Abstract
Histone modification regulates gene expression, and one major regulatory step in this process is the ability of proteins that recognize epigenetic marks to recruit enzymes required to specify transcriptional outcome. Here we show that BRD7 is a component of hSWI–SNF complexes that interacts with PRMT5 and PRC2. Recruitment studies revealed that BRD7 co-localizes with PRMT5 and PRC2 on ‘suppressor of tumorigenecity 7’ (ST7) and retinoblastoma-like protein 2 (RBL2) promoters in patient-derived B cell lines, and that its association with these target genes correlates with hypermethylation of H3R8, H4R3 and H3K27. Furthermore, inhibition of BRD7 expression reduces PRMT5 and PRC2 recruitment to ST7 and RBL2 promoters; however, only ST7 becomes transcriptionally derepressed. Evaluation of the PRMT5- and PRC2-induced epigenetic marks revealed that while H3(Me2)R8, H4(Me2)R3 and H3(Me3)K27 marks are erased from the ST7 promoter, demethylation of RBL2 promoter histones is incomplete. We also show that the arginine demethylase (RDM) JMJD6, which can erase PRMT5-induced H4R3 methylation, and the H3K27-lysine-specific demethylases, KDM6A/UTX and KDM6B/JMJD3, are differentially recruited to ST7 and RBL2. These findings highlight the role played by BRD7 in PRMT5- and PRC2-induced transcriptional silencing, and indicate that recruitment of specific RDMs and KDMs is required for efficient transcriptional derepression.
INTRODUCTION
Transcriptional silencing of tumor-suppressor genes is a common trait of cancer cells, and it is known that there are multiple regulatory steps that can inhibit gene expression. Epigenetic modification of genomic DNA and histones has been tightly linked to chromatin organization and transcriptional regulation, and it has become apparent that there are different classes of proteins involved in the establishment, recognition and maintenance, as well as removal of chromatin marks (1–5). Extensive work has been done over the last decade to characterize enzymes that can modify chromatin, but little has been learned about factors involved in binding histone epigenetic marks and their contribution to gene regulation. One notable example that illustrates the importance of proteins that bind epigenetically modified histones is in the case of Trp-Asp repeat-containing protein 5 (WDR5), a component of mixed lineage leukemia (MLL) complexes, which binds dimethylated H3K4 and facilitates its conversion to triply methylated K4 (6–8). Besides the WD40 motif of WDR5, several other domains have been identified in proteins that exhibit high affinity for epigenetically modified histones. For instance bromodomain, a highly conserved 110 amino acid region, has been shown to preferentially bind acetylated histones and is believed to be involved in mediating protein–protein interactions in both nuclear and cytosolic compartments (9–13).
Among bromodomain-containing proteins that influence gene regulation by promoting protein–protein interactions is BRD7, which was initially identified through a yeast two-hybrid screen as a protein that can interact with the first PDZ (Post-synaptic density protein [PSD95], Drosophila disc large tumor suppressor [DlgA] and zonula occludens-1 protein [zo-1]) domain of protein tyrosine phosphatase-BAS like (PTP-BL). Both northern blot and in situ hybridization analyses showed that BRD7 is ubiquitously expressed in all tissues and at all mouse embryonic stages. Moreover, immunofluorescence experiments indicated that BRD7 is localized predominently in the nucleus, suggesting that it might play a role in signaling events mediated by the PTP-BL multiprotein complex (14). In a separate study, BRD7 was also shown to interact with dishevelled-1 (Dvl-1) and promote β-catenin and TCF4-induced transcription. Further characterization of the BRD7-Dvl-1 interaction indicated that BRD7 enhances Wnt signaling by inducing glycogen synthase kinase-3β (GSK-3β) dephosphorylation at tyrosine 216 and nuclear translocation of β-catenin (15). Therefore, based on these protein–protein interaction studies, a model was proposed in which BRD7 is believed to bring PTP-BL to the Dvl-1/axin/APC/GSK-3β/β-catenin complex where it facilitates GSK-3β dephosphorylation and promotes nuclear translocation of β-catenin.
In addition to BRD7, several transcription factors and histone-modifying enzymes have been shown to contain one or more copies of the bromodomain, and structural studies have clearly demonstrated that bromodomain is a chromatin-targeting module specialized in recognizing acetylated histones (9,11,16). BRD7 can interact with the four core histones, and deletion of its bromodomain abolishes these interactions (17). Because BRD7 binds active chromatin and positively influences Wnt signaling-induced gene transcription, it is believed that its association with target genes occurs only when genes are turned on. However, recent reports have indicated that BRD7 can inhibit expression of E2F3, DP2 and MEK1 (18,19). BRD7 has also been shown to be an integral component of the BRG1-based hSWI–SNF chromatin remodeling complex, and that it is involved in both target gene activation as well as repression in embryonic stem cells (20). Even though BRD7 has been implicated in target gene repression, it is unclear how it contributes to this process.
In this report, we show that BRD7 is a component of PRMT5-containing hSWI–SNF complexes, and we also show using a cell line that stably expresses His-tagged BRD7 (His-BRD7) that subunits of the BRG1 and BRM complexes are tightly associated with BRD7. Interaction of BRD7 with PRMT5-containing hSWI–SNF complexes was also confirmed by glutathione S-transferase (GST) pull-down assays. In addition, we were able to determine that BRD7 is capable of interacting with three core subunits of the polycomb repressor complex 2 (PRC2) including SUZ12. To further understand the relevance of these molecular interactions, we evaluated recruitment of BRD7 and SUZ12 to known PRMT5 and hSWI–SNF target genes. Chromatin immunoprecipitation (ChIP) experiments revealed that BRD7, SUZ12 and PRMT5 co-localize on ST7 and retinoblastoma-like protein 2 (RBL2) promoters, and that their association with these target tumor-suppressor genes correlates with hypermethylation of H3R8, H4R3 and H3K27 in patient-derived mantle cell lymphoma (MCL) and chronic lymphocytic leukemia (CLL) cell lines. Furthermore, when we evaluated the contribution of BRD7 to PRMT5 and PRC2 target gene expression, we found that inhibition of BRD7 expression results in loss of PRMT5 and PRC2 recruitment as well as complete removal of their epigenetic marks at the ST7, but not RBL2 promoter. We have also determined that different histone arginine demethylases (RDMs) and lysine-specific demethylases (KDMs) are involved in transcriptional reactivation of PRMT5 and PRC2 target genes, and that their recruitment differs in a promoter specific manner.
