Abstract
Synthetic small duplex RNAs that are fully complementary to gene promoters can silence transcription in mammalian cells. microRNAs (miRNAs) are endogenous small regulatory RNAs that sequence specifically regulate gene expression. We have developed a computational method to identify potential miRNA target sites within gene promoters. Ten candidate miRNAs predicted to target the human progesterone receptor (PR) gene promoter were tested for their ability to modulate gene expression. Several miRNA mimics inhibited PR gene expression and miR-423-5p, which targets a highly conserved region of the PR promoter, was chosen for detailed analysis. Chromatin immunoprecipitation revealed that the miR-423-5p mimic decreased RNA polymerase II occupancy and increased histone H3 lysine 9 dimethylation (H3K9me2) at the PR promoter, indicative of chromatin-level silencing. Transcriptional silencing was transient, independent of DNA methylation, and associated with recruitment of Argonaute 2 (AGO2) to a non-coding RNA (ncRNA) transcript that overlaps the PR gene promoter. The miR-423-5p mimic also silenced expression of immunoglobulin superfamily member 1 (IGSF1), an additional gene with a predicted target site within its promoter. While additional investigations of endogenous miRNA function will be necessary, these observations suggest that recognition of gene promoters by miRNAs may be a natural and general mechanism for regulating gene transcription.
INTRODUCTION
Small duplex RNAs complementary to gene promoters are potent silencers and activators of target gene expression in mammalian cells (1–12). We and others have demonstrated that these small RNAs sequence specifically recognize non-coding RNA (ncRNA) transcripts that overlap gene promoters (6,12–15). Recognition of the target ncRNA occurs in close proximity to the chromosome and leads to recruitment of Argonaute (AGO) proteins to the ncRNA and to the target gene promoter (13,14,16–18). Modulation of target gene expression occurs at the level of transcription and has been observed in both the presence and absence of DNA methylation (1–7).
Potent and robust transcriptional modulation suggests the existence of an endogenous mechanism that facilitates recognition of gene promoters by small RNAs. AGO proteins implicated in the mechanism of promoter-targeting RNAs are conserved across eukaryotes (19). AGO proteins have been shown to use endogenous small RNAs to direct chromatin modification and regulate transcription in yeast and plants (20–22).
In humans, AGO proteins are well known for their role in microRNA (miRNA) regulation. miRNAs are an endogenous class of short regulatory RNAs ranging in length from 19 to 30 bases. They are generally understood to repress gene expression post-transcriptionally through recognition of sequences within 3′-UTRs of mRNA transcripts (23,24). miRNAs have been reported to regulate transcription in human cells, but much remains to be learned about their mechanism of action (25,26).
We previously performed a genome-wide evaluation of gene promoters and found a significant enrichment of putative miRNA target sites within promoter regions (27). Using the human progesterone receptor (PR) as a model gene we have now identified multiple miRNA mimics that are predicted to target the PR promoter and inhibit PR expression. We further demonstrate that a ncRNA transcribed from the PR promoter serves as the molecular target of these miRNA mimics. Our results indicate that miRNA recognition of gene promoters may be a general mechanism for gene regulation.
MATERIALS AND METHODS
Cell culture
T47D and MCF7 breast cancer cells [American Type Culture Collection (ATCC)] were maintained in RPMI-1640 medium (ATCC) supplemented with 10% (v/v) FBS (Atlanta Biologicals), 0.5% (v/v) nonessential amino acids (Sigma), 10 mM HEPES (Sigma), 1 mM Sodium Pyruvate (Sigma), 0.4 U/ml bovine insulin (Sigma). Cells were cultured at 37°C and 5% CO2.
Cellular delivery of miRNA mimics and siRNAs
RNAiMAX (Invitrogen) was used to deliver small duplex RNAs into T47D or MCF7 cells as per the manufacturer's instructions. For RNA and protein isolation, cells were plated in six-well dishes at densities ranging between 150 and 200 K cells/well. For RNA and chromatin immunoprecipitation, cells were plated in 10 cm2 dishes at a density of 4.5×106 cells/dish. Cells were transfected 48 h after plating. A double transfection protocol was used for AGO reversal experiments. The first transfection was performed as described above (using mismatch-containing duplexes or siRNAs against AGO). After 72 h, cells were dissociated using trypsin and re-seeded in transfection reagent (using MM, miR-423-5p or pre-miR-423 duplexes). Sequences for miRNA mimics and siRNAs are listed in Supplementary Table S1A.
Western blotting
Cells were harvested 5 days post-transfection for protein isolation. Cell pellets were lysed and protein concentrations were quantified by BCA assay (Pierce). Western blots were performed on protein lysates (30 µg/well). Primary antibodies used were α-PR (Cell Signalling Technology) and α-β-actin (Sigma). Protein was visualized with horseradish peroxidase-conjugated α-mouse secondary antibody (Jackson Immunolabs) and Supersignal developing solution (Pierce).
Quantitative PCR
Cells were harvested 72 h post-transfection for RNA isolation. RNA was isolated using TRI Reagent (Sigma) as per the manufacturer's instructions. For each sample, 2 µg of RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). RNA was treated with DNase I (Worthington) prior to reverse transcription. qPCR was performed on an ABI7900 real-time PCR (Applied Biosystems) using iTaq SYBR Green Supermix (Bio-Rad). ABI TaqMan miRNA assays were used to detect miR-423-5p and U6 snRNA as per the manufacturer's instructions. Primers for PR and GAPDH mRNA were supplied by Applied Biosystems. All additional primers were designed using Primer3. Only those primer sets that showed linear amplification over several orders of magnitude were used for quantification. Primers and PCR conditions are listed in Supplementary Table S1B.
