Abstract
The unparalleled growth in the availability of genomic data offers both a challenge to develop orthology detection methods that are simultaneously accurate and high throughput and an opportunity to improve orthology detection by leveraging evolutionary evidence in the accumulated sequenced genomes. Here, we report a novel orthology detection method, termed QuartetS, that exploits evolutionary evidence in a computationally efficient manner. Based on the well-established evolutionary concept that gene duplication events can be used to discriminate homologous genes, QuartetS uses an approximate phylogenetic analysis of quartet gene trees to infer the occurrence of duplication events and discriminate paralogous from orthologous genes. We used function- and phylogeny-based metrics to perform a large-scale, systematic comparison of the orthology predictions of QuartetS with those of four other methods [bi-directional best hit (BBH), outgroup, OMA and QuartetS-C (QuartetS followed by clustering)], involving 624 bacterial genomes and >2 million genes. We found that QuartetS slightly, but consistently, outperformed the highly specific OMA method and that, while consuming only 0.5% additional computational time, QuartetS predicted 50% more orthologs with a 50% lower false positive rate than the widely used BBH method. We conclude that, for large-scale phylogenetic and functional analysis, QuartetS and QuartetS-C should be preferred, respectively, in applications where high accuracy and high throughput are required.
INTRODUCTION
Access to inexpensive, high-throughput genome sequencing has triggered an unprecedented growth in the number of available sequence data. The sequences of more than 1000 prokaryotes are available in public databases and hundreds of bacterial and archaeal genome-sequencing projects are currently underway (1). In parallel, for particular model species, experimental studies are attempting to annotate and decode vast amounts of these genomic data to reveal the molecular functions of genes, their essentiality to a species survival and their connections to the virulence of pathogenic species and human diseases. Unfortunately, the rate at which gene functions are being annotated and decoded through such experimental studies cannot keep pace with today’s high-throughput sequencing capabilities, and this gap is expected to increase in the foreseeable future. Because orthologous genes (i.e. orthologs) across different species often share equivalent molecular functions, orthology detection methods have become pivotal in helping bridge this gap and in hypothesizing gene function in unstudied species. This is achieved by first identifying orthologs between the unstudied and studied species and then transferring knowledge from the annotated genes in the model species to the unstudied species. While the fast accumulation of genomic data offers a challenge to existing orthology detection methods for such large-scale, genome-wide annotations across hundreds, if not thousands, of species, it also offers the opportunity to improve orthology detection accuracy by leveraging the evolutionary evidence accumulated over a much larger set of sequenced genomes.
The essence of orthology detection is to unambiguously distinguish the evolutionary path of two major types of homologous genes (orthologs and paralogs), which evolved from the same ancestral gene but through different evolutionary events: speciation for orthologs and duplication for paralogs (2). In this context, gene duplication is critical for the emergence of new gene functions. While selection pressure oftentimes suppresses the mutation of genes with a particular function, duplication events offer an opportunity for the duplicated genes (i.e. paralogs) to escape such selection pressure and undergo fast mutations that may eventually lead to a new gene function (3,4). Because gene duplication events are both the key feature that defines paralogs and the main cause of their functional differentiation, it should be considered as the critical evolutionary evidence in distinguishing orthologs from paralogs.
One approach to infer and use evolutionary evidence for orthology detection is through reconciliation, i.e. comparison, of gene trees and species trees. Because orthologs originate from speciation events when new species emerge from their common ancestor, it is postulated that a gene tree of orthologous genes should mirror that of their corresponding species tree. Accordingly, topological differences between a gene tree and its corresponding species tree can be used to infer the occurrence of gene duplication events and the existence of paralogs in the gene tree (5). Such tree-based reconciliation approach has led to the development of a number of orthology detection methods, including RAP (5), RIO (6) and Orthostrapper (7). However, when genes of the same species are separated into two sub-trees in a gene tree, gene duplication events may be inferred without the need for tree reconciliation (8,9). Nevertheless, none of these tree-based methods are suitable for large-scale, genome-wide annotations because they require the construction of large gene trees, and often species trees, which is too computationally expensive for practical applications (2,10). The construction of gene trees is also error prone, and techniques for improving their reliability further increase the computational costs (11). Moreover, although tree-based methods are generally considered to be very accurate, a recent comparative study (12) has shown that they yield insignificant improvements over alternative methods that do not require gene tree generation, providing high-throughput orthology detection at modest computational costs. Many of such methods are based on variations of the widely used bi-directional best hit (BBH) method, which simply predicts two genes of two different species as orthologs if they form a BBH pair, i.e. if each of the two genes is the gene in its genome that has the highest sequence similarity (usually measured by the E-value or the bit-score of BLAST searches) with the other gene of the other species. The BBH method, however, is prone to false positive predictions because a paralog can be identified as the BBH of a gene whose true ortholog has been deleted through evolution. In an attempt to reduce such false positive predictions, different methods that exploit evolutionary evidence by making comparisons with the genome of a third species have been proposed (10,13–19).
