGAP are considered together, an RDR of 0.55 is obtained. These results indicate that the Ras family expands almost two times faster than the GEF and GAP families. The expansion rate of GEF and GAP (considering RasGAP and RapGAP together) is very similar. This general trend is maintained consistently in the individual species, except for GEF and RasGAP in Monosiga, and GEF in slime mold and Candida (Supplementary Table S2), where the number of genes in the regulatory families is greater than in Ras.
These findings show a bias in the duplication rate of the Ras family, suggesting that Ras leads the expansion of the module. In accordance with this, in all species except the slime mold, Candida and Monosiga, the number of Ras genes is greater than the number of GEF genes, which is, respectively, greater than the number of GAP genes (Figure 3A).
When we consider the metazoan clade, the Ras switch complexity (measured as the number of genes) is inversely correlated with the phylogenetic distance to humans (Figure 3B and Supplementary Figure S1B). In other words, species closer to humans have more paralog sequences when compared with species phylogenetically distant, evidencing increased complexity in their signaling pathways, with alternative regulatory and functional ramifications. On the other hand, this behavior does not apply when considering other eukaryote clades. Plants, which do not have Ras proteins, or fungi, where the gene numbers have not changed since they diverged about 1500 MYA, are both good examples. On the other hand, D. discoideum, which diverged probably more than 1700 MYA, have a number of genes similar to more complex organisms like insects, which is evidence of complex regulatory pathways probably related to the multicellular stage in the life cycle of this species (26).
Subfamily contribution to the Ras module expansion
Our results indicate a linear expansion of the Ras module correlated to genome protein-coding complexity and evolutionary time. However, some subfamilies may have contributed differently, depending on functional constraints and relevance to speciation events. To examine this possibility, we computed phylogenetic trees for all the families and classified the different groups of genes into subfamilies. This was done taking into account the annotation of known genes, the bootstrap confidence value in the tree, the protein domain composition, and species-dependent annotation (Supplementary Figures S2–S5 for annotated trees of Ras, RasGEF, RasGAP and RapGAP). Subfamilies were supported in most instances by high bootstrap values indicating the statistical reliability of their phylogenetic classification. The number of genes in each subfamily was counted for each clade and the results are shown in Table 1. To avoid errors due to missing sequences in draft genomes, presence in some but not all of the species within a clade, including mammals, birds/reptiles, fish and fungi, is considered a positive. Monosiga, slime mold and yeast sequences frequently showed weak phylogenetic relationships (i.e. low bootstrap values or domain composition similarity, see ‘Materials and Methods’ section) and therefore were difficult to classify into the general subfamilies. Consequently, the number of sequences for these species represented in Table 1 is very low or zero, in spite of the existence of quite a lot of sequences in some cases. This behavior suggests the existence of specific regulatory pathways for the Ras module system in these organisms.
Table 1.
Number of genes in each subfamily per species/clades
| M | R/B | X | F | Ci | Su | I | W | Fu | Mo | Sm | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RAPGAP | |||||||||||
| TSC2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 |
| GARNL1 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| SIPA1 | 4 | 4 | 1 | 4 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| RAP1GAP | 2 | 2 | 1 | 2 | 1 | 0 | 1 | 1 | 0 | 0 | 0 |
| GARNL3 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| RASGAP | |||||||||||
| IQGAP | 3 | 3 | 3 | 3 | 1 | 1 | 0 | 0 | 1 | 1 | 1 |
| NF1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 |
| RASA | 4 | 4 | 3 | 4 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| SYNGAP | 4 | 4 | 1 | 4 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| RASA1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| GAPVD1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| RASGEF | |||||||||||
| PLCE1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| RASGRF | 2 | 2 | 2 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| KNDC1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| RASGEF1 | 3 | 3 | 2 | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 0 |
| RAPGEF1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| RGL | 5 | 4 | 3 | 4 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| RALGPS | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| RAPGEF2/6 | 2 | 2 | 1 | 2 | 1 | 0 | 1 | 1 | 0 | 0 | 0 |
| RAPGEF3/4 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| RAPGEF5 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RAPGEFL1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RASGRP | 4 | 4 | 4 | 3 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| SOS | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 0 |
| RAS | |||||||||||
| RASL10 | 2 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| RASD | 2 | 2 | 2 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| SSR2 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| NKIRAS | 2 | 2 | 2 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| REM | 4 | 4 | 4 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| RGK | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 0 |
| RASL11 | 2 | 2 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RERG | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| RASL12 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| AGAP | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| RERGL | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RHEB | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ERAS | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RAL | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 0 |
| RAP1 | 2 | 2 | 2 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| RAP2 | 3 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| H/K/NRAS | 3 | 3 | 3 | 3 | 0 | 1 | 1 | 1 | 0 | 1 | 0 |
| RRAS | 2 | 2 | 1 | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| MRAS | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| RIT | 2 | 2 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| DIRAS | 3 | 3 | 3 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
M: mammal; R/B: reptile/bird; X: xenopus; F: fish; Ci: ciona; Su: seaurchin; I: insect; W: worm; Fu: fungi; Mo: Monosiga; Sm: slime mold. In bold is highlighted the clades showing a duplication event compared with the parent clades. This table shows only gene sequences consistently classified into the different subgroups, where classification consistency is measured as a combination of bootstrapping value and domain composition. The total number of sequences in each species/clade is presented in Figure 3.