MATERIALS AND METHODS
Plasmid DNA constructs
Plasmid pBABE-puromycin/His-BRD7 was generated by first subcloning a 1.9-kb BRD7 cDNA-encoding amino acids 2–651, which was PCR-amplified from the pCMV6-XL5/BRD7 vector (Origene Technologies Inc.) using forward (5′-CCGCTCGAGGGCAAGAAGCACAAGAAGCACAAG-3′) and reverse (5′-CCGCTCGAGTCAACTTCCACCAGGTCCACACTC-3′) primers that incorporate a XhoI restriction site, into XhoI-digested pET15b vector. Next, the His-tagged BRD7 cDNA was excised out of pET15b/His-BRD7 vector as a ClaI-XbaI fragment and treated with klenow before inserting into SnaBI-linearized pBABE-puromycin. To generate plasmid pBABE-puromycin/Fl-BAF57, the cDNA-encoding full-length BAF57 was PCR amplified from pBS(KS+)/BAF57, which was described previously (21), using a forward primer (5′-CCAGGAATTCATGTCAAAAAGACC-3′) that introduces an EcoRI restriction site, and a reverse primer (5′-CAGGAATTCCTCATTATTTGTCATCGTCGTCCTTGTAGTCTTGTTTTTTCTCATCTTCTGGTATGGG-3′) that introduces a flag epitope tag before a stop codon and EcoRI restriction site. Next, the EcoRI-digested PCR fragment was inserted into the corresponding site of pBABE-puromycin. Plasmid pGEX-2TK/BRD7 (amino acids 2–651), which expresses GST–BRD7 used in GST pull-down assays and antibody production, was constructed by cloning a SmaI-digested 1.9-kb fragment into SmaI-linearized pGEX-2TK. Both forward (5′-TCCCCCGGGGGGCAAGAAGCACAAGAAGCACAAG-3′) and reverse (5′-TCCCCCGGGTCAACTTCCACCAGGTCCACACTC-3′) primers used to amplify BRD7 (amino acids 2–651) included a SmaI restriction site. Plasmids for in vitro expression of PRMT5, mSIN3A and hSWI–SNF subunits, as well as pGEX-2TK/PAH2, which expresses GST fused in frame with the mSIN3A paired amphipathic helix 2 (PAH2) (amino acids 300–404) have been described previously (21). To express MEP50 in vitro, plasmid pBS(KS+)/MEP50 was generated by inserting a 1-kb EcoRI fragment, which was PCR-amplified from a human peripheral blood cDNA library using forward (5′-CCGGAATTCGGCGTCCAGTTTGAGTCTAGGTTG-3′) and reverse (5′-CCGGAATTCGGCAAAGAAGTGGACACTCATGG-3′) primers, into EcoRI-linearized pBS(KS+). Plasmid pBS(KS+)/Fl-BRD7 was constructed by inserting a 2-kb modified BRD7 cDNA fragment, which was generated through a two-step nested PCR-amplification at the 5′-end of the clone using ∼50 bp of the PRMT5 5′-UTR, into pBS(KS+) cut with XbaI. The first PCR amplification was carried out using a 5′ primer (5′-GTGGACAGCGCGAGGAGAAAGATGATGATGGGCAAGAAGCACAAGAAG-3′), which introduces 27 nt of the PRMT5 5′-UTR region (underlined) upstream of the BRD7 start codon, and a 3′ primer (5′-GCTCTAGATCATTTGTCATCGTCGTCCTTGTAGTCACTTCCACCAGGTCCACACTC-3′) designed to include a flag-tag sequence followed by a stop codon and an XbaI restriction site. To further extend the 5′-UTR region of BRD7, we conducted a second PCR amplification using a second 5′ primer (5′-GCTCTAGAGTGATTGGCTACTAGTATCAAGGAATCCCGGCGTGGACAGCGCGAGGAGAAAG-3′), which incorporates an additional 32 nt of the PRMT5 5′-UTR (underlined) and an XbaI restriction site, and the same 3′ primer used above. Plasmids for in vitro expression of the PRC2 subunits, SUZ12, EED, EZH2 and RBAp48 were derived from pFASTBAC vectors that were described previously (22). Each cDNA was excised out of the respective pFASTBAC vector and cloned into pBS(KS+). Plasmid pBS(KS+)/SUZ12 was generated by inserting a 2.2-kb EcoRI fragment into EcoRI-linearized pBS(KS+), pBS(KS+)/EZH2 was constructed by inserting a 2.25-kb BamHI fragment into BamHI-digested pBS(KS+), and pBS(KS+)/RBAp48 was constructed by ligating a 1.4-kb EcoRI–KpnI insert into pBS(KS+) digested with the corresponding restriction enzymes. To induce EED protein expression in bacterial cells, plasmid pGEX-2TK/EED was generated by inserting a 1.3-kb PCR-amplified BamHI–EcoRI fragment, which encodes EED amino acids 2–441, into pGEX-2TK cut with the corresponding restriction enzymes. The full-length EED cDNA sequence was amplified from pCMV6-XL5/EED (Origene Technologies Inc.) using the following forward: 5′-CGCGGATCCTCCGAGAGGGAAGTGTCGACTGCG-3′, and reverse: 5′-CCGGAATTCTTATCGAAGTCGATCGCAGCGCCA-3′ primers. To produce antibodies that recognize SUZ12, plasmid pGEX-2TK/SUZ12 (amino acids 496–737) was constructed by cloning a 0.7-kb SmaI–EcoRI fragment, which was amplified using forward (5′-TCCCCCGGGAGATGGCTCCTATGCAGGAAAT-3′) and reverse (5′-CCGGAATTCTTGTTTTTTGCTCTGTTTTGAAAC-3′) primers, into SamI–EcoRI-linearized pGEX-2TK.
Cell culture and establishment of stable cell lines
HeLa S3, HeLa S3/Fl-BAF45, HeLa S3/Fl-BAF57 and HeLa S3/His-BRD7 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS). When Fl-BAF45, Fl-BAF57 and His-BRD7 expressing cells were grown, puromycin was added at final concentration of 2.5 µg/ml. Normal B cells were isolated from tonsilar tissues obtained from Children’s Hospital through the Cooperative Human Tissue Network as described previously (23). Transformed JeKo and WaC3CD5 cells were cultured in RPMI-1640 supplemented with 10% FBS. To establish HeLa S3/Fl-BAF57 and HeLa S3/His-BRD7 cell lines, 70–80% confluent plates were incubated with 3 ml of retroviral supernatant harvested from Bing cells transfected with either pBABE/Fl-BAF57 or pBABE/His-BRD7 as described previously (24,25). After 2 days, drug-resistant Fl-BAF57 and His-tagged BRD7 cells were selected in the presence of 2.5 µg of puromycin per milliliter. Several puromycin-resistant colonies were grown and positive clones were identified by western blotting using either anti-flag (Santa Cruz) or anti-His (Covance, Inc.) antibodies.
To generate WaC3CD5/shBRD7 cells, 5 × 106 WaC3CD5 cells were electroporated with 15 µg of either control shRNA or a cocktail of four BRD7 specific-shRNA plasmids (QIAGEN), which contain BRD7 specific sequences, in 100 µl of the appropriate reagent using the Amaxa Biosystems nucleofector. Following electroporation, cells were plated in 10 ml of RPMI-1640 supplemented with 10% FBS, and 48 h later puromycin was added at a final concentration of 2.5 µg/ml. To select individual clones that express low levels of BRD7, electroporated cells were diluted to a final concentration of 0.8 cell/100 µl and plated in 96-well plates. Puromycin-resistant clones were expanded in 6-well plates and analyzed by western blotting and real-time RT–PCR to determine the levels of endogenous BRD7.
Purification of Fl-BAF45, Fl-BAF57 and His-BRD7 complexes
Flag tagged hSWI–SNF complexes were purified by incubating 80 mg of nuclear extract from either HeLa S3/Fl-BAF45 or HeLa S3/Fl-BAF57 cells with 1 ml of anti-flag M2 agarose beads as described previously (26). To confirm interaction of BRD7 with hSWI–SNF complexes, 50 mg of extract from His-tagged BRD7 cells was incubated with 1 ml of nickel–nitrilotriacetic acid (Ni–NTA) agarose beads (QIAGEN) for 14 h at 4°C in the presence of 16 U of micrococcal nuclease and 5 mM CaCl2. Extract from His-BRD7 cells was prepared by lysing cells in RIPA buffer [50 mM Tris–HCl (pH 7.5), 250 mM NaCl, 0.5% sodium deoxycholate, 1% NP-40, 0.1% SDS] and diluted 4-fold in buffer A [50 mM Tris–HCl (pH 7.5), 10% glycerol, 2 mM EDTA, 4 mM DTT, 2 mM PMSF] before incubating with Ni–NTA resin, which was prepared by washing with 100 mM NiSO4 solution followed by a wash with buffer B [12.5 mM Tris–HCl (pH 7.5), 62.5 mM NaCl, 0.125% sodium deoxycholate, 10 mM imidazole, 0.25% NP-40, 0.025% SDS]. Next, bound Ni–NTA beads were packed in a column and the flow through was collected and run twice through the column before washing with buffer C [20 mM HEPES–KOH (pH 7.9), 5 mM MgCl2, 10% glycerol, 0.5 mM EDTA, protease inhibitors] supplemented with 500 mM NaCl, followed by another wash with buffer C containing 100 mM NaCl and 8 mM imidazole. Bound proteins were eluted in buffer C containing 100 mM NaCl and 300 mM imidazole, and fractions were analyzed by sodium dodecyl sulfate (SDS)–8% polyacrylamide gel electrophoresis followed by either silver staining or western blotting.
Histone extraction and immunoprecipitation assays
To extract bulk histone from WaC3CD5 and WaC3CD5/shBRD7 cells, ∼1 × 107 cells were washed twice with 10 ml of 1X PBS and resuspended in 1 ml of TEB buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, 0.5% Triton X-100, 0.02% NaN3, 2 mM PMSF). Samples were incubated on ice for 10 min, and cells were collected by centrifugation. Cell pellets were washed with 0.5 ml TEB buffer, resuspended in 250 µl of 0.2 N HCl, and incubated at 4°C for 14 h. Samples were then centrifuged at 14 000 rpm for 10 min at 4°C, and the supernatants containing bulk histones were collected and used to measure the levels of PRMT5- and PRC2-induced methylation marks.
To show that interaction of BRD7 with hSWI–SNF complexes is specific, nuclear extract from HeLa S3/Fl-BAF45 cells and RIPA extract from HeLa S3/His-BRD7 were incubated with either preimmune or immune anti-CBP antibody essentially as described previously (27). After a 4-h incubation at 4°C, 65 µl of a 50% slurry of protein A agarose beads was added to each sample, and the reaction mixtures were incubated at 4°C for 14 h. Beads were collected by centrifugation, washed five times with 0.5 ml of washing buffer [40 mM Tris–HCl (pH 8.0), 180 mM NaCl], and bound proteins were analyzed by western blotting. When HeLa S3 RIPA extract was incubated with either preimmune or immune anti-BRD7 antibody, samples were treated as described above and washed five times with 0.5 ml of washing buffer supplemented with 120 mM NaCl, 0.5% NP-40 and 0.3% sodium deoxycholate. Next, bound endogenous proteins were detected by western blot analysis.
