Abstract
G-quadruplex topologies of telomeric repeat sequences from vertebrates were investigated in the presence of molecular crowding (MC) mimetics, namely polyethylene glycol 200 (PEG), Ficoll 70 as well as Xenopus laevis egg extract by CD and NMR spectroscopy and native PAGE. Here, we show that the conformational behavior of the telomeric repeats in X. laevis egg extract or in Ficoll is notably different from that observed in the presence of PEG. While the behavior of the telomeric repeat in X. laevis egg extract or in Ficoll resembles results obtained under dilute conditions, PEG promotes the formation of high-order parallel topologies. Our data suggest that PEG should not be used as a MC mimetic.
INTRODUCTION
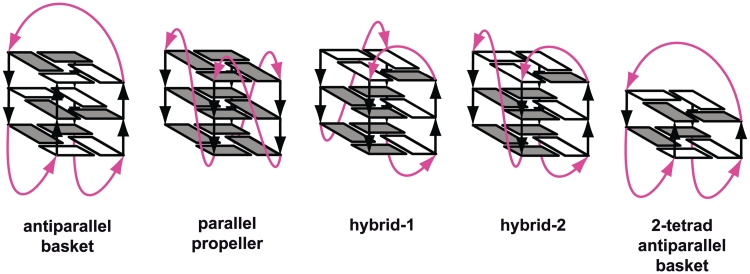
Telomeres are nucleoprotein complexes at the end of linear chromosomes that are essential to prevent improper activation of the DNA damage response. They are also important for protecting chromosomes from nuclease attacks, chromosomal non-homologous end-joining and loss of information during cell division. In vertebrates, telomeric DNA consist of tandem repeats of the hexanucleotide d(TTAGGG)n. Structural investigations have shown that these sequences form G-quadruplexes with various folding topologies under in vitro conditions. G-quadruplex folding topology has been demonstrated to depend on the sequence flanking the core residues, on ions and on the presence of small molecular weight compounds such as polyethylene glycol 200 (PEG). So far, experiments have demonstrated that telomere sequences can adopt four different structures in dilute solution including two hybrid parallel–antiparallel (3+1), one 2- and one 3-tetrad antiparallel folds (Table 1 and Figure 1) (1,2). In addition, studies simulating molecular crowding (MC) using either 40% PEG or 50% ethanol solutions showed that conformational polymorphism exhibited by telomeric DNA in dilute solutions is notably diminished and that telomeric repeats adopt the parallel stranded conformation in the presence of MC (3–5). Based on these and crystallographic data, the parallel G-quadruplex has been suggested to be the biological relevant folding topology of telomeric DNA (4–8). However, recently it was reported that parallel G-quadruplex formation under MC conditions simulated with PEG 200 can rather be explained by the fact that molar concentrations of small molecular weight molecules such as 40% PEG 200 or 50% ethanol cause major dehydration of DNA and that the low hydration state is stabilizing a parallel G-quadruplex conformation (9). In contrast to these conditions, molecules in the interior of a cell are mainly surrounded by large biomolecules such as proteins and nucleic acids and not molar concentrations of small molecular weight molecules (10).
Table 1.
Natural vertebrate telomeric repeat sequences
| Sequence | Name | Main folding topology |
|---|---|---|
| TA(GGGTTA)3G3 | TA-core | hybrid-1 type (3+1)a |
| A(GGGTTA)3G3 | A-core | antiparallela/parallelb |
| A(GGGTTA)3G3T | A-core-T | 2-tetrad antiparallela |
| (GGGTTA)3G3T | core-T | 2-tetrad antiparallela |
| A(GGGTTA)3G3TT | A-core-TT | hybrid-2 type (3+1)a |
| TTA(GGGTTA)3G3TT | TTA-core-TT | hybrid-2 type (3+1)a |
aNMR.
bX-ray.
Figure 1.
Schematic structures of intramolecular G-quadruplexes formed by four-repeat human telomeric sequences. Loops are colored magenta; anti and syn guanines are colored grey and white, respectively.
To determine which in vitro observed quadruplex topology exists in native-like MC environments, we employed NMR spectroscopy to investigate the conformation of different telomeric sequences capable of forming quadruplex structures in cellular extracts (11–13). In particular Xenopus laevis egg extract is a useful mimetic of the cellular environment and has been used in several NMR-based investigations (11,14). Here, we show that the use of PEG leads to a systematic stabilization of a parallel topology of G-quadruplexes regardless of the sequence, while the same sequences adopt different conformations in X. laevis egg extract. In addition, we demonstrate that the stabilization of ligand-favored antiparallel G-quadruplexes by the small chemical ligand TMPyP4 is not influenced under volume exclusion conditions using Ficoll as a large biomolecular mimetic. These data will be important for the search for potential anti-cancer drugs that stabilize the telomeric ends of chromosomes (15–17).