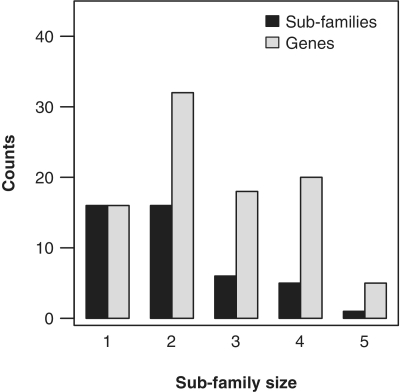
The numeric analysis of Table 1 shows that 16 subfamilies (about 36% of the total) have contributed very little to the module expansion with only a maximum of one gene copy per organism (Figure 4). Among the multigene subfamilies, 36% show two gene copies per species and about 27% are highly expanded subfamilies, which are defined in this work as subfamilies with three or more gene members in any clade. The maximum number of genes per subfamily observed is five (Figure 4). This demonstrates that some subfamilies have contributed differently to the overall duplication rates observed in the Ras module, indicating disparate evolutionary roles and increasing cellular signaling complexity.
Figure 4.
Number of genes and subfamilies versus subfamily size. For each subfamily size class (i.e. maximum number of genes in the subfamily; x-axis), we counted the number of subfamilies with that size (black rectangles), and the number of total sequences in the subfamily (gray rectangles).
Fungi, slime mold and Monosiga genes highly diverge from their Metazoan homologs. IQGAP is the only subfamily whose domain composition and bootstrap values indicate a consistent distribution among all clades. Furthermore, slime mold genes cluster into consistent, independent subfamilies, particularly true for RasGEF, suggesting the exploration of alternative functional pathways not present in the Metazoan clade.
Functional analysis of the expanded subfamilies
To avoid excessive description, only some subfamily expansions and speciation processes involving whole main clades are discussed below. Alternative gene names are provided in parenthesis. Unfortunately, many genes in the Ras switch families show poor functional annotations or are still uncharacterized. Despite the lack of functional information, it seems that many of the gene subfamily expansions and speciation processes in the Ras switch are related to exploration of new functional niches specific to evolution of pluricellular organisms.
For example, the RapGAP subfamily SIPA1 (maximum of four genes in mammals, birds and fish, Table 1), the RasGAP subfamilies such as IQGAP (three genes) and SYNGAP (four genes) and the Ras subfamily DIRAS (three genes) have genetically and functionally expanded presenting genes involved in brain and nervous system development (27–32). On the other hand, the expanded RasGAP subfamily RASA (four genes) and the RasGEF RASGRP (four genes) contain genes involved in the developmental system as well as the adaptive immune system (33–35).
Subfamily expansions linked to clade-specific speciation processes appear in the Ras and RasGEF families (Supplementary Figures S7 and S8). In Ras, the RGK subfamily includes four protein-coding genes: GEM, RRAD (RAD1), REM1 (REM) and REM2; that function as potent inhibitors of voltage-dependent Ca2+ channels, some of them acting as Rho-like cytoskeleton regulators (36). The insect Rgk sequences appear as an independent branch of the animal’s genes (except Caenorhabditis elegans, which is missing), suggesting an independent expansion of the Rgk sequences in insects. However, specific sequence variation, such as absence of a typical motif found in mammalian RGKs [DXWEX in G3; (36)] and lack of strong bootstrap support challenge their classification. The ERAS subfamily in Ras is restricted to the Theria clade (placental mammals and marsupials) and in mice, Eras (also known as HRAS2 in humans) is expressed in embryonic stem cells where it promotes proliferation and tumorigenicity in vitro (37).
Four RasGEF subfamilies show strong evidence of clade speciation processes: RAPGEFL1, RAPGEF5, PLCE1 (PLCE, PPLC) and KNDC1 (RASGEF2, VKIND). RAPGEFL1 and RAPGEF5 seem to be exclusive innovations in the Euteleostomi clade (fish, amphibians, birds and mammals in this work), with RAPGEFL1 proteins being associated with nervous system development (38,39). PLCE1 and KNDC1, also broadly present in the Euteleostomi species, show putative orthologs in the unicellular marine organism M. brevicollis, although with important domain rearrangements in PLCE1. PLCE1 is also present in worm (plc-1) although has been classified into the wrong group. These results point to a selective elimination of PLCE1 and KNDC1 in insects, Ciona and sea urchin ancestor species. PLCE1 may play critical roles in the glomerular development of the kidney, as mutations in PLCE1 are linked to familial nephrotic syndrome (40). In mice, Kndc1 is highly expressed in the cerebellum, where it is restricted to the Purkinje cells (41) and controls dendrite growth by linking Ras with Map2, a protein that is associated with microtubules (42).
Genetic variation of the Ras module orthologs system in mammals
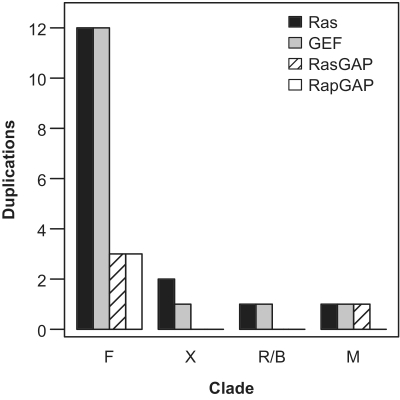
We estimated the number of duplication events presumably driving the appearance of new Ras switch genes in the eukaryote phylogeny (Figure 5). The number of new duplications related to emergence of different clades in the eukaryotic phylogenetic tree shows a drastic reduction after the Euteleostomi split and suggests an early stabilization of the Ras module expansion in the subsequent clades including mammals.
Figure 5.
Number of duplications for Ras, GEF, RasGAP and RapGAP relative to specific clades (F: fish; X: amphibian/xenopus; R/B: reptile/birds; M: mammals).