Chromatin immunoprecipitation and RNA immunoprecipitation
Chromatin immunoprecipitation (ChIP) and RNA immunoprecipitation (RIP) were performed essentially as described (28). Cells were harvested 72 h post-transfection. α-RNA Polymerase II, α-H3K9me2, α-AGO1, α-AGO2, normal rabbit IgG and normal mouse IgG antibodies were supplied by Millipore. For RIP, isolated RNA was treated with DNase I and reverse transcribed. Primers for the PR promoter and GAPDH promoter were designed using Primer3. Only those primer sets that showed linear amplification over several orders of magnitude were used for quantification. Primers and PCR conditions are listed in Supplementary Table S1B.
Methylation specific PCR and bisulphite sequencing
Genomic DNA was extracted using TRI Reagent (Sigma) as per the manufacturer's instructions. For each sample, 2 µg of DNA was diluted into 50 µl of H2O. Single-stranded DNA was created by adding 5.5 µl of 2 M NaOH and incubating at 37°C for 10 min. To each sample, 30 µl of freshly prepared 10 mM hydroquinone (Sigma) and 520 µl of freshly prepared 3 M sodium bisulphite (Sigma) were added. DNA was incubated at 50°C for 16 h. After bisulphite treatment, DNA was purified using the Wizard DNA Clean-Up System (Promega). DNA was then subjected to ammonium acetate precipitation and ethanol washing to remove any remaining impurities. PCR amplification of treated DNA was performed using HotStarTaq DNA polymerase (QIAGEN) as per the manufacturer's instructions. Primers and PCR conditions are listed in Supplementary Table S1B. PCR products were resolved on 3% agarose gel with ethidium bromide. For bisulphite sequencing PCR products were cloned into a PCR-4 Topo vector (Invitrogen) and sequenced (McDermott sequencing core, University of Texas, Southwestern).
Statistical Analysis
Data are presented as means ± standard deviation of three or more independent results. Statistical significance was assessed using a two-tailed unpaired Student's t-test.
RESULTS AND DISCUSSION
miRNAs complementary to the PR promoter inhibit PR expression
We have previously designed small duplex RNAs complementary to the PR promoter and demonstrated that these RNAs silence transcription of the PR gene (4,16). We subsequently showed that the molecular targets of these small RNAs are ncRNA transcripts produced from the PR promoter (14). The PR gene has two major isoforms termed PR-B and PR-A that differ in their transcription start sites (TSS), PR-B being the most upstream. One of the ncRNAs that we characterized is a 2170-base transcript that initiates 1431 bases downstream of the PR-B TSS and is transcribed in the antisense direction through the PR promoter.
In prior studies, we used designed synthetic small duplex RNAs that were fully complementary to the PR promoter. The differences between fully complementary designed RNAs and miRNAs are substantial. miRNAs, which are encoded within the genome, do not require complete complementarity to achieve target recognition. This makes it difficult to accurately predict sequences miRNAs will efficiently target.
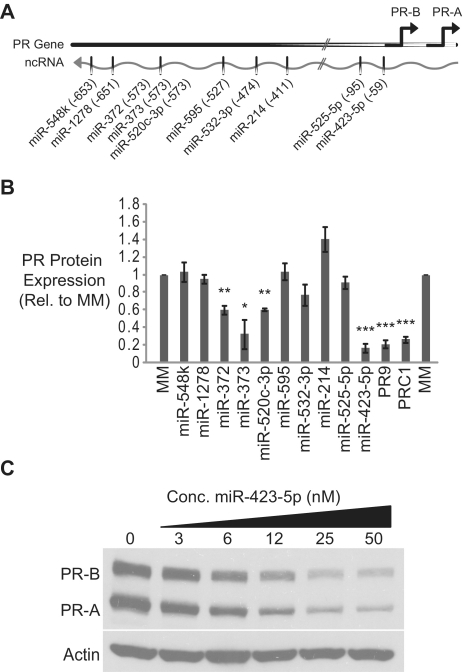
To begin to experimentally determine if miRNAs are capable of recognizing sequences within gene promoters, we obtained sequences for all known human miRNAs from miRBase, the public repository for miRNAs (29). Using an algorithm we developed to identify potential miRNA target sites, we scanned the segment of the ncRNA that overlaps the PR-B promoter for potential miRNA target sites (27). We identified 72 potential miRNA target sites within the analysed region of the ncRNA transcript. We selected 10 miRNAs for experimental validation based on complementarity to their respective target sequences within the PR promoter (Figure 1A and Supplementary Figure S1). We designed miRNA mimics that consisted of the miRNA sequence and a fully complementary RNA carrier strand. The miRNA mimics were transfected into T47D breast cancer cells and PR protein expression was monitored by western blot. PRC1 (an siRNA targeting PR mRNA) and PR9 (a duplex RNA previously shown to target the PR promoter and inhibit PR expression) were used as positive controls. As a negative control we used a mismatched duplex RNA (MM) that does not affect PR expression.
Figure 1.
miRNAs complementary to the PR promoter inhibit PR expression. (A) Schematic of selected miRNAs complementary to the PR promoter. (B) Quantification of three independent experiments measuring reduction of PR protein expression by miRNA mimics. (C) Dose-dependent inhibition of PR by miR-423-5p. miRNA mimics were added to cells at 50 nM unless otherwise noted. Error bars indicate SD (n = 3). P-values were calculated using the two-tailed unpaired Student's t-test with equal variances. *P < 0.05, **P < 0.01, ***P < 0.001.