MATERIALS AND METHODS
Sample preparation
The oligonucleotides d(TAG3(TTAG3)3), d(AG3(TTAG3)3TT), d(G3(TTAG3)3T), d(AG3TG4AG3TG4A) and d(TGAG3TG3TAG3TG3TAA) were purchased from Microsynth (Switzerland). Experiments were carried out with a buffer mimicking the intracellular salt environment [25 mM HEPES (pH = 7.5), 10.5 mM NaCl, 110 mM KCl, 130 nM CaCl2, 1 mM MgCl2, 10% D2O, 1 mM DTT] in the absence or presence of PEG 200 (Fluka) or Ficoll 70 (Sigma-Aldrich). CD and NMR samples were prepared by mixing of G-DNA constructs with egg extract or by mixing or annealing in intra-oocyte buffer supplemented with the indicated PEG 200 or Ficoll 70 concentration. The mixed samples were incubated for 2 h at 37°C prior to measurements.
Xenopus laevis egg extract preparation
Crude cytoplasmic extracts were prepared as described previously (12) with modifications. Briefly, X. laevis oocytes were obtained as described (13). Oocytes were transferred into a petri dish containing 1 µM Progesterone in Ori buffer [5 mM HEPES (pH = 7.6), 110 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2] following an incubation period of at least 12 h. Xenopus eggs were selected and transferred via a glass pipette into a new petri dish and extensively washed with Ori buffer. Subsequently, the petri dish was placed on ice and incubated for 20 min. Cooled eggs were transferred to a pre-cooled petri dish containing a buffer mimicking the salt environment of X. laevis oocytes and eggs (13). Finally, eggs were transferred into an eppendorf tube containing ice-cold intra-oocyte buffer supplemented with 20% instead of 10% D2O. Cells were packed by spinning for 1 min at 400g and the buffer above the eggs was cautiously removed. Subsequently cells were centrifuged for 5 min at 12 000g. On ice, a glass pipette was used to crush eggs to prepare a homogenized egg extract solution. Finally, homogenized eggs were centrifuged for 30 min at 12 000g to obtain the crude inter-phase extract.
CD spectroscopy
CD spectra were collected from 315 to 210 nm on a Jasco J-810 spectrometer using 1-nm bandwidth. The temperature was controlled using a digitized water bath integrated in the instrument. The molar ellipticity, [θ], was calculated from the equation: [θ] = 106 × [θ]obs × C−1 × l−1, where [θ]obs is the ellipticity (mdeg); C is the oligonucleotide molar concentration and l is the optical path length of the cell (cm). Cells with 0.1 and 0.2 cm path lengths and oligonucleotide concentrations of 5 × 10−5 M were used. Scan rates of 50 nm min−1 were used to acquire the data. The spectra were signal-averaged over at least three scans, baseline corrected by subtracting a buffer spectrum and smoothed by the mean value averaging. Each prepared oligo sample, containing 5 × 10−5 M G-DNA, was further used for NMR measurements or supplemented with 100 µM TMPyP4 and incubated at 4°C for 12 h. Melting curves were recorded monitoring ellipticity at 290 nm from 4 to 100°C with a rate of 1°C/min. Ellipticity data points were further fitted (OriginTM) and normalized.
NMR spectroscopy
NMR experiments were performed using a Bruker Avance 700 spectrometer equipped with a cryogenic triple resonance probe at 18°C. The imino protons were observed using the 11-echo pulse sequence (18) with the excitation maximum adjusted to the center of the Hoogsteen imino region (ca. 11.8 ppm). Three millimeter NMR tubes were used for extract measurements with an active sample volume of 180 µl. Samples analyzed via CD were also used for in vitro NMR experiments. The egg extract NMR sample was prepared by mixing 162 µl extract preparation with 18 µl of a G-DNA stock solution leading to final G-DNA concentration of 100 µM. The prepared NMR samples were incubated for 2 h at 37°C prior to the measurements. The background signal from X. laevis egg extract was subtracted from spectra obtained with telomeric constructs.
Native PAGE
Native PAGE analysis was carried out at 4°C and 5 Vcm−1 on a 10% non-denaturing polyacrylamide gel in a buffer containing KCl (110 mM), potassium phosphate (50 mM) and Na2EDTA (1 mM), pH = 7.5. The loading buffer (8 µl, 40% glycerol) was mixed with the DNA samples (8 µl, 50 µM) that prior to mixing were heated to 95°C for 10 min and let slowly cool down to equilibrate. Gels were stained with Toluidine blue O (Sigma).
RESULTS
MC effect on telomeric G-quadruplex formation
In this study, we investigated the following sequences, which represent the currently known conformations observed in the presence of potassium in dilute solution: TAG3(TTAG3)3 ([TA-core]), AG3(TTAG3)3TT ([A-core-TT]) and G3(TTAG3)3 T ([core-T]) (Figure 1 and Table 1).