To reduce bias due to uncertainty in genome assembly and gene prediction in draft genomes, we wanted to confirm the stabilization hypothesis focusing on curated orthologous genes in high-quality annotated mammalian genomes, such as human, mouse and rat. Orthologs in other high-quality annotated organisms, including zebrafish, fly and worm, are also presented for comparison (Table 2). The number and distribution of orthologs indicate very little genetic variation in the mammalian species. DIRAS3 (NOEY2), RGL4 (RGR) and RASA4 (CAPRI, GAPL) are the only three groups over a total of 70 groups of orthologous genes that show variation in the number or distribution of genes within the mammalian sample. This implies that 96% of the Ras switch orthologs system is conserved among all the mammalian species. The observed genetic variations, DIRAS3 and RGL4, correspond actually to duplications related to speciation event as described earlier. Both genes seem to be missing in rodents, but present in primates. There is also evidence for presence of both genes in other mammals, suggesting that these genes may have been lost in the rodent lineage. This co-dependency between DIRAS3 and RGL4 presence/absence suggests that RGL4 might be a regulator of DIRAS3 function. The RASA4 ortholog is duplicated in the human genome and although both genes, RASA4 and RASA4B, map to chromosome 7, RASA4B seems to be a truncated version of RASA4 (and so it is annotated as a pseudogene).
Table 2.
Number of Ras switch homologous genes detected in human, mouse, rat, zebrafish, fly and worm genomes
| Family | Subfamily | Gene | H | M | R | Z | F | W |
|---|---|---|---|---|---|---|---|---|
| Ras | RASL10 | RASL10A | 1 | 1 | 1 | 1 | 1 | 0 |
| RASL10B | 1 | 1 | 1 | 0 | ||||
| RASD | RASD1 | 1 | 1 | 1 | 1 | 1 | 0 | |
| RASD2 | 1 | 1 | 1 | 2 | ||||
| SSR2 | SSR2 | 0 | 0 | 0 | 2 | 2 | 1 | |
| NKIRAS | NKIRAS2 | 1 | 1 | 1 | 1 | 1 | 0 | |
| NKIRAS1 | 1 | 1 | 1 | 1 | ||||
| RGK | REM1 | 1 | 1 | 1 | 1 | 0 | 0 | |
| RRAD | 1 | 1 | 1 | 1 | ||||
| GEM | 1 | 1 | 1 | 1 | ||||
| REM2 | 1 | 1 | 1 | 1 | ||||
| RGK | 0 | 0 | 0 | 0 | 3 | 0 | ||
| RASL11 | RASL11A | 1 | 1 | 1 | 1 | 0 | 0 | |
| RASL11B | 1 | 1 | 1 | 1 | ||||
| RERG | RERG | 1 | 1 | 1 | 1 | 0 | 0 | |
| RASL12 | RASL12 | 1 | 1 | 1 | 1 | 0 | 0 | |
| RERGL | RERGL | 0 | 0 | 1 | 4 | 1 | 0 | |
| RHEB | RHEB | 1 | 1 | 0 | 1 | 1 | 0 | |
| RHEBL1 | 1 | 1 | 1 | 0 | 0 | 0 | ||
| ERAS | ERAS | 1 | 1 | 1 | 0 | 0 | 0 | |
| RAL | RALB | 1 | 1 | 1 | 2 | 1 | 1 | |
| RALA | 1 | 1 | 1 | 2 | ||||
| RAP1 | RAP1B | 2 | 1 | 1 | 1 | 1 | 1 | |
| RAP1A | 1 | 1 | 1 | 1 | ||||
| RAP2 | RAP2C | 1 | 1 | 1 | 1 | 1 | 1 | |
| RAP2A | 1 | 1 | 1 | 1 | ||||
| RAP2B | 1 | 1 | 1 | 1 | ||||
| H/K/NRAS | HRAS | 1 | 1 | 2/1 | 2 | 1 | 1 | |
| KRAS | 1 | 1 | 1 | 0 | ||||
| NRAS | 1 | 1 | 1 | 2 | ||||
| RRAS | RRAS2 | 1 | 1 | 1 | 1 | 1 | ||
| RRAS | 1 | 1 | 1 | 1 | 0 | |||
| MRAS | MRAS | 1 | 1 | 1 | 2 | 0 | 1 | |
| RIT | RIT2 | 1 | 1 | 1 | 0 | 1 | 0 | |
| RIT1 | 1 | 1 | 1 | 1 | ||||
| DIRAS | DIRAS2 | 1 | 1 | 1 | 2 | 1 | 1 | |
| DIRAS1 | 1 | 1 | 1 | 2 | ||||
| DIRAS3 | 1 | 0 | 0 | 0 | 0 | 0 | ||
| RapGAP | TSC2 | TSC2 | 1 | 1 | 1 | 0 | 1 | 0 |
| GARNL1 | C20ORF74 | 1 | 1 | 1 | 1 | 1 | 1 | |
| GARNL1 | 1 | 1 | 1 | 1 | ||||
| SIPA1 | SIPA1L3 | 1 | 1 | 0/1 | 1 | 0 | 1 | |
| SIPA1L1 | 1 | 1 | 1 | 1 | ||||
| SIPA1L2 | 1 | 1 | 1 | 1 | ||||
| SIPA1 | 1 | 1 | 1 | 1 | ||||
| RAP1GAP | GARNL4 | 1 | 1 | 1 | 2 | 1 | 1 | |
| RAP1GAP | 1 | 1 | 1 | 1 | 1 | |||
| GARNL3 | GARNL3 | 1 | 1 | 1 | 1 | 0 | 0 | |
