Abstract
The exact mechanism underlying increases in Sp1 and the physiological consequences thereafter remains unknown. In rat primary cortical neurons, oxygen-glucose deprivation (OGD) causes an increase in H2O2 as well as Sp1 in early ischaemia but apparently does not change mRNA level or Sp1 stability. We hereby identified a longer 5′-UTR in Sp1 mRNA that contains an internal ribosome entry site (IRES) that regulates rapid and efficient translation of existing mRNAs. By using polysomal fragmentation and bicistronic luciferase assays, we found that H2O2 activates IRES-dependent translation. Thus, H2O2 or tempol, a superoxide dismutase-mimetic, increases Sp1 levels in OGD-treated neurons. Further, early-expressed Sp1 binds to Sp1 promoter to cause a late rise in Sp1 in a feed-forward manner. Short hairpin RNA against Sp1 exacerbates OGD-induced apoptosis in primary neurons. While Sp1 levels increase in the cortex in a rat model of stroke, inhibition of Sp1 binding leads to enhanced apoptosis and cortical injury. These results demonstrate that neurons can use H2O2 as a signalling molecule to quickly induce Sp1 translation through an IRES-dependent translation pathway that, in cooperation with a late rise in Sp1 via feed-forward transcriptional activation, protects neurons against ischaemic damage.
INTRODUCTION
Translation occurs through two mechanisms: a cap-dependent pathway and a cap-independent pathway that entails activation of an internal ribosome entry site (IRES) motif in the 5′-UTR region of mRNA (1). Under normal conditions, nearly all of mammalian genes are translated into proteins through the cap-dependent pathway. However, sub-environments including pathogenic infections, heat shock and hypoxia can induce cells to rapidly produce sufficient amounts of proteins through the cap-independent pathway (2–4), activating the IRES to cope with the environment.
Sp1 is a member of an extended family of the DNA-binding proteins that have three zinc-finger motifs that bind guanosine- and cytosine-rich DNA (5–8). This family includes members homologous to Sp1 such as Sp2–Sp4. Sp1 has been implicated in regulating expression of many genes by acting in concert with other factors, including members of the TBP-associated factors family (9), cAMP response element-binding (10,11), nuclear factor-κB (12) and vascular endothelial growth factor receptor-2 (VEGFR-2) (13,14). Sp1 is activated during oxidative stress (15,16) and hypoxia (17,18). However, both pro-survival (19) and pro-death (20) genes are driven by Sp1. In hypoxia, for example, Sp1 activation positively correlates with the CD39 expression that enhances myocardical cell survival (19). However, Sp1 has been speculated to reside in the promoter of the SUR1-regulated NC (Ca-ATP) channel (21) that enhances brain oedema. A recent study also revealed that neuronal injury-inducible genes are synergistically regulated by ATF3, c-Jun and STAT3 through interaction with Sp1 in damaged neurons (20). The exact mechanism underlying the activation or up-regulation of Sp1 during ischaemia, however, remains an open question. Further, whether Sp1 is directly involved in the neuroprotection against hypoxic or middle cerebral artery occlusion (MCAO)/reperfusion insult has not been determined.
Ischaemic insults typically increase the levels of reactive oxygen species (ROS); H2O2 is an early-rising ROS and perhaps has the longest half-life of all ROS. Recent studies suggest, however, that the generation of mitochondrial ROS, especially H2O2, is important for neuroprotection (22,23). In neurons, the hypoxic pre-conditioning protecting against hypoxia seen in the GP×1 transgenic murine brain has been speculated to involve H2O2 (24). In addition, H2O2, but not superoxides, protects mature primary cortical neurons against oxygen-glucose deprivation (OGD) insult (25). Studies have also shown that H2O2 induces HIF-1α through the NF-κB pathway to protect neuronal cells (26). Thus, H2O2 and the Sp1 are implicated in neuronal survival. However, whether and how H2O2 is related to Sp1 remains unknown.
In this study, using primary cortical neurons and the MCAO stroke model in rats, we obtained several novel findings that directly address several important issues. These findings are: (i) the 5′-UTR of Sp1 mRNA contains an IRES motif; (ii) the IRES pathway is activated by H2O2; and (iii) Sp1 acts as an endogenous protector against early ischaemic insult.
MATERIALS AND METHODS
Experimental animals and surgical preparation
Male Sprague-Dawley rats weighing 280–320 g were used. They were housed in group cages of two or three rats each in an air-conditioned vivarium with free access to food and water. Throughout the study, a 12:12-h light/dark cycle was maintained with the lights cycle on at 8 AM. All procedures adhered to the Guidelines for Care and Use of Experimental Animals of the National Cheng-Kung University (Tainan, Taiwan). For surgical preparation, rats (280–320 g) were anaesthetized (i.p.) with 10% chloral hydrate. A 23-gauge stainless steel guide cannula was then stereotaxically implanted into the piriform cortex 2 mm dorsal to the left-middle cerebral artery (MCA). The stereotaxic coordinates were modified for this rat strain (0.2 mm anterior, −5.2 mm lateral and −8 mm ventral from the bregma) (27,28). Another 23-gauge stainless steel guide cannula used for intracerebroventricular administration of 2ME2 was stereotaxically implanted into the lateral ventricle (LV; 0.4 mm posterior, −1.3 mm lateral and −3.5 mm ventral, according to a stereotaxic atlas). The cannula was secured with dental cement and two small screws were inserted into the skull. Animals were housed individually, allowed to recover for 3 days, and divided into drug and vehicle groups before stroke induction. Rats were divided into drug or vehicle groups before induction of stroke. Occlusion of the left MCA was induced in conscious rats by administration of endothelin-1 (ET-1, 240 pmol in 10 μl of saline over 5 min) via a 30-gauge injector that protruded 2 mm beyond the end of the previously implanted guide cannula. The injector was held in place by a poly-tubing cuff and the animal was placed in a clear Plexiglas box for observation during the ET-1 injection. To determine whether the MCA was successfully occluded, regional cerebral blood flow was measured in the cortex supplied by the MCA by continuous laser-Doppler flowmetry (Moor Instruments, Devon, England). At 1 day before stroke induction, a small hole (2 mm posterior and 3.5 mm lateral to the bregma) was drilled and a cannula was fixed to the skull with dental cement. A laser-Doppler flowmetry probe (0.5 mm in diameter) was placed on the dura using the cannula and cortical-regional cerebral blood flow was assessed during the entire surgery to ensure MCA occlusion. Only animals with a regional cerebral blood flow reduction of >75% were included (n = 50). It has been reported that the alteration of cerebral blood flow after ET-1 injection could be recovered within 1 h (29). Sham-operated rats were subjected to the same surgical procedure without MCA occlusion.