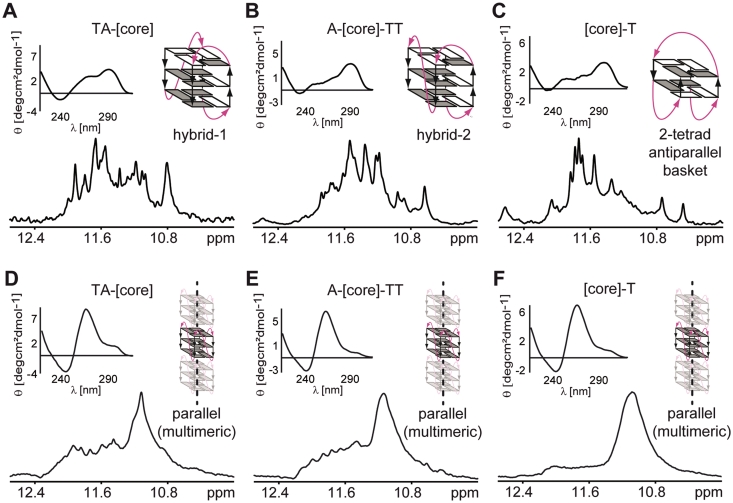
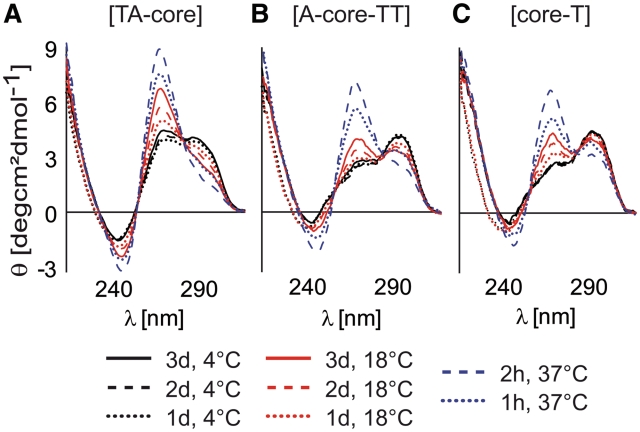
Figure 2A–F shows CD spectra and 1D sections of NMR spectra of the imino region of all sequences in aqueous K+ solution in the absence and presence of 40% PEG. In the K+ solution, the CD and 1D 1H NMR spectra of the [TA-core], [A-core-TT] and [core-T] constructs display characteristic patterns for hybrid-1 and hybrid-2 parallel–antiparallel and 2-tetrad antiparallel G-quadruplex folding topologies, respectively, as shown previously for these sequences in dilute solutions (19–22) (Figure 2A–C). A common feature of all CD spectra is the dominant positive peak at 290 nm which is a typical marker of an antiparallel strand orientation (23). The addition of 40% of PEG results in dramatic changes in both the NMR and CD spectra of all sequences (Figure 2D–F). In the presence of PEG, the positive peak at 290 nm is notably diminished and the resulting CD spectra are dominated by a strong positive peak at 260 nm, which is a typical feature of a parallel G-quadruplex conformation (5). A single broad peak centered at 11.1 ppm dominates the NMR spectra of the PEG containing samples. In accordance with published NMR data (24) and our own reference CD and NMR data of the P19 construct taken from the nuclease-hypersensitivity element III1 from the c-Myc promoter sequence d(AG3TG4AG3TG4A) (25,26) which is known to adopt a parallel topology (Supplementary Figure S1), this peak indicates the formation of highly symmetrical parallel G-quadruplexes. In addition to the broad peak at 11.1 ppm, the NMR spectra of [TA-core] and [A-core-TT] also display several small peaks between 11.5 and 12.4 ppm. Although CD and NMR spectra of all telomeric constructs showed in the presence of PEG typical features of parallel G-quadruplex formation, the lack of spectral resolution in the imino signal region of the NMR spectra was surprising. Other parallel G-quadruplexes were shown to provide well-resolved spectra, for example the extended c-Myc promoter sequence Pu22mut 14/23 d(TGAG3TG3TAG3TG3TAA) (24–27). Lack of spectral resolution and extensive line broadening could be the result of the formation of oligomers. To address this question, we used native PAGE, CD and NMR spectroscopy to compare the behavior of the c-Myc Pu19 and Pu22mut 14/23 constructs. The CD spectra of the Pu19 and Pu22mut 14/23 constructs in dilute conditions were virtually identical (Supplementary Figure S1) indicating the formation of a parallel folding topology. However, comparison of the NMR spectra revealed pronounced differences. While the NMR spectrum of Pu19 showed unresolved, broad imino resonances similar to those of the telomeric repeat constructs in the presence of PEG, the NMR spectrum for Pu22mut 14/23 displayed well resolved imino signals with narrow lines as observed previously (27). To explore whether the lack of resolution is the result of the formation of oligomers we performed native PAGE experiments. These PAGE data indicated that Pu19 migrates significantly slower than Pu22mut 14/23. Slower migration of the shorter Pu19 compared to Pu22mut 14/23 supports the idea of high-order structure formation of Pu19.
Figure 2.
CD spectra and imino regions of 1D 1H 11-echo NMR spectra for [TA-core], [A-core-TT] and [core-T] constructs in intra-oocyte buffer (A–C), and in intra-oocyte buffer supplemented with 40% PEG 200 (D–F) are shown, accompanied by schematic representations of the assumed G-quadruplex conformations. Loops are colored magenta; anti and syn guanines are colored grey and white, respectively.
To investigate if high-order structure formation can also explain the lack of spectral resolution in the NMR spectra of the telomeric repeat constructs in the presence of PEG, we compared the behavior of [core-T] and the Tetrahymena telomeric repeat construct d(G4T4G4T4G4T4G4) in the absence and presence of PEG using native PAGE. As demonstrated by Miyoshi et al. (28), the Tetrahymena telomeric repeat forms high-order structures in the presence of PEG. The addition of 40% PEG 200 resulted in slower migration of both the [core-T] and the Tetrahymena telomeric repeat constructs, indicating the formation of high-order structures (28).