One such approach is based on the use of outgroup species that are located outside of the common clade of the species of interest. For two given genes in two different species, i.e. the target genes, this outgroup approach searches for a gene in an outgroup species such that its sequence similarity with at least one of the target genes is higher than the sequence similarity between the target genes. This provides evidence that the target genes are paralogs because a gene tree formed by three genes (two target genes and one outgroup gene) would not mirror the species tree of the three taxa. Conversely, if such evidence is not found, the target genes are determined to be orthologs. This approach has been used in programs such as INPARANOID (10) and Ortholuge (18), and its drawback is that it requires prior phylogeny knowledge and that the selection of an appropriate outgroup species is not always obvious (for example, which outgroup species should be used to detect orthologs between archaea and bacteria?). An alternative approach that avoids the specification of outgroup species is one where duplication evidence for the two target genes is attained through searches for two genes in a third species. Analysis of the evolution of the four genes from their last common ancestor (LCA) reveals whether a duplication event has occurred during the evolution of the target genes, and, if so, the target genes are determined to be paralogs. The recently developed OMA method exploits this evolutionary concept (14,17,19). However, instead of explicitly inferring and directly using the occurrence of a gene duplication event, its orthology detection procedure, based on heuristic rules, only implies such an occurrence. OMA’s high specificity in a recent comparative study (12) suggests that explicitly inferring and directly using evidence of a gene duplication event may further improve orthology detection.
Another approach to potentially improve orthology detection is to post-process the outputs of an orthology detection method so as to cluster predicted pair-wise orthologs into orthologous groups and consider all genes in the same group as orthologs. Because this process may group together genes that are not pair-wise orthologs or separate out pair-wise orthologs into different groups, it can modify the orthology relationships for some of the genes. In spite of this, the widely used OrthoMCL program, which uses clustering to group together BBH pairs, has achieved very good performance (13,16). However, it is not clear whether clustering consistently improves orthology detection and for what types of applications it should be considered.
In this study, we describe a novel orthology detection method, termed QuartetS, that provides both extremely accurate and high-throughput ortholog predictions for large-scale applications. QuartetS attains accurate predictions to distinguish paralogs from orthologs by explicitly inferring and directly using evolutionary evidence of gene duplication events. This evidence is obtained by identifying the location of a putative duplication event in a quartet gene tree formed by the two target genes and two other genes in a third genome. To reduce the computational time to construct precise quartet trees, we developed a formula that uses pair-wise sequence similarities calculated by BLAST to approximate the location of the putative duplication event in the quartet tree. This allows for effective search of duplication evidence in large genome databases with minimum additional computing time. Large-scale comparative tests, involving 624 bacterial genomes and >2 million genes against BBH, outgroup, OMA and QuartetS followed by clustering, revealed that QuartetS consistently offers advantages in orthology detection.
MATERIALS AND METHODS
QuartetS method
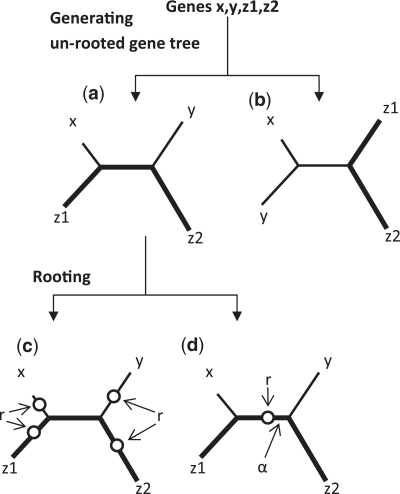
QuartetS determines if two homologous genes x and y of two species X and Y, respectively, are orthologs by searching a genome sequence database for evidence of a gene duplication event that may have occurred along the evolution of the two genes since their LCA. Genes x and y are deemed to be homologs if they form BBH pairs for species X and Y, and evolutionary evidence of potential duplication is provided by two genes z1 and z2 from a third species Z in the database, which, for consistency, we assume to be homologs (although this assumption can be relaxed without detriment). If we ignore horizontal gene transfer, we must infer that z1 and z2 are paralogs originating from a duplication event because they are present in the same species. Central to QuartetS is the observation that if genes x and y have also originated from this same duplication event, they must also be paralogs. Alternatively, if a database search fails to identify a third species Z, where genes x and y originated from the same duplication event inferred by z1 and z2, then x and y are assumed to be orthologs. We can determine if genes x and y have originated from the duplication event implied by z1 and z2 by reconstructing the evolutionary history of the four genes from their LCA and expressing it as an un-rooted quartet gene tree. Figure 1a and b shows the two possible topologies of such an un-rooted tree (when we do not distinguish z1 from z2), where each topology has five branches (four outer branches and one inner branch) linking the four genes, with the length of the branches indicating the evolutionary distance between the genes. The thickened branches highlight the two possible paths between z1 and z2 and indicate the possible locations of a duplication event implied by these genes. Because in the topology depicted in Figure 1b the path between x and y does not overlap with the path between z1 and z2, any duplication event inferred along the z1–z2 path is inconsequential to the relationship between genes x and y. Accordingly, the overlapping inner branch in Figure 1a is the only location in this topology where a duplication event must have occurred, so as to infer that genes x and y must have evolved through it and that they are, therefore, paralogs. If a search for all species Z in the database fails to identify evidence to locate the duplication event in the inner branch, we assumed that genes x and y are orthologs.
Figure 1.
The QuartetS method establishes the homology relationship between two genes x and y from two species X and Y, respectively, by exploiting phylogeny information present in a quartet gene tree formed by these two genes and two paralogous genes z1 and z2 from a third species Z. (a and b) When we do not distinguish z1 from z2, the quartet tree can have two possible topologies, where the thickened branches that highlight the two possible paths between z1 and z2 indicate the possible locations of a duplication event implied by these genes. (a) Genes x and y are paralogs if the duplication event occurs in the inner branch overlapping their path and the path between z1 and z2. (b) Because the path between x and y does not overlap with the path between z1 and z2, any duplication event inferred along the z1–z2 path is inconsequential to the relationship between genes x and y. (c) Rooting the tree can identify the last common ancestor (or root r) in an outer branch, which is not informative. (d) Alternatively, it could identify the root in the inner branch, inferring that x and y are paralogs, where the distance α between r to its nearest inner node provides a measure of the reliability of the estimate for r. We infer that genes x and y are paralogs when α is greater than a specified cutoff value (Ω), with a larger Ω leading to fewer number of paralogs and larger number of orthologs, and vice versa.