| RasGAP | IQGAP | IQGAP1 | 1 | 1 | 1 | 1 | 0 | 0 |
| IQGAP2 | 1 | 1 | 0 | 1 | 0 | 0 | ||
| IQGAP3 | 1 | 1 | 1 | 0 | 0 | 0 | ||
| NF1 | NF1 | 1 | 1 | 1 | 2 | 1 | 0 | |
| RASA | RASA2 | 1 | 1 | 1 | 1 | 1 | 0 | |
| RASA3 | 1 | 1 | 1 | 1 | 0 | |||
| RASA4 | 2 | 1 | 1 | 1 | 0 | 0 | ||
| RASAL1 | 1 | 1 | 1 | 0 | 0 | 0 | ||
| SYNGAP | RASAL2 | 1 | 1 | 1 | 1 | 1 | 0 | |
| DAB2IP | 1 | 1 | 1 | 2 | ||||
| SYNGAP1 | 1 | 1 | 1 | 2 | ||||
| RASAL3 | 1 | 1 | 1 | 0 | ||||
| RASA1 | RASA1 | 1 | 1 | 1 | 2 | 1 | 0 | |
| GAPVD1 | GAPVD1 | 1 | 1 | 1 | 2 | 0 | 0 | |
| RasGEF | PLCE1 | PLCE1 | 1 | 1 | 1 | 0 | 0 | 0 |
| RASGRF | RASGRF2 | 1 | 1 | 1 | 1 | 0 | 0 | |
| RASGRF1 | 1 | 1 | 1 | 1 | ||||
| KNDC1 | KNDC1 | 1 | 1 | 0 | 1 | 0 | 0 | |
| RASGEF1 | RASGEF1A | 1 | 1 | 1 | 0 | 3 | 1 | |
| RASGEF1B | 1 | 1 | 1 | 2 | ||||
| RASGEF1C | 1 | 1 | 1 | 0 | ||||
| RAPGEF1 | RAPGEF1 | 1 | 1 | 0 | 1 | 1 | 1 | |
| RGL | RGL1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| RALGDS | 1 | 1 | 1 | 1 | ||||
| RGL3 | 1 | 1 | 1 | 2 | ||||
| RGL2 | 3/1 | 1 | 1 | 1 | ||||
| RGL4 | 1 | 0 | 0 | 0 | ||||
| RALGPS | RALGPS1 | 1 | 1 | 1 | 1 | 1 | 0 | |
| RALGPS2 | 1 | 1 | 1 | 1 | ||||
| RAPGEF2/6 | RAPGEF6 | 1 | 1 | 1 | 1 | 1 | 1 | |
| RAPGEF2 | 1 | 1 | 1 | 2 | ||||
| RAPGEF3/4 | RAPGEF4 | 1 | 1 | 1 | 3 | 1 | 1 | |
| RAPGEF3 | 1 | 1 | 1 | 1 | 0 | |||
| RAPGEF5 | RAPGEF5 | 1 | 1 | 1 | 2 | 0 | 0 | |
| RAPGEFL1 | RAPGEFL1 | 1 | 1 | 1 | 1 | 0 | 0 | |
| RASGRP | RASGRP3 | 1 | 1 | 1 | 1 | 0 | 1 | |
| RASGRP2 | 1 | 1 | 1 | 1 | ||||
| RASGRP1 | 1 | 1 | 1 | 1 | ||||
| RASGRP4 | 1 | 1 | 1 | 0 | ||||
| SOS | SOS2 | 1 | 1 | 1 | 1 | 1 | 1 | |
| SOS1 | 1 | 1 | 1 | 0 |
H: human; M: mouse; R: rat; Z: zebrafish; F: fly; W: worm. Cases with uncertain number of genes are indicated with ‘/’.
In summary, in the Ras module, duplications of genes within subfamilies are scarce and genetic variation almost non-existent in mammals. Although the pattern of expansion in the Euteleostomi clade suggests an important contribution of 2R and 4R rounds of genome duplication in vertebrates, which would not explain the lack of expansion in the module afterwards. The data suggest crystallization of the Ras module orthologs system in mammals and most likely in the tetrapod clade. This places the maturation of the Ras switch after the Euteleostomi expansion, dated in the Paleozoic era, about 416 MYA (43).
Domain diversity, distribution and organization
Protein domains represent the basic functional and evolutionary units that, when combined in multiple modes (architectures), give sequences with different functional and regulatory properties. Convergent evolution of domain architectures is a rare event (44), representing functional and evolutionary fingerprints. To investigate the role of these evolutionary processes in the Ras module, we computed domain and architecture distribution for all protein sequences detected (Supplementary Figures S6 and S7).
Most Ras sequences are single domain proteins, with very few extra domains appearing in some species (Supplementary Figures S6A and S7A). GEF and GAP proteins, on the other hand, are all multidomain proteins with different architectures (Supplementary Figures S6B–D and S7B–D). RasGEF is the gene family containing the highest diversity of different domains (65 domains), followed by RasGAP (32 domains) and RapGAP (8 domains). Even if the domains in both GAP families are considered together (total of 40 domains), the difference with GEF is still significant.