As already mentioned, the NMR spectra of [TA-core] and [A-core-TT] also show several small and dispersed peaks between 11.5 and 12.4 ppm. These peaks can be attributed to minor populations of monomeric structures co-existing in equilibrium with high-order parallel structures as indicated by the native PAGE data (Supplementary Figure S2). The fact that the imino signals in the region between 11.5 to 12.4 ppm are missing in the NMR spectrum of the [core-T] construct in the presence of PEG suggests that the observed high-order structure formation might depend on the length of the sequences flanking the G-quadruplex core. This explanation is further supported by the effect of 40% PEG on the hybrid-type sequences TTA-[core]-TT and AAA-[core]-AA containing even longer flanking sequences (Supplementary Figure S3) as well as by recently published observations (29).
To analyze whether the observed high-order parallel G-quadruplex formation in the presence of PEG is due to a general MC effect or whether PEG acts via a different mechanism than MC, we simulated MC using Ficoll 70 as an alternative MC mimetic. Ficoll 70 is a neutral, highly branched, high-mass (∼74 kDa), hydrophilic polysaccharide that has been successfully used as an inert MC mimetic in several protein folding studies (10,30–35).
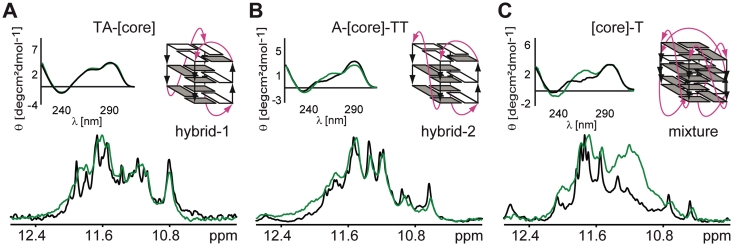
As the intracellular concentration of proteins can reach up to 400 g/l (33), we used 40% Ficoll 70 in order to simulate the MC effect as expected inside living cells. Both CD and NMR spectra of [TA-core] in the presence of 40% Ficoll 70 were virtually identical to those recorded under dilute conditions (Figure 3). These observations indicate that MC as simulated by Ficoll has no effect on folding topology of the [TA-core] construct. While NMR spectra for the [A-core-TT] construct in the absence and presence of 40% Ficoll were very similar as well, the corresponding CD spectra showed minor differences in molar ellipticity at 290 and 260 nm, suggesting a small shift toward a parallel topology (Figure 3). However, this shift was rather small as indicated by the very similar patterns of the corresponding NMR spectra. In contrast, the CD and NMR spectra of [core-T] in the presence of 40% Ficoll were characterized by an increased positive band at 260 nm and a newly appearing broad peak at 11.1 ppm, respectively, suggesting that in the presence of 40% Ficoll the [core-T] construct exists as a mixture of a 2-tetrad antiparallel and a high-order parallel G-quadruplex conformation (Figure 3, Supplementary Figures S1 and S2).
Figure 3.
Overlays of CD spectra and imino regions of 1D 1H 11-echo NMR spectra for [TA-core], [A-core-TT] and [core-T] constructs in intra-oocyte buffer, and in intra-oocyte buffer supplemented with 40% Ficoll are shown, accompanied by schematic representations of the assumed G-quadruplex conformations (A–C). Loops are colored magenta; anti and syn guanines are colored grey and white, respectively.
Considering differences in molecular mass between PEG 200 and Ficoll 70, it is important to note that a 40% w/v of Ficoll 70 solution shows a 1.7 times larger volume exclusion than a 40% v/v PEG 200 solution. To assess the effect of PEG and Ficoll on folding of the telomeric repeats at equivalent volume fractions of these co-solutes (36), we performed CD and NMR experiments for all the telomeric constructs at various concentrations of PEG and Ficoll (Supplementary Figure S4). 40% PEG 200 is expected to have similar volume exclusion as 23% Ficoll 70. As demonstrated in Supplementary Figure S4, the addition of 23% Ficoll had almost no impact on the appearance of both CD and NMR spectra compared to the dilute solution condition for all three telomeric constructs.
