Abstract
The mouse node is a transient early embryonic structure that is required for left-right asymmetry and for generation of the axial midline, which patterns neural and mesodermal tissues. The node is a shallow teardrop-shaped pit that sits at the distal tip of the early headfold (e7.75) embryo. The shape of the node is believed to be important for generation of the coherent leftward fluid flow required for initiation of left-right asymmetry, but little is known about the morphogenesis of the node. Here we show that the FERM domain protein Lulu/Epb4.1l5 is required for left-right asymmetry in the early mouse embryo. Unlike other genes previously shown to be required for left-right asymmetry in the mouse, lulu is not required for specification of node cell identity, for Nodal signaling in the node or for ciliogenesis. Instead, lulu is required for proper morphogenesis of the node and midline. The precursors of the wild-type node undergo a series of rapid morphological transitions. First, node precursors arise from an epithelial-to-mesenchymal transition at the anterior primitive streak. While in the mesenchymal layer, the node precursors form several ciliated rosette-like clusters; they then rapidly undergo a mesenchymal-to-epithelial transition to insert into the outer, endodermal layer of the embryo. In lulu mutants, node precursor cells are specified and form clusters, but those clusters fail to coalesce to make a single continuous node epithelium. The data suggest that the assembly of the contiguous node epithelium from mesenchymal clusters requires a rapid reorganization of apical-basal polarity that depends on Lulu/Epb4.1l5.
Introduction
The mouse embryo node (also called the posterior notochordal plate (PNC) (Blum et al., 2007)) and midline serve as important organizing centers. The role of the node in the establishment of left-right asymmetry has been studied extensively, and the motility of the long cilia in the node is essential to generate a leftward fluid flow that ultimately directs left-side specific expression of genes (Shiratori and Hamada, 2006: Lee and Anderson, 2008). In addition, the axial mesendodermal cells that derive from the node and extend anteriorly provide signals that are important for patterning other tissues in the embryo (Beddington, 1994). Sonic hedgehog (Shh) expressed in the notochord precursors (axial mesendoderm) initiates dorsal-ventral patterning of the overlying neural plate and mediolateral patterning in the flanking somites (Echelard et al., 1993). Lefty1 expressed in the midline cells inhibits Nodal signaling and restricts it to the left side of the embryo, in order to maintain left-right asymmetry (Meno et al., 1998). The anterior-most cells of the midline, the prechordal plate, produce Wnt and Bmp inhibitory signals that are required for development of the forebrain (del Barco Barrantes et al., 2003).
The spatial organization of the node and axial midline is crucial for their patterning roles. It is likely that the proper structure of the mouse node is required to generate the coherent leftward fluid flow that initiates left-right asymmetry (Cartwright et al., 2004). The node, which is derived from the anterior primitive streak (Kinder et al., 2001), is a cluster of ~250–350 cells that assembles on the surface of the distal tip of the e7.75 (early bud - headfold) embryo (Hashimoto et al., 2010). The node is an epithelium of cells with characteristically small apical surfaces and long primary cilia that forms a shallow pit. (This specialized epithelial layer on the embryo surface is often called the “ventral node” or the posterior notochord (PNC); for simplicity, we refer to this structure as the node.) The node epithelium is contiguous with the epithelium of the visceral endoderm, and the visceral endoderm cells that surround the node express Nodal and components of the Notch pathway (Zhou et al., 1993; Krebs et al., 2003).
Fate-mapping studies have shown that the cells that form the axial midline have three different origins. The most anterior midline cells derive from cells at anterior of the primitive streak at e7.0, the cells of the early and mid-gastrula organizer (Kinder et al., 2001). These cells apparently migrate with the mesodermal wings and navigate to the anterior midline to form the prechordal plate and anterior-most axial midline in response to unknown guidance cues. The next-most posterior axial midline cells derive from cells of the node by rearrangement of cells from the relatively wide node to the much narrower axial midline, apparently by convergent extension (Yamanaka et al., 2007; Ybot-Gonzalez et al., 2007). The most posterior midline, the tail notochord, is derived from cells of the node that migrate towards the posterior (Yamanaka et al., 2007).
The cellular and molecular mechanisms that produce the stereotyped organization of the mouse node and axial midline are not well understood. A number of signaling pathways and transcription factors that are essential for specification of cells of the node and axial midline have been identified. The Nodal signaling pathway and the FoxA2 and FoxH1 transcription factors are essential for specification of the node (Ang and Rossant, 1994: Weinstein et al., 1994; Hoodless et al., 2001); Noto and FoxA2 cooperate to specify the trunk notochord (Yamanaka et al., 2007); and Noto defines a transcriptional hierarchy that specifies nodal cilia and some specific aspects of node cells (Beckers et al., 2007). However, little is known about how these transcription factors regulate the morphogenesis of the node and midline.
We previously showed that Lulu (also known as Erythrocyte band 4.1-like 5 (Ebp4.1l5)) is a FERM domain protein that is essential for early mouse development. Null mutations in lulu cause embryonic arrest associated with a set of defects in morphogenesis, including disruption of the organization of the neural plate and failure of the normal gastrulation epithelial-to-mesenchymal transition (Lee et al., 2007; Hirano et al., 2008). Lulu protein, which is localized apically in the epithelia of the epiblast and neural plate (Lee et al., 2007), is required for the reorganization of the actin cytoskeleton that occurs during gastrulation. Lulu is also important for the apical localization of a population of F-actin in the neural plate, and in the absence of Lulu, closure of the neural tube fails completely. Here we show that Lulu is essential for the establishment of left-right asymmetry because it is required for the morphogenesis of the node and midline.
Materials and Methods
Mouse strains
The lulu mutation is an apparent null allele of Epb4.1l5 that was generated by ENU mutagenesis of C57BL/6J mice (Lee et. al., 2007). The GT2 allele of Epb4.1l5 corresponds to BayGenomics line XC282 and was generated in house from ES cells obtained from BayGenomics (Lee et. al., 2007). The nodal-lacZ knock-in allele (Collignon et al., 1996) was a gift from Elizabeth Robertson. The AFP-GFP strain (Kwon et al., 2006) was a gift from A.-K. Hadjantonakis.
Phenotypic analysis
Embryos were dissected in ice-cold PBS. For immunofluorescence, embryos were fixed for 1 hour at room temperature in PBS/4% paraformaldehyde. Embryos were washed after fixation in PBS, and then processed either for cryosectioning or for whole-mount staining. For cryosectioning, embryos were embedded in OCT and sectioned at 8 μm thickness. For antibody staining of both whole-mount and sections, embryos were blocked for 1hr. in PBS/0.1% Triton-X100/1% goat serum at 4°. Primary antibody was diluted in blocking buffer and incubated overnight at 4°; whole mount embryos were incubated on a rocking platform. After incubation, embryos were washed 3 × 20 minutes at room temperature in blocking buffer, and then incubated at RT for 1 hr. in blocking buffer + anti-rabbit secondary conjugated to Alexa-488. TRITC-conjugated phalloidin at 10 U/ml and DAPI were included in the secondary incubation. After the secondary incubation, embryos were washed 3 × 20 min. in blocking buffer, then rinsed 2 times briefly in PBS, and then mounted for imaging. Arl13b antibody (Caspary et al., 2007) was used at 1:2000. Brachyury (T) antibody was a gift from Frank Conlon, and was used at a dilution of 1:1000. Rat anti-E-Cadherin antibody (Sigma) was used at 1:500. TRITC-conjugated phalloidin was used at 10 U/ml.