We could construct a ‘precise’ quartet gene tree through multiple sequence alignments of genes x, y, z1 and z2 and then estimate the location of a putative gene duplication event by rooting the tree, i.e. by identifying the location of the LCA (or the root r) of the four genes in the tree (20). Rooting the tree from the topology in Figure 1a could identify a root r in any one of the four outer branches (Figure 1c), which is not informative for establishing the relationship between genes x and y. Alternatively, it could identify a root r in the inner branch (Figure 1d), which allows us to infer that x and y are paralogs. Due to uncertainties in the construction of gene trees and approximations in the rooting algorithms, we proposed the use of the distance α between the root r to its nearest inner node as a measure of the reliability of the estimate for r. This assumes that the likelihood of correctly estimating the location of the root r is directly proportional to the value of α. Consequently, we could infer that genes x and y were paralogs when α was greater than a specified cutoff value (Ω), with a larger Ω leading to fewer number of paralogs and larger number of orthologs, and vice versa.
We observed, however, that the precise construction of quartet gene trees was still computationally time consuming for large-scale applications. Therefore, we proposed to completely bypass the construction of gene trees and the estimation of the root r and instead used an analytic expression to directly, approximately estimate α. By assuming that the four genes evolved from their LCA with the same mutation rate, i.e. that genes x, y, z1 and z2 are equidistant to r, we can approximately estimate α as follows:
| (1) |
where si,j denotes the sequence similarity of genes i and j computed using BLAST bit-scores [see Supplementary Data for the derivation of Equation (1)]. Note that α is not bounded, with negative values reflecting the case where the root r is located in one of the four outer branches of the tree (Figure 1c) and positive values indicating that r is located in the inner branch (Figure 1d). Therefore, when α > Ω, we determined genes x and y to be paralogs; otherwise, they were assumed to be orthologs. This approximation provides a computationally efficient means to identify orthologs because once we have computed the BBH pairs for all genomes in the database, which involves an all-against-all BLAST search of all genes, the additional computational cost of calculating α in Equation (1) is limited to the time to fetch the values for the six already-computed pair-wise sequence similarities si,j.
Other compared methods
We implemented the BBH method and the outgroup method for comparisons with QuartetS. We identified BBH pairs in a genome sequence database by performing pair-wise all-against-all BLAST searches. Two genes x and y of two different species X and Y, respectively, formed a BBH pair if they possessed the largest sequence alignment score when compared against the sequence alignments of each of these two genes with all other genes of the other one species. In addition, to be considered orthologs, the BBH pairs had to satisfy two conditions: (i) the alignment region had to cover at least 50% of the length of each sequence and (ii) the bit-score of the pair-wise alignment had to exceed a cutoff value (default set to 50, which was equivalent to a 10−5 E-value cutoff in our large-scale evaluations, unless specified otherwise). For the BBH pair computations employed as part of the outgroup and QuartetS methods, we used the same two conditions as the ones described above.
In our implementation of the outgroup method, the orthology relationship inferred by BBH pairs was verified by a set of preselected outgroup species that did not belong to the common clade for which gene orthology was being evaluated. In particular, the method determined that genes x and y were orthologs only if they formed a BBH pair and their sequence similarity was significantly higher than their sequence similarity with any gene z in any outgroup species Z, i.e. only if
| (2) |
where si,j denotes the sequence similarity of genes i and j measured by BLAST bit-scores and Θ represents a cutoff value, with larger values leading to more conservative orthology predictions. The selection of the outgroup species is critical and sometimes difficult, especially for the prediction of orthologs in prokaryotes. As denoted in Equation (2), if the outgroup species Z were distant from species X and Y, then sx,z and sy,z would be small and the method would not be effective in identifying possible false positive predictions. Conversely, if the outgroup species were very close to species X and Y, sequence similarity errors and horizontal gene transfer could yield large sx,z or sy,z, causing the rejection of true orthologs. In our large-scale orthology evaluations for bacterial species, we selected all available sequenced archaea as outgroup species because of their less ambiguous evolutionary relationship with bacteria and fewer horizontal gene transfers with bacteria compared with those among bacterial species.
For post-processing clustering of pair-wise orthologs, we used the Markov Cluster (MCL) program (version 08-213, downloaded from http://micans.org/mcl) previously used by OrthoMCL (13). MCL is an unsupervised clustering algorithm, which clusters genes into orthologous groups, where the size (and therefore the number) of the clusters is controlled by one parameter, the so-called inflation parameter. In general, large inflation parameter values should produce fewer clusters with relatively larger size. We set the inflation parameter to 3 as empirical evaluations with other values did not produce signification changes in the results.
In addition, we compared QuartetS with the recently developed OMA method (19), which also uses two genes in a third genome for orthology prediction. However, unlike QuartetS, which analyzes the evolution of four genes by approximating the reconstruction of a quartet gene tree, OMA uses a set of heuristic rules to predict whether two genes are orthologs. Because the OMA program is not readily available, we downloaded its pre-computed ortholog predictions for prokaryotes from its website (http://omabrowser.org/All.Oct2009/download.html).