Culture of cortical neurons and mixed glial cells
Rat cortical neurons were cultured from post-natal Day 0 (P0) to P1. The cortex was dissected and digested in 7 ml of trypsin (10 U/ml) in phosphate-buffered saline at 37°C for 30 min. After rinsing, the tissue was triturated and filtered through a nylon mesh filter (70 µm; Small Parts, Florida, MI, USA). Cells were plated at a density of 0.5 × 106 cells/cm2 onto a plastic culture plate that was pre-coated with poly-l-lysine (50 µg/ml, Sigma, St Louis, MO, USA) and maintained in Neurobasal-A medium supplemented with 100 U/ml penicillin, 0.1 mg/ml streptomycin, 0.5 mM l-glutamine and 5% fetal borine serum (FBS; Invitrogen, Carlsbad, CA, USA). On the next day, 8 µM cytosine arabinoside (Ara-C; Invitrogen) was added to prevent glial cell growth. The proportion of glial cell was <5% in the total population of primary cortical cultured cells. For primary mixed glial cell culture, cells were plated at 2 × 105 cells/cm2 onto a plastic culture plate coated with poly-l-lysine. Cultures including the U87 glioma cells and A549 lung cancer cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 5% FBS and 100 U/ml penicillin. All of the cells were kept at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
Oxygen-glucose deprivation
In the OGD experiment, primary cortical neurons were transferred into serum-free medium containing 5 mM 2-deoxy-d-glucose (Merck, Darmstadt, Germany) for 1 h. The cells were then placed in an anaerobic chamber that was flushed with a mixture of 95% N2 and 5% CO2. After 30-min incubation, cells were removed from the anaerobic chamber to normal conditions. Having determined that 60-min OGD caused 40% of the neurons to become terminal deoxyuridine-triphosphate nick-end labelling (TUNEL)-positive by 24 h, this duration was used for all further experiments, except when the time-course experiments were conducted.
ROS measurements
Hydrogen peroxide levels in the extracellular medium were measured using a Peroxidetect kit (Sigma). Cultured cortical neurons (0.5 × 106 cells/cm2) were treated as described in figure legends. After treatment, 40 μl of the supernatant was analyzed for H2O2 production according to the manufacturer’s protocol. The absorbance of the samples was obtained at 560 nm and corrected for the absorbance of vehicle. The concentration of peroxides in each of the samples was determined and normalized and expressed as nmol/ml.
RNase protection assays
Briefly, equal amounts of cell lysates were incubated with or without RNase A (10 µg/ml) at 0°C and were then mixed with 600 μl of TRI reagent (Invitrogen, Carlsbad, CA, USA) to extract cellular mRNA. Reverse transcription and real-time polymerase chain reaction (RT–PCR) was performed as described. Primers for the detection of Sp1 5′-UTR were 5′-CTGCAAGGGTCTGATTCTCTA-3′and 5′-AGCTTGTCCACCTTGAACTA-3′. The thermal profile used was initiated at 95°C for 2 min (pre-amplification hot start), followed by 40 cycles of PCR at 95°C for 10 s (denaturation), 56°C for 15 s (annealing) and 72°C for 15 s (extension).
Northern blotting
Equal amounts of total RNA from cells with or without OGD were separated with 0.7% agarose gel and then transfected onto a membrane. After UV cross-linking, the membrane was incubated with the probes, biotin-5′-AAAGGTGAAGTTCGTCGTCCAAC-3′ (Renilla) and biotin-5′-GAGCACGGAAAGACGATG-3′ (Firefly) for 16 h. After three washes to remove any free probe, immunoblotting was performed using anti-streptoavidin antibodies.
Polysomal fractionation
Cultured cells were treated with 100 µg/ml cycloheximide 5 min prior to harvest. They were lysed in polysome lysis buffer containing 100 mM KCl, 5 mM MgCl2, 10 mM HEPES, 1% Triton X-100, 0.5% sodium deoxycholate 100 U/ml RNasin, 100 µg/ml of cycloheximide (Calbiochem) and standard protease inhibitors for 2 min on ice. The total RNA in the samples was determined by measuring the optical density at 260 nm (OD260) and equivalent amounts of RNA were loaded onto a 30% (wt/vol) sucrose solution in polysome lysis buffer containing 500 mM KCl and 20 U/ml RNasin without detergents. Samples were centrifuged at 180 000g (for the average radius of the rotor) for 105 min at 4°C in a Sorvall SW41Ti rotor (Newtown, CT, USA). Fractions of equal volume were collected from the top. RNA in each fraction was extracted using TRI reagent (Invitrogen).
Determination of infarct volume and oedema ratio
After 96 h of MCA occlusion, rats were deeply anaesthetized and decapitated and the brain was cut into 2-mm thick coronal block slices. The brain slices were immersed in 1% solution of 2,3,5-triphenyltetrazolium chloride (TTC) solution in normal saline at 37°C for 30 min and then fixed in 4% paraformaldehyde at 4°C. Brain oedema (brain swelling) was quantified by the ratio of the weight (dry/wet) of the hemisphere. The hemispheres were separated and weighed immediately after removal and again after drying in an oven at 105°C for 24 h.
Statistical analysis
All data from three separate experiments are expressed as mean ± SD. Comparisons among multiple groups were performed using one-way analysis of variance (ANOVA) with appropriate post hoc tests, whereas comparisons between two groups were made using Student’s t-test (StatView 5.01; SAS Institute). Values of P < 0.05 were considered statistically significant.
RESULTS
Focal cerebral ischaemia and OGD induce time-dependent expression of Sp1: in vivo and in vitro studies
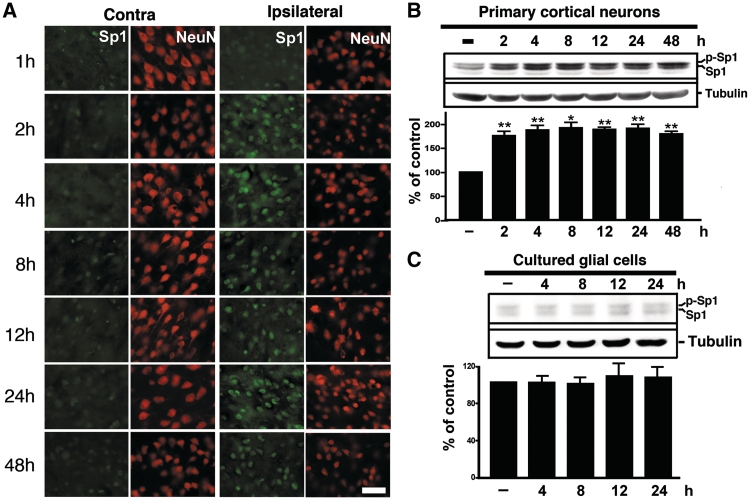
Rats were subjected to the MCAO procedure, and the success of which was verified using behavioural outcomes and Nissl staining for confirmation of hypoxic conditions (Supplementary Figure S1 and Supplementary Videos S1 and S2). Tissues from ischaemic rats were fixed and double-labelled with antibodies against Sp1 and NeuN to examine Sp1 expression in neurons. As illustrated in Figure 1A, Sp1 expression increased after MCAO in the ipsilateral hemispheric neurons but remained unaltered in the contralateral hemisphere. Primary cortical neurons were subjected to OGD in an in vitro ischaemic model. Figure 1B shows that total Sp1 levels (phospho-Sp1 and Sp1) accumulated in primary neurons within 48 h of OGD (ANOVA: F6,14 = 13.57, P < 0.0001). In post hoc analysis, Sp1 expression levels were significantly greater in the OGD group at 2, 4, 8, 12, 24 and 48 h compared with the naïve control group (P < 0.01). These data demonstrate an increase in Sp1 expression from the early period of ischaemia both in vivo and in vitro. OGD treatment did not alter the expression of Sp1 in glial cells (F4,10 = 0.182, P > 0.05; Figure 1C), U87 glioma cells or A549 lung cancer cells (F4,10 = 0.2048, P > 0.05; Supplementary Figure S3) indicating that the OGD-induced up-regulation of Sp1 expression was restricted to neurons.
Figure 1.