Several miRNA mimics inhibited PR protein expression (Supplementary Figure S2A). Quantification of replicate experiments revealed that inhibition by miR-372, miR-373, miR-520c-3p and miR-423-5p was statistically significant (Figure 1B). These results indicate that miRNAs complementary to the PR promoter are capable of inhibiting PR expression. In striking contrast to our prior experience with designed synthetic small RNAs, where introduction of more than one mismatched base between the silencing RNA and its target completely abolished activity, all of the miRNA mimics that inhibited PR expression were extensively mismatched when aligned to the target ncRNA (Supplementary Figure S1). For example, miR-423-5p possesses seven mismatched bases relative to its ncRNA target, including a three base bulge adjacent to the seed sequence. miR-520c-3p also contains seven-mismatched bases relative to the target ncRNA, but with a five base bulge immediately following the seed sequence.
The target sites for the inhibitory miRNA mimics also differ significantly from the sites we have previously targeted with fully complementary small duplex RNAs. In our prior studies, any duplexes that targeted more than 26 bases upstream of the PR-B TSS were inactive (4). However, three of the inhibitory miRNA mimics we identified target more than 500 bases upstream of the TSS. This suggests that small RNAs can target sequences much further upstream of the PR TSS than has previously been appreciated. The most potent mimic, miR-423-5p, targets the region spanning from 59 bases upstream of the PR-B TSS to 37 bases upstream of the TSS. miR-423-5p inhibited PR expression in a dose dependent manner with an IC50 of 7.2 nM (Figure 1C and Supplementary Figure S2B) and was selected as the focus for subsequent experiments.
miR-423-5p inhibits transcription of the PR gene
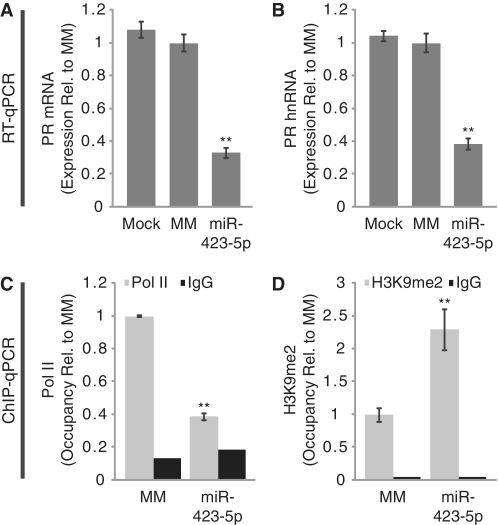
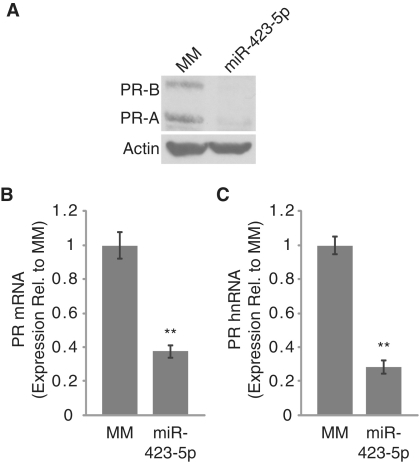
Potent inhibition of PR expression by the miR-423-5p mimic prompted us to investigate the mechanism of silencing. Using quantitative RT–PCR (RT–qPCR) we found that the miR-423-5p mimic reduced PR mRNA levels by >70% (Figure 2A). To evaluate whether decreased mRNA levels are due to reduced transcription, we monitored expression of pre-spliced mRNA, also termed heteronuclear RNA (hnRNA), produced from the PR locus. The miR-423-5p mimic-reduced hnRNA levels by nearly 70% (Figure 2B). We also quantified RNA Polymerase II (Pol II) occupancy at the PR promoter using chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR). Treatment with the miR-423-5p mimic resulted in a >60% decrease in Pol II on the PR promoter as compared with a negative control duplex (Figure 2C). Transcriptional silencing by small RNAs has been associated with induction of repressive histone marks, including dimethylation of histone H3 lysine 9 (H3K9me2) (17). Using ChIP-qPCR we observed a 2.3-fold increase in H3K9me2 following treatment with the miR-423-5p mimic (Figure 2D). Taken together, these results suggest that miR-423-5p represses transcription of the PR gene.
Figure 2.
miR-423-5p inhibits PR transcription. (A and B) RT–qPCR showing inhibition of (A) PR mRNA and (B) PR hnRNA expression by miR-423-5p. (C and D) ChIP-qPCR analysis of (C) RNA Pol II and (D) H3K9me2 occupancy on the PR promoter following treatment with miR-423-5p. miRNA mimics were added to cells at 25 nM for RT–qPCR and 50 nM for ChIP-qPCR. Error bars indicate SD (n = 3). P-values were calculated using the two-tailed unpaired Student's t-test with equal variances. **P < 0.01.
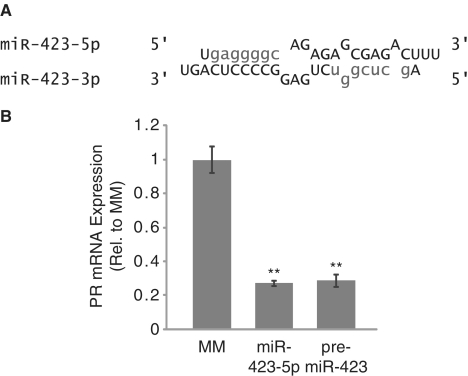
To ensure that inhibition of PR by miR-423-5p was not an artefact of our miRNA mimic design, we designed a pre-miR-423 mimic. The pre-miR-423 mimic retained the structure of endogenous miR-423 after DICER processing (an imperfect duplex RNA consisting of both the miR-423-5p and miR-423-3p sequences) (Figure 3A). Inhibition of PR expression by the pre-miR-423 mimic was indistinguishable from the miR-423-5p mimic (Figure 3B).