MC effect on telomeric G-quadruplex stabilization by TMPyP4
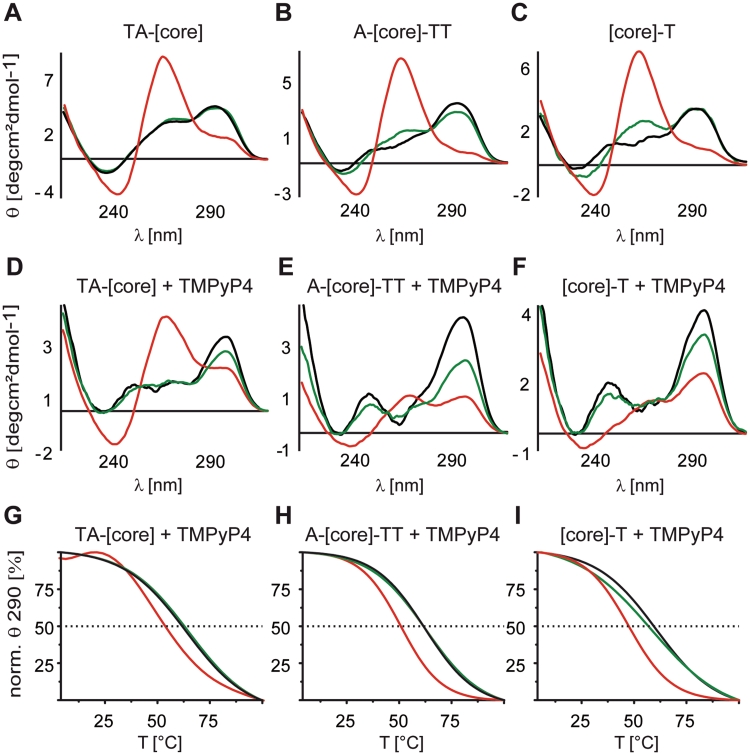
Differences between effects of the two MC mimetics, 40% Ficoll 70 and 40% PEG 200, on G-quadruplex folding topology are further demonstrated by differences in the interaction and stabilization properties of individual telomeric constructs with the G-quadruplex binding ligand TMPyP4. For all telomeric sequences in dilute solution and in the presence of Ficoll TMPyP4 stabilized the antiparallel folding topology (Figure 4D–F, black and green lines). As shown in Figure 4A–C, the addition of PEG leads to a shift of the equilibrium toward a parallel topology for all three sequences. Interestingly, the presence of TMPyP4 significantly suppressed the shift toward a parallel conformation for [A-core-TT] and [core-T] (Figure 4E–F, red lines) while this effect was less pronounced for [TA-core] (Figure 4D, red line). In addition, virtually identical melting temperatures (measured at 290 nm, thus focusing on the antiparallel conformation) were observed for all telomeric constructs in the presence of TMPyP4 when comparing samples in dilute solution with samples in the presence of Ficoll (Figure 4G–I, black and green lines). In contrast, in the presence of PEG and TMPyP4 all telomeric sequences showed a notably reduced melting temperature (Figure 4G–I, red lines). These data again demonstrate that structure and reactivity of G-quadruplexes under simulated MC conditions might be biased by the choice of MC mimetic. At the same time, the data suggest that the previously reported observation of a reduced stabilization of G-quadruplexes by the small chemical ligand TMPyP4 in the presence of PEG (37) is not necessarily due to MC but rather a specific effect of the PEG containing environment leading to distinct G-quadruplex stability (9).
Figure 4.
Overlays of CD spectra of 50 µM [TA-core], [A-core-TT] and [core-T] constructs in intra-oocyte buffer (A–C black lines), supplemented with either 40% PEG 200 (A–C red lines) or 40% Ficoll 70 (A–C green lines) and after the addition of 100 µM TMPyP4 and 12 h sample incubation at 4°C (D–F). Corresponding melting profiles were recorded by measuring the ellipticity at 290 nm, monitoring the stability of antiparallel G-quadruplex conformations of [TA-core], [A-core-TT] and [core-T] in the presence of TMPyP4 (G–I).
Native-like MC effect on telomeric G-quadruplex formation
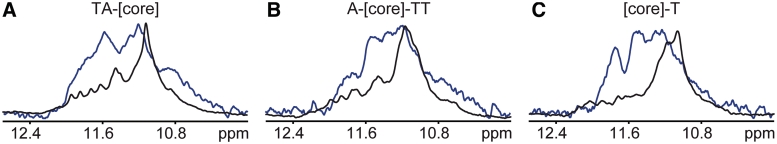
To investigate whether the observed structural changes in the telomeric sequences due to PEG and/or Ficoll are physiologically relevant, we investigated the [TA-core], [A-core-TT] and [core-T] constructs ex vivo in X. laevis egg extract. Due to the fact that the telomeric constructs can only be mixed and not annealed in X. laevis egg extract, various mixing conditions of telomeric constructs were tested in the presence of 40% PEG. All constructs incubated for 2 h at 37°C in the presence of 40% PEG provided virtually the same CD spectra as samples prepared under annealing conditions (Figure 5). Therefore, the NMR measurements aiming to compare the effects of PEG and X. laevis egg extract on the G-quadruplex conformation were performed with samples that were prepared using this protocol. Figure 6 shows a comparison of the imino region of 1D 1H NMR spectra of the telomeric constructs in X. laevis egg extract and in a buffer (simulating the intracellular oocyte salt composition) supplemented with PEG 200. Although, the ex vivo NMR spectra were of lower resolution compared to samples in dilute solution due to inherent sample inhomogeneity and the high viscosity of the egg extract, we clearly recognized differences between the X. laevis egg extract and PEG-containing samples. While the NMR spectra in the presence of PEG are dominated by a single broad peak centered at 11.1 ppm, the spectra in the X. laevis egg extract show a significantly broader chemical shift dispersion (Figure 6). The comparison with the NMR spectra of the PEG-containing samples clearly demonstrates that a parallel G-quadruplex conformation is not the dominant conformation observed in the X. laevis egg extract for any of the studied sequences (Figure 6). These data show that telomeric repeats in the X. laevis egg extract prefer different conformations from those observed in buffers supplemented with PEG or ethanol (3–5) (parallel G-quadruplex).
Figure 5.