Evaluation methods
We employed both function and phylogeny information to evaluate the performance of the orthology detection methods. For the function-based evaluation, we computed the fraction of predicted orthologs (FPOs) as a function of the false positive rate (FPR), with the optimal method producing the maximal number of orthologs with a minimal FPR. In these metrics, we only evaluated the orthology predictions for the gene pairs for which each of the two genes was annotated with a gene function. The FPR was defined as the fraction of the false positive predictions in all evaluated predictions, i.e. the sum of the true positive and false positive predictions, where the predictions were labeled as true positive if both genes had the same function annotation (or at least one function in common, if annotated with multiple functions); otherwise, they were labeled as false positive predictions. The FPO, or coverage, was defined as the ratio of the total number of evaluated predictions (i.e., true positive + false positive) to the total number of evaluated BBH pairs, which was fixed for all methods. Note, however, that we did not use the concept of false negative predictions (observed when two genes annotated with the same function were not predicted to be orthologs), because genes with the same function can also be paralogs.
The premise of phylogeny-based evaluation is that a gene tree constructed using orthologous genes provides a better agreement with its related species tree than a gene tree constructed with paralogs. Thus, the congruence between a gene tree and its related species tree has been used to estimate the quality of orthology predictions (12). However, such a comparison can be complicated because, for a given gene in a given species, different methods may predict different number of orthologs in different species, potentially biasing the congruence toward methods that make fewer predictions involving more closely related taxa. To reduce such biases and allow for a more direct comparison between any two methods, for a given gene, we only evaluated the two sets of orthology predictions when they resulted in the same number of predicted genes in the same set of taxa. When they did not, we discarded the genes in the non-overlapping taxa so as to only compare the overlapping ones. In our evaluations, we used the same species tree and a similar procedure to construct the gene trees as the ones proposed by Altenhoff and Dessimoz (12). To construct gene trees, we first performed multiple sequence alignments using ClustalW (version 1.83, http://www.clustal.org/download/) followed by tree generation with the phyml_3.0.1 program (http://atgc.lirmm.fr/phyml/) using the JTT model with gamma set to 4.
We computed the congruence C of a gene tree and its related species tree as follows:
| (3) |
where Ps is the number of tree partitions shared by the two trees and RF is the Robinson–Foulds metric, which represents the total number of unique partitions in the two trees when compared with each other (21). Congruence C attains a maximum value of 1.0 when the two trees have the exact same topology and a minimum value of 0.0 when the two trees have completely distinct topologies. Both RF and Ps were computed using the treedist program in the PHYLIP software package (version 3.68, http://evolution.genetics.washington.edu/phylip.html).
Evaluated data sets and gold-standard annotations
We performed a large-scale evaluation of QuartetS and compared it with the other methods using prokaryotic genomes in the NCBI RefSeq database (ftp://ftp.ncbi.nih.gov/genomes/Bacteria) in October 2009. Although our analysis was limited to prokaryotes, QuartetS should also be applicable to eukaryotes. Our evaluations involved 624 bacteria of the 949 available prokaryotic genomes, as this was the subset of bacteria whose orthologs had been pre-computed by OMA (19). We also use 66 archaeal genomes of the 949 prokaryotes in the evaluation of the outgroup method. The 624 evaluated genomes belong to 474 distinct bacterial species in 18 phyla, primarily involving Proteobacteria (344) and Firmicutes (114) (see Supplementary Table S1). We used a smaller subset of the prokaryotic genomes to assess the validity of the approximated formula in Equation (1). Because this assessment involved the comparison of QuartetS against precisely constructed quartet gene trees, which is a computationally expensive and time-demanding task, we limited the comparison to 40 well-studied prokaryotes (see Supplementary Table S2) with well-characterized gene function annotations.
For function-based evaluations, we separately used two distinct types of function annotations as gold standards: KEGG Orthology (KO) numbers downloaded from the KEGG database (22) and HAMAP family accession numbers downloaded from the HAMAP databases (23), both in October 2009. For the 624 bacterial genomes used in our large-scale evaluation, which consisted of 2 140 021 protein-coding genes, we assigned 58 888 unique KO numbers to the 967 831 (or 45%) protein-coding genes annotated by KEGG and 1410 unique accession numbers to the 204 663 (or 9.6%) protein-coding genes annotated by HAMAP. Because these function annotations were enriched in the genes forming BBH pairs, a substantial larger fraction of BBH pairs could be evaluated in our orthology predictions (∼79% and ∼19%, respectively, for KEGG and HAMAP). Although the fraction of BBH pair genes annotated by HAMAP was relatively small, it was included in the analysis as a source of more reliable, curated annotations.
RESULTS
Function-based evaluation
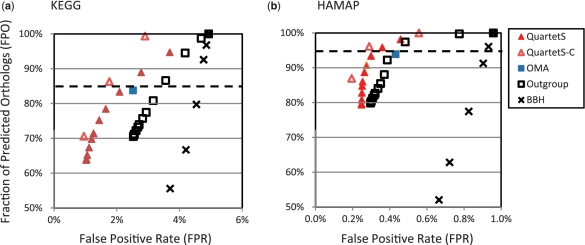
We compared the orthologs predicted by QuartetS with those pre-computed by OMA and those predicted by the outgroup method, clustering (i.e. QuartetS with MCL clustering, termed QuartetS-C), and the simple BBH method. Figure 2 shows the function-based comparisons for the 624 bacterial genomes predicted by these five methods based on different cutoff values (Ω, Θ and bit-scores of 50, 60, 70, 100, 130 and 160 for the BBH method) using KEGG (Figure 2a) and HAMAP (Figure 2b) annotations. The preferred method should yield predictions with high FPO and low FPR, i.e. entries close to the upper left corners of the plots in Figure 2. Although the two annotations produced different values of FPO versus FPR, with the relatively less reliable KEGG annotations producing considerably larger FPRs (possibly attributed to both annotation and prediction errors), Figure 2a and b show similar trends for all five methods, suggesting similar conclusions. The results clearly indicated that all methods significantly outperformed the simple BBH method and that QuartetS-C seemed to be the most effective method. For example, in the KEGG-based evaluation (Figure 2a), for its lowest FPR of 3.70%, the BBH method yielded an FPO of only 55.6%. This is in sharp contrast with both QuartetS and QuartetS-C, which, for Ω = 30, yielded FPRs of ∼1.9% with ∼85% FPO, essentially predicting 50% more orthologs with a 50% lower FPR. This is a remarkable improvement in algorithmic efficiency when we consider that QuartetS consumed <0.5% additional computational time than the BBH method. Figure 2a and b show that QuartetS is consistently and significantly superior to the outgroup method and slightly superior to OMA, which is represented by only one entry in each plot corresponding to its pre-computed results. Comparisons of QuartetS with QuartetS-C suggest that post-processing clustering can improve the overall performance of orthology detection, predicting more orthologs with fewer false positives. This is supported by the analysis of Chen et al. (24), who reported improved performance when BBH pairs were grouped into clusters using the OrthoMCL program.