Protein identification by mass spectrometry and western blot analysis
Protein mixtures purified by anti-flag affinity chromatography were concentrated into a single 1-mm wide band by electrophoresing through an SDS ‘4% stacking gel’ until entering the ‘10% separating gel’. Next, the gel was stained with Coommassie blue and the stacked band was excised out. In-gel tryptic digests were subjected to a micro-clean-up procedure on a 2-µl bed-volume of Poros 50 R2 reversed-phase beads, which was packed in an Eppendorf gel-loading tip before samples were analyzed using a QSTAR-XL hybrid quadrupole time-of-flight mass spectrometer (AB/MDS Sciex) equipped with a NanoSpray ion source (AB/MDS Sciex) (28). Peptide mixtures (in 20 µl) were loaded onto a trapping guard column (0.3 × 5-mm PepMap C18 100 cartridge from LC Packings) using a Tempo nano MDLC system (Applied Biosystems, Inc.) at a flow rate of 20 µl/min. A detailed description of the capillary liquid chromatography coupled to tandem mass spectrometry (LC–MS/MS) analysis has recently been published (29).
Initial protein identifications from MS/MS fragment ion data were done using the Mascot search engine (Matrix Science, version 2.3.01) (30) and the NCBI (National Library of Medicine, National Institutes of Health) and IPI (International Protein Index, EBI, Hinxton, UK) databases. One missed cleavage site is allowed, precursor ion mass tolerance = 0.4 Da, fragment ion mass tolerance = 0.4 Da, peptide modifications are allowed for Met-oxide, Cys-acrylamide and using significance threshold score P < 0.001. Unique peptide and percent sequence coverages for all identified proteins are exported to Excel for further analysis.
To visualize purified proteins, fractions containing Fl-BAF45 or His-BRD7 complexes were separated on an SDS–8% polyacrylamide gel and detected by western blotting using enhanced chemiluminescence reagents according to the manufacturer’s recommendations (GE Healthcare). Antibodies raised against hSWI–SNF subunits and PRMT5 have been described previously (21,24,27). Anti-mSIN3A, anti-flag, anti-JMJD6 and anti-CBP antibodies were purchased from Santa Cruz, and anti-His antibody was purchased from Covance. Rabbit polyclonal antibodies raised against GST–BRD7 (amino acids 2–651) and GST–SUZ12 (amino acids 496–737) were generated by Covance. Anti-H3(Me2)R8 and anti-H4(Me2)R3 have been described previously (23,27,31), while anti-H3(Me3)K27, anti-UTX were purchased from Abcam. Anti-JMJD3 was purchased from Millipore.
GST protein expression and pull-down assays
Expression of GST, GST-PAH2A, GST-BRD7 and GST-EED was carried out essentially as described previously (32). Bacterial strains carrying pGEX-2TK plasmids for expression of either GST or GST fusion proteins were induced with 1.5 mM IPTG for 4 h at 37°C. To purify induced proteins, 500 µg of bacterial extract was incubated with 30 µl of a 50% slurry of GST beads on ice for 30 min. Bound GST beads were washed three times with buffer STE-100 (20 mM Tris–HCl [pH 7.6], 5 mM MgCl2, 100 mM NaCl, 1 mM EDTA) supplemented with 0.5% NP-40 and 1% bovine serum albumin (BSA), and blocked for 4 h at 4°C in 250 µl of STE-100 containing 1 mg of uninduced bacterial extract per ml, 1% BSA, 0.5% Carnation milk and 100 µg of ethidium bromide per ml. Next, 35S-labeled proteins were synthesized using the TNT-coupled rabbit reticulocyte lysate as specified by the manufacturer (Promega), and ∼9 × 104 cpm of each in vitro-translated protein was incubated with immobilized GST or GST fusion proteins at 4°C for 16 h. Samples were washed three times with 200 µl of STE-100 supplemented with 50 mM NaCl, 1% BSA and 0.5% Carnation milk, and twice with STE-100 before the retained proteins were analyzed by SDS–polyacrylamide gel electrophoresis followed by autoradiography.
ChIP and ChIP-re-ChIP assays
Cross-linked chromatin was prepared as described previously (21), except that after sonication, samples were digested with 1 unit of MNase per milliliter at 37°C for 20 min before adding 200 µl of stop buffer [100 mM Tris–HCl (pH 8.6), 0.45% SDS, 2.5% Triton X-100, 5 mM EDTA (pH 8.0), protease inhibitors]. Chromatin was analyzed by agarose gel electrophoresis to ensure that DNA fragment sizes do not exceed 500 bp, and ChIP assays were carried out as described previously (23,31). Chromatin samples were pre-cleared with 30 µl of protein A sepharose beads, which were treated overnight with blocking buffer (0.2 mg of salmon sperm DNA per ml, 0.5 mg of BSA per ml), before incubation with specific antibodies. To amplify ST7, RBL2 and ST5 promoter sequences, pre-immune and immune antibodies were incubated with pre-cleared chromatin from control and transformed B cells at 4°C for 5 h. Next, nucleoprotein complexes were immunoprecipitated by adding pre-blocked protein A sepharose beads at 4°C for 16 h. Samples were then washed extensively and bound nucleoprotein complexes were eluted in 200 µl of elution buffer [50 mM Tris–HCl (pH 8.0), 10 mM EDTA, 1% SDS] at 65°C for 20 min. After elution, samples were incubated at 65°C for 12 h to reverse cross-links, and treated with buffer D (0.4 mg of proteinase K per milliliter, 0.06 mg of yeast tRNA per milliliter) for 2 h at 37°C, followed by phenol and chloroform extraction. Samples were resuspended in 40 µl of Tris–EDTA (pH 8.0) supplemented with 0.2 µg of RNase A/µl, and incubated at 37°C for 30 min before performing real time PCR using the following gene-specific primer sets and probes: ST7 (forward, 5′-CCACTTGGCCTTCTCTTTC-3′; reverse, 5′-GGTCCCTACAAGTGGCTTT-3′; ABI probe, 5′-6FAM-CCCTCGCGTTCTGGGTCCATT-MGBNFQ-3′), RBL2 (forward, 5′-ATTTTTGGCCCCCTTGAA-3′; reverse, 5′-GCACCCGTAGTCTTGAGCAC-3′; Roche universal probe no. 3), ST5 (forward, 5′-CGCCACGAAAGGTCAGAG-3′; reverse, 5′-CTTAAGCTCCGATACCT GCTG-3′; Roche universal probe no. 17), GAS1 (forward: 5′-GCGACAGTGAGCCTCTCC-3′; reverse: 5′-GATCTGGTCCCGCTCTCC-3′; Roche universal probe no. 77) and GAS2 (forward: 5′-CTTTTCCTCCCCACGTATCC-3′; reverse: 5′-GGAGTAAAGAAGCACAGGTCTTG-3′; Roche universal probe no. 13).
For ChIP-re-ChIP assays, immunoprecipitated nucleoprotein complexes were first eluted twice with 75 µl of elution buffer containing 20 mM dithiothreitol (DTT) at room temperature for 15 min. The eluates were combined, and 600 µl of ChIP dilution buffer [100 mM Tris–HCl (pH 8.6), 5 mM EDTA] was added to each sample. Next, the second immunoprecipitation was conducted by adding the appropriate antibody and pre-blocked protein A sepharose beads. Samples were washed, and bound nucleoprotein complexes were eluted and processed as described above.
Reverse transcription (RT) real-time PCR
To measure the BRD7, ST7, RBL2 and ST5 mRNA levels, ∼1 µg of total RNA was reverse transcribed in a 20-µl reaction containing 2.5 µM random hexamer primers and Taqman Reverse Transciption reagents (Applied Biosystems, Inc). Next, real-time-PCR was carried out using the TaqMan system in a 10-µl reaction as described previously (23). Primer sets and probes used to detect BRD7 and its target genes were as follows: BRD7 (forward, 5′-CAACCTATGGGGAAGACTCTG-3′; reverse, 5′-TCTGCCATGACATACGGATAA-3′; Roche universal probe no. 89), ST7 (forward, 5′-CCCCT AATTGCTTCCTCTACC-3′; reverse, 5′-CCAAGAGAATATAAGCAGTTGCAC-3′; Roche universal probe no. 14), RBL2 (forward, 5′-TTGTTGGGTGCTTTTTATATATGC-3′; reverse, 5′-TTTCCATAAACTAAGTCCAAAGCA-3′; Roche universal probe no. 62) and ST5 (forward, 5′-TTTTTGTGGCAGATAAGCTCAG-3′; reverse, 5′-GGAGAAGGGGTAGAGCAAGG-3′; Roche universal probe no. 78). To normalize mRNA levels, we measured the levels of 18 S rRNA in both control and BRD7 knockdown cell lines using 1X pre-mixed 18 S primer/probe set (Applied Biosystems, Inc).
Statistical analysis
Results were expressed as the means ± standard deviations unless otherwise specified. Paired t-tests were used to generate P-values for comparisons between two groups.