CD spectra of 5 × 10−6 M [TA-core], [A-core-TT] and [core-T] constructs mixed with 40% PEG at different temperatures and incubated for various periods of time. In studies employing synthetic polymers as MC mimetics, re-annealing of DNA constructs in the presence of MC agents is typically used to accelerate the MC promoted DNA refolding. However, re-annealing of DNA constructs in X. laevis egg extracts cannot be performed as it would lead to thermal denaturation of proteins that are dominant contributing factors to the native MC environment. Here, we show that the CD spectra of telomeric constructs incubated in the presence of PEG for 2 h at 37°C are essentially the same as those obtained for the telomeric constructs re-annealed in the presence of PEG (cf. Figure 2D–F). To avoid any bias from differences in sample preparations, the telomeric repeat constructs were incubated in X. laevis egg extract and 40% PEG 200 for 2 h at 37°C prior to acquisition of NMR spectra (see Figure 6).
Figure 6.
Overlays of imino regions of 1D 1H 11-echo NMR spectra of the telomeric constructs in X. laevis egg extract (A–C blue lines) and in intra-oocyte buffer supplemented with 40% PEG 200 (A–C black lines). In the presence of PEG, several small and dispersed peaks can be observed between 11.5 and 12.4 ppm in the NMR spectra of [TA-core] and [A-core-TT] constructs (A and B black lines). These peaks can be attributed to low-level populated alternative conformation(s) co-existing in equilibrium with the dominant parallel high-order structures based on the analysis native PAGE data (Supplementary Figures S1 and S2).
DISCUSSION
While the resolution of the NMR spectra is sufficient for revealing differences in the general topology of the same sequence either in X. laevis egg extract or in PEG containing samples, the resolution of 1D 1H NMR spectra is too low for a more detailed characterization. At this level of resolution, the NMR spectra of all three telomeric repeat constructs acquired in X. laevis egg extracts seem to combine features of spectra obtained in dilute solutions and those obtained in the presence of PEG (Figure 6) suggesting that telomeric repeats might co-exist under native conditions as an equilibrium mixture of antiparallel, hybrid and parallel topologies. This explanation is further supported by the recent observation of co-existing hybrid and parallel G-quadruplexes in dilute solutions for G-rich stretches of the hTERT promoter (24). Our data show that while the high-order parallel G-quadruplex folding topology is dominant in a PEG containing environment, the equilibrium is dramatically shifted in favor of monomeric antiparallel and/or hybrid conformations in Ficoll or cellular extracts.
The presented comparison of the effects of PEG 200, Ficoll 70 and X. laevis egg extract on the conformation of G-quadruplexes and their stabilization by small chemical ligands highlights two main issues associated with the use of MC mimetics. First, the results depend on the choice of the particular MC mimetic and might therefore not be directly related to MC but could be based on for example dehydration effects (9). Second, the observed effects of MC mimetics might not necessarily resemble a physiologically relevant situation in vivo. The fact that true macromolecular crowding results from the addition of high molecular weight co-solutes such as proteins and nucleic acids that occupy and exclude volume inside cells (10,38) suggests that currently established approaches to investigate the effect of MC on structure and binding ability of potential drugs, which are based on the use of small molecular weight MC mimetics such as PEG 200, should be generally reconsidered.
Recent findings that stabilization of G-quadruplexes can inhibit the activity of telomerase, an enzyme responsible for maintenance of telomeric DNA and being over-activated in ∼80–85% of cancer cells and primary tumors, have stimulated intense interest in exploring small molecular ligands having potential to stabilize G-quadruplexes as a new class of anticancer drugs. The suggestion that the parallel G-quadruplex topology is the biologically relevant fold of telomeric DNA, has encouraged studies aiming at identification and rational design of small ligands selectively targeting parallel G-quadruplexes. Our results reported here and recently published data (9) show that the parallel G-quadruplex topology, formed under low hydration-conditions such as in the presence of molar concentrations of PEG and ethanol or in the crystalline state might not be the physiologically prevalent conformation and implicates that rational design of small molecular ligands stabilizing G-quadruplexes should be redirected toward ligands selectively targeting antiparallel and hybrid type G-quadruplex folding topologies.
Since its introduction in 1974 (39), PEG has become the most frequently used MC mimetic for nucleic acids. Many recent studies reported a dramatic effect of MC as simulated by PEG on the structure of both DNA and RNA (4,8,22,38,40–43). PEG simulated MC was found to strongly modulate stability and promote formation of non-canonical motifs such as parallel duplexes, G-quadruplexes, i-motifs or H-DNA, for example (5,40,44–48). All these observations provide the impression that MC is a crucial modulator and determinant of nucleic acid structure in vivo. However, our data acquired in Ficoll and cellular extracts indicate that the observed structural effects in the presence of PEG might not necessarily relate to the physiological relevant situation and might be caused by other mechanisms than MC (9). Instead more detailed analysis of the conformation of G-quadruplexes using cellular extracts or investigations in living cells (13,49,50) will be necessary to study the influence of MC on nucleic acid structure.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Center for Biomolecular Magnetic Resonance (BMRZ), the Cluster of Excellence Frankfurt (Macromolecular Complexes), the Fond der Chemischen Industrie (FCI). Funding for open access charge: DFG funding.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Access to Research Infrastructures is gratefully acknowledged.