Figure 2.
Function-based evaluations of the different orthology detection methods, involving 624 bacterial genomes and >2 million genes. Each entry represents the results corresponding to a given cutoff value, except for OMA, where there is only one entry corresponding to its pre-computed results. (a) Evaluations using KEGG protein function annotations. (b) Evaluations using HAMAP protein function annotations. The preferred method should yield predictions with a high fraction of predicted orthologs (FPO) and a low false positive rate (FPR), i.e. predictions close to the upper left corners of the plots. Entries close to the horizontal dashed line correspond to cutoff values for the different methods that predict similar number of orthologs as the ones in OMA.
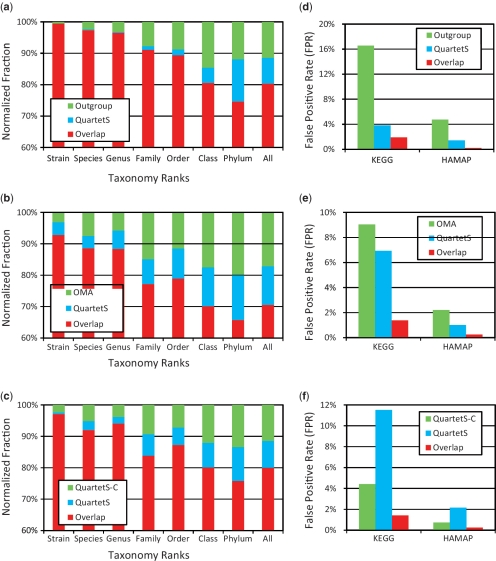
To evaluate the coverage of the different prediction methods, we performed pair-wise comparisons for the 624 bacterial genomes between QuartetS and each of the three methods: outgroup, OMA and QuartetS-C. The cutoff values used by each of the methods in these comparisons are those associated with the horizontal dashed lines in Figure 2a and b. Figure 3a–c shows the three pair-wise comparisons for the 624 genomes when all genomes were compared as one group (rightmost bar) as well as when the comparisons were performed within different granularity levels, each representing distinct evolutionary relationships based on seven bacterial taxonomy ranks, ranging from the closest relationship (i.e. strain) to the most remote relationship (i.e. phylum). For example, for the bacterial strain evaluations (leftmost bar), we compared the predicted orthologs for each of the genes comprising the different strains of a given species and aggregated the results of each such comparisons for all species. We compared the fractions of unique and overlapping orthology predictions made by each of the two compared methods for the eight different granularity levels, each normalized to the total number of corresponding predictions. Figure 3a–c shows that, overall, when we compared QuartetS with each of the other three methods, the fraction of overlapping orthology predictions ranged from 70% to 80% and that the overlap tended to decrease as we moved up along the taxonomy ranks from the inter-strain comparisons (92–99% overlap) to the inter-phylum comparisons (65–75% overlap). The reduced overlap suggests that the evolutionary information extracted from the different methods tended to diverge as the basis for the orthology comparisons included more remotely related organisms, likely also increasing the prediction error in each method. This also reflects the challenges in detecting orthologs for organisms that have long and complex evolutionary history. Figure 3a–c also shows that QuartetS consistently produced slightly smaller coverage than the other methods, as we could not identify ‘optimal’ cutoff values for each of the three methods that produced exactly the same coverage.
Figure 3.
Function-based pair-wise comparisons between QuartetS and each of the three methods, outgroup, OMA and QuartetS with clustering (QuartetS-C), using the cutoff values associated with the horizontal dashed lines in Figure 2. (a–c) Comparisons for the 624 genomes when all genomes were compared as one group (rightmost bar) as well as when the comparisons were performed within different granularity levels, each representing distinct evolutionary relationships based on seven bacterial taxonomy ranks, ranging from the least remote relationship (i.e. strain) to the most remote (i.e. phylum). (d–f) False positive rates (FPRs) for the unique and overlapping predictions using KEGG- and HAMAP-based function annotations.
Figure 3d–f shows the FPRs for the unique and overlapping predictions between each of the three pair-wise comparisons for the 624 bacterial genomes using KEGG- and HAMAP-based function annotations. As might be expected, the overlapping predictions yielded the lowest FPR in each of the six comparisons. The observed FPRs of the unique predictions support the findings presented in Figure 2, showing even more sizeable differences between the methods. Figure 3d–f shows that QuartetS significantly outperformed the outgroup method (e.g. 3.83% versus 16.56% for KEGG annotations) and slightly outperformed OMA (e.g. 6.93% versus 9.03% for KEGG annotations) and that its performance could be further improved through post-processing clustering in QuartetS-C (e.g. 11.52% versus 4.42% for KEGG annotations and 2.15% versus 0.74% for HAMAP annotations). Supplementary Figure S1a–d shows the pair-wise comparisons between QuartetS-C and each of the outgroup and OMA methods, strongly supporting the potential advantages of clustering to improve orthology predictions for gene function annotation.