RESULTS
BRD7 is associated with PRMT5-containing hSWI–SNF chromatin remodeling complexes
We have previously shown by mass spectrometry of individually excised and stoichiometric gel bands that affinity purified flag-tagged hSWI–SNF complexes contain protein arginine methyltransferase PRMT5, and we have also confirmed this interaction by immunoprecipitation of endogenous complexes purified by conventional chromatography (21,27). To further characterize the subunit composition of hSWI–SNF complexes, protein mixtures purified by anti-flag affinity chromatography were concentrated into a single 1-mm wide band by electrophoresing through an SDS–10% polyacrylamide gel. After staining with Coomassie blue, the 1-mm wide band was excised out and analyzed by mass spectrometry. Protein identification revealed that in addition to the common BRG1 and BRM-associated factors (BAFs), PRTM5-containing hSWI–SNF complexes purified from two distinct HeLa cell lines, which express either Fl-BAF45 or Fl-BAF57, included novel subunits such as BRD7 (Supplementary Table S1).
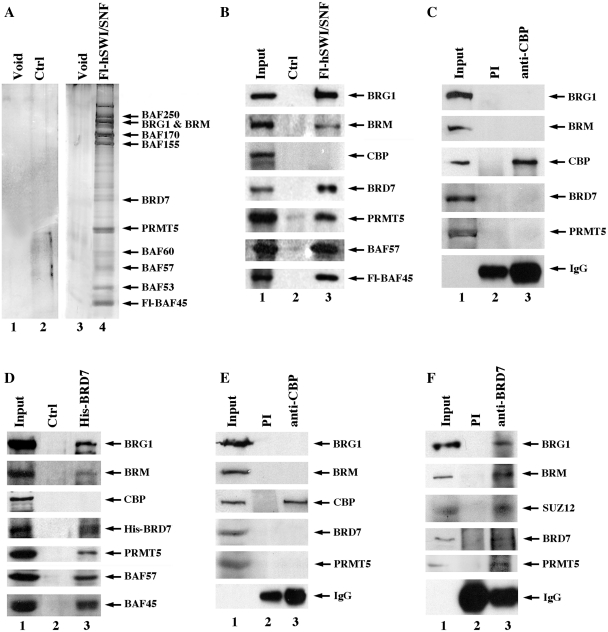
In light of the recent findings, which show that BRD7 is a bromodomain-containing protein capable of interacting with the four core histones and is involved in transcriptional repression of target genes (17,20), we sought to examine and characterize its association with PRMT5-containing hSWI–SNF complexes (Figure 1). Nuclear extracts from either control HeLa S3 or HeLa S3 cells that express Fl-BAF45 were incubated with anti-flag M2 affinity gel, and after extensive washing the retained proteins were eluted with a 20-fold molar excess of flag peptide. Consistent with the mass spectrometry results, silver staining and western blot analysis of affinity-purified Fl-BAF45 complexes revealed the presence of characteristic hSWI–SNF subunits such as BRG1, BRM, BAF57, BAF45 and PRMT5 (Figure 1A and B). Using a highly specific anti-BRD7 antibody, we were able to confirm the presence of BRD7 in purified Fl-hSWI–SNF fractions but not in control HeLa S3 fractions. When we tested for the presence of CREB-binding protein (CBP), we were unable to detect it in affinity-purified Fl-BAF45 fractions. Similarly, when we checked immunopurified anti-CBP fractions using antibodies that can recognize BRG1, BRM, BRD7 and PRMT5, there was no signal detected (Figure 1C), indicating that association of BRD7 with hSWI–SNF is specific.
Figure 1.
BRD7 co-purifies with PRMT5-containing hSWI/SNF complexes. (A) Nuclear extracts from either control (Ctrl) HeLa S3 (lanes 1 and 2) or HeLa S3/Fl-BAF45 cells (lanes 3 and 4) were purified by affinity chromatography using anti-flag M2 agarose beads, and 5 µl of void and peak fractions was analyzed by silver staining. (B) Western blot analysis was performed on 15 µl of affinity-purified control HeLa S3 (lane 2) and flag-tagged hSWI/SNF complexes (lane 3) using the indicated antibodies. The input lane shows expression of hSWI–SNF subunits in 20 µg of HeLa S3/Fl-hSWI–SNF nuclear extract. (C) Nuclear extract (250 µg) from HeLa S3/Fl-BAF45 cells was immunoprecipitated using either preimmune (PI) (lane 2) or immune anti-CBP antibody (lane 3), and western blot analysis was conducted using the indicated antibodies. Input represents 20 µg of HeLa S3/Fl-BAF45 nuclear extract. (D) RIPA extract from either control HeLa S3 or HeLaS3/His-BRD7 cells were incubated with Ni–NTA agarose beads, and after extensive washing ∼15 µl of affinity-purified control HeLa S3 (lane 2) or His-BRD7 fraction (lane 3) was analyzed by western blotting as described in (B). Input shows levels of BRD7-associated proteins in 20 µg of HeLa S3 RIPA extract. (E) HeLa S3/His-BRD7 RIPA extract was immunoprecipitated with either PI (lane 2) or anti-CBP antibody (lane 3), and proteins were detected by western blotting using the indicated antibodies. (F) Approximately 250 µg of HeLa S3 RIPA extract was immunoprecipitated with PI (lane 2) and immune anti-BRD7 antibody (lane 3), and BRD7-associated proteins were detected by western blot analysis.
To further verify interaction of BRD7 with PRMT5-containing hSWI–SNF complexes, we established a stable HeLa cell line that expresses His-tagged (His) BRD7 and proceeded to affinity-purify BRD7 and its associated partners. Both silver staining and western blot analysis showed that BRG1, BRM, BAF57, BAF45 and PRMT5 co-purify with His-BRD7 (Figure 1D, data not shown). In stark contrast, BRD7 and hSWI–SNF subunits were not detected in control HeLa S3 fractions. Moreover, western blot analysis revealed that CBP was absent in His-BRD7 affinity-purified fractions, and that components of hSWI/SNF were not detected in anti-CBP immunoprecipitates (Figure 1D and E). These results suggest that reciprocal interaction of BRD7 with hSWI/SNF complexes is highly specific.
To rule out the possibility that interaction of BRD7 with PRMT5–hSWI–SNF complexes is not a direct result of overexpressed subunits, we immunoprecipitated endogenous BRD7 complexes and checked for the presence of BRG1, BRM, SUZ12 and PRMT5 (Figure 1F). All four subunits were detected in anti-BRD7 immunoprecipitates but not in control preimmune fractions, suggesting that association of BRD7 with hSWI–SNF and SUZ12 can be detected at the endogenous level.
BRD7 directly interacts with hSWI–SNF subunits, PRMT5 and MEP50
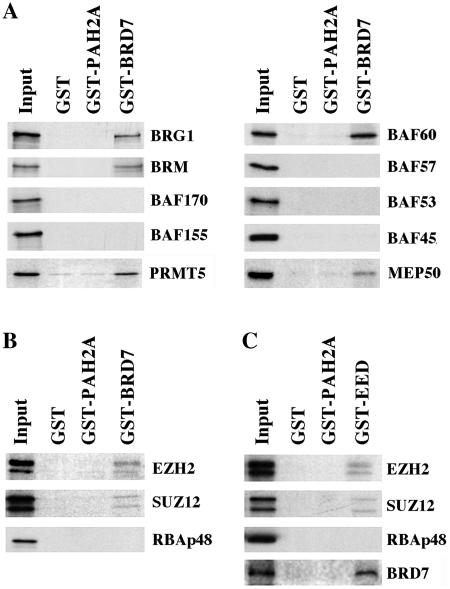
Having found that BRD7 co-purifies with PRMT5-containing hSWI–SNF complexes by affinity chromatography on two distinct columns, we wanted to ascertain the specificity of their interaction by identifying the subunits that mediate interaction with BRD7. GST pull-down assays were carried out using bacterially expressed GST-full length BRD7 and individually in vitro translated hSWI–SNF subunits. When GST–BRD7 was incubated with 35S-labeled hSWI–SNF subunits, BRG1, BRM, PRMT5 and BAF60 were able to interact with BRD7, but not with control GST or GST–PAH2A (Figure 2A). A specific interaction was also observed between BRD7 and MEP50, a protein known for its ability to interact with PRMT5 and components of the PRC2 complex (33).
Figure 2.
BRD7 can directly interact with hSWI/SNF and PRC2 components. (A and B) Equal amounts (1–2 µg) of GST, GST-PAH2 or GST-BRD7 were immobilized on glutathione agarose beads, and incubated with 35S-labeled hSWI–SNF and PRC2 subunits. After extensive washing the retained proteins were detected by autoradiography. The input lane represents 10% of the total amount of protein used in each reaction. (C) Similar amounts of GST, GST-PAH2 or GST-EED were bound to glutathione agarose beads before adding 35S-labeled PRC2 subunits and BRD7. Samples were processed and detected as described in A.