For whole-mount node imaging, whole embryos were mounted immediately after staining in molten 1% low-melting agarose dissolved in PBS, on Mattek coverslip-dishes. Embryos were oriented vertically, with the distal tip of the embryo as close to the coverslip as possible. Embryos were imaged by confocal microscopy using a Leica TCS AOBS SP2 inverted stand confocal microscope. Alternatively, the distal tip of the embryo was removed and flat-mounted on a slide using Vectashield mounting media, and then scanned using a Leica TCS AOBS SP2 upright stand confocal microscope. Cryosections were mounted in Vectashield and scanned using both Leica TCS AOBS SP2 microscopes. Confocal datasets were analyzed using the Volocity software suite (Improvision).
For in situ hybridization, embryos were fixed overnight at 4° in PBS/4% PFA. For XGal staining, embryos were fixed for 15 min. at room temperature in 0.2% glutaraldehyde. Whole-mount in situ hybridization and X-Gal staining were performed using standard protocols. Samples for SEM were fixed overnight in 2.5% glutaraldehyde, then processed and observed according to standard procedures using a Zeiss Supra 25 Field Emission Scanning Electron Microscope.
Results
Left-right asymmetry is randomized in lulu mutants
lulu mutant embryos arrest at approximately e9.0, prior to many of the morphological manifestations of left-right asymmetry (Lee et al., 2007). In particular, morphogenesis of the heart is abnormal in lulu embryos, so it was not possible to determine whether there were defects in left-right asymmetry based on morphology of the mutant hearts. Nevertheless, molecular markers of left-right asymmetry are asymmetrically expressed at e8.5, when lulu embryos are healthy. Nodal-lacZ (Collignon et al., 1996) and lefty2 are normally expressed exclusively in the left lateral plate mesoderm at early somite stages. In contrast, the vast majority of lulu embryos expressed the nodal-lacZ transgene inappropriately: 11/20 mutants expressed the transgene bilaterally; 7/20 showed no transgene expression in the lateral plate mesoderm; 2/20 showed weak left-sided expression only (Fig. 1A, B). Similarly, all three lulu mutants examind expressed lefty2 bilaterally (Fig. 1C).
Fig. 1.
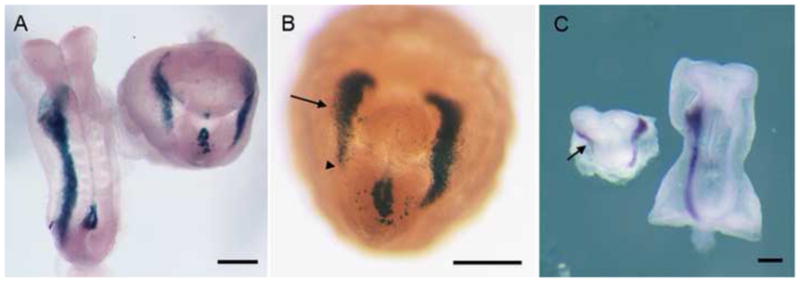
Left-right asymmetry is disrupted in lulu mutant embryos. (A) Dorsal views of nodal-lacZ expression in e8.5 wild-type (left) and lulu mutant embryos (right). Nodal-lacZ is expressed in the left lateral plate mesoderm (LPM) in wild type, but is expressed bilaterally in the mutant. (B) Ventral view of another lulu mutant embryo, showing Nodal-lacZ expression in both the left and right (arrow) LPM. The expression in the right LPM does not extend as far posteriorly (arrowhead) as in the left LPM. Note the abnormal expression of nodal-lacZ in the region of the mutant node in (A) and (B). (C) lefty2 RNA expression is bilateral in lulu (left; ventral view) (seen in 3/3 mutant embryos examined), but is confined to the left LPM in wild type (right; dorsal view). As with Nodal-lacZ, lefty expression in the right LPM (arrow) does not extend as far posteriorly as in the left LPM. Scale bars = 150μm.
Lulu is required for organization of the embryonic node and midline
Left-right asymmetry depends on signals generated in the node that are propagated to the left lateral plate and maintenance of left-sided signals depends on an intact axial midline. Markers of the node and midline were expressed in lulu embryos, but the spatial organization of the midline structures was abnormal. As previously reported, Brachyury (T) is expressed continuously along the wild-type e8.5 midline, but was discontinuous in lulu embryos (Lee et al., 2007). We found that the expression of the midline markers Shh and FoxA2 was also discontinuous in lulu embryos (Fig. 2A–D). Midline markers were expressed robustly in the anterior-most midline, at a position corresponding to the prechordal plate, but expression at more posterior positions was patchy and varied among lulu embryos. The node becomes a distinct structure at the distal tip of the wild-type embryo by e7.5, and node cells express T, Shh and FoxA2, as well as specific markers of the node such as Foxj1 (Fig. 2E). These markers were expressed at the distal tip of lulu embryos, although the expression was often weaker than in wild type (Fig. 2B, D, F).
Fig. 2.
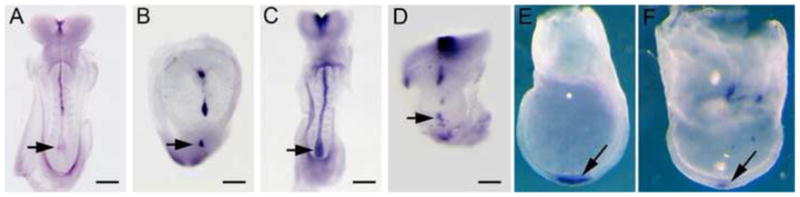
Abnormal expression of midline and node markers in lulu embryos, assayed by in situ hybridization. (A, B) Shh expression in e8.5 wild-type (A) and lulu (B) embryos. (C, D) FoxA2 expression in e8.5 wild-type (C) and lulu (D) embryos. Expression of both Shh and FoxA2 is discontinuous in the lulu midline. (E, F) FoxJ1 expression is expressed in a lateral view of an e7.5 wild-type (E) and a frontal view of a lulu (F) node arrows, but the expression level is lower in the mutant. Arrows point out the node. Scale bars = 150μm in A–D.
The disruption of the structure of the lulu node was more clearly revealed by the pattern of expression of the nodal-lacZ transgene. Prior to its expression in the left lateral plate, nodal is expressed in the endodermal crown cells that surround the node. The expression pattern of nodal-lacZ revealed that the organization of the nodal-expressing cells around the node was abnormal in lulu embryos (Fig. 1A, B). The pattern of nodal-lacZ expression varied among lulu embryos, but it was never expressed in the wild-type pattern (Fig. 3). In some embryos, the nodal-lacZ expression domain collapsed towards the midline. In other embryos, there were two or more rings of nodal-lacZ expression, as if more than one node had initiated.
Fig. 3.