Phylogeny-based evaluation
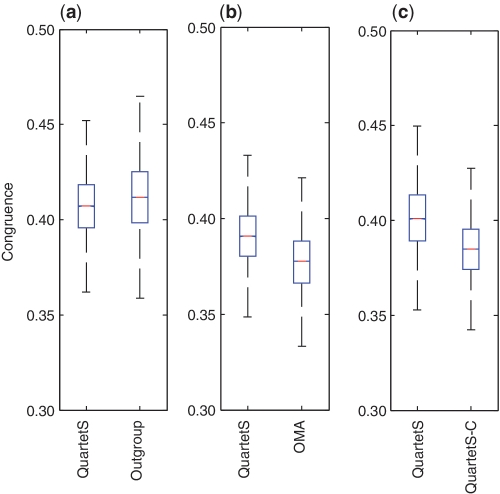
In the phylogeny-based evaluations, we compared the congruence of gene trees constructed for a set of predicted orthologs with a fixed bacterial species tree representing eight different taxa (Supplementary Figure S2), following the procedure proposed by Altenhoff and Dessimoz (10). Consistent with these taxa, we randomly selected 120 000 genes from the corresponding subset of the 624 bacterial genomes and performed pair-wise comparisons for orthologs predicted by QuartetS and each of the outgroup, OMA, and QuartetS-C methods. Figure 4 shows the box plot results for the three pair-wise comparisons. The congruence of QuartetS is slightly different for the different comparisons because, as discussed in the ‘Materials and Methods’ section, to reduce biases between each pair of compared methods, the basis for comparisons was limited to the same number of predicted genes in the same set of taxa, which varied for each pair-wise comparison. The figure indicates that, overall, congruence C in Equation (3) fluctuated ∼0.400 for the different methods, with the outgroup method having the largest standard deviation and range and QuartetS-C the smallest. The congruence of QuartetS was slightly lower than that of the outgroup method but slightly higher than that for OMA and QuartetS-C. The moderate improvement of QuartetS compared with OMA is consistent with the function-based evaluations, while the results of the other two comparisons are inconsistent. This highlights the fundamental differences of the different methods, their common underpinning with function- and phylogeny-based evaluation metrics and the potential limitations of these metrics in evaluating orthology prediction methods.
Figure 4.
Phylogeny-based pair-wise comparisons involving 120 000 genes from a subset of the 624 bacterial genomes. We used box plots to compare the congruence of a pre-specified species tree (12) with gene trees constructed by orthologous genes predicted by QuartetS and (a) outgroup, (b) OMA and (c) QuartetS-C. Higher congruence implies better orthology predictions.
Effect of QuartetS approximation
To avoid the computationally expensive, time-demanding task of constructing precise quartet gene trees to identify the exact location of the LCA (or root) in the tree and acquire homology (paralogy versus orthology) evidence, QuartetS uses an approximated formula [Equation (1)]. Therefore, we evaluated the effect of this approximation on the prediction accuracy of QuartetS by comparing its predictions against those obtained with precisely constructed quartet gene trees, termed QuartetT, for 40 well-studied prokaryotes (Supplementary Table S2). We performed an identical set of analysis as the ones discussed earlier and found that, as expected, QuartetT consistently outperformed QuartetS in both the function- and phylogeny-based evaluations (Supplementary Figures S3 and S4). However, such improvements were marginal. Considering the high-throughput gains provided by QuartetS (we found QuartetT to be >170-fold more computational demanding than QuartetS), we believe that the observed performance reduction is an acceptable tradeoff.
DISCUSSION
One of the challenges of orthology detection methods is to provide both extremely accurate and high-throughput ortholog predictions for large-scale applications. At one extreme, methods predicated on evolutionary evidence deduced from phylogeny trees are considered the most accurate. These methods identify orthologous and paralogous relationships by reconciling gene and species trees, thus requiring the construction of precise trees of both types (5–7). Given the computational requirements for constructing precise trees, they are not practical for large-scale applications (2,10). At the other extreme, methods predicated on BBHs using sequence similarity through standard BLAST searches can provide high-throughput orthology detection at modest computational costs. However, the lack of an evolutionary underpinning limits their prediction accuracy. In theory, their accuracy could be improved through the use of more stringent BLAST cutoff values (bit-scores or E-values); however, in practice, while the use of more stringent cutoff values can provide modest improvements, as shown here (Figure 2), they also significantly reduce the fraction of predicted orthologs.
To overcome the weaknesses of these approaches while capturing their strengths, we developed a novel orthology detection method that balances the tradeoff between prediction accuracy and high throughput. QuartetS attains accurate predictions by exploiting evolutionary evidence extracted from quartet gene trees formed by the two genes of interest and two genes from a third genome, for all available genomes. The analysis of quartet gene trees reveals if a gene duplication event, inferred by the two genes of the third genome, has occurred along the evolution of the two genes of interest, indicating a paralog relationship. Otherwise, the genes are assumed to be orthologs. As evolutionary evidence is extracted from all available sequenced genomes, we expect that the accuracy of QuartetS will continue to increase over time as more organisms are sequenced. QuartetS attains computational efficiency by approximating the precise construction and analysis of quartet gene trees through an analytic expression based on pair-wise sequence similarities using BLAST. Together, this strategy produced an acceptable tradeoff between accuracy and high throughput: the computational cost is <0.5% larger than that obtained with the widely used BBH method and the prediction accuracy is roughly akin to that obtained with precisely constructed gene trees (Supplementary Figures S3 and S4).