Since BRD7 had previously been shown to interact with the four core histones and we have found that it interacts with MEP50, we examined whether BRD7 could interact with components of the PRC2 complex. GST pull-down assays showed that both SUZ12 and EZH2 specifically interacted with GST–BRD7, while RBAp48 failed to form a complex with BRD7 (Figure 2B). Because we have been unable to express the fourth PRC2 subunit in vitro, we generated a GST–EED fusion protein and performed GST pull-down assays using in vitro translated and 35S-labeled BRD7 (Figure 2C). As expected, GST-immobilized EED was able to interact with components of the PRC2 complex including SUZ12 and EZH2, but not RBAp48. Similarly, GST–EED was also able to interact with BRD7 suggesting that multiple interactions occur between BRD7 and subunits of the PRC2 complex.
BRD7 co-localizes with PRMT5, PRC2 and their epigenetic marks on ST7 and RBL2 promoters in transformed lymphoma and leukemia cell lines
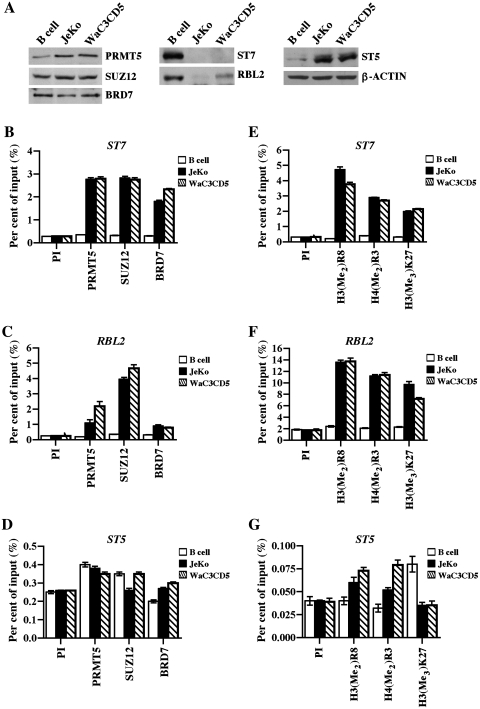
We have previously determined that PRMT5 is involved in transcriptional repression of ST7 and RBL2 in transformed B cells (23,31). To investigate the role played by BRD7 in PRMT5 target gene expression and to further explore the relationship between BRD7, PRMT5 and PRC2, we monitored their proteins levels and recruitment to the promoter region of known target genes in normal B cells as well as transformed patient-derived MCL (JeKo) and B-CLL (WaC3CD5) cell lines (Figure 3). Western blot analysis revealed that while PRMT5 protein levels were increased in transformed B cells, expression of SUZ12 and BRD7 proteins was unaffected (Figure 3A). When we measured the levels of target genes, we found that unlike ST5, expression of ST7 and RBL2 proteins was significantly reduced in JeKo and WaC3CD5 cells. Consistent with our previous findings, ChIP experiments showed that PRMT5 is enriched 4- to 9.7-fold at the ST7 and RBL2 promoters in both JeKo (P < 10−4 for ST7 and P < 10−3 for RBL2) and WaC3CD5 (P < 10−3 for ST7 and P = 0.0018 for RBL2) cell lines compared to B cells (Figure 3B and C). To determine if PRC2 and BRD7 are recruited to PRMT5 target genes, we assessed their association with the ST7 and RBL2 promoters. When an antibody that recognizes SUZ12, an integral component of the PRC2 repressor complex, was used to immunoprecipitate cross-linked chromatin from either normal or transformed B cells, we noticed that there was a 9.4- to 16-fold increase in its recruitment to the ST7 (P < 10−4 in JeKo and P < 10−3 in WaC3CD5) and RBL2 (P < 10−4 in JeKo and P < 10−3 in WaC3CD5) promoters in both transformed B cell lines compared to normal B cells (Figure 3B and C).
Figure 3.
PRMT5, SUZ12 and BRD7 co-localize on ST7 and RBL2 promoters. (A) RIPA extracts from normal CD19+ B cells or transformed JeKo MCL and WaC3CD5 B-CLL cell lines were analyzed by western blotting using the indicated antibodies. Anti-β-ACTIN was used to show equal loading. (B) Cross-linked chromatin from either normal B cells or transformed JeKo and WaC3CD5 cells was subjected to immunoprecipitation using control PI, anti-PRMT5, anti-SUZ12 or anti-BRD7 antibody, and the immunoprecipitated DNA was analyzed by real-time PCR using primer sets and probes specific for ST7 (amplicon 1: from –102 to +6) (B), RBL2 (amplicon 1: from –85 to –16) (C) and control ST5 (amplicon 1: from –118 to –59) (D). (E–G) ChIP assays were performed using either PI or immune antibodies that can specifically recognize PRMT5-induced methylation marks, H3(Me2)R8 and H4(Me2)R3 or PRC2-induced H3(Me3)K27, and real-time PCR was performed to detect ST7, RBL2 and ST5 promoter sequences as described in (B–D). Enrichment was measured as per cent of input. ChIP experiments were performed twice and PCR reactions were carried out in triplicate.
Similarly, interaction of BRD7 with the ST7 and RBL2 promoters was enhanced 3- to 8-fold in JeKo (P = 10−3 for ST7 and P < 10−4 for RBL2) and WaC3CD5 (P = 0.0265 for ST7 and P < 10−2 for RBL2) cell lines compared to normal B cells (Figure 3B and C), indicating that PRMT5, SUZ12 and BRD7 co-exist on common target genes. To verify if association of PRMT5, SUZ12 and BRD7 is unique to ST7 and RBL2, we tested their interaction with the promoter region of ‘ST5′ (Figure 3D). Real-time PCR analysis of immunoprecipitated DNA showed that there was no enrichment of PRMT5, SUZ12 and BRD7 at the ST5 promoter in both normal and transformed B cells. To further confirm our findings, we evaluated recruitment of PRMT5, SUZ12 and BRD7 at different ST7, RBL2, and ST5 promoter sites using a distinct set of primers (Supplementary Figure S1). Consistent with our current results, recruitment of PRMT5 was enhanced 2- to 5-fold (P < 10−3) at the ST7 promoter and 4.7- to 8-fold (P < 10−3) at the RBL2 promoter in both JeKo and WaC3CD5 cells. Similarly, binding of SUZ12 was enriched 3- to 5-fold (P < 10−3) and 6- to 13-fold (P < 10−4) at the ST7 and RBL2 promoters, respectively. Interaction of BRD7 with target promoters was also increased 3- to 5-fold (P < 10−4) at the ST7 promoter and 3.5- to 7-fold (P < 10−3) at the RBL2 promoter, while association of all three chromatin binding proteins with the ST5 promoter was unchanged. These results indicate that enhanced association of PRMT5, SUZ12 and BRD7 with the ST7 and RBL2 promoters is highly specific.
We have already established that PRMT5 symmetrically methylates histone residues H3R8 and H4R3 at target promoters, and we have also shown that methylation of H3K4 is missing at these promoters (23,31). To confirm our previous findings, we measured the levels of PRMT5-induced marks (Figure 3E–G). When anti-H3(Me2)R8 and anti-H4(Me2)R3 antibodies were used to evaluate the methylation status of promoter histones H3 and H4, we found that methylation of H3R8 was increased 11.5- to 15-fold (P = 0.0044 in JeKo and P < 10−3 in WaC3CD5), while methylation of H4R3 was increased 8- to 9-fold (P < 10−4 in JeKo and P < 10−3 in WaC3CD5) at the ST7 promoter in cancer cells compared to normal B cells (Figure 3E). Methylation of H3R8 and H4R3 was also enhanced 6- to 7-fold at the RBL2 promoter in JeKo [P = 0.0031 for H3(Me2)R8 and P < 10−3 for H4(Me2)R3] and WaC3CD5 [P < 10−4 for H3(Me2)R8 and P = 0.0024 for H4(Me2)R3] cells (Figure 3F). Consistent with our results using the anti-PRMT5 antibody, there was no enrichment of PRMT5-induced epigenetic marks at the ST5 promoter (Figure 3G). Because ChIP studies have shown that SUZ12 is co-recruited to PRMT5 target genes, we investigated whether the PRC2-induced H3(Me3)K27 epigenetic mark is enriched at the ST7 and RBL2 promoters (Figure 3E–G). ChIP analysis showed that methylation of H3K27 was enhanced 6-fold at the ST7 promoter (P = 0.0004 in JeKo and P = 0.0002 in WaC3CD5), and 4- to 5-fold at the RBL2 promoter (P = 0.0006 in JeKo and P = 0.0002 in WaC3CD5) in transformed B cells. No significant enrichment of methylated H3K27 was noticed at the ST5 promoter.