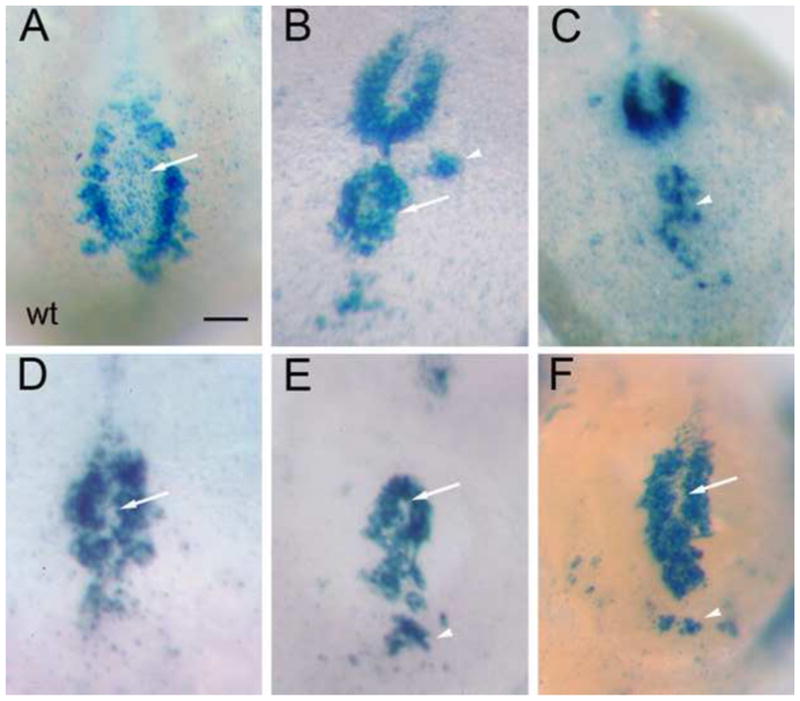
Variable disruption of node morphology in lulu mutant embryos, as revealed by the pattern of nodal-lacZ expression. Ventral views of wild type (A) and lulu nodes (B–F), in which node shape is outlined by the expression of nodal-lacZ in the crown cells. Anterior is to the top. Node shape is highly variable in the mutants; defects include loss or reduction of the node pit (arrows, D–F, compare to arrow in A), ectopic crown cells (arrowheads, B, C, E, F), and an apparently duplicated node (arrow, B). All images are at the same magnification; scale bar = 20 μm.
Lulu was expressed at high levels in the outer (ventral) epithelial layer of the node, based on expression of a gene trap allele of lulu that fuses most of the FERM domain to β-galactosidase (luluGT2) (Lee et al., 2007). The fusion protein was expressed in most cell types in the embryo (Fig. 4), but was expressed at relatively high levels in cells of the e8.0 node (Fig. 4B), consistent with a direct role of the Lulu protein within the cells of the node.
Fig. 4.
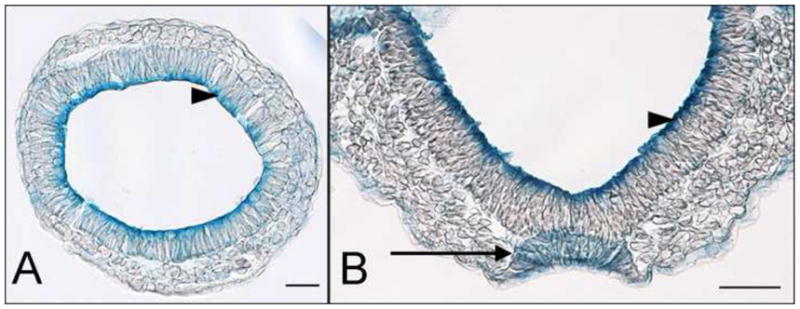
Expression of lulu in the node. (A, B) The LuluGT2-β-gal fusion protein (blue) is enriched apically in the epiblast epithelium (arrowheads) at both transverse sections of e7.5 embryos (A) and frontal sections of headfold stage embryos (B) that are heterozygous for the gene trap allele. Localization of the gene trap fusion protein parallels the localization of the endogenous Lulu/Epb4.1l5 protein (Lee et al., 2007). At the headfold stage (B), the epithelium of the node has assembled (arrow). The LuluGT2-β-gal fusion protein is present throughout these cells, rather than being enriched at the apical surface.
The lulu node fails to assemble correctly
The nodal-lacZ expression pattern indicated that the crown cells associated with the node were not organized correctly in lulu embryos. We therefore examined the cellular organization of the node at higher resolution. Cells of the node have two clear morphological hallmarks: each cell has a single long cilium and the apical surfaces of the cells are strongly constricted (Lee and Anderson, 2008). As a result, node cells can be recognized even when not properly organized into the node pit.
The apical constrictions of node cells are readily visualized by staining of F-actin with phalloidin. The apically constricted cells of the wild-type node form a stereotyped teardrop-shaped array (Fig. 5). The disrupted organization of the cells within the node was apparent in phalloidin-stained lulu embryos. The organization of the apically constricted node cells varied among lulu embryos, but the cells never formed a single well-organized field. In many cases, multiple patches of apically constricted cells were seen at the distal tip of the embryo, at the position where the node should develop (Fig. 5B). The apically constricted cells in ectopic clusters had long cilia, as visualized by staining with the cilia marker Arl13b (Caspary et al., 2007) (Fig. 5C).
Fig. 5.
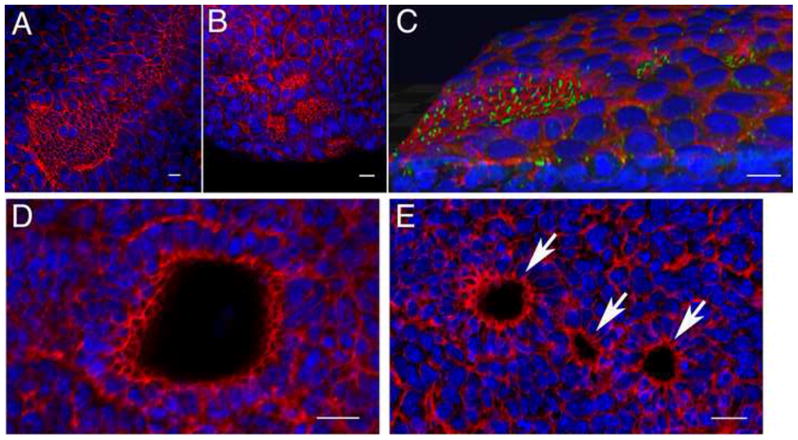
Abnormal cellular organization of the lulu node. Phalloidin (red) and DAPI (blue) staining revealed that multiple node pits form in lulu embryos. (A) Phalloidin staining of the distal surface of an e7.75 wild-type embryo shows the organization of the cells with small apical surfaces in the node and midline, compared to the large surfaces of the surrounding squamous cells of the endoderm. Anterior is to the upper right. (B) At the same stage in lulu embryos, cells with small apical surfaces are found in several separate clusters; this is an extreme example of the disruptions of the node in lulu embryos. Anterior is to the upper left. (C, D, E) Phalloidin: red. DAPI: blue. (C) 3D rendering of an e7.75 lulu embryo reveals that ectopic node cells form pits. Several clusters of cells with long cilia (marked by expression of Arl13b in green) are present outside the main node region; even the small clusters are depressed from the surface of the endoderm in small pits. Anterior to the left. See also Supplemental Movie 1. (D) A confocal section parallel to the surface of the wild-type node highlights the pit of the node, seen here as the empty circle surrounded by cells with small apical surfaces. (E) A similar confocal section through the node region of a lulu embryo shows three separate pits (arrows). Scale bars = 25μm.
Each cluster of lulu cells appeared to initiate the morphogenetic movements that create the node pit. Three-dimensional renderings of confocal images of the lulu node indicated that the clusters of ectopic ciliated cells formed small pits (Fig. 5C; Supplemental Movie 1). Optical sections parallel to the surface confirmed that there was a single large pit in the wild-type node, whereas similar sections of the distal region of lulu embryos often showed more than one pit (Fig. 5D, E).