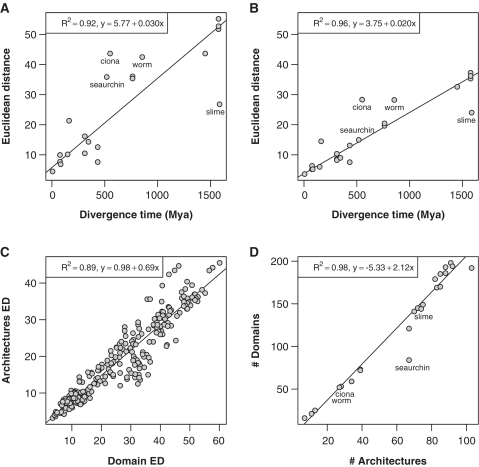
Domain and architecture counts for all the Ras switch families were concatenated in two independent domain and architecture occurrence profiles for each of the 24 eukaryotic species (Supplementary Figures S8 and S9). The ED between domain and architecture profiles was computed to assess closeness between species. Figure 6A and B show domain and architecture ED versus divergence times (MYA) from humans. Both plots suggest a linear tendency and show that species closely related to humans tend to share domain and architecture composition compared with more distant species, indicating an important role of domain fusion and rearrangement in the divergence process driving to speciation. Domain and architecture composition are dependent variables, and species with similar domain composition share similar architectures, as shown by a strong linear correlation between domain and architecture profiles in Figure 6C. In a similar way, the total number of domains versus architectures also displays strong linear correlation (Figure 6D).
Figure 6.
Comparison of species similarity profiles. (A and B) ED versus divergence time from humans (MYA) for domains and architectures, respectively. (C) Domain versus architecture profile’s EDs among all species. (D) Number of domains versus number of architectures. Outliers (Ciona, slime mold, seaurchin and worm) were removed from the linear fits in subfigures A, B and D, and the corresponding species points labeled. Fitted regression lines and R2 values are also shown for all the plots.
Comparison between occurrence profiles from human, rat and mouse shows high domain and architecture composition conservation in the Ras switch protein system (ED values below 10 in Figure 6A–C). These results reveal that the Ras switch orthologs system is conserved in mammals beyond the canonical Ras, GAP and GEF domains, and includes the sets of domains present in the Ras switch protein architectures.
As expected, the GEF family contains the highest number of different domains and architectures, with 65 architectures, followed by RasGAP (32), RapGAP (8) and Ras (8) (Supplementary Figure S9). The different architectures reflect (and in a sense define) gene subfamily membership (Supplementary Figures S2–S5) and therefore potentially contain information about functional differences between family members. Domain and architecture distribution shows an effect of species-specific divergence for species outside the metazoan clade. For example, fungi and D. discoideum present some specific domains, in particular in the RasGEF family, not present in the Metazoan clade. This also happens in C. elegans, insects, sea urchin and Monosiga, indicating species-specific divergent evolution of the Ras regulatory network. These results indicate that some species or clade-specific functional variation have been obtained by different domain rearrangements.
Protein architectures in the Ras and RapGAP families do not show any common Pfam domains with the RasGAP or RasGEF families. On the other hand, some RasGAP and RasGEF proteins share up to 9 out of 97 total Pfam domains present in both families (e.g. PDZ, PH, IQ, C2, etc.). Considering that many of these domains are involved in basic regulatory processes, lack of common domains among the different Ras switch families suggests high independence in their molecular mechanisms.
Role of alternative splicing in the Ras module functional expansion
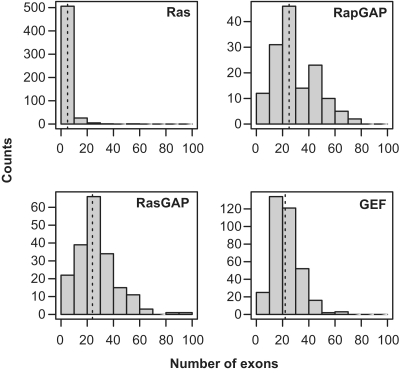
Earlier we considered gene duplication as the main evolutionary process involved in generation of sequence diversity (Figure 3). We showed that domain and architecture composition complexity increased positively in correlation to divergence times (Figure 6). However, changes in domain composition and architecture appear to be the effect of alternative evolutionary processes, like domain fusion and exon shuffling, which work in concert with gene duplication. This architecture variability may be exploited to generate sequence diversity at the transcript level by using alternative splicing. Alternative splicing generates different RNA sequences from a unique DNA template by removing selected exons during the splicing of introns (45,46). Indeed, alternative splicing seems to play an important role in the Ras module functional expansion and diversification (47,48), and provides an alternative hypothesis to explain the gap in gene number between Ras and the other families. To assess this possibility, we measured the number of exons and transcript variants and estimated the potential of the different gene families to increase functionality by alternative splicing (Figure 7 and Table 3).
Figure 7.
Distribution of exon counts within each gene family.
Table 3.
Percentage of human genes with a corresponding number of splice variant (number of SV), extracted from the ASTD database (URL: www.ebi.ac.uk/astd/) for Ras, RapGAP, RasGAP and RasGEF
| No. of SV | Ras | RapGAP | RasGAP | RasGEF |
|---|---|---|---|---|
| 1 | 27 | 17 | 0 | 6 |
| 2 | 33 | 33 | 60 | 6 |
| 3 | 13 | 17 | 20 | 47 |
| 4 | 13 | 33 | 20 | 12 |
| 5 | 7 | 0 | 0 | 6 |
| 6 | 0 | 0 | 0 | 12 |
| 7 | 7 | 0 | 0 | 6 |
| 8 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 6 |
| Total (N) | 15 | 6 | 10 | 17 |
| Missing (%) | 58 | 33 | 29 | 43 |
The percentage is computed relative to the total number of genes with splicing information (total, N). The percentage of detected genes in this work with missing information is also indicated (missing %).