Figure 3.
Inhibition of PR by pre-miR-423. (A) Structure of pre-miR-423 duplex (seed sequences shown in lower case). (B) RT–qPCR showing inhibition of PR mRNA by pre-miR-423. Error bars indicate SD (n = 3). P-values were calculated using the two-tailed unpaired Student's t-test with equal variances. **P < 0.01.
In parallel to miR-423-5p, we tested additional inhibitory miRNA mimics miR-372, miR-373 and miR-520c-3p for their effects on expression of PR mRNA. All three mimics reduced PR mRNA expression by > 70% (Supplementary Figure S2C). The mimics also reduced levels of PR hnRNA expression (Supplementary Figure S2D). The ability of multiple promoter-targeting miRNA mimics to inhibit PR expression suggests that recognition of gene promoter sequences may be a general property of miRNAs.
Sequence analysis of the miR-423-5p target site within the PR promoter
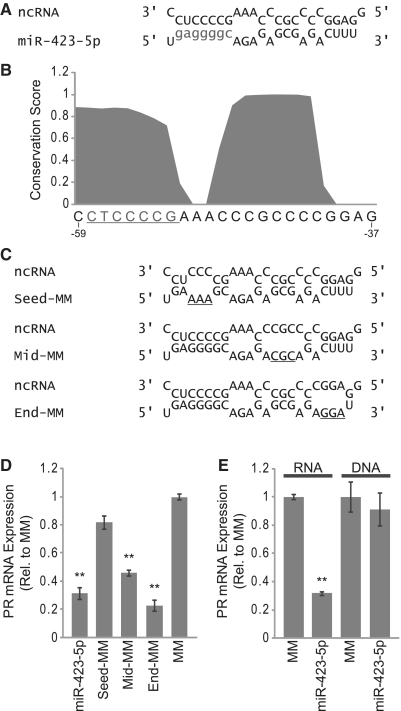
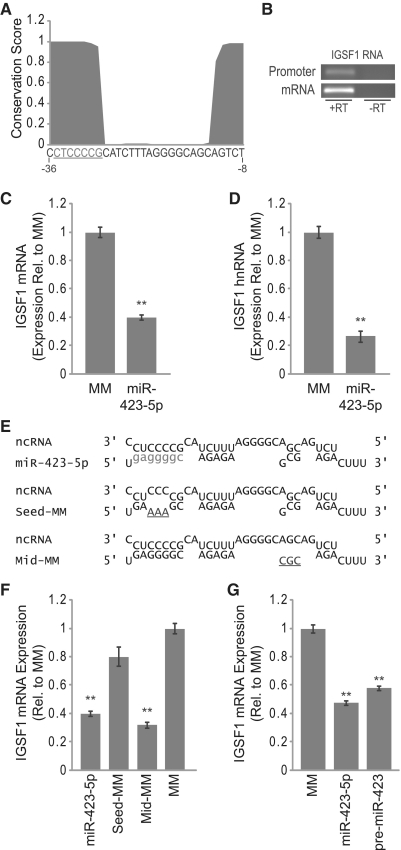
miR-423-5p has a high degree of complementarity to an antisense transcript that overlaps the PR gene promoter (Figure 4A). The region of complementarity spans from 59 bases upstream of the PR TSS through 37 bases upstream of the TSS. PhastCons analysis of the multiz alignment of 44 vertebrate species revealed that this region of the PR promoter is highly conserved, implying that the target sequence is of biological importance (Figure 4B).
Figure 4.
Inhibition of PR by miR-423-5p requires seed sequence complementarity. (A) Alignment of miR-423-5p with ncRNA overlapping the PR promoter (seed sequence shown in lower case). (B) PhastCons conservation analysis of the miR-423-5p target site within the PR promoter (seed sequence target site underlined, sequence listed 3′ to 5′). (C) Alignment of mutant miR-423-5p mimics with ncRNA overlapping the PR promoter (mismatched bases underlined). (D) RT–qPCR measuring PR mRNA expression following treatment with mutant miR-423-5p mimics. (E) RT–qPCR measuring PR mRNA expression following treatment with a DNA analog of miR-423-5p. Duplexes were added to cells at 25 nM. Error bars indicate SD (n = 3). P-values were calculated using the two-tailed unpaired Student's t-test with equal variances. **P < 0.01.
A major determinant of miRNA target recognition is complete complementarity between the miRNA seed sequence (bases 2–8 of the mature miRNA) and the target RNA sequence. One component of the algorithm, we used to identify candidate miRNAs was the requirement for seed sequence complementarity with the ncRNA overlapping the PR promoter. Interestingly, we observed that the entire miR-423-5p seed sequence target site is highly conserved within the PR promoter. In addition to seed sequence complementarity, we noted two additional regions of complementarity between miR-423-5p and the ncRNA (Figure 4A).
To determine the importance of each segment of complementarity on gene silencing, we designed mutant miRNA mimics that contained three-base changes in either the seed sequence (Seed-MM), the middle region of complementarity (Mid-MM) or the terminal region of complementarity (End-MM) (Figure 4C). We used RT–qPCR to measure expression of PR mRNA following treatment with each mutant miRNA mimic. Mutating the seed sequence abolished silencing activity while mutating the middle or end regions of complementarity had little or no effect on silencing, respectively (Figure 4D). Identical results were obtained when evaluating the effects of these mutant mimics on PR protein expression (Supplementary Figure S3A and B). These results are consistent with known targeting properties of miRNAs and demonstrate that the seed sequence of miR-423-5p is a major determinant of target recognition.