Scanning electron microscopy studies have shown that the wild-type node assembles during the six hour period between the late streak to zero bud stages (~e7.5-e7.75). At this stage, clusters of ciliated cells with small apical surfaces appeared on the surface of the embryo near its distal tip, separated by squamous cells of the visceral endoderm (Sulik et al., 1994; Lee and Anderson, 2008). By e7.75, there was a single field of ciliated cells with small apical surfaces at the distal tip of the wild-type embryo (Fig. 6A–C). Similar to the results of immunofluorescent staining, SEM views showed that ectopic clusters of ciliated cells were present in the distal region of the lulu embryo (Fig. 6D–E). Thus all the analysis shows that the defect in the lulu embryo was not in acquisition of node cell identity, but the formation of a single coherent node.
Fig. 6.
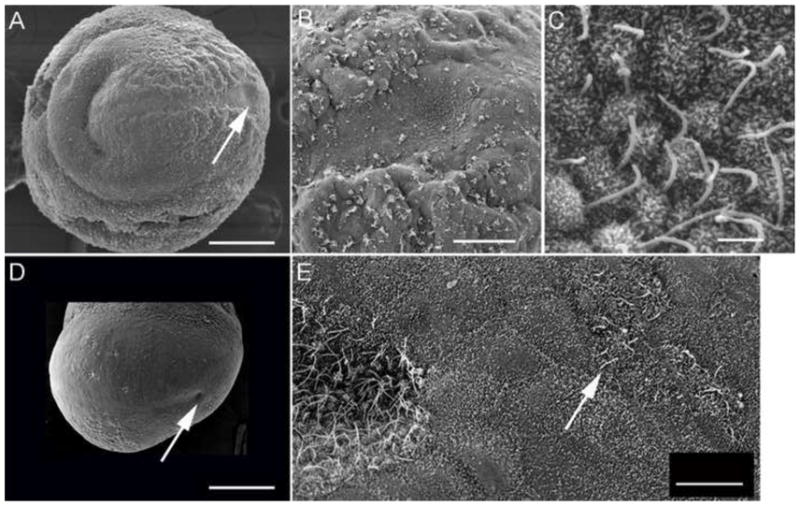
Scanning electron micrographs of wild type (A, B, C) and lulu mutant (D, E) embryos, e7.75. Anterior is to the left in all panels. The node is located distally on the ventral surface of the embryo (arrow, A). A higher magnification view (B) shows the node is visible as a concave pit of cells with small apical surfaces, surrounded by squamous visceral endoderm cells; node cells have long cilia (C). (D) lulu mutant, lateral/ventral view. This node in this lulu embryo (arrow) is approximately half the diameter of the wild-type node, and is less deep. (D) A higher magnification view shows that cilia on the lulu node appear normal. Ectopic cells with long, node-like are found to the posterior of the node (arrow), surrounded by cells of the visceral endoderm. Scale bars in A, C = 200 μm; B = 50 μm; C = 2 μm; E = 10 μm.
Lulu disrupts the coalescence of clustered pre-node cells
Node cells arise from the anterior primitive streak at the late streak stage (Kinder et al., 2001), and go through a series of rapid morphological transitions to generate the node. Node precursors undergo an epithelial-to-mesenchymal transition as they move out of the epiblast layer of the streak. At the late streak stage, an accumulation of cells between the epithelial layers of the epiblast and the endoderm can be seen at the anterior primitive streak that is likely to correspond to the node precursors (Fig. 7A). Unlike most cells that have transited the primitive streak, these pre-node cells express high levels of E-cadherin on all aspects of their surfaces. Shortly thereafter, the same cells undergo a mesenchymal-to-epithelial transition as they incorporate into the endodermal layer to generate the outer epithelial layer of the node. Optical cross sections of E-cadherin stained embryos reveal that there is an organization within the mesenchymal node precursors: several rosette-like structures composed of 10–20 cells with clustered apical surfaces are present between the epiblast and the endoderm layers of the pre-node embryo (Fig. 7B, 7C). These clustered cells near the distal tip of the embryo between the epiblast and the visceral endoderm layers already have the long cilia characteristic of nodal cilia (Fig. 8B). Within a few hours, all the node cells are visible on the surface of the embryo, and the mesenchymal-to-epithelial transition is complete (Fig. 7D).
Fig. 7.
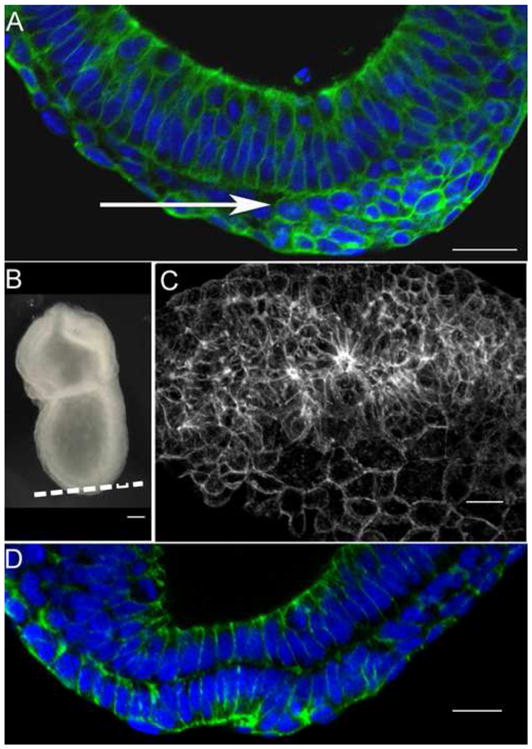
Sequential epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions produce the wild-type node. E-cadherin: green. DAPI: blue. (A) At the late streak stage, cells of the presumptive node accumulate at the anterior primitive streak between the epithelial layers of the epiblast and endoderm (arrow), after having delaminated from the epiblast layer (above the mesenchymal cells in this image). These cells express E-cadherin (green) around their circumferences. (B) A late streak embryo, indicating the plane of the optical section in (C). (C) E-cadherin expression (white) reveals that the presumptive node cells between the epiblast and endoderm layers form several large three-dimensional rosette-like structures in the presumptive node region, where E-cadherin is enriched at the center of each rosette. (D) Within a few hours, the cells of the presumptive node have completed the mesenchymal-to-epithelial transition. The node cells have incorporated into the outer layer of the embryo and express apical E-cadherin. The node has formed its characteristic pit at the distal tip of embryo. Scale bars in A, C, D = 25 μm; in D = 50 μm.
Fig. 8.
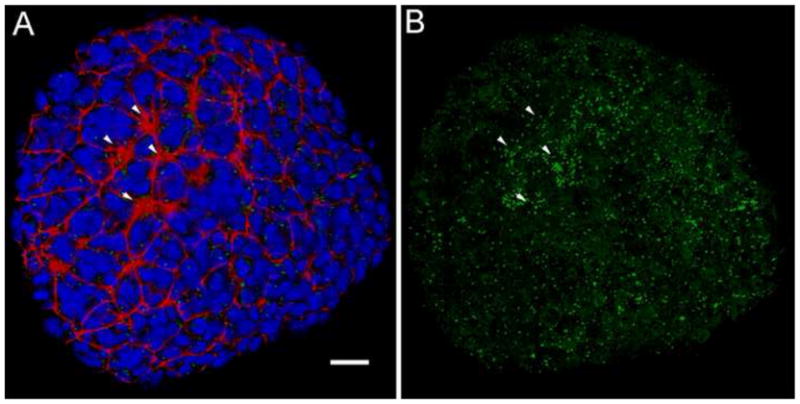
Groups of ciliated presumptive node cells are present below the endodermal layer in the wild-type embryo. (A, B) 3D rendering of a ventral view of an e7.25 wild-type embryo, early in the process of node morphogenesis. (A) Red = phalloidin; blue =DAPI; green = Arl13b. Several clusters of cells with small apical surfaces are visible on surface of the embryo (arrowheads). At this stage, the nuclei between the clusters belong to the visceral endoderm (Kwon et al., 2008). (B) Same embryo as in (A), Arl13b channel only, to visualize cilia. Clusters of long cilia are seen at the position of the clusters seen in phalloidin staining (arrowheads); additional cells with long cilia are found beneath the endoderm (compare to (A)). Note that short cilia are also broadly distributed on endoderm cells. Scale bar = 25 μm.