Hypoxia/ischaemia-induced Sp1. (A) Double immunostaining of Sp1 and NeuN at various time points of recovery from MCAO was performed. Brain sections were obtained at various time points after MCAO and were subsequently probed with anti-Sp1 and anti-NeuN antibodies. HIF-1α was detected using Alexa 488 (green) while NeuN was detected using Alexa 568 (red). Panels show images taken from the cortex of non-ischemic and ischemic hemispheres. Scale bar, 50 µm. (B and C) The time course of Sp1 expression induced by OGD treatment in primary neurons or glial cells was assessed. Total lysates of primary neurons (B) or glial cells (C) were obtained at various time points following OGD challenge. Sp1 expression was detected by performing immunoblotting with anti-Sp1 antibodies using tubulin as an internal control. All experiments were independently carried out in triplicate and were expressed as a percentage of the levels in the naïve controls. Statistical analysis was carried out using the one-way ANOVA with appropriate post hoc tests. **P < 0.01 versus the naïve control group.
H2O2-induced Sp1 expression
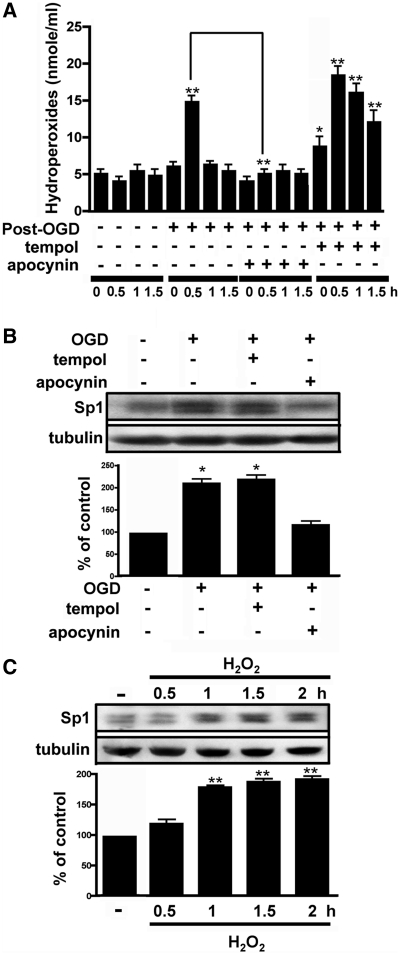
An earlier report indicated that Sp1 is induced by oxidation and that Sp1 is an anti-death transcription factor in cortical neurons (16). We examined here whether free radicals or ROS regulate Sp1 expression after OGD treatment. First, we determined whether OGD could induce H2O2 in primary neurons. As shown in Figure 2A, the concentration of H2O2 increased at 0.5 h after OGD. ANOVA showed a main effect of the group (F15,32 = 42.51, P < 0.0001). Post hoc analysis revealed that at 0.5 h the expression level was significantly greater in the OGD group than in the naïve control group (P < 0.01). Primary neurons that were treated with the NADPH oxidase inhibitor apocynin harboured lower levels of H2O2 0.5 h after OGD as compared to treatment with OGD alone (Figure 2A). Pre-treatment with the superoxide dismutase mimetic tempol increased H2O2 levels. In Newman–Keuls post hoc comparison, the naïve control group differed from neurons that were treated with tempol 1 h before OGD at 0 (P < 0.05), 0.5 (P < 0.01), 1 (P < 0.01) and 1.5 h (P < 0.01). Notably, apocynin treatment blocked the OGD-induced increase in Sp1, and tempol treatment led to higher Sp1 levels (F3,8 = 53.11, P < 0.0001; Figure 2B). In Newman–Keuls post hoc comparison, we also observed differences between the naïve control, OGD and OGD plus tempol groups (P < 0.05). The naïve control and OGD plus apocynin groups did not differ (P > 0.05). Furthermore, we administered H2O2 directly to neuronal cultures and found that it significantly increased Sp1 expression (F4,10 = 89.24, P < 0.0001; Figure 2C), and in Newman–Keuls post hoc comparison, we found differences between the naïve control and the 1-, 1.5- and 2-h time points (P < 0.01). No significant difference was observed between the naïve control and the 0.5-h time point (P > 0.05). Collectively, these results suggest that OGD-induced Sp1 expression involves H2O2. Notably, the OGD-induced rise in H2O2 was transient, peaking at 0.5 h and stabilizing thereafter (Figure 2A).
Figure 2.
Role of H2O2 in Sp1 expression. (A) Primary neurons treated with either normoxia, OGD, OGD with apocynin or OGD with tempol. Total lysates were prepared at the 0.5-, 1- and 1.5-h time points after the recovery from OGD. The concentration of H2O2 was measured as described in Materials and Methods. (B) Cultured cortical neurons were pre-treated with either apocynin or tempol 1 h prior to OGD treatment. Further, the cells were harvested 4 h after treatment, immunoblotting was performed with anti-Sp1 antibodies and tubulin was used as an internal control. The Sp1 expression level was quantified in three independent experiments. (C) Total lysates of the primary neurons were obtained at various time points following H2O2 treatment. Sp1 expression was detected by western blotting using anti-Sp1 antibodies and using tubulin as an internal control. The values represent the mean ± SD. In (B and C), statistical analysis was carried out using the one-way ANOVA with appropriate post hoc tests. *P < 0.05, **P < 0.01 versus the naïve control group.
H2O2 and OGD increase Sp1 translational efficiency at the early stage and Sp1 transcriptional activity at the late period
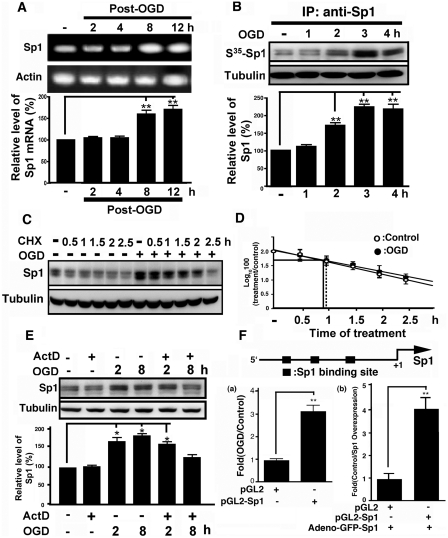
We performed RT–PCR to measure Sp1 mRNA levels in primary neurons to determine the mechanism by which OGD up-regulates Sp1 expression. Total mRNA was extracted from cultured cortical neurons at various time points following OGD treatment. Results revealed that Sp1 mRNA increased after OGD treatment for 8 and 12 h (P < 0.01; Figure 3A). ANOVA showed a main effect of the group (F4,11 = 32.04, P < 0.0001), and by Newman–Keuls post hoc comparison, we observed differences between the naïve control and the 8- and 12-h (P < 0.01) time points. Next, we investigated whether Sp1 protein synthesis increased after OGD treatment. After adding 35S-methionine to the culture medium, we observed that the expression of 35S-methionine-labelled Sp1 expression was significantly up-regulated following OGD treatment (F4,10 = 47.08, P < 0.0001; Figure 3B). Results revealed significant differences between the naïve control and treated groups at the 2, 3 and 4 h (P < 0.01) time points. No significant differences were observed between the naïve control and the 1 h groups (P > 0.05). We also examined protein stability during OGD-induced Sp1 expression by incubating OGD-treated primary neurons and controls with cycloheximide. Cell lysates were prepared and analysed at various time points after cycloheximide treatment. The Sp1 degradation did not differ significantly between OGD-treated neurons and controls (Figure 3C and D). Finally, primary neurons were pretreated with actinomycin D before OGD treatment to inhibit transcriptional activity. As shown in Figure 3E, Sp1 remained up-regulated at 2 h in the OGD-treated neurons even in the presence of actinomycin D; at 8 h, however, this increase vanished (P > 0.05). These results suggest that the up-regulation of Sp1 at the 2 h after OGD correlates with enhanced translation of existing Sp1 mRNAs, whereas OGD-induced Sp1 increases in Sp1 at later time points (i.e. 8 h) are under transcriptional control.