Another challenge is the lack of a factual ‘gold standard’ to truly evaluate orthology detection methods. However, similar to previous studies (12,25), we used both function- and phylogeny-based metrics, each having its own limitations. On the one hand, function-based evaluation inherently assumes that only orthologous genes share equivalent functions, while it is known that paralogs, in particular those originated from more recent duplication events, can also share the same functions (2). Thus, this assumption leads to the incorrect evaluation of some false positive predictions. On the other hand, phylogeny-based evaluation assumes that the rate of evolution among different specifies is equivalent to those of their corresponding genes, generally approximated by pair-wise gene sequence similarity scores; however, there exists sufficient evidence to challenge such assumption (26). Thus, this assumption leads to incorrect evaluation of some true positive predictions. Nevertheless, we used these two metrics to perform large-scale evaluations, involving 624 bacterial genomes and >2 million genes, to compare QuartetS with four other methods: BBH, outgroup, OMA and QuartetS followed by clustering (i.e. QuartetS-C).
The function-based evaluations indicated that QuartetS-C consistently achieved the best performance, followed by the QuartetS, OMA, outgroup and BBH methods (Figure 2). Each of the methods significantly improved orthology detection beyond the simple BBH method, with QuartetS and QuartetS-C producing >50% additional predictions with 50% lower FPRs than the BBH method. The comparisons between QuartetS-C and QuartetS suggest that, based on gene function, clustering can improve orthology detection by increasing the number of predictions while seemingly reducing the FPR (Figures 2, 3c and f). This new insight suggests that post-processing through clustering should be favored when the purpose of orthology detection is to infer gene functions. QuartetS consistently performed slightly better than OMA. This improvement was even more apparent when we compared the FPRs of the unique predictions inferred by each method (Figure 3e), although in this case QuartetS predicted slightly fewer orthologs. QuartetS more significantly outperformed the outgroup method (Figures 2 and 3d). We attribute this success, in part, to the larger set of reference third genomes from which QuartetS draws evolutionary evidence from, i.e. while outgroup references are limited to those clades outside the compared genomes, QuartetS extracts evidence from all available genome sequences.
The phylogeny-based metric provided a separate, independent means to comparatively evaluate QuartetS with the other methods and to highlight the dependency of the performance of some of the methods on the evaluation metric. Overall, each of the three pair-wise comparisons between QuartetS and outgroup, OMA, and QuartetS-C provided similar results, with congruence fluctuating ∼0.400 for the different methods (Figure 4). Although the pair-wise comparisons showed only slight improvements of one method over the other, only one of the three pair-wise rankings [QuartetS (0.390) versus OMA (0.378)] matched those observed in the function-based metric, whereas the other two [QuartetS (0.408) versus outgroup (0.413) and QuartetS (0.400) versus QuartetS-C (0.385)] produced the reverse ranking. This is attributed to many factors. First, the phylogeny-based evaluations involved only 120 000 genes of the >2 million genes evaluated with the function-based metric, and these evaluations were performed for one fixed bacterial species tree, covering a subset of the taxa of the 624 studied bacterial genomes (10). Nevertheless, we believe that the number of genes and species (514) covered in this analysis was sufficiently large as to not drastically alter the nature of the results and that, given the complexity in performing phylogeny-based analysis, a more comprehensive evaluation involving a much larger set of genes and comparisons against multiple species trees would be overwhelming. Second, the phylogeny-based evaluation, which measures the congruence between a gene tree formed by the predicted orthologs and a species tree, favors phylogeny-based approaches such as the one in the outgroup method. The outgroup method requires that two orthologous genes have higher sequence similarity with each other than with a third gene in an outgroup species. This is equivalent to requiring that the tree formed by the three genes (the two orthologs and the gene in a third species) be congruent to the tree formed by the corresponding three species (the two concerned species and the outgroup species). This supports the slight improvement of the outgroup method over QuartetS (Figure 4). However, given the marginal nature of the improvement and the potential difficulty in identifying appropriate, if not optimal, outgroup species (for example, which outgroup should be used to detect orthologs between archaea and bacteria?), we recommend the use of QuartetS even if the intent of orthology detection is to establish phylogenetic relationships. Finally, function-based metrics artificially inflate the performance of clustering methods. The principle underlying gene clustering inherently assumes a transitive orthology relationship, i.e. if gene x is an ortholog of gene y and gene y is an ortholog of gene z, then clustering may infer that genes x and z are orthologs, although there may be no evidence to support such an inference. When all three genes happen to have the same function, function-based evaluations will incorrectly score this unsupported orthology relationship between genes x and z as a true prediction, whereas phylogeny-based evaluations will detect such false positive prediction. This supports our observation that QuartetS-C outperformed QuartetS in the function-based evaluations but not in the phylogeny-based evaluations. While this bias of function-based metrics for clustering methods is acceptable if the intent of orthology detection is to predict gene function, it may not be adequate if the intent is to establish evolutionary relationships.
Consistent with the function-based evaluation, QuartetS also outperformed OMA in the phylogeny-based evaluation. Although the improvement is modest, it is notable because OMA has been, arguably, deemed to be the most specific method to date (12). We attribute the superior ability of QuartetS to discriminate between orthologs and paralogs to its explicit attempt to identify and directly use the occurrence of gene duplication events along the evolution of the gene pairs being evaluated and the soundness of the approximation used to analyze quartet gene trees.