A higher number of exons indicate more potential to generate functional diversity using alternative splicing (49). The distribution of the number of exons in the Ras module shows a significant bias. Ras genes have a highly skewed distribution with an average around five exons per gene, but with some sequences with very large values. GEF and GAP distributions show higher variance, indicating more variability in the functional landscape. The number of exons is larger than in Ras, with median values of 25 for RapGAP, 24 for RasGAP and 22 for RasGEF (Figure 7).
This suggests that GEF and GAP proteins have on average higher potential than Ras proteins to generate sequence diversity using alternative splicing. Direct evidence comes from the distribution of human alternative splicing sequences from the ASTD database (Table 3). The results indicate that the Ras family has the highest percentage of genes of all families with just one splice variant (27%, Table 3). RapGAP genes show two maximums of 33% of genes with two and four splice variants. The RasGAP family shows no genes with one splice variant, have the highest percentage of genes with two variants (60%), and present the remaining 40% of genes with three or more alternative splicing variants. RasGEF genes show the most significantly skewed distribution towards multiple splice variants categories. The RasGEF family has the highest percentage of genes with three variants (47%), and shows 36% of genes distributed between four and nine variant categories. These results are consistent, although they are not taking into account genes with missing alternative splicing information (Ras: 8%, RapGAP: 33%, RasGAP: 29% and RasGEF: 43%).
Overall, GEF (in particular) and GAP proteins show higher potential than Ras to generate functional diversity by alternative splicing. Consequently, although gene duplication is the main evolutionary force driving functional expansion of the Ras family, alternative splicing has a considerable role in the GEF and GAP families. This may be an important mechanism to compensate for the disparity in the GEF and GAP family sizes, compared with Ras.
CONCLUSIONS
An evolutionary genetic model for the Ras module expansion
The Ras network works as a critical regulatory module in many signaling pathways. Given the minimal number of genes present in the fungi species, we can assume that the original Ras module originated at some point with at least one member of the Ras, GEF and GAP families. In a compatible scenario to explain the Ras module expansion the bulk sequence pool is generated by gene duplication, a genetic mechanism with a more prominent role in the Ras family (Figure 3). This genetic expansion has accompanied the overall increment in coding genome size in the Eukaryotic kingdom (Figure 3). The observed behavior appears very robust given that many processes have affected the organization of genomic material in some of the studied organisms (e.g. 2R genome duplication leading to vertebrates, the fish-specific duplication, large-scale losses in Drosophila, C. elegans and Saccharomycotina).
Far from being a homogeneous phenomenon, the Ras module expansion is the overall result of uneven gene duplication rates of the different generated subfamilies within the Ras module families (Figure 4). This uneven expansion is likely the result of speciation-specific functional requirements, such as the immune and nervous systems. Some of the relevant expanded subfamilies are seldom studied (e.g., the SIPA subfamily), limiting the functional analysis of these subfamilies and suggesting that further efforts are necessary to disentangle their functional implications. In this sense, our findings are useful to direct future experimental efforts to study some of these subfamilies.
Together with gene duplication, other processes have contributed to generate functional divergence linearly correlated to the speciation processes in Eukaryote evolution. Domain fusion, which includes exogenous domains, and exon shuffling, which alters the internal protein domain architecture (Figure 7), both play important roles in the GEF and GAP families.
Although domain and architecture variation is not a source of functional expansion by itself, this domain diversity in protein architectures can be used at the transcript level to expand functional complexity by alternative splicing. This is another mechanism with a prominent role in the Ras module to expand functionality, particularly used by GEF and GAP genes. By these means, transcript variants could eventually fill the gap in the number of genes between Ras and its GAP and GEF regulators observed in the genomes of higher eukaryotes.
Genetic expansion in the Ras module has decreased after the evolution of the Euteleostomi clade (fish, amphibians, birds and mammals in this work) (Figure 5). In addition, similar protein architectures and domain composition suggests that the Ras switch is practically crystallized into a genetically and functionally stable system of orthologs in the mammalian clade after the Euteleostomi expansion, dated in the Paleozoic era, about 416 MYA (43). This implies that the molecular and functional mechanisms performed by these orthologs are conserved among mammal species, supporting the use of mammalian models, such as rat or mouse, to guide functional and biomedical studies of the Ras switch components in humans.
Gene duplication and alternative splicing have been the main genetic processes for generating Ras switch genetic expansion, while sequence divergence together with domain fusion and rearrangement have been the main sources of Ras module speciation. As a result of theses genetic mechanisms, the Ras switch has become a complex system with about 70 sets of orthologous genes in mammals.
The deep differences in protein architectures and domain composition observed among the Ras switch regulatory families in many species, with almost complete absence of any shared domains, suggest highly functional specificity and pathway independency regulating and propagating Ras signaling transduction. This molecular function independency of the Ras switch families creates a network with a ‘decision making’ topology, creating the possibility to integrate, through a homologous GEF molecular mechanism, different input signals into Ras paralogous proteins. In this network, Ras proteins function as central nodes conducting the signal flow through the bifurcation in the GAP signal propagation (RasGAP and RapGAP no homologous domains).