Figure 3.
Mechanism underlying the induction of Sp1 expression by OGD challenge. (A) Primary neurons were exposed to OGD challenge and total RNA was isolated at various time points after the treatment. The mRNA expression of Sp1 was analyzed by RT–PCR. The results are expressed as arbitrary mRNA units and have been normalized to GAPDH expression. (B) At various time points following OGD treatment, the neurons were incubated with a medium containing 35S-methionine for 1 h. Whole-cell lysates were analysed by immunoprecipitation with anti-Sp1 antibodies followed by autoradiography. (C) Primary neurons were treated with cycloheximide (100 μM) in the absence or presence of OGD treatment. The samples were analysed by immunoblotting with anti-Sp1 antibodies using tubulin as an internal control. (D) The level of Sp1 was plotted on logarithmic scales and lines of best fit were calculated for cycloheximide-treated (shaded bar) and cycloheximide plus OGD-treated (open bar) cells. Error bars show SD. (E) Primary neurons were treated with actinomycin D (1μg/ml) in the absence or presence of OGD treatment. The samples were analysed by immunoblotting with anti-Sp1 antibodies using tubulin as an internal control. (F) Sp1 promoter was inserted into pGL3 containing the luciferase gene, and then transfected into primary cells with or without OGD and adeno-GFP-Sp1 virus infection for 24 h to determine luciferase activity. Statistical analysis was carried out using Student’s t-test. *P < 0.05 versus the naïve control group. In (A, B, D and E), all experiments were carried out independently in triplicate and were expressed as a percentage of the levels in the naïve controls. Statistical analysis was carried out using the one-way ANOVA with appropriate post hoc tests. *P < 0.05, **P < 0.01 versus the naïve control group.
Although the rise in H2O2 after OGD peaked at 0.5 h (Figure 2A), the Sp1 levels peaked at 2 h and plateaued thereafter (Figure 1B). Thus, Sp1 from H2O2-induced early spike might bind to the Sp1 promoter to stimulate transcription of Sp1. This model is indirectly supported by the finding that apocynin also blocks Sp1 levels at late time points by preventing H2O2 formation (Figure 2B). Thus, to confirm that binding of Sp1 to the Sp1 promoter stimulated its transcription, the Sp1 promoter was inserted into a pGL3 plasmid that contained the luciferase gene and was examined for luciferase activity under OGD conditions. As shown in Figure 3F, Sp1 transcription was induced in the late stage of OGD (P < 0.01; Student’s t-test). Thus, during early ischaemic insult, Sp1 induction may be attributed to increased translational activity, whereas late Sp1 accumulation may be primarily due to increased transcriptional activity. To confirm that Sp1 can auto-regulate its own promoter activity in neuron cells, Sp1 was expressed by adeno-GFP-Sp1 infection in neuron primary cells, and the promoter activity of Sp1 was studied (Figure 3F, panel b). The data obtained indicated that Sp1 promoter-driven luciferase activity increased significantly upon Sp1 overexpression. Further, these results suggest that ROS influence the Sp1 up-regulation during early ischaemic insult.
Increased ribosome recruitment in a cap-independent manner and the IRES pathway contributes to H2O2-induced increases in Sp1 translational efficiency
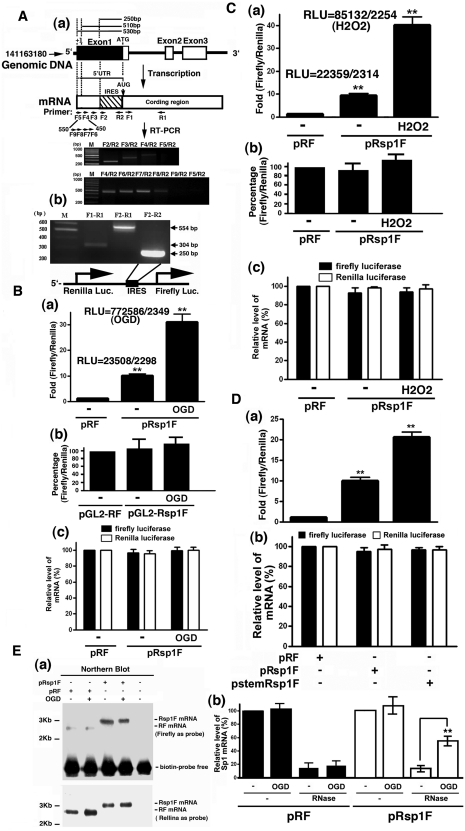
Because Sp1 expression increases within 2 h of OGD, the Sp1 5′-UTR might contain an IRES. The IRES Omnibus WebServer was used to predict the location of a conserved 250-bp IRES motif upstream of the Sp1 coding region. A typical IRES that forms a Y-shift IRES motif (Supplementary Figure S4) was identified in this region. However, because the transcriptional initiation site (+1) and the 5′-UTR of rat Sp1 mRNA have not been fully determined, we speculate that a longer 250-bp 5′-UTR might exist in the 5′ region of rat Sp1 mRNA. To test this possibility, several primers were designed and RT–PCR was performed to estimate the 5′-UTR length. As shown in Figure 4A, a longer 5′-UTR (>510 bp but <530 bp) was thus identified (Figure 4A). Based on this data, we can prove that this IRES motif, the 1st–250th bp from the translational initiation codon, is really located within the 5′-UTR that contains >510 bp. To determine whether the predicted IRES could increase Sp1 mRNA translational efficiency, the Sp1 5′-UTR containing the IRES was inserted into a bicistronic reporter plasmid that harboured Renilla luciferase and firefly luciferase genes. The resulting construct (pRsp1F) was used to determine the effect of the predicted IRES on firefly luciferase expression by transient transfection into cultured cortical neurons. As shown in Figure 4B and C, the Sp1 5′-UTR increased the firefly luciferase–Renilla luciferase ratio by 10-fold and 30-fold without or with OGD or H2O2 treatment in primary neurons, suggesting that the Sp1 5′-UTR uses the IRES to promote translation (Figure 4B and C, panel a). In addition, to rule out the possibility that this region of the 5′-UTR may contain the promoter activity, two internal controls were used to prove the conclusion shown here. First, we constructed a pRsp1F-promoterless plasmid which deleted the SV40 promoter and to study the luciferase activity of Renilla and firefly (Figure 4B and C, panel b). The luciferase activity of Renilla and firefly were nearly abolished. Second, the ratio of Renilla and firefly luciferase activity was studied under OGD or H2O2 treatment (Figure 4B and C, panel c). Data indicated that there was no significant variation between Renilla and firefly luciferase activity. These data imply that the 5′-UTR of Sp1 inserted in the bicistronic plasmid has no promoter activity. To verify that the increased firefly luciferase codons were not translated, but due to the re-initiation of ribosomes that failed to dissociate from the mRNA after encountering the stop codon of the upstream Renilla luciferase, a palindromic sequence was inserted into the transcriptional start site to form a strong stem–loop structure adjacent to the 5′-cap, which inhibits cap-dependent translation (30). Compared with the pRF group, relative firefly luciferase activity increased significantly in the pStemRSp1F and pRSp1F groups (Figure 4D), indicating that the translation of the firefly luciferase does not need the 5′-cap on Renilla mRNA. Conversely, IRES activities are modulated by proteins called IRES trans-activating factors (ITAFs) (31,32). We used the mRNA of the transfected plasmids, pRF and pRsp1F, to perform RNase protection assay, and then detected the region between Renilla and firefly luciferase mRNA by RT-PCR assay to determine whether this IRES region is protected during OGD treatment. As shown in Figure 4E, a Northern blot assay using firefly luciferase-coding sequence (634th–656th bp) and Renilla luciferase-coding sequence (1462th–1479th bp) labelled with biotin as a probe was carried out to detect the bicistronic mRNA (Figure 4E, panel a). Data indicated that only full-length bicistronic mRNA containing the Renilla and firefly luciferase mRNA was observed, not individual Renilla or firefly luciferase mRNA (Figure 4E, panel a). The total RF and Rsp1F mRNA expression did not alter after 2 h of OGD treatment, but nearly all of the RF mRNA was abolished upon RNAse treatment with or without OGD. However, more Rsp1F mRNA shielded from RNAse degradation under OGD condition indicates that the IRES motif forms a Y-shift against RNAse activity (F3,8 = 177.4, P < 0.0001) (Figure 4E, panel b). These results suggest that the Sp1 5′-UTR indeed contains a conserved sequence that promotes translational efficiency through a cap-independent pathway (i.e. the IRES pathway) and that the Sp1 IRES is activated by ROS specifically H2O2.