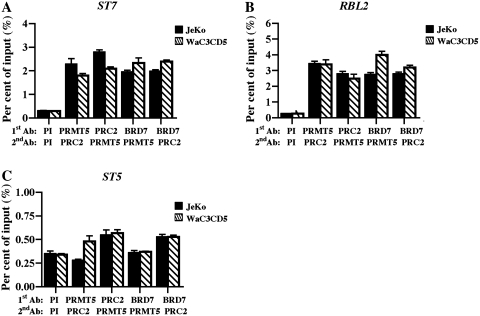
To ensure that BRD7, PRMT5 and SUZ12 are recruited to the same nucleosomes in the promoter region of ST7 and RBL2 target genes, we performed ChIP-re-ChIP experiments on cross-linked and MNase-treated chromatin from transformed B cells (Figure 4). Our findings indicate that when anti-PRMT5 is used first to immunoprecipitate chromatin from either JeKo or WaC3CD5 cells, SUZ12 is associated with the immunoprecipitated PRMT5 nucleoprotein complexes as evidenced by its 6- to 13-fold enrichment at the ST7 (P = 0.0058 for JeKo and P = 0.0009 for WaC3CD5) and RBL2 (P < 10−4 for both cancer cell lines) promoters (Figure 4A and B). When the same experiment was carried out using the anti-SUZ12 antibody first, association of PRMT5 with the ST7 (P = 0.0002 for JeKo and P < 10−4 for WaC3CD5) and RBL2 (P = 0.0006 for JeKo and P < 10−4 for WaC3CD5) promoters was enhanced to the same extent (7- to 10-fold). More importantly, when we examined binding of BRD7 in combination with either PRMT5 or SUZ12, we found that BRD7 is tightly associated with both proteins at the ST7 (6- to 8-fold enrichment, P < 10−4) and RBL2 (10- to 15-fold enrichment, P < 10−4) promoters. Analysis of BRD7, PRMT5 and SUZ12 recruitment to the ST5 promoter region did not show any enrichment. Collectively, these results indicate that BRD7, PRMT5 and SUZ12 physically co-exist in the promoter region of target tumor suppressor genes.
Figure 4.
PRMT5, PRC2 and BRD7 co-exist on the same nucleosomes in the promoter region of ST7 and RBL2. (A–C) ChIP assays were performed using cross-linked and MNase-treated chromatin from either JeKo or WaC3CD5 cells, and after nucleoprotein complexes were first immunoprecipitated (first Ab) with the indicated antibodies, they were released in the presence of 20 mM DTT. Next, a second round of immunoprecipitation (second Ab) was carried out using either PI, anti-PRMT5 or anti-SUZ12 antibodies, and real-time PCR was conducted as described in Figure 3. This experiment was performed twice in triplicate.
Having found that BRD7 is in complex with PRMT5-containing hSWI–SNF, we sought to evaluate recruitment of one of its core subunits, BRG1, to PRMT5 and SUZ12 target genes (Supplementary Figure S2). BRG1 binding to the ST7 promoter was increased 5- to 8-fold (P < 10−3), while its recruitment to the RBL2 promoter was enriched 4- to 5-fold (P < 10−4) in transformed B cells. Consistent with its ability to interact with BRD7 and PRMT5, interaction of BRG1 with ST7 and RBL2 promoters was enhanced to the same extent using a second set of primers (Supplementary Figure S2B and D). Moreover, there was no noticeable change in BRG1 recruitment to the ST5 promoter. These results suggest that BRG1–hSWI–SNF is recruited to the same target promoters as PRMT5, SUZ12 and BRD7.
BRD7 knockdown results in transcriptional derepression of ST7 but not RBL2
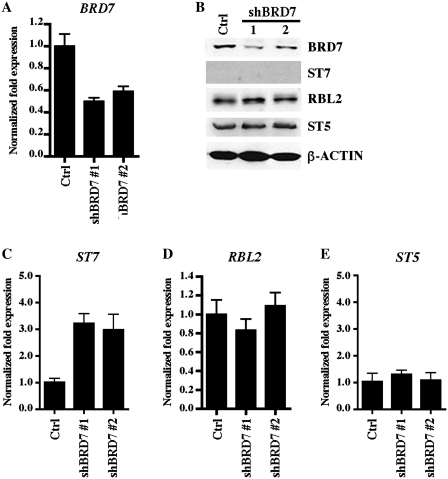
To gain more insight into the role played by BRD7 in epigenetic regulation of PRMT5 and PRC2 target genes, we established WaC3CD5 stable cell lines where BRD7 levels were reduced via concomitant expression of four BRD7-specific shRNAs, and measured the mRNA and protein levels of PRMT5 and PRC2 target genes (Figure 5). Real time RT–PCR showed that BRD7 mRNA was knocked down 50–60% in most WaC3CD5/shBRD7 stable cell lines examined including clones 1 and 2 (P < 10−3), and western blot analysis revealed that BRD7 protein levels were reduced 3- to 4-fold (Figure 5A and B). We have previously shown that reducing expression of PRMT5 triggers transcriptional derepression of ST7 and RBL2 in JeKo, Raji and WaC3CD5 cell lines (23,31). Therefore, to determine if reduced expression of BRD7 impacts transcription of PRMT5 and PRC2 target genes, we measured the ST7 and RBL2 mRNA levels in both control and BRD7 knockdown cells (Figure 5C and D). Our results revealed that while ST7 is transcriptionally derepressed 2–4-fold (P < 10−4) in WaC3CD5/shBRD7 stable cell lines clones 1 and 2, RBL2 mRNA levels are unaffected, indicating that there are different molecular determinants involved in their regulation. Furthermore, when we measured the levels of ST7 and RBL2 proteins in control and BRD7 knockdown cell lines clones 1 and 2, we found that RBL2 protein expression was unaffected, whereas expression of ST7 protein was suppressed (Figure 5B), suggesting that there might be post-transcriptional mechanisms involved in its regulation.
Figure 5.
BRD7 knockdown triggers transcriptional derepression of ST7, but not RBL2. (A) Levels of BRD7 mRNA were measured by RT–PCR using 1 µg of total RNA from either control or BRD7 knockdown cell lines. Levels of 18 S were used as an internal control. (B) RIPA extract (40 µg) from either control WaC3CD5 or BRD7 knockdown WaC3CD5 cell lines clones 1 and 2 were analyzed by western blotting using anti-BRD7, anti-ST7, anti-RBL2, anti-ST5 or anti-β-ACTIN antibody. (C–E) Levels of ST7, RBL2 and ST5 mRNA were determined by real-time RT–PCR as in Figure 5A. Graphs show normalized changes in expression for each gene relative to control WaC3CD5 cells using 18S as an internal control.
Recruitment of PRMT5 and SUZ12 to ST7 and RBL2 target promoters is compromised in BRD7 knockdown cell lines
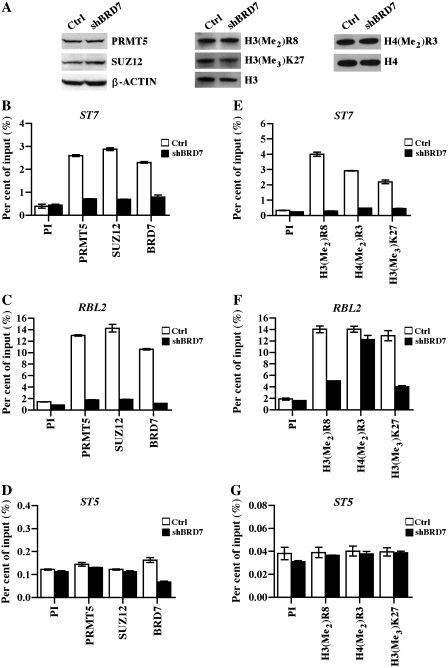
To uncover the underlying cause for the lack of RBL2 transcriptional derepression in BRD7 knockdown cells, we considered the possibility that association of PRMT5 and SUZ12 with the RBL2 promoter might be preserved in the absence of BRD7. Therefore, we carried out ChIP experiments using cross-linked chromatin from control WaC3CD5 and WaC3CD5/shBRD7 cells, and we also examined the levels of PRMT5 and SUZ12 proteins as well as their epigenetic marks (Figure 6). Western blot analysis showed that expression of PRMT5, SUZ12 and global levels of their induced methylation marks were unaffected in BRD7 knockdown cells (Figure 6A). As expected, recruitment of PRMT5, SUZ12 and BRD7 to the ST7 promoter was enhanced 5.8- to 7.3-fold (P < 10−4 for PRMT5 and BRD7, and P < 10−3 for SUZ12) in control WaC3CD5 cells, whereas binding of these proteins was completely abolished in BRD7 knockdown cells (Figure 6B). Interestingly, when we examined association of PRMT5, SUZ12 and BRD7 with the RBL2 promoter, we discovered that their binding was enriched 7.5- to 10-fold (P < 10−4 for PRMT5, P = 0.0018 for SUZ12 and P < 10−3 for BRD7) in control cells, and reduced significantly in WaC3CD5/shBRD7 cells (Figure 6C), implying that the presence of BRD7 is essential for PRMT5 and PRC2 recruitment. Interaction of PRMT5, SUZ12 and BRD7 with the ST5 promoter was unaffected in both control and test cell lines (Figure 6D), further confirming the importance played by BRD7 in PRMT5 and SUZ12 recruitment to their target genes. We have also monitored binding of PRMT5, SUZ12 and BRD7 to target promoters using a second and distinct set of primers, and we have found similar results (Supplementary Figure S3).