Some Ras switch proteins are known to lack activity (e.g. Ras proteins missing GTPase activity) or interact with and regulate other members of the Ras superfamily (e.g. RasGEF proteins that activate Rho) (12,50). Although these are important aspects of Ras evolution, this work focuses on a global analysis of these gene families, and their remarkable coordinated expansion. The particular evolutionary pathways that each individual gene followed are not explicitly included in our study, although it is implicitly covered by the divergent nature of some of the described subgroups. Ras switch proteins may avoid cross-communication between paralogous genes by diversifying their tissue and cellular location and gene expression timing in complex organisms. However, there is also the possibility that different sets of Ras switch orthologs co-localize and integrate their signal propagation in ‘decision making’ network topologies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Science and Innovation Ministry through the Ramón y Cajal program (RYC-2007-01649), the Plan Nacional project (SAF-2009-09839–Subprograma MED), and the ENFIN NoE EU project (to J.A.G.R.) Programa I3P de la Red de Bioinformática del CSIC (pre-doctoral fellowship), Comunidad de Madrid 08.5/0042/2003 and GR/SAL/0382/2004, MEC BFI2002-00489 and Red de Centros RCMN (C03/08) and by a postdoctoral fellowship from the Japanese Society for the Promotion of Science (JSPS) (to D.D.); CIBERER, which is an initiative of the Instituto de Salud Carlos III, and Spanish Plan Nacional project (SAF2008-02522 to F.S.J.). Funding for open access charge: Spanish Science and Innovation Ministry through the Spanish Plan Nacional projects SAF2009-09839 and SAF2008-02522.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Miguel A. Medina for his help in revising the manuscript.
REFERENCES
- 1.Brambilla R, Gnesutta N, Minichiello L, White G, Roylance AJ, Herron CE, Ramsey M, Wolfer DP, Cestari V, Rossi-Arnaud C, et al. A role for the Ras signalling pathway in synaptic transmission and long-term memory. Nature. 1997;390:281–286. doi: 10.1038/36849. [DOI] [PubMed] [Google Scholar]
- 2.Manabe T, Aiba A, Yamada A, Ichise T, Sakagami H, Kondo H, Katsuki M. Regulation of long-term potentiation by H-Ras through NMDA receptor phosphorylation. J. Neurosci. 2000;20:2504–2511. doi: 10.1523/JNEUROSCI.20-07-02504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 4.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 5.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci. STKE. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao GH, Shoarinejad F, Jin F, Golemis EA, Yeung RS. The tuberous sclerosis 2 gene product, tuberin, functions as a Rab5 GTPase activating protein (GAP) in modulating endocytosis. J. Biol. Chem. 1997;272:6097–6100. doi: 10.1074/jbc.272.10.6097. [DOI] [PubMed] [Google Scholar]
- 7.Hu KQ, Settleman J. Tandem SH2 binding sites mediate the RasGAP-RhoGAP interaction: a conformational mechanism for SH3 domain regulation. EMBO J. 1997;16:473–483. doi: 10.1093/emboj/16.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burbelo PD, Miyamoto S, Utani A, Brill S, Yamada KM, Hall A, Yamada Y. p190-B, a new member of the Rho GAP family, and Rho are induced to cluster after integrin cross-linking. J. Biol. Chem. 1995;270:30919–30926. doi: 10.1074/jbc.270.52.30919. [DOI] [PubMed] [Google Scholar]
- 9.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Ranea JA, Valencia A. Distribution and functional diversification of the ras superfamily in Saccharomyces cerevisiae. FEBS Lett. 1998;434:219–225. doi: 10.1016/s0014-5793(98)00967-3. [DOI] [PubMed] [Google Scholar]
- 11.Vernoud V, Horton AC, Yang Z, Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003;131:1191–1208. doi: 10.1104/pp.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- 13.Bernards A. Ras superfamily and interacting proteins database. Methods Enzymol. 2006;407:1–9. doi: 10.1016/S0076-6879(05)07001-1. [DOI] [PubMed] [Google Scholar]
- 14.Hubbard T, Andrews D, Caccamo M, Cameron G, Chen Y, Clamp M, Clarke L, Coates G, Cox T, Cunningham F, et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donoghue PC, Benton MJ. Rocks and clocks: calibrating the Tree of Life using fossils and molecules. Trends Ecol. Evol. 2007;22:424–431. doi: 10.1016/j.tree.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Blair Hedges S, Kumar S. Genomic clocks and evolutionary timescales. Trends Genet. 2003;19:200–206. doi: 10.1016/S0168-9525(03)00053-2. [DOI] [PubMed] [Google Scholar]
- 17.Hedges SB. The origin and evolution of model organisms. Nat. Rev. Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 18.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 19.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2005. [Google Scholar]
- 20.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howe K, Bateman A, Durbin R. QuickTree: building huge Neighbour-Joining trees of protein sequences. Bioinformatics. 2002;18:1546–1547. doi: 10.1093/bioinformatics/18.11.1546. [DOI] [PubMed] [Google Scholar]
- 22.Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koscielny G, Le Texier V, Gopalakrishnan C, Kumanduri V, Riethoven JJ, Nardone F, Stanley E, Fallsehr C, Hofmann O, Kull M, et al. ASTD: The Alternative Splicing and Transcript Diversity database. Genomics. 2009;93:213–220. doi: 10.1016/j.ygeno.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 24.The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 25.Ohno S. Evolution by Gene Duplication. London: Springer; 1970. [Google Scholar]