REFERENCES
- 1.Dai J, Carver M, Yang D. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phan AT. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renciuk D, Kejnovska I, Skolakova P, Bednarova K, Motlova J, Vorlickova M. Arrangements of human telomere DNA quadruplex in physiologically relevant K+ solutions. Nucleic Acids Res. 2009;37:6625–6634. doi: 10.1093/nar/gkp701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xue Y, Kan ZY, Wang Q, Yao Y, Liu J, Hao YH, Tan Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 6.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 7.Miyoshi D, Sugimoto N. Molecular crowding effects on structure and stability of DNA. Biochimie. 2008;90:1040–1051. doi: 10.1016/j.biochi.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Zhang DH, Fujimoto T, Saxena S, Yu HQ, Miyoshi D, Sugimoto N. Monomorphic RNA G-quadruplex and polymorphic DNA G-quadruplex structures responding to cellular environmental factors. Biochemistry. 2010;49:4554–4563. doi: 10.1021/bi1002822. [DOI] [PubMed] [Google Scholar]
- 9.Miller MC, Buscaglia R, Chaires JB, Lane AN, Trent JO. Hydration is a major determinant of the G-quadruplex stability and conformation of the human telomere 3′ sequence of d(AG(3)(TTAG(3))(3)) J. Am. Chem. Soc. 2010;132:17105–17107. doi: 10.1021/ja105259m. [DOI] [PubMed] [Google Scholar]
- 10.Pielak GJ, Miklos AC. Crowding and function reunite. Proc. Natl Acad. Sci. USA. 2010;107:17457–17458. doi: 10.1073/pnas.1013095107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selenko P, Serber Z, Gadea B, Ruderman J, Wagner G. Quantitative NMR analysis of the protein G B1 domain in Xenopus laevis egg extracts and intact oocytes. Proc. Natl Acad. Sci. USA. 2006;103:11904–11909. doi: 10.1073/pnas.0604667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serber Z, Selenko P, Hänsel R, Reckel S, Lohr F, Ferrell JE, Jr, Wagner G, Dötsch V. Investigating macromolecules inside cultured and injected cells by in-cell NMR spectroscopy. Nat. Protoc. 2006;1:2701–2709. doi: 10.1038/nprot.2006.181. [DOI] [PubMed] [Google Scholar]
- 13.Hänsel R, Foldynova-Trantirkova S, Lohr F, Buck J, Bongartz E, Bamberg E, Schwalbe H, Dötsch V, Trantirek L. Evaluation of parameters critical for observing nucleic acids inside living Xenopus laevis oocytes by in-cell NMR spectroscopy. J. Am. Chem. Soc. 2009;131:15761–15768. doi: 10.1021/ja9052027. [DOI] [PubMed] [Google Scholar]
- 14.Selenko P, Frueh DP, Elsaesser SJ, Haas W, Gygi SP, Wagner G. In situ observation of protein phosphorylation by high-resolution NMR spectroscopy. Nat. Struct. Mol. Biol. 2008;15:321–329. doi: 10.1038/nsmb.1395. [DOI] [PubMed] [Google Scholar]
- 15.Hampel SM, Sidibe A, Gunaratnam M, Riou JF, Neidle S. Tetrasubstituted naphthalene diimide ligands with selectivity for telomeric G-quadruplexes and cancer cells. Bioorg. Med. Chem. Lett. 2010;20:6459–6463. doi: 10.1016/j.bmcl.2010.09.066. [DOI] [PubMed] [Google Scholar]
- 16.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Neidle S. Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010;277:1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- 18.Sklenar V, Bax A. Spin-echo water suppression for the generation of pure-phase two-dimensional Nmr-spectra. J. Magn. Reson. 1987;74:469–479. [Google Scholar]
- 19.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai J, Carver M, Punchihewa C, Jones RA, Yang D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim KW, Amrane S, Bouaziz S, Xu W, Mu Y, Patel DJ, Luu KN, Phan AT. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martino L, Pagano B, Fotticchia I, Neidle S, Giancola C. Shedding light on the interaction between TMPyP4 and human telomeric quadruplexes. J. Phys. Chem. B. 2009;113:14779–14786. doi: 10.1021/jp9066394. [DOI] [PubMed] [Google Scholar]
- 23.Kypr J, Kejnovska I, Renciuk D, Vorlickova M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim KW, Lacroix L, Yue DJ, Lim JK, Lim JM, Phan AT. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 2010;132:12331–12342. doi: 10.1021/ja101252n. [DOI] [PubMed] [Google Scholar]
- 25.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 26.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatzakis E, Okamoto K, Yang D. Thermodynamic stability and folding kinetics of the major G-quadruplex and its loop isomers formed in the nuclease hypersensitive element in the human c-Myc promoter: effect of loops and flanking segments on the stability of parallel-stranded intramolecular G-quadruplexes. Biochemistry. 2010;49:9152–9160. doi: 10.1021/bi100946g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyoshi D, Karimata H, Sugimoto N. Drastic effect of a single base difference between human and tetrahymena telomere sequences on their structures under molecular crowding conditions. Angew. Chem. Int. Ed. Engl. 2005;44:3740–3744. doi: 10.1002/anie.200462667. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Dai J, Veliath E, Jones RA, Yang D. Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: insights into the interconversion of human telomeric G-quadruplex structures. Nucleic Acids Res. 2010;38:1009–1021. doi: 10.1093/nar/gkp1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, He H, Li S. Effect of Ficoll 70 on thermal stability and structure of creatine kinase. Biochemistry. 2010;75:648–654. doi: 10.1134/s0006297910050160. [DOI] [PubMed] [Google Scholar]