In lulu mutants, there are normal numbers of node cells (an average of 220 ciliated cells in lulu compared to 200 in wild type littermates), node cells form cilia of normal length (4.2±1.0 μm in lulu; 3.5±0.6μm in wild-type), and clustered node cells constrict their apical surfaces (Fig. 5B, 5C, 6E). However, the early clusters of node precursors do not resolve into a single field, and several groups of node cells remain separated in the surface layer of most mutant embryos.
Like the morphogenesis of the node, organization of the anterior mesendoderm also involves clusters of apically constricted ciliated cells that coalesce to form a narrow stripe of apically constricted cells on the midline (Fig. 5A). The midline of the embryo has more than one origin. Cells of the trunk notochord (caudal to the second somite) derive from the cells of the node, while more anterior midline cells (the anterior head process and prechordal plate) derive from cells of the early/mid gastrula organizer (Kinder et al., 2001; Yamanaka et al., 2007). Although somite formation is blocked in lulu embryos, the mutants arrest at the equivalent of the about two-somite stage, so the midline cells in lulu embryos most probably derive from the early and mid-gastrula organizer. In lulu mutants, there are midline cells that express midline markers, but, as in the node, these cells did not coalesce to make a single midline (Fig. 2).
Coordination of cell behaviors between the visceral endoderm and the node/midline
During the formation of the node and midline, cells derived from the primitive streak insert into the visceral endoderm layer that surrounds the e7.5 embryo. An AFP-GFP transgene (Kwon et al., 2006), which is expressed in the cells of the visceral endoderm (VE) at this stage, makes it possible to visualize the behavior of the VE cells. At approximately e7.5, cells of the definitive endoderm begin to insert into the outer VE layer, causing the AFP-GFP+ VE cells to become dispersed over most of the embryo (Kwon et al., 2008). At the same stage, VE cells remain covering the region of the primitive streak and form a single-cell file surrounding the node and midline (Kwon et al., 2008; Fig 9A). The same general pattern was retained in lulu embryos: AFP-GFP+ VE cells became dispersed over most of the embryo and were retained over the primitive streak. In the region of the node and midline, the arrangement of AFP-GFP+ VE cells in lulu embryos paralleled the disorganization of the node and axial mesendoderm cells (Fig. 9B, C). The most striking feature of the mutant AFP-GFP+ VE cells was that they continued to organize around mutant node cells. For example, in the embryo shown in Fig. 9B, there appeared to be two contiguous node-like regions, as defined by two oval regions that express T (Fig. 9B′), and AFP-GFP+ cells aligned between the two nodes (Fig. 9B″, arrow).
Fig. 9.
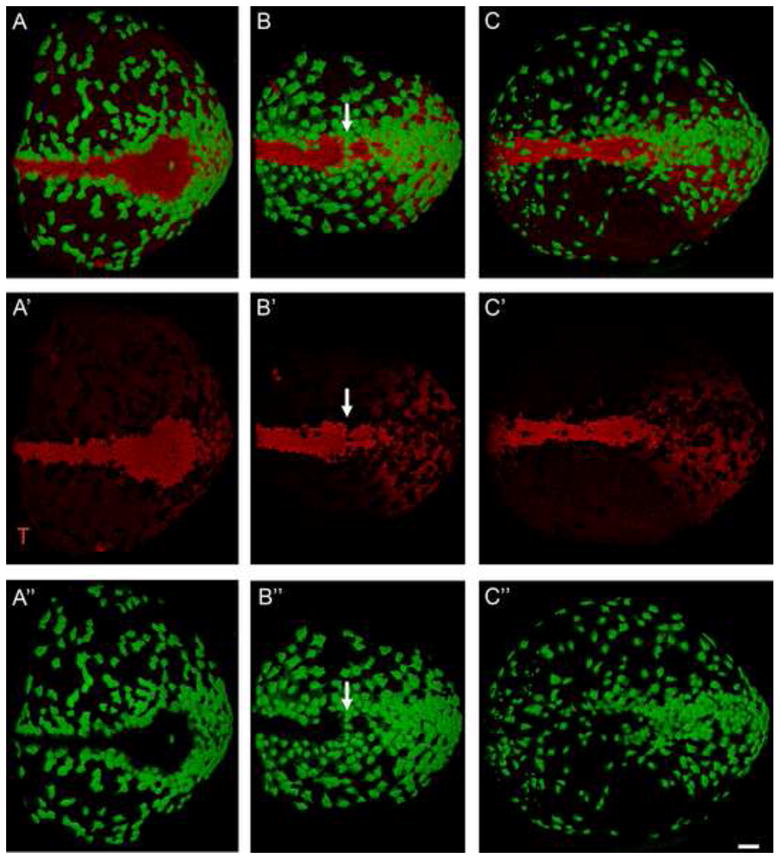
The organization of the node and midline is mirrored by the organization of cells of the visceral endoderm. Green = AFP-GFP, marks the visceral endoderm. Red = T; expressed in the node, axial midline and primitive streak. Anterior to the left; posterior (the primitive streak) to the right. (A) The axial midline of the wild-type headfold (e7.75) embryo. The formation of the node and midline is complete at this stage. Most visceral endoderm cells (green) have dispersed by this stage, except for those lying over the primitive streak (right). At this stage, a single row of visceral endoderm cells surrounds the node and midline. (A′) T channel only shows the shape of the node and midline. (A″) GFP channel shows the arrangement of visceral endoderm cells around the wild-type node and midline. (B and C) lulu mutant embryos; (B′ and C′) T channel only; (B″ and C″) GFP channel only. (B). In this lulu embryo the node appears to be pinched off into two adjacent clusters of node cells, and AFP+ cells align between the two node-like regions (arrow). (C) In this mutant, the node is reduced in size. A few AFP-GFP+ visceral endoderm cells are present over the axial midline of this mutant embryo and others lie over the posterior node. All images are at the same magnification; scale bar = 50 μm.
Discussion
Roles of lulu/Epb4.15 in left-right asymmetry
Many mouse genes have been shown to be required for the generation of left-right asymmetry, but most appear to regulate either ciliogenesis or Nodal signaling. Cilia are required to generate a leftward fluid flow across the node; mutations that disrupt ciliogenesis, cilia motility or the calcium signaling activated by nodal flow lead to randomization of laterality, frequently accompanied by bilateral expression of Nodal in the lateral plate (Lee and Anderson, 2008; Basu and Brueckner, 2008). Mutations in the Nodal signaling pathway have direct effects on signaling from the node to the lateral plate mesoderm (Brennan et al., 2002; Saijoh et al., 2003). Other signaling pathways are also important in left-right asymmetry: activity of the Hedgehog pathway in the lateral plate mesoderm is required to propagate Nodal signaling in the lateral plate (Tsiairis and McMahon, 2009) and Fgf signaling is required for laterality in the mouse, perhaps through regulation of Nodal signaling or cilia length (Meyers and Martin, 1999; Neugebauer et al., 2009). Other genes required for left-right asymmetry (e.g. T, Tbx6, Dll1, Wnt3a) (King et al., 1998; Hadjantonakis et al., 2008; Przemeck et al., 2003; Nakaya et al., 2005) are thought to act, at least in part, through regulation of Nodal signaling.