Figure 4.
The Sp1 5′-UTR contains an IRES. (A) Primers were designed in the different regions of the Sp1 gene including the coding and non-coding region. Total RNA was extracted for preparing the cDNA, which was used as a template for PCR. PCR products were analysed by gel and then inserted into the biscistronic plasmid between the Renilla luciferase gene and firefly luciferase gene. (B and C) Primary neurons were transiently transfected with pRsp1F or pRF (panel a) and pGL2-RF or pGL2-Rsp1F (panel b). Thirty hours post-transfection, the cells were subjected to OGD (B) or H2O2 (C). The value of firefly luciferase activity was divided by that of Renilla luciferase activity and was then finally normalized with that of the pRF only. Real RLU values are shown here. The mRNA levels of firefly luciferase and Renilla luciferase were measured by performing real-time RT–PCR and served as the internal control (B and C, panel c). Statistical significance of the difference between the normoxic and treatment groups was tested using the one-way ANOVA with appropriate post hoc tests. *P < 0.05, **P < 0.01 versus the pRF in the normoxia group. (D) Effect of a 5′ stem–loop on relative expression of the downstream reporter enzyme. Primary neurons were transfected with pRF, pRsp1F or pStemRsp1F. Thirty hours post-transfection, cells were harvested to detect the luciferase activity, and then the IRES activities are represented as ratios of firefly luciferase to Renilla luciferase. The ratios are graphed relative to the ratio obtained with pRF, which was given a value of 1. The data shown are mean (±SEM) of triplicate samples from each of three independent experiments. (E) pRF or pRsp1F were transfected into primary neurons for 12 h, and then were subjected to OGD or were left under control conditions for 2 h before harvesting. Total RNA was extracted and separated with 0.7% gel in a northern blot assay by using the firefly luciferase-coding sequence (1462th–1479th bp) and renilla luciferase-coding sequence (634–656th bp) labelled with biotin as a probe. Finally, the streptavidin and anti-streptavidin were used to find the mRNA signal (panel a). Total lysates of primary neurons were then incubated with RNase (10 µg/ml) at 0°C and the remaining 5′-UTR of the Sp1 IRES region in each group, and the RF mRNA and Rsp1F mRNA was measured by real-time RT–PCR (panel b). These levels were expressed as a percentage of those of the naive controls. Statistical analysis was performed using one-way ANOVA with appropriate post hoc tests. *P < 0.05, **P < 0.01 versus the naive control group.
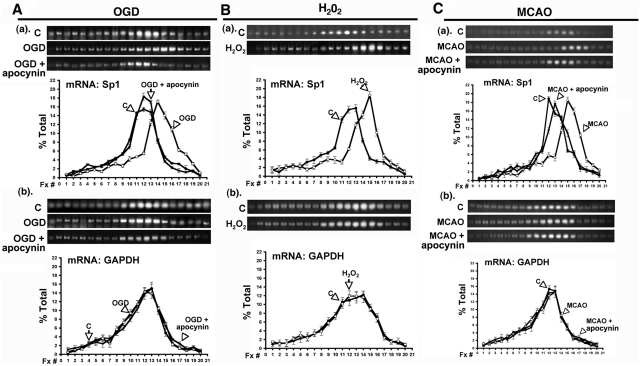
To determine whether Sp1 translational efficiency is altered after OGD or H2O2 treatment, we examined the effects of OGD and H2O2 on Sp1 mRNA distribution on polysomes. We used a linear 30% sucrose solution containing 500 mM KCl to prevent any non-translational interactions between mRNAs and ribosomes. Under normoxic conditions, distribution of Sp1 mRNA on polysomes showed a peak in fraction 12 (Figures 5A–C). OGD led to a selective increase in the number of ribosomes that were associated with Sp1 transcripts, distributing them to the larger polysome fractions 14 and 15 (Figure 5A, panel a). In contrast, glyceraldehydes-3-phosphate dehydrogenase (GAPDH) mRNA remained primarily in polysome fractions 12 and 13 during normoxia and OGD (Figure 5A, panel b). Treatment with apocynin suppressed the OGD-induced shift in the distribution of Sp1 transcripts to a larger polysome fraction (Figure 5A, panel a) but had a minimal effect on the distribution of GAPDH transcripts after OGD (Figure 5A, panel b). Moreover, H2O2 led to an increase in the number of ribosomes that were associated with Sp1 mRNA, distributing them to polysome fractions, 14, 15 and 16 (Figure 5B, panel a) but did not affect the ribosome distribution on GAPDH mRNA (Figure 5B, panel b). Notably, the same effect was observed when ribosomes and Sp1 mRNAs from the brains of MCAO rats were assayed in the same manner. MCAO rats showed an increase in the number of ribosomes that were associated with Sp1 mRNA with more polysomes present in fractions 15, 16 and 17 (Figure 5C, panel a), whereas no such changes were seen for GAPDH mRNA (Figure 5C, panel b). Thus, these results indicate that in vitro OGD and in vivo MCAO raise intracellular ROS concentrations, especially that of H2O2, thus stimulating Sp1 translational efficiency of Sp1 through the IRES-dependent pathway to increase Sp1.
Figure 5.