- 31.Hong J, Gierasch LM. Macromolecular crowding remodels the energy landscape of a protein by favoring a more compact unfolded state. J. Am. Chem. Soc. 2010;132:10445–10452. doi: 10.1021/ja103166y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dhar A, Samiotakis A, Ebbinghaus S, Nienhaus L, Homouz D, Gruebele M, Cheung MS. Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding. Proc. Natl Acad. Sci. USA. 2010;107:17586–17591. doi: 10.1073/pnas.1006760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christiansen A, Wang Q, Samiotakis A, Cheung MS, Wittung-Stafshede P. Factors defining effects of macromolecular crowding on protein stability: an in vitro/in silico case study using cytochrome c. Biochemistry. 2010;49:6519–6530. doi: 10.1021/bi100578x. [DOI] [PubMed] [Google Scholar]
- 34.Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin J, Hartl FU. The effect of macromolecular crowding on chaperonin-mediated protein folding. Proc. Natl Acad. Sci. USA. 1997;94:1107–1112. doi: 10.1073/pnas.94.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spink CH, Chaires JB. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry. 1999;38:496–508. doi: 10.1021/bi9820154. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z, Zheng KW, Hao YH, Tan Z. Reduced or diminished stabilization of the telomere G-quadruplex and inhibition of telomerase by small chemical ligands under molecular crowding condition. J. Am. Chem. Soc. 2009;131:10430–10438. doi: 10.1021/ja9010749. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Li C, Pielak GJ. Effects of proteins on protein diffusion. J. Am. Chem. Soc. 2010;132:9392–9397. doi: 10.1021/ja102296k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laurent TC, Preston BN, Carlsson B. Conformational transitions of polynucleotides in polymer media. Eur. J. Biochem. 1974;43:231–235. doi: 10.1111/j.1432-1033.1974.tb03404.x. [DOI] [PubMed] [Google Scholar]
- 40.Zheng KW, Chen Z, Hao YH, Tan Z. Molecular crowding creates an essential environment for the formation of stable G-quadruplexes in long double-stranded DNA. Nucleic Acids Res. 2010;38:327–338. doi: 10.1093/nar/gkp898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei C, Wang J, Zhang M. Spectroscopic study on the binding of porphyrins to (G(4)T(4)G(4))4 parallel G-quadruplex. Biophys. Chem. 2010;148:51–55. doi: 10.1016/j.bpc.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 42.Zhou J, Wei C, Jia G, Wang X, Feng Z, Li C. Human telomeric G-quadruplex formed from duplex under near physiological conditions: spectroscopic evidence and kinetics. Biochimie. 2009;91:1104–1111. doi: 10.1016/j.biochi.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Arora A, Maiti S. Stability and molecular recognition of quadruplexes with different loop length in the absence and presence of molecular crowding agents. J. Phys. Chem. B. 2009;113:8784–8792. doi: 10.1021/jp809486g. [DOI] [PubMed] [Google Scholar]
- 44.Karimata H, Miyoshi D, Sugimoto N. Structure and stability of DNA quadruplexes under molecular crowding conditions. Nucleic Acids Symp. Ser. 2005:239–240. doi: 10.1093/nass/49.1.239. [DOI] [PubMed] [Google Scholar]
- 45.Miyoshi D, Matsumura S, Nakano S, Sugimoto N. Duplex dissociation of telomere DNAs induced by molecular crowding. J. Am. Chem. Soc. 2004;126:165–169. doi: 10.1021/ja036721q. [DOI] [PubMed] [Google Scholar]
- 46.Miyoshi D, Nakamura K, Tateishi-Karimata H, Ohmichi T, Sugimoto N. Hydration of Watson-Crick base pairs and dehydration of Hoogsteen base pairs inducing structural polymorphism under molecular crowding conditions. J. Am. Chem. Soc. 2009;131:3522–3531. doi: 10.1021/ja805972a. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura K, Karimata H, Ohmichi T, Miyoshi D, Sugimoto N. Effects of cosolutes on the thermodynamic stability of parallel DNA duplex and triplex. Nucleic Acids Symp. Ser. 2007:167–168. doi: 10.1093/nass/nrm084. [DOI] [PubMed] [Google Scholar]
- 48.Rajendran A, Nakano S, Sugimoto N. Molecular crowding of the cosolutes induces an intramolecular i-motif structure of triplet repeat DNA oligomers at neutral pH. Chem. Commun. 2010;46:1299–1301. doi: 10.1039/b922050j. [DOI] [PubMed] [Google Scholar]
- 49.Serber Z, Corsini L, Durst F, Dötsch V. In-cell NMR spectroscopy. Methods Enzymol. 2005;394:17–41. doi: 10.1016/S0076-6879(05)94002-0. [DOI] [PubMed] [Google Scholar]
- 50.Krstić I, Hänsel R, Romainczyk O, Engels JW, Dötsch V, Prisner TF. In-cell Long Range Distance Measurements on Nucleic Acids by Pulsed EPR Spectroscopy. Angew. Chem. Int. Ed. Engl. 2011 doi: 10.1002/anie.201100886. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.