It has been reported that specific cellular RNAs are capable of sequestering DNA binding transcription factors (30). To test whether miR-423-5p sequesters-specific protein factors that normally recognize the conserved DNA sequence within the PR promoter, we designed a small duplex DNA corresponding to the miR-423-5p sequence. The miR-423-5p DNA duplex did not affect PR mRNA expression, indicating that transcriptional silencing is specific to small RNAs and most likely not a result of sequestering putative DNA binding proteins (Figure 4E).
Mechanism of miRNA-induced transcriptional silencing
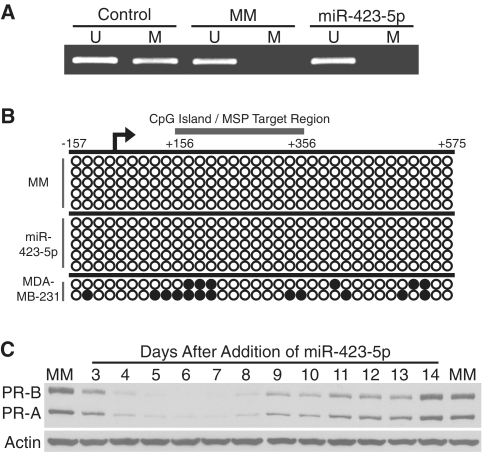
Small RNA-induced transcriptional silencing has been reported in both the presence and absence of DNA methylation (1–7). There is a CpG island near the PR-B TSS that has the potential to become methylated. We examined the methylation state of this CpG island using bisulphite treatment followed by methylation specific PCR (MSP). As a positive control we used genomic DNA isolated from MDA-MB-231 breast cancer cells where the PR promoter is methylated. Treatment with the miR-423-5p mimic did not induce DNA methylation at the PR promoter (Figure 5A). Bisulphite sequencing confirmed the lack of DNA methylation throughout the PR promoter (Figure 5B). Consistent with the absence of DNA methylation, silencing of PR protein expression by miR-423-5p was transient, lasting ∼7–8 days before beginning to return to normal levels (Figure 5C). The mechanism of transcriptional silencing by miR-423-5p at the PR promoter does not appear to involve induction of permanent epigenetic changes.
Figure 5.
Mechanism of miR-423-5p-induced silencing of PR. (A) Methylation specific PCR of the PR promoter after treatment with miR-423-5p. U, unmethylated. M, methylated. (B) Bisulphite sequencing of the PR promoter following addition of miR-423-5p. Open circles, unmethylated CpG. Closed circles, methylated CpG. Circles represent individual CpG dinucleotides. (C) Timecourse of PR inhibition by miR-423-5p. miRNA mimics were added to cells at 25 nM.
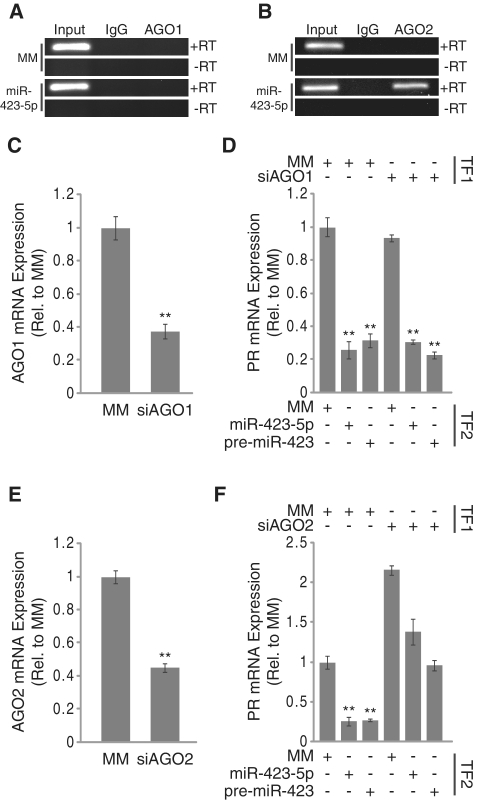
We have previously demonstrated that synthetic small duplex RNAs targeting the PR promoter recruit AGO2 to the antisense ncRNA transcript overlapping the PR promoter (16,18). Likewise, other groups have reported the involvement of AGO1 in small RNA-induced transcriptional gene silencing (17). We used RIP to evaluate both AGO1 and AGO2 occupancy on the ncRNA following treatment with the miR-423-5p mimic. RIP revealed no association between AGO1 and the ncRNA under any treatment condition (Figure 6A). Conversely, treatment with the miR-423-5p mimic resulted in association of AGO2 with the ncRNA transcript (Figure 6B). We used RT–qPCR to determine if recruitment of AGO2 affected expression of the ncRNA overlapping the PR promoter. Addition of the miR-423-5p mimic resulted in a 30% decrease in expression of the ncRNA suggesting that the miRNA moderately destabilized the target ncRNA (Supplementary Figure S5). These results indicate that the ncRNA that overlaps the PR promoter is a target for the miR-423-5p mimic.
Figure 6.
Inhibition of PR expression by miR-423-5p is mediated by AGO2. (A and B) RIP analysis of (A) AGO1 and (B) AGO2 recruitment to the ncRNA overlapping the PR promoter by miR-423-5p. (C) RT–qPCR analysis of AGO1 mRNA expression following treatment with siRNA against AGO1. (D) RT–qPCR analysis of PR mRNA inhibition by miR-423-5p following AGO1 knockdown. (E) RT–qPCR analysis of AGO2 mRNA expression following treatment with siRNA against AGO2. (F) RT–qPCR analysis of PR mRNA inhibition by miR-423-5p following AGO2 knockdown. TF1 = transfection 1, TF2 = transfection 2. siRNAs and miRNA mimics were added to cells at 25 nM for AGO reversal experiments and 50 nM for RIP. Error bars indicate SD (n = 3). P-values were calculated using the two-tailed unpaired Student's t-test with equal variances. **P < 0.01.