Ribosomal distribution with respect to Sp1 mRNA during OGD or H2O2 treatment. (A) Panel a shows the effect of normoxia, OGD and OGD with apocynin on the polysomes distribution in cultured cortical neurons with respect to Sp1 mRNAs. GAPDH mRNA served as an internal control (A panel b). (B) Panel a shows the effect of H2O2 in cultured cortical neurons on polysomes distribution with respect to Sp1 mRNA, with GAPDH mRNA serving as an internal control (B, panel b). (C) Panel a shows the effect of MCAO in rats on polysomes distribution with respect to Sp1 mRNA and GAPDH mRNA was used as an internal control (C, panel b). RNA in each fraction was extracted and the product of RT–PCR was presented. The results are shown as a percentage of the specific mRNA in each fraction, where the total amount of the specific mRNA in all fractions was set at 100%. The data shown are mean (±SEM) of triplicate samples from each of three independent experiments.
Protective effect of Sp1 following OGD- or MCAO-induced brain damage
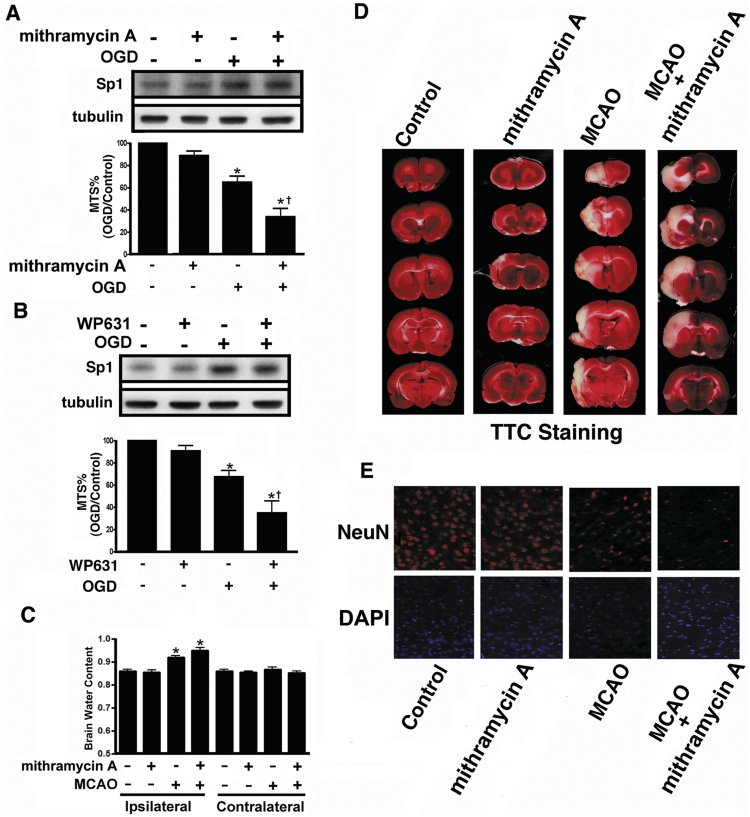
The potential neuroprotective effects of Sp1 were examined in vitro using primary cortical neurons and in vivo using the MCAO stroke model in rats by blocking Sp1 binding to DNA with mithramycin A or WP631. Further, the Sp1 short hairpin RNA (shRNA) was used to knock-down Sp1 in primary neurons to examine Sp1 function in neuronal survival. As shown in Figure 6A and B, 24-h OGD in primary cortical neurons reduced cell viability. After mithramycin A or WP631 treatment, cell viability did not decrease significantly compared with OGD treatment alone; indicating that Sp1 induction during OGD protects cells. Next, we examined whether endogenous Sp1 confers protection against MCAO-induced brain injury by injecting mithramycin A into the injured regions of ischaemic brains 0.5 h after MCAO. Brain oedema, as indicated by increased brain/water content, was typically observed 48 h after the MCAO procedure. The water content in the MCAO ipsilateral hemispheres increased significantly compared with the sham-control group (F7,16 = 20.48, P < 0.0001; Figure 6C). Mithramycin A administered 0.5 h after MCAO further increased the mean water content of the ipsilateral hemispheres relative to those in the MCAO group. Brain water content in the contralateral hemispheres did not differ between the MCAO and MCAO-mithramycin A groups (Figure 6C). Furthermore, we measured the extent of damage 4 days after MCAO using 2,3,5-triphenyl tetrazolium chloride. Cerebral infarction in the rat brains of each group is shown in Figure 6D. The white areas indicate infarction. Severe infarction was observed in all of the MCAO rats. Administration of mithramycin A 0.5 h after MCAO significantly increased infarct volume compared to treatment with MCAO alone (Figure 6D). Conversely, in immunohistochemistry using anti-NeuN antibodies, administration of mithramycin A, 0.5 h after MCAO decreased the number of NeuN-positive neurons in ipsilateral hemispheres compared to treatment with MCAO alone (Figure 6E).
Figure 6.
Effects of Sp1 inhibitors on brain injury after hypoxia/ischaemia challenge. (A and B) Primary neurons were treated with mithramycin A (A) or WP631 (B), followed by OGD treatment. Viability of the cultured cortical neurons was assessed by MTS assay in which Sp1 expression level was used as an internal control. Experiments were carried out independently in triplicate. The values that were normalized to the protein content are provided as the mean ± SD (n = 3) and are expressed as percent viability relative to untreated cells. Statistical analysis was carried out using one-way ANOVA with appropriate post hoc tests. *P < 0.05 versus the naïve control group; †P < 0.01 versus the OGD group. (C) Hemispheres were separated and weighed immediately after removal and again after drying in an oven at 105°C for 24 h. Data are presented as mean ± SD (n = 4). (D) Infarct area was evaluated by TTC staining. Representative sections from the indicated treatments after 4 day of recovery are shown. (E) Brain sections were obtained 4 day after MCAO recovery, probed with anti-NeuN antibodies and detected with Alexa 568 (red). The nucleus was stained with 4′, 6-diamidino-2-phenylindole (DAPI; blue). Scale bar, 50 μm. Statistical analysis was carried out using one-way ANOVA with appropriate post hoc tests. *P < 0.05 versus the sham-control group.
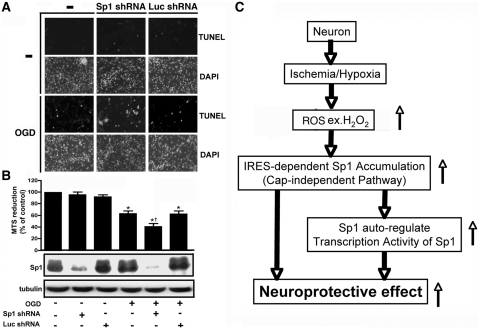
Finally, on TUNEL staining of Sp1 knock-down primary neurons, the number of apoptotic cells was higher in the Sp1 shRNA plus OGD group than in the OGD group (Figure 7A). In addition, Sp1 knock-down by shRNA attenuated cell viability compared with OGD treatment alone (Figure 7B). Differences in cell viability were observed between the naïve control, OGD, OGD plus Sp1 shRNA and the OGD plus luciferase shRNA groups (control shRNA) (Figure 7B). However, under conditions in which OGD was not induced, cell viability did not differ between the control, Sp1 shRNA and luciferase shRNA groups (P > 0.05). Thus, Sp1 apparently regulates cell survival only under hypoxic/ischaemic conditions.
Figure 7.