In contrast to these previously characterized mutations, the failure of lulu mutants to establish left-right asymmetry appears to be due to a disruption of morphogenesis of the node and midline. Although lulu embryos arrest before handedness of the visceral organs would be apparent and have independent defects in heart morphogenesis that would prevent the detection of defects in heart looping (Lee et al., 2007), the symmetrical expression of nodal and lefty2 in the lateral plate mesoderm (Fig. 1) demonstrate that lulu mutants fail to establish correct laterality. Both Nodal signaling and formation of nodal cilia are required for establishment of left-right asymmetry, but both appear to be normal in lulu embryos. A loss of Nodal signaling would lead to a failure to express nodal in the node or lateral plate mesoderm, but nodal is expressed in both regions of lulu embryos. Homologues of Lulu/Epb4.1l5 have been shown to regulate the activity of Crumbs in Drosophila and zebrafish, and siRNA and shRNA experiments have implicated Crumbs proteins in mammalian ciliogenesis (Fan et al., 2004; Fan et al., 2007); however Lulu/Epb4.1l5 is not required for ciliogenesis, as nodal cilia of normal length are present at the distal tip of the headfold stage lulu embryos (Fig. 5C, 6E). Instead, we conclude that Lulu/Epb4.1l5 is required for left-right asymmetry because it is required for cells to organize a correctly structured node and midline.
Coalescence of clusters of node precursors is required for formation of the wild-type node
Formation of the epithelial layer of the node on the outer surface of the mouse embryo is an interesting and unusual process in which a population of cells undergoes rapid, sequential epithelial-to-mesenchymal and then mesenchymal-to-epithelial transitions, such that the new epithelium has the opposite polarity to the epithelium of origin. Cells of the node derive from the anterior primitive streak of the late streak stage embryo (~e7.5) (Beddington, 1981; Kinder et al., 2001). As in other regions of the primitive streak, these cells undergo an epithelial-to-mesenchymal transition as they move from the epithelial layer of the epiblast into the mesenchymal layer between the epiblast and the endoderm. Cells in more proximal regions of the streak rapidly down-regulate E-cadherin, and that down-regulation is essential for migration of the nascent mesoderm away from the primitive streak (Burdsal et al., 1993; Ciruna and Rossant, 2001). In contrast, the node precursors at the primitive streak apparently never down-regulate E-cadherin from the time they are in the epiblast until they assemble into the node; instead, when cells move out of the primitive streak, E-cadherin is redistributed so it that is not restricted to an apical surface and instead is uniformly distributed on the cell surface (Fig. 7A). While still in the layer between the epiblast and endoderm, the precursors of the node begin to grow long primary cilia and assemble into several rosettes-like clusters (Fig. 7C). Each rosette appears to have higher levels of E-cadherin at the center, suggesting that the center of the rosette has an apical identity. These separate rosettes then coalesce to form the single epithelium of the node, which has the opposite apical-basal polarity of the epiblast epithelium from which they derived a few hours earlier (Fig. 7D). Similar rosettes of Noto-expressing node cells were reported by Yamanaka et al (2007), although those authors did not observe the coalescence of the rosettes or describe how they insert into the endodermal layer.
In the wild-type embryo, the separated rosette-like structures emerge on to the embryo surface as they insert as groups into the endodermal layer. They first appear as clusters of apically-constricted ciliated cells in SEM images at the zero bud stage (Sulik et al., 1994; Lee and Anderson, 2008) and as a sheet of ciliated cells below the endoderm layer (Fig. 8B). At the end of morphogenesis of the node and midline, a single row of visceral endoderm cells outlines these midline structures (Kwon et al., 2008; Fig. 9A).
The assembly of the mouse node epithelium from multiple rosette-like structures has a number of parallels with the morphogenesis of Kupffer’s vesicle, the organ of laterality in the zebrafish embryo (Amack et al., 2007; Oteíza et al., 2008). Like the node, Kupffer’s vesicle (KV) is a ciliated epithelium that generates a fluid flow required to initiate left-right asymmetry in the fish. Like the node, the KV epithelium arises as the result of a mesenchymal-to-epithelial transition (Amack et al., 2007) that goes through an intermediate of several epithelial rosette like-structures that resolve to form the single KV (Amack et al., 2007; Oteíza et al., 2008).
Lulu/Epb4.1l5 is required for coalescence of node precursor clusters
In the lulu mutant, cells derived from the anterior streak are specified to have a node identity and have most properties of normal node cells. lulu mutant node cells express node markers, such as Foxj1; they form long, apical primary cilia, they change cell shape and acquire a small apical surface, and clusters of cells form concave depressions. The lulu mutant cells fail, however, to coalesce into a single node field. Although the morphology of the node varies among mutant embryos, there are always node-like cells on the surface of the embryo that are outside the primary node region.
A few other mouse mutants have been described that cause abnormal morphogenesis of the node and midline. For example, in several Notch pathway mutants (Dll1, Baf60c; Rbpj) loss of left-right asymmetry is correlated with the presence of some endoderm cells over the node and/or loss of expression of midline markers (Przemeck et al., 2003; Takeuchi et al., 2007). The presence of endoderm cells over the node could reflect a delay in development, as endoderm cells normally move off the node over time, or it could be due to a breakdown in communication between the node and visceral endoderm. However, the disruptions in left-right asymmetry in these mutants have been attributed to loss of Nodal expression in the node (Krebs et al., 2003; Przemeck et al., 2003; Takeuchi et al., 2007), which is not the case in lulu mutants.
In the zebrafish, two Tbox transcription factors, Brachyury/no tail and Tbx16/spadetail, are required for morphogenesis of Kuppfer’s vesicle. In particular, Tbx16/spadetail appears to be required for coalescence of the epithelial rosettes that give rise to the KV. The mouse homologue of Tbx16/spadetail appears to be Tbx6, and left-right asymmetry is disrupted in mouse Tbx6 mutants, but that defect appears to be the result of defects in cilia structure and Notch signaling rather than morphogenesis of the node (Hadjantonakis et al., 2008). However, the relatively intense expression of lulu in the node (Fig. 4B) suggests that it could be a target of Tbox transcription factors that promote morphogenesis of the organs of laterality.
Loss of Lulu/Epb4.1l5 affects the behavior of both the cells of the node and the adjacent visceral endoderm cells. Mutant cells with node identity form ectopic clusters (Fig. 5C, E; Fig. 6C; Fig 9B′) and, in parallel, the surrounding nodal-expressing mutant visceral endoderm cells are found in ectopic locations, often in ectopic rings (Fig. 2B, C, E). In the regions where the lulu node and midline cells are organized more normally, most endoderm cells move away from the midline and outline the ectopic node and midline cells (Fig. 9B″, C″). These findings highlight the coupling of the cell behaviors in these two tissues. Because of the roles of Lulu/Epb4.1l5 in epithelial reorganization (Lee et al., 2007), we suggest that the defect in lulu node morphogenesis may be intrinsic to the node precursor cells, but future genetic mosaic experiments will be required to test this possibility.