Figure 6.
Recruitment of PRMT5 and PRC2 to ST7 and RBL2 promoters is reduced in BRD7 knockdown cells. (A) Whole-cell extracts (40 µg) from control or BRD7 knockdown cells were analyzed by western blotting using anti-PRMT5, anti-SUZ12 or anti-β-ACTIN. To detect PRMT5- and PRC2-induced epigenetic marks, 5 µg of hydrochloric acid-extracted histones were analyzed by western blotting using anti-H3(Me2)R8, anti-H3(Me3)K27 or anti-H4(Me2)R3 antibody. Both anti-H3 and anti-H4 were used to show equal loading. Chromatin from control WaC3CD5 and BRD7 knockdown cells was immunoprecipitated with either control PI, anti-PRMT5, anti-SUZ12 or anti-BRD7 antibody, and the purified DNA was analyzed by RT–PCR to determine enrichment of ST7 (B), RBL2 (C) and ST5 (D) promoter sequences. (E–G) Both arginine and lysine methylation marks are reduced at the ST7, but not RBL2 promoter in BRD7 knockdown cells. ChIP assays were carried out using PI, anti-H3(Me2)R8, anti-H4(Me2)R3 or anti-H3(Me3)K27 antibody, and target promoter sequences were detected as in Figure 3. The data points in each graph show the average from two independent experiments.
To assess if reduced recruitment of PRMT5 and PRC2 is accompanied by a decrease in H3R8, H4R3 and H3K27 methylation, we measured the levels of all three epigenetic marks at the ST7 and RBL2 promoters (Figure 6E and F). ChIP analysis showed that methylation of histones H3R8 and H4R3 is highly enriched (7- to 13-fold, P < 10−4) at the ST7 promoter in control WaC3CD5 cells; however, methylation of ST7 promoter histones H3 and H4 is dramatically reduced in WaC3CD5/shBRD7 cells (Figure 6E). Similarly, when we checked the levels of H3R8, H4R3 and H3K27 methylation at the RBL2 promoter in control and BRD7 knockdown cells, we found that all three epigenetic marks were increased 7- to 8-fold (P < 10−4) in control WaC3CD5 cells. We also discovered that while the levels of H3(Me2)R8 and H3(Me3)K27 were decreased 2.4- to 2.8-fold (P < 10−4) in BRD7 knockdown cells, methylation of H4R3 was unaffected (Figure 6F). No changes in histone methylation were detected at the ST5 promoter (Figure 6G). Together, these results demonstrate that BRD7 plays an important role in PRMT5 and PRC2 recruitment, and that complete removal of PRMT5- and PRC2-induced epigenetic marks is important for efficient transcriptional derepression of their target genes.
Both RDM and KDM activities are recruited to PRMT5 and PRC2 target genes in BRD7 knockdown cells
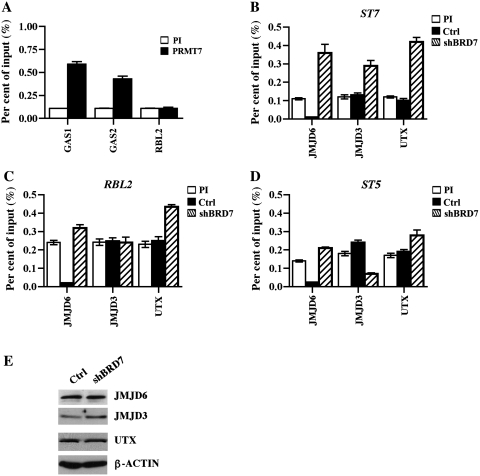
Having found that symmetric methylation of H4(Me2)R3 is unaffected at the RBL2 promoter in BRD7 knockdown cells, and knowing that there is only one other type II protein arginine methyltransferase, PRMT7, capable of H4R3 symmetric methylation (34), we tested if PRMT7 was associated with the RBL2 promoter in WaC3CD5 cells (Figure 7A). Our results showed that even though PRMT7 is heavily recruited to other tumor suppressor genes such as growth arrest specific (GAS) genes 1 and 2, there was a complete lack of PRMT7 binding to RBL2, suggesting that PRMT5 is the only histone arginine methyltransferase involved in symmetric methylation of H4R3 at the RBL2 promoter.
Figure 7.
Arginine and lysine-specific demethylases are differentially recruited to the ST7 and RBL2 promoters. (A) PRMT7 is not involved in transcriptional regulation of RBL2 in WaC3CD5 cells. Cross-linked chromatin from WaC3CD5 cells was immunoprecipitated using either PI or anti-PRMT7 antibody, and GAS1, GAS2 and RBL2 promoter sequences were detected by real-time PCR using gene-specific primer sets and probes. This experiment was repeated twice and real-time PCR was carried out in triplicate. ChIP assays were performed on cross-linked chromatin from control WaC3CD5 and BRD7 knockdown cells using either PI, anti-JMJD6, anti-JMJD3 or anti-UTX antibody. Immunoprecipitated DNA was amplified by real-time PCR to determine enrichment of each demethylase at the ST7 (B), RBL2 (C) and ST5 (D) promoters. (E) RIPA extracts (40 µg) from control WaC3CD5 and WaC3CD5/BRD7 knockdown cells were analyzed by western blotting using the indicated antibodies. Anti-β-ACTIN was used to show equal loading.
Because recruitment of PRMT5 is abolished in the absence of BRD7, we reasoned that demethylation of H4(Me2)R3 might be the limiting step for RBL2 transcriptional derepression. Therefore, we sought to identify and study recruitment of enzymes capable of erasing PRMT5-induced epigenetic marks. Recent work has shown that demethylation of specific lysine residues is intimately involved in transcriptional regulation, cellular differentiation and disease (1,2,5). Similarly, there are enzymes such as peptidylarginine deiminase 4 (PADI4), which is capable of converting methylarginine to citrulline (35,36), and Jumonji C domain-containing protein 6 (JMJD6), which represents a novel class of iron and 2-oxoglutarate-dependent dioxygenase enzymes, that can erase mono- and di-methylarginine marks (37).
Based on the recent findings, which showed that JMJD6 demethylates histones H3R2 and H4R3 (37), we checked whether it is recruited to PRMT5 target genes (Figure 7B–D). ChIP experiments showed that binding of JMJD6 to the ST7 promoter was enhanced 3.3-fold (P < 10−4) in BRD7 knockdown cells compared to control WaC3CD5 cells (Figure 7B). When we evaluated JMJD6 binding to the promoter regions of RBL2 and control ST5, we found no significant enrichment (1.3- to 1.5-fold, P < 10−3) (Figure 7C and D). We have shown that recruitment of PRC2 and the levels of its epigenetic mark are altered at the ST7 and RBL2 promoters in BRD7 knockdown cells (Figure 6). Therefore, we examined the binding status of two H3(Me3)K27-specific demethylases, KDM6A/UTX and KDM6B/JMJD3, at the ST7 and RBL2 promoters in both control and BRD7 knockdown cells (Figure 7B and C). Our results showed that binding of KDM6A/UTX and KDM6B/JMJD3 is augmented 2.4- to 3.5-fold (P < 10−4) at the ST7 promoter. Binding of both H3(Me3)K27-specific demethylases differed at the RBL2 promoter in that only KDM6A/UTX was bound to the promoter region (1.9-fold enrichment, P < 10−4). To rule out the possibility that differences in recruitment of demethylases arise from changes in their protein levels, we performed western blot analysis (Figure 7E). Expression of all three demethylases was unaltered in control and BRD7 knockdown cells. Taken together, these results indicate that both arginine and lysine demethylases are involved in transcriptional regulation of PRMT5 and PRC2 target genes.
DISCUSSION
Regulation of gene expression via histone post-translational modification has been the focus of many studies, and understanding how various epigenetic constellations impact gene transcription has become very important, especially as more histone marks are being identified and implicated in both transcriptional activation as well as repression. Studies by various groups have shown that there are different molecular effectors capable of recognizing and binding specific histones marks; however, their mode of action remains obscure. For instance, recent work by Xhemalce and Kouzaridez showed that acetylation of H3K4, a mark generally associated with gene activation, plays a major role in the formation of repressive heterochromatin in Schizosaccharomyces pombe by promoting a switch in binding of HP1-like proteins to methylated H3K9 (38). This finding redefines the notion that there is a strict code of histone modifications that specifies a particular transcriptional outcome, and suggests that both activation and repression-specific marks could play dual roles during gene transcription. Another example of the complexity of mechanisms used to regulate chromatin structure comes from studies where WDR5, a component of MLL complexes that binds methylated H3K4, can be found in association with the H3(Me3)K27-specific demethylase KDM6A/UTX (39), indicating that histone-binding proteins can interact with both histone methyltransferases as well as histone demethylases to promote either gene activation or repression.