Effect of shRNA targeting Sp1 on cellular apoptosis after ischaemic challenge. (A) shRNA targeting Sp1 or luciferase was transfected into primary cortical neurons 30 h before OGD. TUNEL staining was performed 96 h after OGD challenge. A TUNEL-positive cell counting analysis was determined under each condition. Scale bars, 50 μm. (B) Sp1 levels in OGD-treated or OGD-untreated primary neurons were knocked-down by Sp1-shRNA. Viability of the cultured cortical neurons was assessed by the MTS assay, for which Sp1 expression levels were used as an internal control. All experiments were carried out independently in triplicate. The values that were normalized to the protein content are provided as mean ± SD (n = 3) and are expressed as percent viability relative to the untreated cells. Statistical analysis was carried out using the one-way ANOVA with appropriate post hoc tests. *P < 0.05 versus the naïve control group; †P < 0.05 versus OGD plus luciferase shRNA group. (C) The model proposed here was used to elucidate the accumulation of Sp1 and its role under ischaemic conditions.
DISCUSSION
This study described novel findings addressing several important issues with regard to neuronal survival. The early rise in H2O2 during OGD/ischaemic insult might be used by neurons as a signalling mechanism to elicit endogenous neuroprotective activity in neurons. The 5′-UTR of Sp1 is longer than expected and contains an IRES. H2O2 can activate the Sp1 IRES in neurons and increase Sp1 translation. Ischemia-activated Sp1 is neuroprotective. Finally, the early rise in Sp1 auto-activates the Sp1 promoter in a forward manner increasing late transcription of Sp1 (Figure 7C).
As a ROS, H2O2 is recognized as a pro-death molecule, because it reacts with superoxide anion to form hydroxyl radicals or reacts with nitric oxide to form peroxynitrate. Several reports have indicated that H2O2 is neuroprotective (16), however, our in vitro and in vivo results suggest that H2O2 is used by neurons as a signalling molecule to trigger endogenous neuroprotective mechanisms. We observed that neurons respond to H2O2 and use it to activate the IRES region of Sp1 to mediate a relatively early and rapid rise in Sp1 through cap-independent translation. However, it is still not clear why H2O2 activates Sp1 translation only in neurons and not in glial cells. Perhaps neurons and glial cells have disparate ITAFs that respond differently to H2O2, but more experiments are needed to confirm this hypothesis. Regarding the Sp1 IRES, specifically the 5′-UTR of Sp1, although much is known about this important transcription factor, the exact length of its 5′-UTR has not been really established. To this end, we identified a much longer 5′-UTR of Sp1 and discovered that it contains an IRES region, confirming our hypothesis that H2O2 activates Sp1 translation through a fast mechanism (i.e. via the IRES system rather than the cap-operated system). H2O2 activates the IRES of Sp1 in neurons, enabling them to respond quickly to ischaemic attack.
Sp1 is known to be pro-apoptotic and pro-survival, and the neuronal injury-inducible gene is synergistically regulated by ATF3, c-Jun and STAT3 through the interaction with Sp1 in damaged neurons but the detailed function of Sp1 in ischaemic insult or stroke is less well understood (20,21). Using both inhibitors that block Sp1 binding to DNA and using shRNAs against Sp1, we demonstrated that Sp1 is neuroprotective both in vitro and in vivo. As discussed, the early rise in H2O2 induces an increase in Sp1 through a fast translational system involving the IRES. In this regard, we observed that the upstream promoter of Sp1 contains a G/C-rich region that serves as an Sp1-binding motif to enhance Sp1 transcription. Based on our data, we propose a model in which H2O2 causes an early rise in Sp1 that auto-activates the Sp1 promoter in a forward manner, causing a subsequent rise in Sp1 during the course of ischaemic insult. We speculate that the rises in Sp1 during the early or late phases protect neurons against ischaemia-induced injuries. Based on a bioinformatics screen of gene expression under hypoxic conditions (GEO accession no. GSE63), it is notable that most genes activating during hypoxic conditions are regulated by Sp1 and are related to cell survival (Supplementary Figure S2). In contrast, other genes such as cyclooxygenase-2 and caveolin-1 (33) are also activated by Sp1 and regulate the pathology of neurodegenerative diseases, including Huntington and Alzheimer disease (34–36). Exactly which genes are activated by Sp1 and how they are related to neuronal survival requires further investigation.
In conclusion, we have observed several novel findings and have shown that the ROS H2O2 plays a critical pro-survival role in neurons during ischaemic insult by rapidly causing an early rise in the transcription factor Sp1 through an IRES-dependent translational mechanism. Thus, agents that increase H2O2 such as tempol or other agents that artificially activate the IRES system may constitute new, effective therapeutic interventions against ischaemic insult and stroke.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Cheng-Kung University project of the Program for Promoting Academic Excellence and Developing World Class Research Centers, National Science Council, Taiwan (NSC 97-2320-B-006-016-MY3 and NSC 97-2311-B-006-002-MY3); Intramural Research Program, National Institute on Drug Abuse, National Institutes of Health, Department of Health and Human Services, USA. Funding for open access charge: NSC, Taiwan.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Blaszczyk L, Dutkiewicz M, Ciesiolka J. [Translation of eukaryotic mRNA in a cap-independent mode] Postepy Biochem. 2007;53:400–412. [PubMed] [Google Scholar]
- 2.Dinkova TD, Zepeda H, Martinez-Salas E, Martinez LM, Nieto-Sotelo J, de Jimenez ES. Cap-independent translation of maize Hsp101. Plant J. 2005;41:722–731. doi: 10.1111/j.1365-313X.2005.02333.x. [DOI] [PubMed] [Google Scholar]
- 3.Bornes S, Prado-Lourenco L, Bastide A, Zanibellato C, Iacovoni JS, Lacazette E, Prats AC, Touriol C, Prats H. Translational induction of VEGF internal ribosome entry site elements during the early response to ischemic stress. Circ. Res. 2007;100:305–308. doi: 10.1161/01.RES.0000258873.08041.c9. [DOI] [PubMed] [Google Scholar]
- 4.Conte C, Riant E, Toutain C, Pujol F, Arnal JF, Lenfant F, Prats AC. FGF2 translationally induced by hypoxia is involved in negative and positive feedback loops with HIF-1alpha. PLoS ONE. 2008;3:e3078. doi: 10.1371/journal.pone.0003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs MR, Kadonaga JT, Bell SP, Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 6.Lania L, Majello B, De Luca P. Transcriptional regulation by the Sp family proteins. Int. J. Biochem. Cell. Biol. 1997;29:1313–1323. doi: 10.1016/s1357-2725(97)00094-0. [DOI] [PubMed] [Google Scholar]
- 7.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y, Hwang CK, Junn E, Lee G, Mouradian MM. ZIC2 and Sp3 repress Sp1-induced activation of the human D1A dopamine receptor gene. J. Biol. Chem. 2000;275:38863–38869. doi: 10.1074/jbc.M007906200. [DOI] [PubMed] [Google Scholar]