- 26.Wilkins A, Insall RH. Small GTPases in Dictyostelium: lessons from a social amoeba. Trends Genet. 2001;17:41–48. doi: 10.1016/s0168-9525(00)02181-8. [DOI] [PubMed] [Google Scholar]
- 27.Maruoka H, Konno D, Hori K, Sobue K. Collaboration of PSD-Zip70 with its binding partner, SPAR, in dendritic spine maturity. J. Neurosci. 2005;25:1421–1430. doi: 10.1523/JNEUROSCI.3920-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, McNulty DE, Marler KJ, Lim L, Hall C, Annan RS, Sacks DB. IQGAP1 promotes neurite outgrowth in a phosphorylation-dependent manner. J. Biol. Chem. 2005;280:13871–13878. doi: 10.1074/jbc.M413482200. [DOI] [PubMed] [Google Scholar]
- 29.Briggs MW, Sacks DB. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep. 2003;4:571–574. doi: 10.1038/sj.embor.embor867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Briggs MW, Sacks DB. IQGAP1 as signal integrator: Ca2+, calmodulin, Cdc42 and the cytoskeleton. FEBS Lett. 2003;542:7–11. doi: 10.1016/s0014-5793(03)00333-8. [DOI] [PubMed] [Google Scholar]
- 31.Rumbaugh G, Adams JP, Kim JH, Huganir RL. SynGAP regulates synaptic strength and mitogen-activated protein kinases in cultured neurons. Proc. Natl Acad. Sci. USA. 2006;103:4344–4351. doi: 10.1073/pnas.0600084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang SN, Huang CB, Yang CH, Lai MC, Chen WF, Wang CL, Wu CL, Huang LT. Impaired SynGAP expression and long-term spatial learning and memory in hippocampal CA1 area from rats previously exposed to perinatal hypoxia-induced insults: beneficial effects of A68930. Neurosci. Lett. 2004;371:73–78. doi: 10.1016/j.neulet.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Guo J, Dzhagalov I, He YW. An essential function for the calcium-promoted Ras inactivator in Fcgamma receptor-mediated phagocytosis. Nat. Immunol. 2005;6:911–919. doi: 10.1038/ni1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebinu JO, Stang SL, Teixeira C, Bottorff DA, Hooton J, Blumberg PM, Barry M, Bleakley RC, Ostergaard HL, Stone JC. RasGRP links T-cell receptor signaling to Ras. Blood. 2000;95:3199–3203. [PubMed] [Google Scholar]
- 35.Pasvolsky R, Feigelson SW, Kilic SS, Simon AJ, Tal-Lapidot G, Grabovsky V, Crittenden JR, Amariglio N, Safran M, Graybiel AM, et al. A LAD-III syndrome is associated with defective expression of the Rap-1 activator CalDAG-GEFI in lymphocytes, neutrophils, and platelets. J. Exp. Med. 2007;204:1571–1582. doi: 10.1084/jem.20070058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Correll RN, Pang C, Niedowicz DM, Finlin BS, Andres DA. The RGK family of GTP-binding proteins: regulators of voltage-dependent calcium channels and cytoskeleton remodeling. Cell. Signal. 2008;20:292–300. doi: 10.1016/j.cellsig.2007.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi K, Mitsui K, Yamanaka S. Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature. 2003;423:541–545. doi: 10.1038/nature01646. [DOI] [PubMed] [Google Scholar]
- 38.Kawasaki H, Springett GM, Toki S, Canales JJ, Harlan P, Blumenstiel JP, Chen EJ, Bany IA, Mochizuki N, Ashbacher A, et al. A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc. Natl Acad. Sci. USA. 1998;95:13278–13283. doi: 10.1073/pnas.95.22.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ichiba T, Hoshi Y, Eto Y, Tajima N, Kuraishi Y. Characterization of GFR, a novel guanine nucleotide exchange factor for Rap1. FEBS Lett. 1999;457:85–89. doi: 10.1016/s0014-5793(99)01012-1. [DOI] [PubMed] [Google Scholar]
- 40.Jefferson JA, Shankland SJ. Familial nephrotic syndrome: PLCE1 enters the fray. Nephrol. Dial. Transplant. 2007;22:1849–1852. doi: 10.1093/ndt/gfm098. [DOI] [PubMed] [Google Scholar]
- 41.Mees A, Rock R, Ciccarelli FD, Leberfinger CB, Borawski JM, Bork P, Wiese S, Gessler M, Kerkhoff E. Very-KIND is a novel nervous system specific guanine nucleotide exchange factor for Ras GTPases. Gene Expr. Patterns. 2005;6:79–85. doi: 10.1016/j.modgep.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 42.Huang J, Furuya A, Furuichi T. Very-KIND, a KIND domain containing RasGEF, controls dendrite growth by linking Ras small GTPases and MAP2. J. Cell Biol. 2007;179:539–552. doi: 10.1083/jcb.200702036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benton MJ, Donoghue PC. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- 44.Gough J. Convergent evolution of domain architectures (is rare) Bioinformatics. 2005;21:1464–1471. doi: 10.1093/bioinformatics/bti204. [DOI] [PubMed] [Google Scholar]
- 45.Sharp PA. Splicing of messenger RNA precursors. Science. 1987;235:766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- 46.Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 47.Bernards A, Haase VH, Murthy AE, Menon A, Hannigan GE, Gusella JF. Complete human NF1 cDNA sequence: two alternatively spliced mRNAs and absence of expression in a neuroblastoma line. DNA Cell Biol. 1992;11:727–734. doi: 10.1089/dna.1992.11.727. [DOI] [PubMed] [Google Scholar]
- 48.Guil S, de La Iglesia N, Fernandez-Larrea J, Cifuentes D, Ferrer JC, Guinovart JJ, Bach-Elias M. Alternative splicing of the human proto-oncogene c-H-ras renders a new Ras family protein that trafficks to cytoplasm and nucleus. Cancer Res. 2003;63:5178–5187. [PubMed] [Google Scholar]
- 49.Kopelman NM, Lancet D, Yanai I. Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat. Genet. 2005;37:588–589. doi: 10.1038/ng1575. [DOI] [PubMed] [Google Scholar]
- 50.Bernards A, Settleman J. GEFs in growth factor signaling. Growth Factors. 2007;25:355–361. doi: 10.1080/08977190701830375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.