In this study, we have identified BRD7 as a component of the PRMT5–hSWI–SNF complex, and using stable cell lines that express epitope-tagged BRD7 complexes we have shown that PRMT5 and hSWI–SNF subunits co-purify with His-BRD7. We have also determined that BRD7 interacts with specific hSWI–SNF subunits including BRG1, BRM and BAF60, as well as PRMT5 and MEP50. Moreover, we have found that BRD7 can also interact with components of the PRC2 repressor complex in vitro. Our previous work showed that hSWI–SNF-associated PRMT5 and its epigenetic marks are enriched at the promoter region of ST7 and RBL2 tumor suppressor genes (23,31). Therefore, we investigated the involvement of BRD7 and PRC2 in PRMT5 target gene regulation. Both ChIP and ChIP-re-ChIP analyses indicated that BRD7 and PRC2 co-localize with PRMT5 on ST7 and RBL2 target promoters in vivo. To gain a better understanding of the role played by BRD7 in ST7 and RBL2 transcriptional regulation, we knocked down its expression and measured the ST7 and RBL2 mRNA levels. Our findings clearly show that reducing expression of BRD7 triggers transcriptional derepression of ST7 as evidenced by the increase in ST7 mRNA levels in BRD7 knockdown cells; however, transcription of RBL2 was not affected even though recruitment of both PRMT5 and PRC2 was compromised in BRD7 knockdown cells.
To determine if lack of recruitment of PRMT5 and PRC2 could result in a decrease in methylation of histones H3 and H4, we used ChIP assays to measure the levels of PRMT5 and PRC2-induced epigenetic marks at the ST7 and RBL2 promoters in both control as well as BRD7 knockdown cells. In accordance with the real-time RT–PCR results (Figure 5C), we discovered that H3(Me2)R8, H4(Me2)R3 and H3(Me3)K27 epigenetic marks were completely removed from the ST7 promoter region. When we checked the promoter region of RBL2, we found that while methylation of H3R8 and H3K27 was reduced in the absence of BRD7, symmetric methylation of H4R3 was unaffected, raising the possibility that another type II PRMT might be involved in RBL2 transcriptional regulation. To date the only other type II PRMT known for its ability to symmetrically methylate H4R3 is PRMT7 (34). Therefore, we tested its association with the RBL2 promoter in WaC3CD5 cells. Our results indicated that even though PRMT7 was recruited to two other tumor suppressor genes, GAS1 and GAS2, its association with the RBL2 promoter was not increased.
Since it has previously been shown that lysine methylation marks are removed by specific KDMs, and recent studies have indicated that histone arginine methylation can be erased by Jumonji C domain-containing RDMs, we checked if there was altered recruitment of lysine and arginine demethylases to the ST7 and RBL2 promoters.
ChIP analysis of H3(Me3)K27-specific demethylases showed that while both KDM6A/UTX and KDM6B/JMJD3 were efficiently recruited to the ST7 promoter, only KDM6A/UTX was associated with RBL2. In addition, recruitment studies of the H4(Me2)R3-specific demethylase JMJD6 revealed that its association with the ST7 promoter is highly enriched in BRD7 knockdown cells, while its binding to the RBL2 promoter is lacking in these cells. These data are in complete agreement with the ChIP results obtained using antibodies specific to PRMT5- and PRC2-induced methylation marks (Figure 7). Although it is not clear if binding of BRD7 to PRMT5 and PRC2 target genes precludes recruitment of JMJD6, KDM6A/UTX and KDM6B/JMJD3, our results provide important clues about the function of BRD7 in recruiting PRMT5 and PRC2 repressors, and suggest that transcriptional activation of repressed genes does not only require release of enzymes involved in inducing specific epigenetic marks, but also efficient recruitment of different combinations of lysine and arginine-specific demethylases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute grants (CA116093 and CA101956 to S.S.); National Cancer Institute Cancer Center support grant (P30 CA08748 to P.T.). Funding for open access charge: NCI/CA116093.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank K. Helin for kindly providing pFASTBAC vectors for the expression of PRC2 subunits, J. Sudaram, L. Wang and S. Pal for technical help, and members of the laboratory for critical reading of the article.
REFERENCES
- 1.Cloos PAC, Christensen J, Agger K, Helin K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 3.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 4.Pal S, Sif S. Interplay between chromatin remodelers and protein arginine methyltransferases. J. Cell Physiol. 2007;213:306–315. doi: 10.1002/jcp.21180. [DOI] [PubMed] [Google Scholar]
- 5.Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 6.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3K4 methyltransferase MLL1 and the H4K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 7.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 8.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 9.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 10.Haynes SR, Dollard C, Winston F, Beck S, Trowsdale J, Dawid IB. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 12.Jeanmougin F, Wurtz JM, Douarin BL, Chambon P, Losson R. The bromodomain revisited. Trends Biochem. Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 13.Winston F, Allis CD. The bromodomain: a chromatin-targeting module. Nat. Struct. Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- 14.Cuppen E, Ham MV, Pepers B, Wieringa B, Hendriks W. Identification and molecular characterization of BP75, a novel bromodomain-containing protein. FEBS Lett. 1999;459:291–298. doi: 10.1016/s0014-5793(99)01191-6. [DOI] [PubMed] [Google Scholar]
- 15.Kim S, Lee J, Park J, Chung J. BP75, bromodomain-containing Mr 75,000 protein, binds disheveled-1 and enhances Wnt signaling by inactivating glycogen synthase kinase-3β1. Cancer Res. 2003;63:4792–4795. [PubMed] [Google Scholar]
- 16.Zhang Q, Chakravarty S, Ghersi D, Zeng L, Plotnikov AN, Sanchez R, Zhou MM. Biochemical profiling of histone binding selectivity of the yeast bromodomain family. PLoS ONE. 2010;5:e8903. doi: 10.1371/journal.pone.0008903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kzhyshkowska J, Rusch A, Wolf H, Dobner T. Regulation of transcription by the heterogeneous nuclear ribonucleoprotein E1B-AP5 is mediated by complex formation with the novel bromodomain-containing protein BRD7. Biochem. J. 2003;371:385–393. doi: 10.1042/BJ20021281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J, Nie J, Hao B, Li L, Xing G, Wang Z, Zhou Y, Sun Q, Li G, et al. Ceap/BLOS2 interacts with BRD7 and selectively inhibits its transcription-suppressing effect on cellular proliferation-associated genes. Cell Signalling. 2008;20:1151–1158. doi: 10.1016/j.cellsig.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Ma J, Zhang BC, Li XL, Shen SR, Zhu SG, Xiong W, Liu HY, Huang H, Zhou M, et al. BRD7, a novel bromodomain gene, inhibits G1-S progression by transcriptionally regulating some important molecules involved in ras/MEK/ERK and Rb/E2F pathways. J. Cell Physiol. 2004;200:89–98. doi: 10.1002/jcp.20013. [DOI] [PubMed] [Google Scholar]
- 20.Kaeser MD, Aslanian A, Dong MQ, Yates JR, III, Emerson B. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J. Biol. Chem. 2008;283:32254–32263. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, Tempst P, Sif S. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 2003;23:7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasini D, Bracken AP, Jensen MR, Denchi EL, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S. Low levels of miR-92 b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007;26:3558–3569. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Baiocchi RA, Pal S, Mosialos G, Caligiuri M, Sif S. The BRG1- and hBRM-associated factor BAF57 induces apoptosis by stimulating expression of the cylindromatosis tumor suppressor gene. Mol. Cell. Biol. 2005;25:7953–7965. doi: 10.1128/MCB.25.18.7953-7965.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erdjument-Bromage H, Lui M, Lacomis L, Grewal A, Annan RS, MacNulty DE, Carr SA, Tempst P. Micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. 1998;A826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
- 29.Li W, You L, Cooper J, Schiavon G, Pepe-Caprio A, Zhou L, Ishii R, Giovannini M, Hanemann CO, Long SB, et al. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiqutin ligase CRL4 DCAF1 in the nucleus. Cell. 2010;140:477–490. doi: 10.1016/j.cell.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perkins DN, Pappin DJ, Creasy DM, Cotrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Pal S, Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol. Cell. Biol. 2008;28:6262–6277. doi: 10.1128/MCB.00923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 33.Furunno K, MasaTsugu T, Sonada M, Sasazuki T, Yamamoto K. Association of polycomb group SUZ12 with WD-repeat protein MEP50 that binds histone H2A selectively in vitro. Biochem. Biophys. Res. Commun. 2006;345:1051–1058. doi: 10.1016/j.bbrc.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Jelinic P, Stehle JC, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, et al. Human PAD4 regulated histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 37.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 38.Xhemalce B, Kouzarides T. A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev. 2010;24:647–652. doi: 10.1101/gad.1881710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Croce LD, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.