- 9.Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, Mouradian MM, Young AB, Tanese N, Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 10.Liu YW, Tseng HP, Chen LC, Chen BK, Chang WC. Functional cooperation of simian virus 40 promoter factor 1 and CCAAT/enhancer-binding protein beta and delta in lipopolysaccharide-induced gene activation of IL-10 in mouse macrophages. J. Immunol. 2003;171:821–828. doi: 10.4049/jimmunol.171.2.821. [DOI] [PubMed] [Google Scholar]
- 11.Nucifora FC, Jr, Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson VL, et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 12.Fridmacher V, Kaltschmidt B, Goudeau B, Ndiaye D, Rossi FM, Pfeiffer J, Kaltschmidt C, Israel A, Memet S. Forebrain-specific neuronal inhibition of nuclear factor-kappaB activity leads to loss of neuroprotection. J. Neurosci. 2003;23:9403–9408. doi: 10.1523/JNEUROSCI.23-28-09403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meissner M, Stein M, Urbich C, Reisinger K, Suske G, Staels B, Kaufmann R, Gille J. PPARalpha activators inhibit vascular endothelial growth factor receptor-2 expression by repressing Sp1-dependent DNA binding and transactivation. Circ. Res. 2004;94:324–332. doi: 10.1161/01.RES.0000113781.08139.81. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka T, Kanai H, Sekiguchi K, Aihara Y, Yokoyama T, Arai M, Kanda T, Nagai R, Kurabayashi M. Induction of VEGF gene transcription by IL-1 beta is mediated through stress-activated MAP kinases and Sp1 sites in cardiac myocytes. J. Mol. Cell. Cardiol. 2000;32:1955–1967. doi: 10.1006/jmcc.2000.1228. [DOI] [PubMed] [Google Scholar]
- 15.Lee J, Kosaras B, Aleyasin H, Han JA, Park DS, Ratan RR, Kowall NW, Ferrante RJ, Lee SW, Ryu H. Role of cyclooxygenase-2 induction by transcription factor Sp1 and Sp3 in neuronal oxidative and DNA damage response. FASEB J. 2006;20:2375–2377. doi: 10.1096/fj.06-5957fje. [DOI] [PubMed] [Google Scholar]
- 16.Ryu H, Lee J, Zaman K, Kubilis J, Ferrante RJ, Ross BD, Neve R, Ratan RR. Sp1 and Sp3 are oxidative stress-inducible, antideath transcription factors in cortical neurons. J. Neurosci. 2003;23:3597–3606. doi: 10.1523/JNEUROSCI.23-09-03597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee M, Bikram M, Oh S, Bull DA, Kim SW. Sp1-dependent regulation of the RTP801 promoter and its application to hypoxia-inducible VEGF plasmid for ischemic disease. Pharm. Res. 2004;21:736–741. doi: 10.1023/b:pham.0000026421.09367.b3. [DOI] [PubMed] [Google Scholar]
- 18.Miki N, Ikuta M, Matsui T. Hypoxia-induced activation of the retinoic acid receptor-related orphan receptor alpha4 gene by an interaction between hypoxia-inducible factor-1 and Sp1. J. Biol. Chem. 2004;279:15025–15031. doi: 10.1074/jbc.M313186200. [DOI] [PubMed] [Google Scholar]
- 19.Eltzschig HK, Kohler D, Eckle T, Kong T, Robson SC, Colgan SP. Central role of Sp1-regulated CD39 in hypoxia/ischemia protection. Blood. 2009;113:224–232. doi: 10.1182/blood-2008-06-165746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiryu-Seo S, Kato R, Ogawa T, Nakagomi S, Nagata K, Kiyama H. Neuronal injury-inducible gene is synergistically regulated by ATF3, c-Jun, and STAT3 through the interaction with Sp1 in damaged neurons. J. Biol. Chem. 2008;283:6988–6996. doi: 10.1074/jbc.M707514200. [DOI] [PubMed] [Google Scholar]
- 21.Simard JM, Chen M, Tarasov KV, Bhatta S, Ivanova S, Melnitchenko L, Tsymbalyuk N, West GA, Gerzanich V. Newly expressed SUR1-regulated NC(Ca-ATP) channel mediates cerebral edema after ischemic stroke. Nat. Med. 2006;12:433–440. doi: 10.1038/nm1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emerling BM, Platanias LC, Black E, Nebreda AR, Davis RJ, Chandel NS. Mitochondrial reactive oxygen species activation of p38 mitogen-activated protein kinase is required for hypoxia signaling. Mol. Cell. Biol. 2005;25:4853–4862. doi: 10.1128/MCB.25.12.4853-4862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzy RD, Hoyos B, Robin E, Chen H, Liu L, Mansfield KD, Simon MC, Hammerling U, Schumacker PT. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Sheldon RA, Aminoff A, Lee CL, Christen S, Ferriero DM. Hypoxic preconditioning reverses protection after neonatal hypoxia-ischemia in glutathione peroxidase transgenic murine brain. Pediatr. Res. 2007;61:666–670. doi: 10.1203/pdr.0b013e318053664c. [DOI] [PubMed] [Google Scholar]
- 25.Furuichi T, Liu W, Shi H, Miyake M, Liu KJ. Generation of hydrogen peroxide during brief oxygen-glucose deprivation induces preconditioning neuronal protection in primary cultured neurons. J. Neurosci. Res. 2005;79:816–824. doi: 10.1002/jnr.20402. [DOI] [PubMed] [Google Scholar]
- 26.Bonello S, Zahringer C, BelAiba RS, Djordjevic T, Hess J, Michiels C, Kietzmann T, Gorlach A. Reactive oxygen species activate the HIF-1alpha promoter via a functional NFkappaB site. Arterioscler. Thromb. Vasc. Biol. 2007;27:755–761. doi: 10.1161/01.ATV.0000258979.92828.bc. [DOI] [PubMed] [Google Scholar]
- 27.Macrae IM, Robinson MJ, Graham DI, Reid JL, McCulloch J. Endothelin-1-induced reductions in cerebral blood flow: dose dependency, time course, and neuropathological consequences. J. Cereb. Blood Flow Metab. 1993;13:276–284. doi: 10.1038/jcbfm.1993.34. [DOI] [PubMed] [Google Scholar]
- 28.Sharkey J, Ritchie IM, Kelly PA. Perivascular microapplication of endothelin-1: a new model of focal cerebral ischaemia in the rat. J. Cereb. Blood Flow Metab. 1993;13:865–871. doi: 10.1038/jcbfm.1993.108. [DOI] [PubMed] [Google Scholar]
- 29.Bogaert L, Scheller D, Moonen J, Sarre S, Smolders I, Ebinger G, Michotte Y. Neurochemical changes and laser Doppler flowmetry in the endothelin-1 rat model for focal cerebral ischemia. Brain Res. 2000;887:266–275. doi: 10.1016/s0006-8993(00)02959-0. [DOI] [PubMed] [Google Scholar]
- 30.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 31.Holcik M, Korneluk RG. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holcik M, Gordon BW, Korneluk RG. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell. Biol. 2003;23:280–288. doi: 10.1128/MCB.23.1.280-288.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dasari A, Bartholomew JN, Volonte D, Galbiati F. Oxidative stress induces premature senescence by stimulating caveolin-1 gene transcription through p38 mitogen-activated protein kinase/Sp1-mediated activation of two GC-rich promoter elements. Cancer Res. 2006;66:10805–10814. doi: 10.1158/0008-5472.CAN-06-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu Z, Norflus F, Singh B, Swindell MK, Buzescu R, Bejarano M, Chopra R, Zucker B, Benn CL, DiRocco DP, et al. Sp1 is up-regulated in cellular and transgenic models of Huntington disease, and its reduction is neuroprotective. J. Biol. Chem. 2006;281:16672–16680. doi: 10.1074/jbc.M511648200. [DOI] [PubMed] [Google Scholar]
- 35.Zhai W, Jeong H, Cui L, Krainc D, Tjian R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 36.Santpere G, Nieto M, Puig B, Ferrer I. Abnormal Sp1 transcription factor expression in Alzheimer disease and tauopathies. Neurosci. Lett. 2006;397:30–34. doi: 10.1016/j.neulet.2005.11.062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.