The initial studies of Epb4.1l5 homologues in Drosophila and zebrafish (Yurt and Mosaic Eyes) emphasized the role of these proteins in the regulation of apical Crumbs complexes (Laprise et al., 2006; Hsu et al., 2006; Gosens et al., 2007). However, recent studies have shown that Drosophila Yurt and mammalian Lulu/Epb4.1l5 are also important for the stabilization of the basolateral domain of epithelial cells (Laprise et al., 2009). A role for Lulu in establishment of the basolateral domain is supported by the similarity of the global morphology of lulu embryos and those of FAK, Paxillin and Fibronectin mutants, which should lack normal focal adhesions (Furuta et al., 1995; Hagel et al., 2002; George et al., 1993). Although the Paxillin mutant phenotype has not been characterized in depth, the similar time of arrest and global morphology of the lulu and Paxillin mutants is especially provocative, as the region of Lulu/Epb4.l15 C-terminal to the FERM domain has been shown to bind Paxillin (Hirano et al., 2008).
Mammalian genomes encode approximately 50 FERM proteins, most of which have not been studied. Lulu/Epb4.1l5, and its homologues Drosophila Yurt and zebrafish Mosaic eyes, are members of the FERM-FA subfamily, which includes ~14 mammalian genes (Tepass, 2009). The mouse protein most closely related to Lulu/Epb4.1l5 is Epb4.1ll4b (Ehm2), which was first identified based on its upregulation in a metastatic melanoma cell line (Hashimoto et al., 1996) and is expressed in prostate epithelium (Wang et al., 2006). Although the expression and function of Epb4.1l4b in the mouse embryo has not been studied, it will be interesting to test whether it cooperates with Lulu in early embryonic morphogenesis. FERM-FA proteins, including the founding member of the family, Band 4.1, associate with spectrin, and our previous studies suggest that Lulu is required for organization the membrane-associated actin cytoskeleton, as actin-associated proteins including β2-spectrin are mislocalized in the lulu neuroepithelium.
Lulu/Epb4.1l5 is required for the gastrulation epithelial-to-mesenchymal transition (Lee et al., 2007; Hirano et al., 2008), and we propose that the defect in node morphogenesis in lulu embryos may be due to defects in the reorganization of the actin cytoskeleton during the MET that generates the node epithelium. We note that while the LuluGT2-β-gal fusion protein is localized apically in the epithelium of the epiblast, like the Lulu protein itself (Figure 4; Lee et al., 2007), LuluGT2-β-gal was not restricted to apical regions of the node, and instead was present throughout those cells. We suggest that the fractured node of lulu embryos reflects the importance of Lulu/Epb4.1l5 in the rapid reorganizations of apical-basal polarity during the sequential EMT and MET required for formation of the node.
Supplementary Material
Acknowledgments
We thank Kat Hadjantonakis for the AFP-GFP mice and for helpful discussions. We thank Nina Lampen at the MSKCC Electron Microscopy Core Facility, and the staff of the MSKCC Molecular Cytology Core Facility, for their expert assistance during SEM and confocal imaging. IM was supported by a long-term EMBO post-doctoral fellowship and a Starr Foundation fellowship. The work was supported by NIH grant HD035455 to KVA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amack JD, Wang X, Yost HJ. Two T-box genes play independent and cooperative roles to regulate morphogenesis of ciliated Kupffer’s vesicle in zebrafish. Dev Biol. 2007;310:196–210. doi: 10.1016/j.ydbio.2007.05.039. [DOI] [PubMed] [Google Scholar]
- Ang SL, Rossant J. HNF–3β is essential for node and notochord formation in mouse development. Cell. 1994;78:561–74. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Basu B, Brueckner M. Cilia multifunctional organelles at the center of vertebrate left-right asymmetry. Curr Top Dev Biol. 2008;85:151–74. doi: 10.1016/S0070-2153(08)00806-5. [DOI] [PubMed] [Google Scholar]
- Beckers A, Alten L, Viebahn C, Andre P, Gossler A. The mouse homeobox gene Noto regulates node morphogenesis, notochordal ciliogenesis, and left right patterning. Proc Natl Acad Sci USA. 2007;104:15765–70. doi: 10.1073/pnas.0704344104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddington SP. An autoradiographic analysis of the potency of embryonic ectoderm in the 8th day postimplantation mouse embryo. J Embryol Exp Morphol. 1981;64:87–104. [PubMed] [Google Scholar]
- Beddington RS. Induction of a second neural axis by the mouse node. Development. 1994;120:613–20. doi: 10.1242/dev.120.3.613. [DOI] [PubMed] [Google Scholar]
- Blum M, Andre P, Muders K, Schweickert A, Fischer A, Bitzer E, Bogusch S, Beyer T, van Straaten HW, Viebahn C. Ciliation and gene expression distinguish between node and posterior notochord in the mammalian embryo. Differentiation. 2009;75:133–46. doi: 10.1111/j.1432-0436.2006.00124.x. [DOI] [PubMed] [Google Scholar]
- Brennan J, Norris DP, Robertson EJ. Nodal activity in the node governs left-right asymmetry. Genes Dev. 2002;16:2339–44. doi: 10.1101/gad.1016202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdsal CA, Damsky CH, Pedersen RA. The role of E-cadherin and integrins in mesoderm differentiation and migration at the mammalian primitive streak. Development. 1993;118:829–44. doi: 10.1242/dev.118.3.829. [DOI] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–78. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Cartwright JH, Piro O, Tuval I. Fluid-dynamical basis of the embryonic development of left-right asymmetry in vertebrates. Proc Natl Acad Sci USA. 2004;101:7234–9. doi: 10.1073/pnas.0402001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Collignon J, Varlet I, Robertson EJ. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 1996;381:155–8. doi: 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- del Barco Barrantes I, Davidson G, Gröne HJ, Westphal H, Niehrs C. Dkk1 and noggin cooperate in mammalian head induction. Genes Dev. 2003;17:2239–44. doi: 10.1101/gad.269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–30. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- Fan S, Hurd TW, Liu CJ, Straight SW, Weimbs T, Hurd EA, Domino SE, Margolis B. Polarity proteins control ciliogenesis via kinesin motor interactions. Curr Biol. 2004;14:1451–61. doi: 10.1016/j.cub.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Fan S, Fogg V, Wang Q, Chen XW, Liu CJ, Margolis B. A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin beta interactions. J Cell Biol. 2007;178:387–98. doi: 10.1083/jcb.200609096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y, Ilić D, Kanazawa S, Takeda N, Yamamoto T, Aizawa S. Mesodermal defect in late phase of gastrulation by a targeted mutation of focal adhesion kinase, FAK. Oncogene. 1995;11:1989–95. [PubMed] [Google Scholar]
- George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119:1079–91. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- Gosens I, Sessa A, den Hollander AI, Letteboer SJ, Belloni V, Arends ML, Le Bivic A, Cremers FP, Broccoli V, Roepman R. FERM protein EPB41L5 is a novel member of the mammalian CRB-MPP5 polarity complex. Exp Cell Res. 2007;313:3959–70. doi: 10.1016/j.yexcr.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis AK, Pisano E, Papaioannou VE. Tbx6 regulates left/right patterning in mouse embryos through effects on nodal cilia and perinodal signaling. PLoS One. 2008;3:e2511. doi: 10.1371/journal.pone.0002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel M, George EL, Kim A, Tamimi R, Opitz SL, Turner CE, Imamoto A, Thomas SM. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol Cell Biol. 2002;22:901–15. doi: 10.1128/MCB.22.3.901-915.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Shindo-Okada N, Tani M, Takeuchi K, Toma H, Yokota J. Identification of genes differentially expressed in association with metastatic potential of K–1735 murine melanoma by messenger RNA differential display. Cancer Res. 1996;56:5266–71. [PubMed] [Google Scholar]