CONCLUSIONS
The gap created by the unprecedented growth in the availability of sequenced genomic data and the inability to rapidly annotate and decode such information experimentally creates opportunities for computational orthology detection methods that can balance accuracy and high throughput. In this study, we developed a novel method that meets these requirements by explicitly searching, in a computationally efficient manner, for gene duplication evolutionary evidence to differentiate paralogs from orthologs. As such evidence increases with sequenced data, we expect the accuracy of the proposed QuartetS method to continuously improve. Based on our large-scale evaluation of bacterial genomes and comparisons of QuartetS with widely used and leading orthology detection methods, we conclude that QuartetS should be preferred when the intent of orthology detection is to infer phylogenetic relationships and that grouping the QuartetS predictions into clusters optimizes gene function predictions and should be preferred in this case.
AVAILABILITY
The source code and executables for QuartetS are available at http://www.bhsai.org/downloads/quartets/quartets.tar.gz. The QuartetS-C results are available at http://www.bhsai.org/downloads/quartets/quartets_prokaryotes_orthologs.tar.gz.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the U.S. DoD High Performance Computing Modernization Program, under the High Performance Computing Software Applications Institutes Initiative. Funding for open access charge is the same as the funding for the performed research.
Conflict of interest statement. The opinions and assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the U.S. Army or the U.S. Department of Defense. This paper has been approved for public release with unlimited distribution.
Supplementary Material
REFERENCES
- 1.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2010;38:D346–D354. doi: 10.1093/nar/gkp848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 3.Ohta T. Evolution by gene duplication revisited: differentiation of regulatory elements versus proteins. Genetica. 2003;118:209–216. [PubMed] [Google Scholar]
- 4.Serres MH, Kerr AR, McCormack TJ, Riley M. Evolution by leaps: gene duplication in bacteria. Biol. Direct. 2009;4:46. doi: 10.1186/1745-6150-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dufayard JF, Duret L, Penel S, Gouy M, Rechenmann F, Perriere G. Tree pattern matching in phylogenetic trees: automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics. 2005;21:2596–2603. doi: 10.1093/bioinformatics/bti325. [DOI] [PubMed] [Google Scholar]
- 6.Zmasek CM, Eddy SR. RIO: analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinformatics. 2002;3:14. doi: 10.1186/1471-2105-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollich V, Storm CE, Sonnhammer EL. OrthoGUI: graphical presentation of Orthostrapper results. Bioinformatics. 2002;18:1272–1273. doi: 10.1093/bioinformatics/18.9.1272. [DOI] [PubMed] [Google Scholar]
- 8.van der Heijden RT, Snel B, van Noort V, Huynen MA. Orthology prediction at scalable resolution by phylogenetic tree analysis. BMC Bioinformatics. 2007;8:83. doi: 10.1186/1471-2105-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pryszcz LP, Huerta-Cepas J, Gabaldon T. MetaPhOrs: orthology and paralogy predictions from multiple phylogenetic evidence using a consistency-based confidence score. Nucleic Acids Res. 2010;39:e32. doi: 10.1093/nar/gkq953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 11.Salter LA, Pearl DK. Stochastic search strategy for estimation of maximum likelihood phylogenetic trees. Syst. Biol. 2001;50:7–17. [PubMed] [Google Scholar]
- 12.Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput. Biol. 2009;5:e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dessimoz C, Cannarozzi G, Gil M, Margadant D, Roth A, Schneider A, Gonnet GH. OMA, a comprehensive, automated project for the identification of orthologs from complete genome data: Introduction and first achievements. Compar. Genomics. 2005;3678:61–72. [Google Scholar]
- 15.Alexeyenko A, Tamas I, Liu G, Sonnhammer EL. Automatic clustering of orthologs and in paralogs shared by multiple proteomes. Bioinformatics. 2006;22:e9–e15. doi: 10.1093/bioinformatics/btl213. [DOI] [PubMed] [Google Scholar]
- 16.Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dessimoz C, Boeckmann B, Roth AC, Gonnet GH. Detecting non-orthology in the COGs database and other approaches grouping orthologs using genome-specific best hits. Nucleic Acids Res. 2006;34:3309–3316. doi: 10.1093/nar/gkl433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fulton DL, Li YY, Laird MR, Horsman BG, Roche FM, Brinkman FS. Improving the specificity of high-throughput ortholog prediction. BMC Bioinformatics. 2006;7:270. doi: 10.1186/1471-2105-7-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roth AC, Gonnet GH, Dessimoz C. Algorithm of OMA for large-scale orthology inference. BMC Bioinformatics. 2008;9:518. doi: 10.1186/1471-2105-9-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huelsenbeck JP, Bollback JP, Levine AM. Inferring the root of a phylogenetic tree. Syst. Biol. 2002;51:32–43. doi: 10.1080/106351502753475862. [DOI] [PubMed] [Google Scholar]
- 21.Robinson DF, Foulds LR. Comparison of phylogenetic trees. Math. Biosci. 1981;53:131–147. [Google Scholar]
- 22.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2009;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lima T, Auchincloss AH, Coudert E, Keller G, Michoud K, Rivoire C, Bulliard V, de Castro E, Lachaize C, Baratin D, et al. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 2009;37:D471–D478. doi: 10.1093/nar/gkn661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen F, Mackey AJ, Vermunt JK, Roos DS. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS ONE. 2007;2:e383. doi: 10.1371/journal.pone.0000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hulsen T, Huynen MA, de Vlieg J, Groenen PM. Benchmarking ortholog identification methods using functional genomics data. Genome Biol. 2006;7:R31. doi: 10.1186/gb-2006-7-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henz SR, Huson DH, Auch AF, Nieselt-Struwe K, Schuster SC. Whole-genome prokaryotic phylogeny. Bioinformatics. 2005;21:2329–2335. doi: 10.1093/bioinformatics/bth324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.