To further evaluate the necessity of AGO proteins for inhibiting PR by the miR-423-5p mimic we used siRNAs to knock down expression of either AGO1 or AGO2 prior to miRNA mimic addition (Figure 6C and E). Depletion of AGO1 had no effect on PR expression or the inhibition of PR by miR-423-5p (Figure 6D). Knockdown of AGO2 caused a 2-fold increase in PR expression (Figure 6F). While we routinely observe a 75% decrease in PR expression by miR-423-5p, PR expression was only reduced by 35% in an AGO2 depleted background. AGO2 expression was required for inhibition by both the miR-423-5p mimic and the pre-miR-423 mimic suggesting that our mimic design did not bias the AGO isoform used for inhibition of PR expression (Figure 6D and F).
Effects of miR-423-5p inhibition on PR expression
Our approach in this investigation has been to screen for miRNAs that are capable of silencing transcription using miRNA mimics as a proof-of-principle for miRNA-induced transcriptional silencing and subsequently characterize the mechanism of action. An alternative approach would be to inhibit endogenously expressed miRNAs and evaluate the effects on expression of genes that contain predicted target sites within their promoters. This is more complicated because in addition to the computational target predictions, knowledge about endogenous miRNA expression is required and the endogenous control pathway must be present in the cell types used for the experiment. Furthermore, the tools available for inhibiting miRNAs are not as well characterized as those for mimicking their function. However, miR-423-5p is expressed at detectable levels in T47D cells so we evaluated whether the endogenous miRNA was regulating PR expression.
To inhibit the function of endogenous miR-423-5p, we designed an antisense 2′-O-methyl RNA that was fully complementary to the miRNA (anti-miR). As controls we also designed 2′-O-methyl RNAs consisting of either mismatched (MM) or scrambled (SCR) derivatives of the anti-miR sequence. RT–qPCR confirmed that addition of the anti-miR significantly reduced detection of miR-423-5p (Supplementary Figure S4A). Inhibition of miR-423-5p did not affect PR mRNA expression (Supplementary Figure S4B). This result is not particularly surprising as T47D cells express high basal levels of the PR gene and it is unlikely that the gene is under miRNA-mediated repression in this cell line.
The PR gene is expressed at low basal levels in MCF7 breast cancer cells and we reasoned that it might be a better cell line for studying the effects of endogenous miRNAs on PR expression. Using RT–qPCR, we were able to detect miR-423-5p expression in MCF7 cells and used the anti-miR to inhibit the function of the endogenous miRNA (Supplementary Figure S4C). Addition of the anti-miR had no effect on PR mRNA expression (Supplementary Figure S4D). These data suggest that while miR-423-5p is capable of silencing transcription of the PR gene, the endogenous miRNA is not performing this function in the cell lines we tested and that a physiological role for miR-423-5p in PR regulation, if any, will be found in other cell types or under other physiologic conditions.
Generality of miRNA-induced transcriptional silencing
The potency of miR-423-5p-induced silencing of the PR gene in T47D cells led us to test the effect of synthetic miR-423-5p on PR expression in MCF7 cells. MCF7 cells express PR at low basal levels as compared with T47D cells (9). We transfected miR-423-5p into MCF7 cells and monitored PR expression. Western blot revealed that the miR-423-5p mimic inhibited PR protein expression (Figure 7A). Using RT–qPCR, we observed a >60% reduction in PR mRNA expression (Figure 7B). In addition, the miR-423-5p mimic resulted in a >70% reduction in PR hnRNA levels (Figure 7C). These data demonstrate that miRNA-induced transcriptional silencing of PR is readily observed in two human cell lines with differing basal levels of PR expression.
Figure 7.
Inhibition of PR expression by miR-423-5p in MCF7 cells. (A) Western analysis of PR expression following treatment with miR-423-5p in MCF7 cells. (B and C) RT–qPCR showing inhibition of (B) PR mRNA and (C) PR hnRNA expression by miR-423-5p in MCF7 cells. miRNA mimics were added to cells at 25 nM. Error bars indicate S.D. (n = 3). P-values were calculated using the two-tailed unpaired Student's t-test with equal variances. **P < 0.01.
To further establish the generality of miRNA-induced transcriptional silencing, we searched for additional genes with miR-423-5p target sites within their promoters. Sequence analysis revealed a miR-423-5p target site within a conserved region of the immunoglobulin superfamily member 1 (IGSF1) gene promoter (Figure 8A). miR-423-5p is complementary to a putative promoter-overlapping ncRNA transcribed in the antisense direction relative to the IGSF1 gene at a region spanning from 36 to 8 bases upstream of the IGSF1 TSS. IGSF1 is expressed at very low levels in T47D cells; however, RT–PCR readily detected expression of IGSF1 mRNA in MCF7 cells (Figure 8B). We also detected expression of an RNA species overlapping the IGSF1 promoter that included the miR-423-5p target sequence (Figure 8B).
Figure 8.
Inhibition of IGSF1 expression by miR-423-5p. (A) PhastCons conservation analysis of the miR-423-5p target site within the IGSF1 promoter (seed sequence target site underlined, sequence listed 3′ to 5′). (B) RT–PCR detection of IGSF1 promoter-overlapping ncRNA and mRNA expression. (C and D) RT–qPCR showing inhibition of (C) IGSF1 mRNA and (D) IGSF1 hnRNA expression by miR-423-5p. (E) Alignment of miR-423-5p and mutant miR-423-5p mimics with ncRNA overlapping the IGSF1 promoter (seed sequence show in lower case, mismatched bases underlined). (F) RT–qPCR analysis of IGSF1 mRNA expression following treatment with mutant miR-423-5p mimics. (G) RT–qPCR analysis of IGSF1 mRNA expression following treatment with pre-miR-423 mimic. Duplexes were added to cells at 25 nM. Error bars indicate SD (n = 3). P-values were calculated using the two-tailed unpaired Student's t-test with equal variances. **P < 0.01.