- Hirano M, Hashimoto S, Yonemura S, Sabe H, Aizawa S. EPB41L5 functions to post-transcriptionally regulate cadherin and integrin during epithelial-mesenchymal transition. J Cell Biol. 2008;182:1217–30. doi: 10.1083/jcb.200712086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoodless PA, Pye M, Chazaud C, Labbé E, Attisano L, Rossant J, Wrana JL. FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes Dev. 2001;15:1257–71. doi: 10.1101/gad.881501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Willoughby JJ, Christensen AK, Jensen AM. Mosaic Eyes is a novel component of the Crumbs complex and negatively regulates photoreceptor apical size. Development. 2006;133:4849–59. doi: 10.1242/dev.02685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinder SJ, Tsang TE, Wakamiya M, Sasaki H, Behringer RR, Nagy A, Tam PP. The organizer of the mouse gastrula is composed of a dynamic population of progenitor cells for the axial mesoderm. Development. 2001;128:3623–34. doi: 10.1242/dev.128.18.3623. [DOI] [PubMed] [Google Scholar]
- King T, Beddington RS, Brown NA. The role of the Brachyury gene in heart development and left-right specification in the mouse. Mech Dev. 1998;79:29–37. doi: 10.1016/s0925-4773(98)00166-x. [DOI] [PubMed] [Google Scholar]
- Krebs LT, Iwai N, Nonaka S, Welsh IC, Lan Y, Jiang R, Saijoh Y, O’Brien TP, Hamada H, Gridley T. Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes Dev. 2003;17:1207–12. doi: 10.1101/gad.1084703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon GS, Fraser ST, Eakin GS, Mangano M, Isern J, Sahr KE, Hadjantonakis AK, Baron MH. Tg(Afp-GFP) expression marks primitive and definitive endoderm lineages during mouse development. Dev Dyn. 2006;235:2549–58. doi: 10.1002/dvdy.20843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon GS, Viotti M, Hadjantonakis AK. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev Cell. 2008;15:509–20. doi: 10.1016/j.devcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprise P, Beronja S, Silva-Gagliardi NF, Pellikka M, Jensen AM, McGlade CJ, Tepass U. The FERM protein Yurt is a negative regulatory component of the Crumbs complex that controls epithelial polarity and apical membrane size. Dev Cell. 2006;11:363–74. doi: 10.1016/j.devcel.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprise P, Lau KM, Harris KP, Silva-Gagliardi NF, Paul SM, Beronja S, Beitel GJ, McGlade CJ, Tepass U. Yurt, Coracle, Neurexin IV and the Na(+),K(+)-ATPase form a novel group of epithelial polarity proteins. Nature. 2009;459:1141–5. doi: 10.1038/nature08067. [DOI] [PubMed] [Google Scholar]
- Lee JD, Anderson KV. Morphogenesis of the node and notochord: the cellular basis for the establishment and maintenance of left-right asymmetry in the mouse. Dev Dyn. 2008;237:3464–76. doi: 10.1002/dvdy.21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JD, Silva-Gagliardi NF, Tepass U, McGlade CJ, Anderson KV. The FERM protein Epb4.1l5 is required for organization of the neural plate and for the epithelial-mesenchymal transition at the primitive streak of the mouse embryo. Development. 2007;134:2007–16. doi: 10.1242/dev.000885. [DOI] [PubMed] [Google Scholar]
- Meno C, Shimono A, Saijoh Y, Yashiro K, Mochida K, Ohishi S, Noji S, Kondoh H, Hamada H. lefty–1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell. 1998;94:287–97. doi: 10.1016/s0092-8674(00)81472-5. [DOI] [PubMed] [Google Scholar]
- Meyers EN, Martin GR. Differences in left-right axis pathways in mouse and chick: functions of FGF8 and SHH. Science. 1999;285:403–6. doi: 10.1126/science.285.5426.403. [DOI] [PubMed] [Google Scholar]
- Nakaya MA, Biris K, Tsukiyama T, Jaime S, Rawls JA, Yamaguchi TP. Wnt3a links left-right determination with segmentation and anteroposterior axis elongation. Development. 2005;132:5425–36. doi: 10.1242/dev.02149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer JM, Amack JD, Peterson AG, Bisgrove BW, Yost HJ. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature. 2009;458:651–4. doi: 10.1038/nature07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oteíza P, Köppen M, Concha ML, Heisenberg CP. Origin and shaping of the laterality organ in zebrafish. Development. 2008;135:2807–13. doi: 10.1242/dev.022228. [DOI] [PubMed] [Google Scholar]
- Przemeck GK, Heinzmann U, Beckers J, Hrabé de Angelis M. Node and midline defects are associated with left-right development in Delta1 mutant embryos. Development. 2003;130:3–13. doi: 10.1242/dev.00176. [DOI] [PubMed] [Google Scholar]
- Saijoh Y, Oki S, Ohishi S, Hamada H. Left-right patterning of the mouse lateral plate requires nodal produced in the node. Dev Biol. 2003;256:160–72. doi: 10.1016/s0012-1606(02)00121-5. [DOI] [PubMed] [Google Scholar]
- Shiratori H, Hamada H. The left-right axis in the mouse: from origin to morphology. Development. 2006;133:2095–104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- Sulik K, Dehart DB, Iangaki T, Carson JL, Vrablic T, Gesteland K, Schoenwolf GC. Morphogenesis of the murine node and notochordal plate. Dev Dyn. 1994;201:260–78. doi: 10.1002/aja.1002010309. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Lickert H, Bisgrove BW, Sun X, Yamamoto M, Chawengsaksophak K, Hamada H, Yost HJ, Rossant J, Bruneau BG. Baf60c is a nuclear Notch signaling component required for the establishment of left-right asymmetry. Proc Natl Acad Sci USA. 2007;104:846–51. doi: 10.1073/pnas.0608118104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Katuri V, Dillner A, Mishra B, Deng CX, Mishra L. Disruption of transforming growth factor-beta signaling in ELF beta-spectrin-deficient mice. Science. 2003;299:574–7. doi: 10.1126/science.1075994. [DOI] [PubMed] [Google Scholar]
- Tepass U. FERM proteins in animal morphogenesis. Curr Opin Genet Dev. 2009;19:357–67. doi: 10.1016/j.gde.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Tsiairis CD, McMahon AP. An Hh-dependent pathway in lateral plate mesoderm enables the generation of left/right asymmetry. Curr Biol. 2009;19:1912–7. doi: 10.1016/j.cub.2009.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cai Y, Penland R, Chauhan S, Miesfeld RL, Ittmann M. Increased expression of the metastasis-associated gene Ehm2 in prostate cancer. Prostate. 2006;66:1641–1652. doi: 10.1002/pros.20474. [DOI] [PubMed] [Google Scholar]
- Weinstein DC, Ruiz i Altaba A, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JE., Jr The winged-helix transcription factor HNF-3β is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Tamplin OJ, Beckers A, Gossler A, Rossant J. Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Dev Cell. 2007;13:884–96. doi: 10.1016/j.devcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Ybot-Gonzalez P, Savery D, Gerrelli D, Signore M, Mitchell CE, Faux CH, Greene ND, Copp AJ. Convergent extension, planar-cell-polarity signalling and initiation of mouse neural tube closure. Development. 2007;134:789–99. doi: 10.1242/dev.000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sasaki H, Lowe L, Hogan BL, Kuehn MR. Nodal is a novel TGF-beta-like gene expressed in the mouse node during gastrulation. Nature. 1993;361:543–7. doi: 10.1038/361543a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