We transfected the miR-423-5p mimic into MCF7 cells and monitored IGSF1 expression using RT–qPCR. We observed a 60% reduction in IGSF1 mRNA expression after treatment with miR-423-5p (Figure 8C). In addition, miR-423-5p caused a >70% reduction in IGSF1 hnRNA levels (Figure 8D). We next tested the effects of our previously designed mutant miR-423-5p mimics on IGSF1 mRNA expression (Figure 8E). Just as we observed with PR, silencing of IGSF1 by miR-423-5p required seed sequence complementarity to the predicted target RNA (Figure 8F). Also consistent with our previous results, expression of IGSF1 was significantly inhibited by pre-miR-423 (Figure 8G). These results underscore the potential of single miRNAs to target multiple gene promoters and regulate transcription.
Transcriptional regulation by miRNAs
miRNAs are powerful regulators of gene expression that function by recognizing complementary RNA sequences. The vast majority of studies on miRNAs have focused on their ability to target sequences within mRNA. Here we show that miRNAs can also recognize ncRNAs overlapping gene promoters and regulate transcription. We have identified multiple promoter-targeting miRNA mimics that inhibit PR expression, suggesting that transcriptional regulation may be a general property of miRNAs. We have also characterized one miRNA mimic in detail and demonstrated that it can target additional gene promoters, further supporting the possibility that recognition of gene promoters by miRNAs may be a general mechanism of gene regulation.
In this study, we have used exogenously added miRNA mimics and have demonstrated their ability to act as potent transcriptional regulators. We chose the mimic approach because we anticipated that the mechanism of transcriptional regulation would be complex. Using miRNA mimics complementary to the PR promoter allowed us to take advantage of our previous detailed characterization of transcription at the PR locus and established assays for studying the regulation PR expression. As a result, we identified several miRNA mimics that inhibit PR expression with potencies that are similar to those previously observed with our designed fully complementary RNAs.
While the addition of exogenous miR-423-5p resulted in potent inhibition of PR transcription, we did not observe a role for endogenous miR-423-5p in regulating PR expression in the two cell lines used in this study (Supplementary Figure S4). It is possible that there is an expression threshold that must be surpassed by miRNAs before they can function as transcriptional regulators. This would be consistent with a recent study in the moss Physcomitrella patens showing that miRNAs can induce transcriptional silencing, but only under conditions where the specific miRNAs become highly expressed (31). Identifying endogenous miRNAs that target gene promoters remains an important goal and will require more complex screening approaches covering many genes, cell types and miRNAs. The mechanistic data provided here should assist in designing those experiments.
Our findings here differ from our previous observations of transcriptional silencing by designed small RNAs. For example, our earlier reports focused on the region spanning just 50 bases upstream of the PR-B TSS and no duplexes that targeted more than 26 bases upstream of the TSS were active. However, three of the inhibitory miRNA mimics we have identified here target more than 500 bases upstream of the TSS (Figure 1A). This report is also the first time we have used duplex RNAs with two imperfectly complementary RNA strands and observed gene silencing (Figure 3A and B). This result demonstrates that small duplex RNAs do not require full complementarity to be processed by the RNAi machinery for transcriptional silencing.
Perhaps the most striking observation in our current study is the degree of imperfect complementarity between the inhibitory miRNA mimics and their target ncRNA. In our prior studies, we designed small duplex RNAs that were fully complementary to the PR promoter and the activity of these silencing RNAs was completely abolished when duplexes contained more than one mismatched base from their target sequence (4). In contrast, all of the inhibitory miRNA mimics we identified were extensively mismatched relative to their target sequence (Supplementary Figure S1).
Small RNAs with imperfect complementarity to their targets face challenges that differ substantially from those that have full complementarity. miRNAs with imperfect complementarity likely have greatly reduced affinity for their target sequences (as compared with fully complementary RNAs) which would increase their difficulty achieving potent and selective binding to targets. To overcome this hurdle, at least in the case of traditional miRNA targeting, 3′-UTRs containing miRNA binding sites with poor complementarity often have multiple miRNA binding sites within the same UTR to promote cooperativity (32). The miRNAs identified in our study, in contrast, are potent inhibitors of transcription despite having several mismatched bases with their target sequences and only one target site within the ncRNA. This result implies that the rules for miRNA target recognition within promoter regions may differ with respect to target site multiplicity and cooperativity.
There have been previous reports of miRNA mimics targeting gene promoters in human cells. The first report of a promoter-targeting miRNA, from Place et al. (25), showed that introduction of exogenous miR-373 induced expression of two genes that contain promoter regions with complementarity to the miRNA. Subsequently, Kim et al. (26) reported that miR-320 targets its own genomic location in cis and silences transcription of an adjacent gene, POLR3D through an AGO1-dependent mechanism. Our data differ significantly from these previous results. Because miR-320 targets its own genomic location, it inherently has full complementarity to its target sequence. We demonstrate that miRNAs do not require full complementarity to recognize their targets within gene promoters and silence transcription. We also show that gene silencing can be achieved by miRNAs that target gene promoters in trans and that a single miRNA can silence transcription of multiple genes. Finally, whereas miR-320 was reported to recruit AGO1 to its target, we find that miRNAs with incomplete complementarity to their targets require AGO2, suggesting the potential for different silencing mechanisms.
Our work provides insights into the mechanism of miRNA-induced transcriptional silencing that were not reported in previous studies. We show that synthetic miRNAs can recruit AGO2 to ncRNA transcripts that overlap their target gene promoter, implicating recognition of ncRNA transcripts in the mechanism of promoter-targeting miRNAs (Figure 6B). In our model system, recognition of the target ncRNA by the miRNA mimic results in moderate reduction in expression of the ncRNA (Supplementary Figure S5). We do not believe that the reduction in ncRNA expression is a major factor in the silencing mechanism. In support of this conclusion, we have previously shown that depletion of the target ncRNA using an RNase H based approach (which is independent of the RNAi machinery) does not affect PR expression (14). Instead, we believe that AGO2 is recruiting other protein factors to the PR promoter that affect transcription of the PR gene. For example, we observed an increase in H3K9 dimethylation following treatment with miR-423-5p suggesting that some of these factors may be chromatin-modifying enzymes (Figure 2D).
Transcriptome studies have revealed that over 70% of gene promoters are overlapped by ncRNA transcripts (33). One function of these ncRNAs may be to serve as targets for miRNAs, which would be consistent with our previous observation that gene promoter sequences are enriched with potential miRNA target sites (27). Transcriptome studies also suggest that over 90% of the human genome is transcribed into RNA, raising the possibility that additional non-coding regions of the genome could serve as targets for miRNAs (33). In support of this hypothesis, we have recently demonstrated that synthetic small duplex RNAs can target regions beyond the 3′-terminus of protein coding genes and regulate transcription of the upstream gene (34). These small RNAs recognize ncRNA transcripts that overlap gene termini in a manner similar to promoter-targeting small RNAs and subsequently regulate transcription through long-range chromosome interactions.
miRNAs have been understood to regulate gene expression at the post-transcriptional level through recognition of 3′-UTRs within mRNA transcripts. While our demonstration of transcriptional silencing involves miRNA mimics rather than endogenous miRNAs, our results are robust and are consistent with the hypothesis that miRNAs can function to recognize ncRNA transcripts that overlap gene promoters. Sequence specific recognition of gene promoters by miRNAs may complement protein transcription factors. In addition, the ability of small RNAs to rapidly evolve specificity for new sequences would have evolutionary advantages.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIGMS 77253 to D.R.C.); The Robert A. Welch Foundation (I-1244); Alnylam Pharmaceuticals; NIH Pharmacological Sciences Training Grant (GM07062 to S.T.Y.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki K, Shijuuku T, Fukamachi T, Zaunders J, Guillemin G, Cooper D, Kelleher A. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J. RNAi Gene Silencing. 2005;1:66–78. [PMC free article] [PubMed] [Google Scholar]
- 3.Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat. Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy D, Shames DS, Minna JD, Corey DR. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat. Chem. Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 5.Pulukuri SM, Rao JS. Small interfering RNA directed reversal of urokinase plasminogen activator demethylation inhibits prostate tumor growth and metastasis. Cancer Res. 2007;67:6637–6646. doi: 10.1158/0008-5472.CAN-07-0751. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Napoli S, Pastori C, Magistri M, Carbone GM, Catapano CV. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J. 2009;28:1708–1719. doi: 10.1038/emboj.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl Acad. Sci. USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Place RF, Jia ZJ, Pookot D, Dahiya R, Li LC. Antitumor effect of dsRNA-induced p21(WAF1/CIP1) gene activation in human bladder cancer cells. Mol. Cancer Ther. 2008;7:698–703. doi: 10.1158/1535-7163.MCT-07-2312. [DOI] [PubMed] [Google Scholar]
- 11.Huang V, Qin Y, Wang J, Wang X, Place RF, Lin G, Lue TF, Li LC. RNAa is conserved in mammalian cells. PLoS ONE. 2010;5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc. Natl Acad. Sci. USA. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA. Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris KV. RNA-directed transcriptional gene silencing and activation in human cells. Oligonucleotides. 2009;19:299–306. doi: 10.1089/oli.2009.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat. Struct. Mol. Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 17.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 18.Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 20.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 21.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 22.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ambros V. The evolution of our thinking about microRNAs. Nat. Med. 2008;14:1036–1040. doi: 10.1038/nm1008-1036. [DOI] [PubMed] [Google Scholar]
- 25.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl Acad. Sci. USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Younger ST, Pertsemlidis A, Corey DR. Predicting potential miRNA target sites within gene promoters. Bioorg. Med. Chem. Lett. 2009;19:3791–3794. doi: 10.1016/j.bmcl.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watts JK, Yu D, Charisse K, Montaillier C, Potier P, Manoharan M, Corey DR. Effect of chemical modifications on modulation of gene expression by duplex antigene RNAs that are complementary to non-coding transcripts at gene promoters. Nucleic Acids Res. 2010;38:5242–5259. doi: 10.1093/nar/gkq258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebralidze A, Wang Y, Petkova V, Ebralidse K, Junghans RP. RNA leaching of transcription factors disrupts transcription in myotonic dystrophy. Science. 2004;303:383–387. doi: 10.1126/science.1088679. [DOI] [PubMed] [Google Scholar]
- 31.Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W. Transcriptional control of gene expression by microRNAs. Cell. 2010;140:111–122. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 32.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gingeras TR. Origin of phenotypes: genes and transcripts. Genome Res. 2007;17:682–690. doi: 10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- 34.Yue X, Schwartz JC, Chu Y, Younger ST, Gagnon KT, Elbashir S, Janowski BA, Corey DR. Transcriptional regulation by small RNAs at sequences downstream from 3′ gene termini. Nat. Chem. Biol. 2010;6:621–629. doi: 10.1038/nchembio.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.